Abstract
Botanical dietary supplements and herbal remedies are widely used for health promotion and disease prevention. Due to the high chemical complexity of these natural products, it is essential to develop new analytical strategies to guarantee their quality and consistency. In particular, the precise characterization of multiple botanical markers remains a challenge. This study demonstrates how a combination of computer-aided spectral analysis and 1D quantitative 1H NMR spectroscopy (qHNMR) generates the analytical foundation for innovative means of simultaneously identifying and quantifying botanical markers in complex mixtures. First, comprehensive 1H NMR profiles (fingerprints) of selected botanical markers were generated via 1H iterative Full Spin Analysis (HiFSA) with PERCH. Next, the 1H fingerprints were used to assign specific 1H resonances in the NMR spectra of reference materials, enriched fractions and crude extracts of Ginkgo biloba leaves. These 1H fingerprints were then used to verify the assignments by 2D NMR. Subsequently, a complete purity and composition assessment by means of 1D qHNMR was conducted. As its major strengths, this tandem approach enables the simultaneous quantification of multiple constituents without the need for identical reference materials, the semi-quantitative determination of particular sub-classes of components, and the detection of impurities and adulterants.
INTRODUCTION
During the last decade, the use of Complementary and Alternative Medicine (CAM) practices and products has become increasingly popular in Western countries. In particular, the use of Botanical Dietary Supplements (BDSs) and herbal remedies to improve health and treat different diseases and ailments has become a common practice among the general public. However, as the popularity of BDSs grows, concern about the quality of commercially available products also increases.1–8 Considering the relatively complex chemical composition of herbal preparations, which may contain tens, hundreds, or even more bioactive and non-bioactive constituents in a wide concentration range, both qualitative and quantitative information becomes essential to ensure the consistency, safety and efficacy of these materials.
Quality control of BDSs and herbal medicines is typically carried out using a combination of modern separation techniques such as gas and liquid chromatography (GC, LC) with sensitive analytical techniques including ultraviolet-visible absorption spectroscopy (UV/vis), evaporative light scattering detection (ELSD) or mass spectrometry (MS).9–13 In order to obtain a detailed profile of the chemical markers in each sample (e.g., identify and quantify main, characteristic, and bioactive constituents), all these methods require a set of well-characterized reference materials that are identical to the target analytes, henceforth called identical reference materials. In the case of MS-based detection, additional standards are required for internal calibration. Accordingly, experimental design frequently includes multiple sample preparation steps, such as serial dilutions of stock solutions, addition of standards and filtration, thus making this approach not only laborious and time-consuming, but also particularly dependent on the availability and quality of phytochemical reference materials. As a consequence, while the quantification of major, known constituents can certainly be achieved, the analysis of minor and unknown compounds is often not feasible.
Because the biological effect of herbal preparations has been recurrently ascribed to the complex interaction between many of their constituents,14–16 there is a growing interest in the development of methods for the simultaneous quantification of multiple botanical markers, i.e., multi-target standardization and normalization, in raw materials and finished products. Due to its non-destructive and universal nature, quantitative Nuclear Magnetic Resonance (qNMR)17,18 measurements represent an interesting option in this regard. Typically used for the structural analysis of pure organic compounds, NMR has evolved into a powerful technique for mixture analysis,19 and even though its quantitative applications have been known for decades, qNMR has received renewed interest only in recent years due to the development of high-field NMR systems with concomitant high sensitivity. Publication of several reports on the relatively low uncertainty and high accuracy of qNMR methods add strong support to this field of application.17,20–26 However, interpretation of qNMR data of complex mixtures is challenging due to frequent signal overlap, especially in the 1H NMR domain, as a result of its narrow chemical shift range.
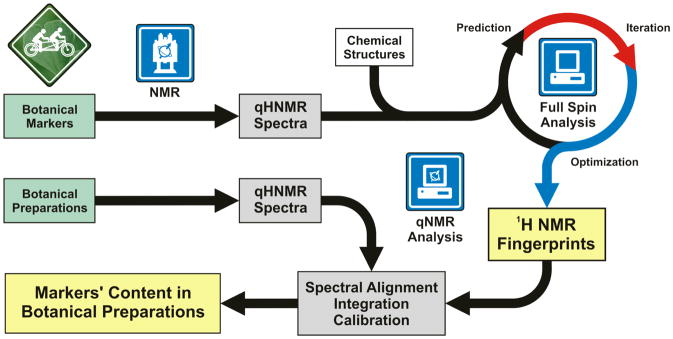
In this report, we describe a new tandem approach to the evaluation of complex botanical mixtures using qualitative and quantitative 1H NMR spectroscopy in combination with computer-assisted spectral analysis (Scheme 1). Precise NMR profiles of botanical markers, generated by means of 1H iterative Full Spin Analysis (HiFSA) using PERCH NMR software,27,28 were aligned with the experimental NMR data in order to facilitate the identification of individual constituents. The high-resolution fingerprints resulting from HiFSA contain all 1H chemical shifts (δ) and spin-spin coupling constants (J) and were used to unequivocally assign each botanical marker in 1D 1H NMR spectra of complex botanical mixtures. In addition, the identity of the markers was verified using their characteristic cross peaks patterns in 2D 1H,1H-COSY experiments. After the markers had been unambiguously identified in the quantitative 1H NMR (qHNMR) spectra of the botanical preparations, molar ratios were readily calculated from the integrals of their characteristic 1H resonances. Furthermore, absolute quantification was achieved using different calibration approaches and without the need for identical reference materials.
Scheme 1.
Flowchart of the proposed botanical standardization scheme, which utilizes a tandem of qNMR and computer-aided full spin analysis.
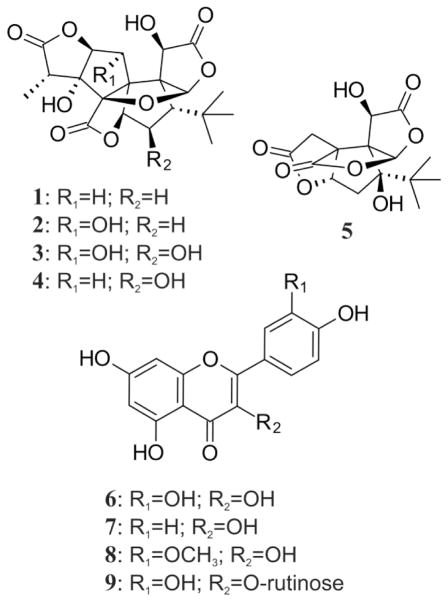
As an example of the application of this methodology, here we evaluate the composition of different preparations from Ginkgo biloba leaves. Ginkgo, also known as Maiden hair tree, has been used for centuries in Traditional Chinese Medicine (TCM) and is currently one of the top-selling BDSs worldwide.29 Due to their antioxidant, neuroprotective and cardioprotective effects, Ginkgo leaf extracts are used to treat or help prevent memory impairments and circulatory disorders.30,31 In fact, a standardized preparation (EGb761) has been the subject of a number of clinical trials.32,33 The pharmacological effects of Ginkgo leaf extracts are attributed to two classes of compounds (Chart 1), which were chosen as botanical markers for qHNMR analysis: the structurally unique Ginkgo terpene trilactones (TTLs) and the flavonoids. The first group includes the ginkgolides A, B, C and J (1–4), which are known antagonists of the platelet-activating factor receptor (PAFR), and bilobalide (5), a potent neuroprotective agent. The second group is considered responsible for the antioxidant properties of Ginkgo extracts and comprises a large number of flavonol derivatives, primarily mono-, di- and triglycosides, with quercetin (6), kaempferol (7) and isorhamnetin (8) as the main aglycones. In addition, the content of rutin (9), one of the major flavonol glycosides in Ginkgo biloba, was evaluated in order to test the applicability of this approach to the analysis of this class of compounds in complex botanical mixtures.
Chart 1.
Ginkgo biloba markers examined in this study.
RESULTS AND DISCUSSION
Solvent Selection for qHNMR Analysis
The choice of a suitable deuterated solvent is a key step in NMR method development and depends on three main factors: the solubility characteristics of the analyte, the overall signal dispersion of the resulting NMR spectrum, and the quality of the commercial grade solvent. A number of solvent systems have been used in previous NMR approaches to the quantification of Ginkgo chemical markers,34–37 ranging from neat acetone-d6 to ternary mixtures such as acetone-d6/pyridine-d5/methanol-d4 (18:6:1). For the present study, we chose DMSO-d6 as the NMR solvent because it fulfills both the solubility and dispersion requirements. The use of DMSO-d6 to analyze Ginkgo TTLs was suggested in the first qHNMR report by van Beek et al.,34 and successfully applied by Nakanishi et al., who added a small amount of acetic acid to standardize 1H chemical shifts.35 Although DMSO-d6 has a number of well-known weaknesses such as mild oxidizing power, high viscosity, hygroscopicity, and relatively high melting and boiling points, it offers three important advantages for identification and quantification purposes: (i) exchangeable 1H resonances can be detected and included in the qualitative analysis;38 (ii) the signal of the residual protonated solvent, DMSO-d5, can be calibrated against a known standard and used for internal calibration;39 and (iii) DMSO-d5 resonates in a relatively non-overlapped region of the 1H NMR spectra of Ginkgo preparations.
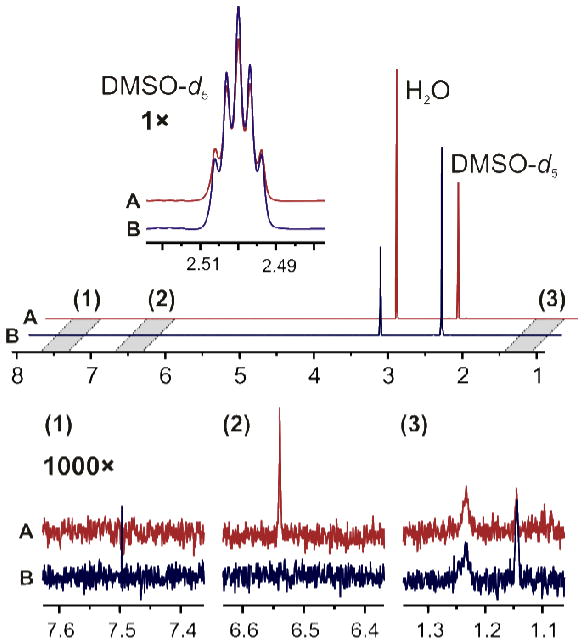
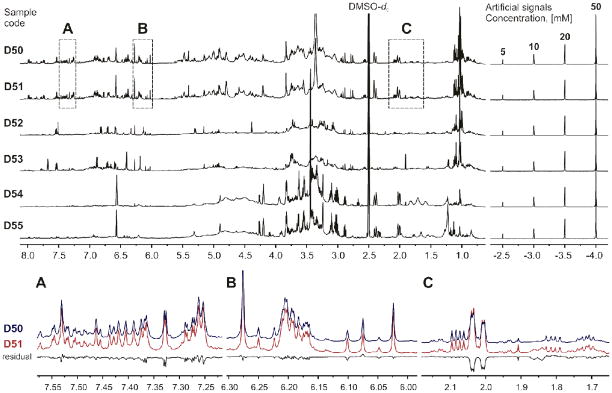
In order to evaluate the purity of the deuterated solvents used in this study, several batches of DMSO-d6 were subjected to NMR analysis. Two of them were identified as suitable and subsequently analyzed in depth. In both cases, only traces of impurities were found. However, differences in the impurity profiles of both DMSO-d6 batches were observed (Figure 1), thus emphasizing the importance of background analysis and subtraction before qHNMR analysis is conducted. Further analysis also revealed significant differences in the total intensity of the residual solvent signal. Therefore, the DMSO-d5 content in each lot was determined by calibration against serial dilutions of caffeine24 and dimethyl sulfone (DMSO2).40 The calibration curves exhibited excellent linearity (R2 > 0.999) over the concentration range explored (0.3–50 mM), and the results confirmed the differences in the degree of deuteration with the amount of residual DMSO-d5 being 78 mM in lot A and 110 mM in lot B. Moreover, resulting from the excellent linearity of the NMR response, subsequent quantitative analyses could be performed via a single-point calibration using an accurate, gravimetrically prepared sample within the evaluated concentration range.
Figure 1.
NMR analysis of two DMSO-d6 batches: A (top) and B (bottom).
Full Spin Analysis of Ginkgo Chemical Markers
One requirement for the unequivocal identification of known chemical structures by NMR is a reliable and complete set of characteristic spin parameters, δ and J, for each target analyte. This equally applies for pure compounds and mixture constituents. In the case of Ginkgo biloba, a plethora of spectroscopic information has been published over the last 40 years.41 In fact, 1H NMR data of the TTLs in DMSO-d6 have been described in several reports,42–44 including one of the seminal works on the structure of the ginkgolides by Nakanishi.45 A similar scenario exists for the flavonoids, since all four compounds have been found in numerous plants. However, a thorough inspection of the available NMR data revealed that several parameters must be updated, while others need to be specified. For example, a number of reported chemical shifts are only estimates, while coupling constants are often not reported due to signal overlap. Moreover, proton signal multiplicities are sometimes omitted, or only described as multiplets, m. This situation impedes the comprehensive interpretation of the NMR data, thus preventing the use of the full set of 1H NMR signals to identify the botanical markers in complex mixtures. As a result, qHNMR analysis is frequently performed using only a small number of the characteristic NMR signals, e.g., the H-12 singlets of Ginkgo TTLs. This makes it difficult to accommodate situations where the measured intensities (e.g., integrals) are altered due to signal overlap or baseline distortion, which is frequently the case in samples with complex matrices such as crude extracts.
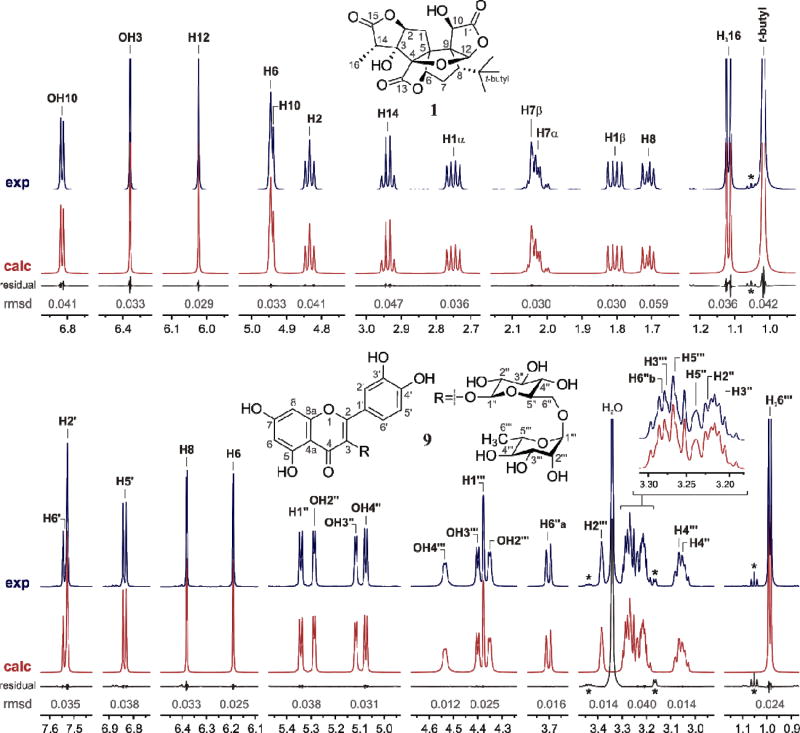
In order to generate a complete, reproducible 1H NMR profile (1H fingerprint) for each botanical marker, a supervised46 1H iterative full spin analysis (HiFSA) was conducted with PERCH. Molecular structures of the selected markers were imported into PERCH and used as the starting point to analyze each discrete spin system and predict the basic NMR parameters (δ, J). Next, the predicted NMR parameters were optimized through spin system calculations with the PERCHit iterator, which systematically adjusted all the δ, J-couplings and line widths values to replicate the experimental 1H NMR data (Figure 2). The outcome of this iterative fitting process is a detailed description of all the 1H NMR signals, together with their corresponding assignments (see Supporting Information). In other words, the 1H fingerprints generated by HiFSA with PERCH provide an unequivocal interpretation of the 1H NMR spectra of pure botanical markers.
Figure 2.
Examples of 1H fingerprints of selected markers, ginkgolide A (1) and rutin (9), resulting from HiFSA with PERCH: comparison of experimental (exp, DMSO-d6, 600 MHz) and calculated (calc) 1H NMR spectra with residuals after complete iteration. Small but significant residuals (*) denotes the presence of residual ethanol in the samples (0.24% and 0.33% w/w, respectively).
qHNMR Analysis of Ginkgo Preparations
Taking into account that HiFSA-generated 1H fingerprints can mimic the 1H NMR spectra of botanical markers, the identification of these constituents in Ginkgo preparations may be simplified to a comparison between these characteristic profiles and the experimental qHNMR data using spectral alignment or, even better, curve fitting procedures. Once the constituents have been identified using their 1H fingerprints, quantitative analysis can immediately follow and be accomplished by measuring the corresponding integrals using NMR processing software. In order to test the applicability of this approach, a qHNMR analysis of different Ginkgo materials, ranging from high-purity isolates to crude extracts, was conducted. Molar proportions between the different components in the samples were obtained through the relative (100%) qNMR method,47,48 while absolute quantification was initially achieved using the residual solvent signal as internal calibrant39 and verified by the internal standard method. Anticipating a low signal dispersion and extensive spectral overlap in crude extracts, two external calibration methods were implemented: the first involved the generation of artificial signals,49,50 which were calibrated using gravimetrically prepared samples of caffeine and DMSO2, while the second relied on the use of a 1H NMR spectrum of caffeine/DMSO2 as calibrant, acquired under identical experimental conditions, plus tuning, matching and 90° pulse width optimization.24,51 All the qHNMR experiments were acquired using a number of scans required to reach a signal-to-noise ratio (S/N) of at least 200:1, which ensures an uncertainty level below 1% for the integration procedure.22,47 Preliminary results on the validation of the tandem approach to qHNMR showed that PERCH is able to identify and align 1H NMR even for signals with S/N of 10:1 (twenty times below the quantification threshold used in this study) with a total intensity root-mean-square deviation (RMSD) below 0.1% (see Supporting Information). In addition, the limits of detection (LOD) and quantification (LOQ) of the qNMR method were calculated according to IUPAC and ACS definitions,52 using serial dilutions of caffeine and DMSO2. As a result, the LOD and LOQ for one proton equivalent were established at 23 ± 4 μM and 73 ± 14 μM, respectively.
When the newly developed tandem approach was applied to purified compounds, the identification of the main constituent and potential common impurities became straightforward. The qHNMR analysis confirmed the high purity (>98.5% w/w) of the commercially available Ginkgo TTLs. In addition, traces of organic solvents, commonly ethanol (Figure 2), and structurally related compounds were identified as the main impurities. For example, 2 contained 0.43% of 1. The presence of these impurities should be considered when these isolates are selected as reference materials for LC-based analysis or used in biological tests, especially taking into account the known synergistic effects of the ginkgolides in vitro.14,15 With regard to the Ginkgo flavonoids, their purity determination by qHNMR led to values over 90% w/w in all cases, with small amounts of related flavonols among the known impurities. For instance, 6 contained 1.87% of 7, 1.53% of 8, and 0.14% of 9.
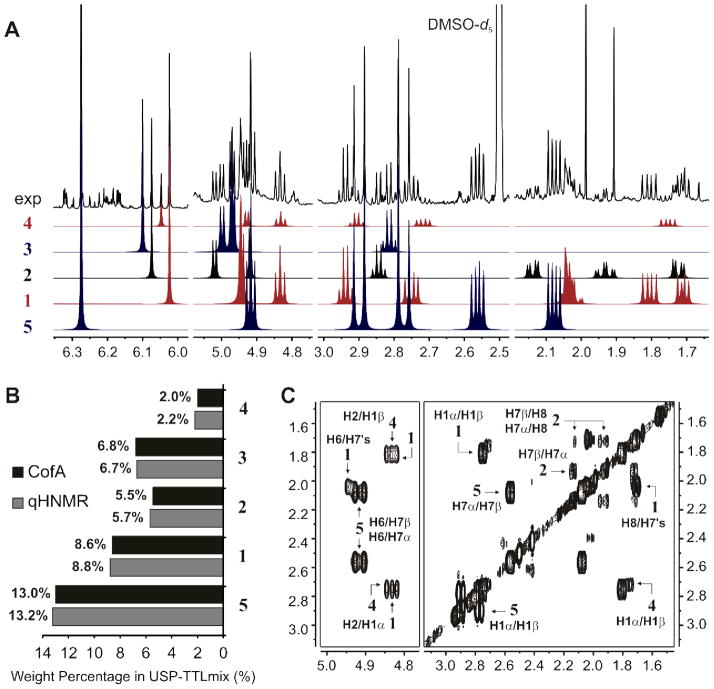
In a first attempt to evaluate the composition of mixtures of Ginkgo chemical markers, a USP-certified mixture of TTLs (USP-TTLmix) was analyzed using the described qHNMR method. The analysis of highly defined reference materials provides the best scenario to test the suitability of the new analytical approach in samples with complex matrices. At the same time, this enables comparison with results from the Certificate of Analysis (CofA), which were obtained by the manufacturer using an LC-ELSD method, specifically developed and validated for the quantification of terpene lactones in Ginkgo biloba.53 When the 1H fingerprints of the TTLs were aligned with the qHNMR spectrum of USP-TTLmix, an efficient and simultaneous identification of the five TTLs was accomplished (Figure 3). The identity of the TTLs was verified by 2D NMR through characteristic cross peaks patterns in 1H,1H-COSY experiments. Furthermore, parallel quantification of TTLs in Mnova showed that qHNMR results were in excellent agreement with the CofA. In fact, the capability of the qHNMR method to replicate weight percentages determined by a well-established and validated LC-ELSD method supports the validity of both approaches. While, in our experience, this is not always the case when using orthogonal methodology for purity evaluation, this observation supports the choice of Ginkgo biloba as a case for building a new analytical approach.
Figure 3.
qHNMR analysis of USP-TTLmix. (A) Experimental (exp, DMSO-d6, 600 MHz) qHNMR spectrum and intensity-adjusted subspectra of ginkgolide A (1), ginkgolide B (2), ginkgolide C (3), ginkgolide J (4) and bilobalide (5) as determined by PERCH iteration of individual 1H fingerprints. (B) qHNMR results and comparison with those provided in the Certificate of Analysis (CofA), which were obtained using an established LC-ELSD method. (C) Identification of key cross peaks in a magnitude-mode 1H,1H-COSY experiment.
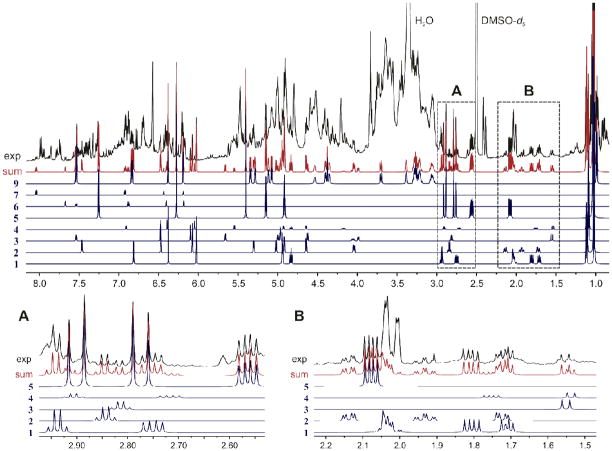
Taking into account the previous results, six crude extracts obtained from different commercial suppliers were evaluated. In general, for certain BDSs, there has been a tendency to use groups of phytoconstituents rather than individual compounds as targets for standardization and quantitation. In the case of Ginkgo biloba, it is widely accepted that the total content of one or two classes of compounds such as the percentage of flavonol glycosides and/or terpenes is reported. However, when the 1H fingerprints of the five TTLs and the four flavonols were aligned with the qHNMR data of the Ginkgo preparations, the content of all nine individual botanical markers can be determined at once. The qHNMR results, which are summarized in Table 1 and visualized in Figure 4, reveal noticeable differences in the composition of the samples. Only D50 and D51, two different batches from the same manufacturer, showed comparable profiles (Figure 4), which would be anticipated considering the (apparent) similarity of their NMR spectra. The difference spectrum generated by arithmetic subtraction of the two qHNMR spectra (residual, Figure 4) reveals that significant differences exist in the intensities of the signals, including those of the target analytes. These small differences are quantitatively reflected in Table 1. The total TTL content in all six samples varied from ~10% w/w in D50 to less than 1% w/w in D55, a fresh Ginkgo leaf extract, with 5 as the most abundant compound of this particular sub-class. The extracts D52 and D53, which according to their CofAs contained at least 6% of TTLs, were found to contain a total of 6.77 and 8.24 % w/w TTLs, respectively, while simultaneously providing a precise measure of each individual compound. This indicates that the qHNMR tandem approach is fit for both multi-target and compound class standardization of BDSs, without the need for identical reference materials. The qHNMR analysis also showed a high variability in the content of flavonoids between samples. D52 and D53, from the same manufacturer, provide a particularly illustrative example: the former contains much 9 (~12% w/w) while the latter shows a remarkably high concentration of the corresponding aglycone, 6 (>7% w/w), presumably due to hydrolysis. Further qNMR analysis indicated the presence of small amounts of the aglycones 6 and 7 in the remaining samples, whereas compound 8 was not detected under the current experimental conditions.
Table 1. Variability of selected botanical markers in commercially available Ginkgo biloba preparations.
Weight percentage of compounds 1–9 in six Ginkgo samples according to qHNMR measurements. All signals used for quantification exhibited signal-to-noise ratios (S/N) 200:1. As indicated by the differences in the individual concentrations between samples D50 and D51 (Δ), even apparently small differences in the qHNMR spectra (see regions A, B, C in Figure 4) reflect significant variation in the content of the phytoconstituents.
| Ginkgo biloba markers | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| concentration, % (w/w) | ||||||||||
| sample | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Σa |
| D50 | 2.34 | 1.92 | 1.15 | 0.68 | 4.03 | 0.73 | 0.38 | ∅b | 3.33 | 14.56 |
| D51 | 2.70 | 1.60 | 1.32 | 0.93 | 3.95 | 0.52 | 0.31 | ∅ | 3.05 | 14.38 |
| Δc | −0.36 | +0.32 | −0.17 | −0.25 | +0.08 | +0.21 | +0.07 | ∅ | +0.28 | +0.18 |
| D52 | 1.14 | 0.81 | 1.54 | 0.68 | 2.60 | 0.96 | 0.23 | ∅ | 11.91 | 19.87 |
| D53 | 1.74 | 0.77 | 1.76 | 1.12 | 2.85 | 7.14 | 0.37 | ∅ | 3.86 | 19.61 |
| D54 | 0.06 | 0.09 | 0.13 | 0.04 | 0.36 | 0.02 | <0.01 | ∅ | 0.70 | 1.40 |
| D55 | 0.07 | 0.09 | 0.14 | 0.03 | 0.36 | 0.04 | <0.01 | ∅ | 0.65 | 1.38 |
Σ denotes the total weight percent of compounds 1–9.
∅ denotes values below the calculated limit of detection (LOD).
Δ denotes the differences in concentration between samples D50 and D51 (Δ = D50 – D51).
Figure 4.
Top: 1H NMR spectra of Ginkgo biloba commercial preparations (DMSO-d6, 600 MHz), including the artificial, calibrated signals used in one of the quantification methods. Bottom: Comparison of Ginkgo preparations D50 (blue) and D51 (red). Although the two preparations exhibit very similar NMR profiles upon visual inspection, the difference spectrum (residual) shows that small but significant differences exist between the two materials which can be quantitated (see Table 1).
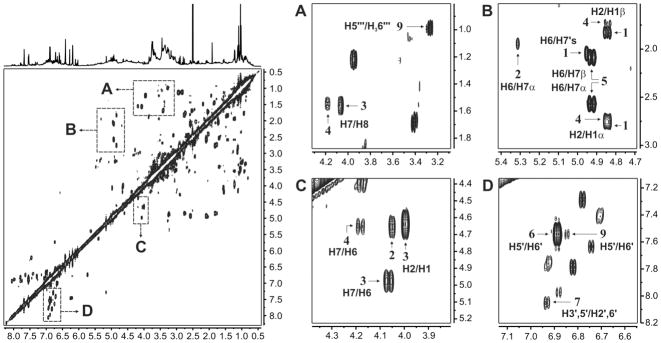
Figure 5 shows how the target analytes were identified in the 1D qHNMR spectra of Ginkgo preparations using their 1H fingerprints. The integrals were computationally adjusted to match the corresponding integration areas in the experimental data. At the same time, δ values were adjusted to account for slight variations in the relative position of the signals. In DMSO-d6 solutions, these chemical shift differences (Δδ) are commonly within a ±0.01 ppm range, which gives additional support to the use of this solvent for qHNMR analysis. Considering that the fitting process is essentially an intensity-driven alignment of the individual 1H fingerprints, this approach enables a rapid identification of botanical markers in relatively high concentration, e.g., compounds 6 and 9 in samples D53 and D52, respectively. The process also allows an efficient identification of suitable signals for quantification in relatively clear regions of the 1H NMR spectrum. Furthermore, as all the integrals were adjusted according to their relative number of protons, the individual contributions of the target analytes to the total area of crowded spectral regions can be projected. However, it is important to keep in mind that the accuracy of the fitting procedures is strongly dependent on the signal-to-noise ratio. As a result, the accuracy of the major constituents is better than that of the minor ones. Depending on the degree of spectral overlap, attempts to fit markers found in very small concentrations can lead to erroneous integral assessments, with differences up to 20%. Nevertheless, although extensive spectral overlap might complicate the identification of specific compounds in 1D NMR spectra, these difficulties can be overcome using 2D NMR experiments such as the 1H,1H-COSY (Figure 6). This approach was particularly useful in the detection of 9 in Ginkgo extracts as most 1H resonances of the rutinose moiety are located in the crowded spectral region between 3.0 and 4.0 ppm.
Figure 5.
Experimental (exp) qHNMR spectrum of sample D50 (DMSO-d6, 600 MHz) and intensity-adjusted subspectra (in blue) of ginkgolide A (1), ginkgolide B (2), ginkgolide C (3), ginkgolide J (4), bilobalide (5), quercetin (6), kaempferol (7) and rutin (9). The subspectra were determined by PERCH iteration of individual 1H fingerprints. The total contribution of the eight markers (sum) is shown in red.
Figure 6.
Identification of ginkgolide A (1), ginkgolide B (2), ginkgolide C (3), ginkgolide J (4), bilobalide (5), quercetin (6), kaempferol (7) and rutin (9) in a commercial Ginkgo preparation (sample D53) using a magnitude-mode 1H,1H-COSY experiment (DMSO-d6, 600 MHz).
After the identification of the target analytes was completed, the simultaneous quantification of these constituents was performed using a suitable calibration method. In this study, four different methods for internal or external calibration were tested. The traditional internal standard method allowed a direct calculation of molar ratios between each botanical marker and the calibrant, but it requires the addition of a known amount of standard to each sample. Additionally, applications to increasingly complex botanical samples are difficult due to potential signal overlap and interactions between botanical constituents and the added standard. The residual solvent method provides a valid alternative to the internal standard method, because the amount of internal calibrant added is strictly related to the volume of deuterated solvent used for the analysis, thereby avoiding the introduction of errors associated with an additional sample preparation step. However, as shown in previous sections, the DMSO-d5 content of each solvent batch needs to be determined, and although the residual protonated signal is located in a relatively clear region of the spectra, signal overlap still can represent a significant source of error.
The external calibration methods are intended to enable the absolute quantification through precise measurement of the integral(s) of the calibrant, while minimizing the effects of signal overlap and chemical interaction. One option is the artificial signal method, which uses calibrated signals that can be placed in any clear, flat region of the qHNMR spectrum of the botanical sample. However, this approach requires the calibration of the artificial signals against the 1H NMR spectrum of a known standard, or in other words, the application of the second external calibration method, developed by Walter et al., i.e., the comparison of two 1H NMR datasets acquired under well-controlled, quantitative conditions.24 Essentially, the two methods are different approaches to exploit the linearity, stability and reproducibility of modern NMR systems. When the direct comparison of two qHNMR datasets was used, molar ratios between botanical constituents and the calibration standard were readily calculated using the absolute values of the integrals in both spectra. On the other hand, a number of additional steps were needed to calculate the same molar ratios by the artificial signal method, including the generation, calibration and arithmetic addition of the artificial signals to the experimental data.
In general, the four calibration methods provide comparable results, with the level of precision increasing in a concentration-dependent manner. The qHNMR analysis of highly-pure isolates and major constituents in Ginkgo preparations yielded results with <2% error, while maximum deviations up to 20% were found for minor components (<0.1% w/w) in crude extracts. In this context, it should be kept in mind that the qHNMR approach enables multi-target standardization in one experiment and for many target compounds at very different abundance levels. Considering these observations, the choice of a suitable calibration method for qNMR will depend on the specific characteristics of each sample and the signal dispersion of the corresponding 1H NMR spectrum. Among the calibration methods described above, we recommend the direct comparison of qHNMR spectra because of its simplicity and flexibility.24 This approach not only allows the qNMR analysis of intact botanical preparations, but also can be adapted to situations in which some acquisition parameters, e.g., relaxation delay, number of scans or receiver gain, must be adjusted according to the characteristics of the analyte. This way, absolute quantification can be performed by simply re-acquiring the qHNMR spectrum of the gravimetrically prepared standard sample under the new acquisition conditions.
Concluding Remarks
The qualitative and quantitative assessment of botanical materials is a crucial step in the verification of product efficacy, safety and batch-to-batch consistency. Taking into account the dependence of LC-based techniques on the availability and quality of identical reference materials, the application of NMR for non-targeted screening represents a practical alternative. The analysis of Ginkgo biloba preparations demonstrates the applicability of the described qNMR method for the simultaneous qualitative and quantitative characterization of multiple constituents in botanical samples. The use of individual 1H fingerprints, generated by full spin analysis, was the key step to the unambiguous identification of the selected markers. These complete 1H NMR profiles, used as characteristic 1H resonances in 1D spectra and as cross peaks patterns in 2D experiments, provide high specificity for the discrimination of target analytes, even in the frequently crowded spectra of complex botanical mixtures. Strategies to also quantify botanicals markers using 2D 1H,1H-COSY experiments are currently being developed by our group.
Once the individual constituents were identified, a parallel quantification was performed using the same qHNMR dataset. This qNMR method is capable of detecting variations in the amount of specific botanical markers, which might be dependent of the geographical location, the time of harvest, the extraction protocol, or the storage conditions. Furthermore, qHNMR analysis can indicate potential adulteration of Ginkgo preparations, e.g., the addition of relatively inexpensive rutin or quercetin to increase the total content of flavonoids.54,55 In addition, it cannot be overstated that the individual botanical markers are only required once, when generating the 1H fingerprints and not for any future analysis. Consequently, the qNMR method does not require the use of identical reference materials for calibration. Accordingly, available identical reference compounds can be preserved for further examination in biological assays or LC-MS analysis, which is still necessary when the target analytes fall below the qNMR limit of detection. Additional aspects of quantitative calibration and validation of the qHNMR method will be subject of a forthcoming report.
In summary, the use of HiFSA and qHNMR in tandem is capable of identifying and quantifying nine Ginkgo marker compounds simultaneously. The present study demonstrates that this approach can be successfully applied over a fairly wide concentration range, i.e., from micromolar to tens of millimolar, to a wide variety of Ginkgo preparations, extending from pure isolates to crude extracts. While this confirms the initial hypothesis of the study, a detailed examination of the qHNMR spectra offers a new challenge: there are still numerous 1H resonances that need to be assigned (Figures 4–6). In some cases, these signals could be associated with unidentified components. The generation of additional 1H fingerprints for known Ginkgo constituents will certainly increase the number of compounds that can be simultaneously identified, while Multivariate Data Analysis (MDA)56–59 will be necessary to improve our understanding of full 1H NMR profiles of mixtures. Nonetheless, the flexibility and convenience of using different calibration approaches depending on the characteristics of each sample, together with the simplicity of sample preparation and the high reproducibility under easy-to-setup conditions, makes qHNMR a particularly strong tool for fast, routine quality control of botanical preparations. Application of this methodology to other important BDSs in the U.S. market including Trifolium pratense, Actaea racemosa (formerly Cimicifuga racemosa) and Camellia sinensis is currently underway.
EXPERIMENTAL SECTION
Materials
Ginkgo terpene trilactones and flavonols were purchased from Indofine Chemical Company Inc. (Hillsborough, NJ). The Certified Ginkgo Terpene Lactones Reference Standard was kindly provided by the United States Pharmacopeial Convention Inc. (Rockville, MD). Ginkgo enriched fractions and crude extracts were provided by Naturex Inc. (South Hackensack, NJ), Vitality Works Inc. (Albuquerque, NM), and Indena S.A.S. (Tours, France). Hexadeuterodimethyl sulfoxide (DMSO-d6, D 99.9%) was obtained from Cambridge Isotope Laboratories Inc. (Andover, MA). Caffeine (>99%) and dimethyl sulfone (DMSO2, >98%) were purchased from Sigma-Aldrich Inc. (St. Louis, MO). All samples were used for NMR analyses without further purification.
Sample Preparation
NMR samples of pure botanical markers were prepared by precisely weighing 1–5 mg of each marker directly into 5 mm, 7″ standard NMR tubes (part no. XR-55-7, Norell Inc., Landisville, NJ) using a Mettler Toledo XS105 Dual Range analytical balance calibrated with Alloy 8 stainless steel precision weights (Henry Troemner LLC, Thorofare, NJ), followed by the addition of 600 μL of DMSO-d6 using a Pressure-Lok gas syringe (VICI Precision Sampling Inc., Baton Rouge, LA) for accurate volume delivery (597 ± 5 μL, n = 15, DMSO). NMR samples of Ginkgo reference materials, enriched fractions and crude extracts were prepared following the same procedure, except that a higher amount of material was used (20–25 mg, precisely weighed).
NMR Spectroscopy
NMR measurements were recorded at 600.13 MHz on a Bruker AVANCE spectrometer equipped with a 5 mm TXI cryoprobe. The 1H chemical shifts (δ) are expressed in ppm with reference to the residual solvent signal of DMSO-d6 (DMSO-d5, 2.500 ppm relative to the TMS scale) and coupling constants (J) are given in Hertz. The 1D 1H NMR spectra were acquired at 298 K under quantitative conditions using a 90° single-pulse experiment (Bruker pulprog: zg). The 90° pulse width for each sample was determined by prorating the measured 360° pulse width (p90 = ¼ × p360). The receiver gain was set to 16 for all 1H NMR measurements. A total of 64 transients were acquired with a relaxation delay of 60 s, which is more than five times the longest T1 observed. The spectral width and the acquisition time were set to 17,985.61 Hz and 4.0 s, respectively, and the resulting number of data points was 143,882. The probes were frequency tuned and impedance matched before each sample run. Off-line 1D and 2D NMR data processing was performed using Mnova software (v.6.0.2, MestreLab Research S.L., A Coruña, Spain). For qHNMR analysis, the following processing scheme was used: a mild Lorentzian-to-Gaussian window function (line broadening = −0.3 Hz, Gaussian factor = 0.05) was applied, followed by zero filling to 256k acquired data points before Fourier transformation. The digital resolution after zero-filling was 0.069 Hz/pt (0.11 ppb/pt at 600.13 MHz). After manual phasing, a fifth order polynomial baseline correction was applied. For 1H full spin analysis, 1D NMR data was processed with NUTS software (v.201004, Acorn NMR Inc., Las Positas, CA) using the scheme previously described, except that a different Lorentzian-to-Gaussian window function for resolution enhancement (line broadening = −1.0 Hz, Gaussian factor = 0.10) was used. The 2D 1H,1H-COSY experiments (Bruker pulprog: cosygpqf) were acquired in magnitude mode with 2k data points in F2 and 256 increments in F1, with a spectral width of 8400 Hz in each dimension (acquisition time = 0.5 s). A total of eight transients were collected with a relaxation delay of 1.0 s. The data were zero filled to 4k data points in F2, and linear predicted to 2k and zero filled to 4k data points in F1. Non-shifted sine-bell window functions were applied to both dimensions before double Fourier transformation, followed by a third order polynomial baseline correction.
1H NMR Full Spin Analysis
PERCH NMR software (v.2010.1, PERCH Solutions Ltd., Kuopio, Finland) was used for full spin analysis. The resolution-enhanced 1H NMR spectra were imported into PERCH as JCAMP-DX files and subjected to baseline correction, peak picking and integration. The X-ray crystal structures of 4 (CCDC ID 183040)60 and 6 (in complex with bovine xanthine oxidase, PDB ID 3NVY) were used as templates to build the molecular models of the Ginkgo TTLs and flavonols, respectively. After geometry optimization (GO) and molecular dynamics (MD) simulations, 1H NMR parameters in DMSO-d6 were predicted using the PERCH Molecular Modeling System (MMS). After a manual examination of the 1H assignments, the calculated 1H chemical shifts, signal line widths, and J-couplings were refined with PERCHit, using both the Integral-transform (D) mode and the Total-line-fitting (T) mode, until an excellent agreement between the observed and simulated spectra was reached (iteration convergence with a total intensity RMSD below 0.1%) to yield the 1H fingerprints of marker compounds 1–9. Finally, the 1H assignments were verified using 2D 1H,1H-COSY experiments.
Qualitative and Quantitative NMR Analysis
For parallel identification of the nine target analytes, their 1H fingerprints were combined into a single .PMS text file (see Supporting Information) and imported into PERCH PMS module, together with the qHNMR spectra of each of the Ginkgo preparations. The nine 1H fingerprints were simultaneously fitted to the NMR spectra of the preparations using PERCHit in D mode, thus identifying the compounds present in each sample. In those cases where PERCH iteration failed to complete the spectral fitting due to insufficient signal dispersion, botanical markers were fitted using 1D spectral alignment and 2D 1H,1H-COSY analysis in Mnova.
Absolute quantification was carried out in Mnova using qNMR scripts developed and provided by C. Peng (MestreLab Research S.L.). The qNMR scripts allow the precise definition of integration ranges for the target analytes and the calibration standard, as well as the automatic calculation of molar concentrations. The qHNMR analysis can be performed with a single sample (qNMR_Basic script) or in batch-mode (APqNMR script). Caffeine and DMSO2 were used as calibration standards in all cases; their purity was evaluated by the 100% qNMR method and water content was assessed by Karl Fischer titration. The results were found to be in excellent agreement with the certificates of analysis provided by the manufacturer [DMSO2: 99.4% (CofA); 99.3% (qHNMR), <0.09% H2O; caffeine: 99.5% (CofA); 98.7% (qHNMR), <0.9% H2O]. For the residual solvent calibration method, stock solutions of caffeine and DMSO2 in DMSO-d6 at nine concentrations, 0.3 to 50 mM, were used to generate calibration curves. Synthetic NMR signals were created in Mnova’s Spin Simulation module and calibrated against a gravimetrically prepared solution of both standards in DMSO-d6 (caffeine 25.2 mM; DMSO2 24.6 mM). The same standard sample was used to acquire the 1H NMR spectrum used in the direct external calibration method.
As a general procedure for quantification, integrated signal areas in 1D NMR experiments were measured by defining a narrow region of at least five times the full-width at half-height (FWHH) of the 1H NMR signals, excluding the 13C satellites. When signal overlap precluded the direct measurement of integrals, line shape fitting was performed with Fityk software (v.0.9.7, http://fityk.nieto.pl/)61 using Voigt-type functions and the Levenberg-Marquardt algorithm for non-linear least-squares fitting.
Supplementary Material
Acknowledgments
The authors thank Dr. J.B. McAlpine for his very valuable comments and suggestions during the manuscript preparation. We are also thankful to Dr. B. Ramirez for his support in the NMR facility at the UIC Center for Structural Biology (CSB), and Dr. C. Peng (MestreLab Research S.L.) for his assistance with Mnova’s qNMR scripts. The support and kind hospitality of M. Niemitz as well as Dr. S.-P. Korhonen (PERCH Solutions Ltd.) during a training visit of J.G.N. to their facilities is gratefully acknowledged. The authors thank the United States Pharmacopeial Convention Inc. (USP), Naturex Inc., Vitality Works Inc., and Indena S.A.S. for kindly providing the Ginkgo biloba preparations analyzed in this study. We are also grateful to the following individuals for helpful discussions of various NMR topics: Drs. K. Colson and J. Hicks (Bruker BioSpin Corp.); I.W. Burton as well as Drs. J.A. Walter and T.K. Karakach (Institute for Marine Biosciences, National Research Council of Canada). The website development efforts of J. Wrenn and E.T. Lin as part of the qreference.com project are also acknowledged. The authors also thank an anonymous reviewer for helpful comments and suggestions regarding qHNMR validation.
The present research work was financially supported by the National Institute of Health (NIH) through grant RC2 AT005899, awarded by the National Center for Complementary and Alternative Medicine (NCCAM). T.G. was partially supported as recipient of a generous USP Fellowship. The authors want to express their sincere gratitude and appreciation to Dr. P.G.W. Gettins for his support and key role in the construction of the UIC CSB via the NIH grant P41 GM068944, awarded by the National Institute of General Medical Sciences (NIGMS).
Footnotes
Contains 1D 1H NMR spectra, 2D 1H,1H-COSY experiments and 1H fingerprints of the botanical markers. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Elvin-Lewis M. J Ethnopharmacol. 2001;75:141–164. doi: 10.1016/s0378-8741(00)00394-9. [DOI] [PubMed] [Google Scholar]
- 2.Cardellina JH., II J Nat Prod. 2002;65:1073–1084. doi: 10.1021/np0200515. [DOI] [PubMed] [Google Scholar]
- 3.Rapaka RS, Coates PM. Life Sci. 2006;78:2026–2032. doi: 10.1016/j.lfs.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 4.van Breemen RB, Fong HHS, Farnsworth NR. Chem Res Toxicol. 2007;20:577–582. doi: 10.1021/tx7000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betz JM, Fisher KD, Saldanha LG, Coates PM. Anal Bional Chem. 2007;389:19–25. doi: 10.1007/s00216-007-1342-8. [DOI] [PubMed] [Google Scholar]
- 6.van Breemen RB, Fong HHS, Farnsworth NR. Am J Clin Nutr. 2008;87:509S–513S. doi: 10.1093/ajcn/87.2.509S. [DOI] [PubMed] [Google Scholar]
- 7.Stoney CM, Coates PM, Briggs JP. J Am Med Assoc. 2008;300:1995–1996. doi: 10.1001/jama.2008.557. [DOI] [PubMed] [Google Scholar]
- 8.Phillips MM, Rimmer CA, Wood LJ, Lippa KA, Sharpless KE, Duewer DL, Sander LC, Betz JM. J AOAC Int. 2011;94:803–814. [PMC free article] [PubMed] [Google Scholar]
- 9.Smillie TJ, Khan IA. Clin Pharmacol Ther. 2010;87:175–186. doi: 10.1038/clpt.2009.287. [DOI] [PubMed] [Google Scholar]
- 10.Mattioli L, Cangi F, Ghiara C, Burico M, Maidecchi A, Bianchi E, Ragazzi E, Bellotto L, Seraglia R, Traldi P. Metabolomics. 2011;7:437–445. [Google Scholar]
- 11.Rimmer CA, Howerton SB, Sharpless KE, Sander LC, Long SE, Murphy KE, Porter BJ, Putzbach K, Rearick MS, Wise SA, Wood LJ, Zeisler R, Hancock DK, Yen JH, Betz JM, NguyenPho A, Yang L, Scriver C, Willie S, Sturgeon R, Schaneberg B, Nelson C, Skamarack J, Pan M, Levanseler K, Gray D, Waysek EH, Blatter A, Reich E. Anal Bional Chem. 2007;389:179–196. [Google Scholar]
- 12.van Beek TA, Montoro P. J Chromatogr A. 2009;1216:2002–2032. doi: 10.1016/j.chroma.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Tang D, Yang D, Tang A, Gao Y, Jiang X, Mou J, Yin X. Anal Bional Chem. 2010;396:3087–3095. doi: 10.1007/s00216-010-3536-8. [DOI] [PubMed] [Google Scholar]
- 14.Wagner H, Ulrich-Merzenich G. Phytomedicine. 2009;16:97–110. doi: 10.1016/j.phymed.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Wagner H. Fitoterapia. 2011;82:34–37. doi: 10.1016/j.fitote.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Junio HA, Sy-Cordero AA, Ettefagh KA, Burns JT, Micko KT, Graf TN, Richter SJ, Cannon RE, Oberlies NH, Cech NB. J Nat Prod. 2011;74:1621–1629. doi: 10.1021/np200336g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauli GF. Phytochem Anal. 2001;12:28–42. doi: 10.1002/1099-1565(200101/02)12:1<28::AID-PCA549>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Pauli GF, Jaki BU, Lankin DC. J Nat Prod. 2005;68:133–149. doi: 10.1021/np0497301. [DOI] [PubMed] [Google Scholar]
- 19.Novoa-Carballal R, Fernandez-Megia E, Jiménez C, Riguera R. Nat Prod Rep. 2011;28:78–98. doi: 10.1039/c005320c. [DOI] [PubMed] [Google Scholar]
- 20.Al-Deen TS, Hibbert DB, Hook JM, Wells RJ. Accred Qual Assur. 2004;9:55–63. [Google Scholar]
- 21.Jancke H, Malz F, Haesselbarth W. Accred Qual Assur. 2005;10:421–429. [Google Scholar]
- 22.Malz F, Jancke H. J Pharm Biomed Anal. 2005;38:813–823. doi: 10.1016/j.jpba.2005.01.043. [DOI] [PubMed] [Google Scholar]
- 23.Rizzo V, Pinciroli V. J Pharm Biomed Anal. 2005;38:851–857. doi: 10.1016/j.jpba.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 24.Burton IW, Quilliam MA, Walter JA. Anal Chem. 2005;77:3123–3131. doi: 10.1021/ac048385h. [DOI] [PubMed] [Google Scholar]
- 25.Pauli GF, Jaki BU, Lankin DC. J Nat Prod. 2007;70:589–595. doi: 10.1021/np060535r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webster GK, Marsden I, Pommerening CA, Tyrakowski CM. Appl Spectros. 2010;64:537–542. doi: 10.1366/000370210791211655. [DOI] [PubMed] [Google Scholar]
- 27.Laatikainen R, Niemitz M, Malaisse WJ, Biesemans M, Willem R. Magn Reson Med. 1996;36:359–365. doi: 10.1002/mrm.1910360306. [DOI] [PubMed] [Google Scholar]
- 28.Laatikainen R, Niemitz M, Weber U, Sundelin J, Hassinen T, Vepsäläinen J. J Magn Reson A. 1996;120:1–10. [Google Scholar]
- 29.Nakanishi K. Bioorg Med Chem. 2005;13:4987–5000. doi: 10.1016/j.bmc.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Strømgaard K, Nakanishi K. Angew Chem Int Ed. 2004;43:1640–1658. doi: 10.1002/anie.200300601. [DOI] [PubMed] [Google Scholar]
- 31.Mahadevan S, Park Y. J Food Sci. 2008;73:R14–R19. doi: 10.1111/j.1750-3841.2007.00597.x. [DOI] [PubMed] [Google Scholar]
- 32.Gardner CD, Taylor-Piliae RE, Kiazand A, Nicholus J, Rigby AJ, Farquhar JW. J Cardiopulm Rehabil Prev. 2008;28:258–265. doi: 10.1097/01.HCR.0000327184.51992.b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeKosky ST, Williamson JD, Fitzpatrick AL, Kronmal RA, Ives DG, Saxton JA, Lopez OL, Burke G, Carlson MC, Fried LP, Kuller LH, Robbins JA, Tracy RP, Woolard NF, Dunn L, Snitz BE, Nahin RL, Furberg CD. J Am Med Assoc. 2008;300:2253–2262. doi: 10.1001/jama.2008.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Beek TA, van Veldhuizen A, Lelyveld GP, Piron I, Lankhorst PP. Phytochem Anal. 1993;4:261–268. [Google Scholar]
- 35.Lichtblau D, Berger JM, Nakanishi K. J Nat Prod. 2002;65:1501–1504. doi: 10.1021/np0201974. [DOI] [PubMed] [Google Scholar]
- 36.Li CY, Lin CH, Wu CC, Lee KH, Wu TS. J Agric Food Chem. 2004;52:3721–3725. doi: 10.1021/jf049920h. [DOI] [PubMed] [Google Scholar]
- 37.Agnolet S, Jaroszewski JW, Verpoorte R, Staerk D. Metabolomics. 2010;6:292–302. doi: 10.1007/s11306-009-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charisiadis P, Exarchou V, Troganis AN, Gerothanassis IP. Chem Commun. 2010;46:3589–3591. doi: 10.1039/b927256a. [DOI] [PubMed] [Google Scholar]
- 39.Pierens GK, Carroll AR, Davis RA, Palframan ME, Quinn RJ. J Nat Prod. 2008;71:810–813. doi: 10.1021/np8000046. [DOI] [PubMed] [Google Scholar]
- 40.Wells RJ, Cheung J, Hook JM. Accred Qual Assur. 2004;9:450–456. [Google Scholar]
- 41.van Beek TA. Bioorg Med Chem. 2005;13:5001–5012. doi: 10.1016/j.bmc.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 42.Weinges K, Hepp M, Jaggy H. Liebigs Ann Chem. 1987:521–526. [Google Scholar]
- 43.Roumestand C, Perly B, Hosford D, Braquet P. Tetrahedron. 1989;45:1975–1983. [Google Scholar]
- 44.Llabres G, Baiwir M, Sbit M, Dupont L. Spectrochim Acta A. 1989;45:1037–1045. [Google Scholar]
- 45.Nakanishi K. Pure Appl Chem. 1967;14:89–113. doi: 10.1351/pac196714010089. [DOI] [PubMed] [Google Scholar]
- 46.Although PERCH’s Automatic Consistency Analysis (ACA) is able to create full 1H NMR profiles without user intervention, the analysis of all the possible assignment combinations (solutions) can take an important amount of CPU time. In order to reduce the number of solutions to be considered, full spin analysis was performed manually using PERCH’s shell.
- 47.Malz F. In: NMR Spectroscopy in Pharmaceutical Analysis. 1. Holzgrabe U, Wawer I, Diehl B, editors. Chapter 2. Elsevier; Amsterdam: 2008. pp. 43–62. [Google Scholar]
- 48.Pauli GF, Jaki BU, Lankin DC, Walter JA, Burton IW. In: Bioactive Natural Products: Detection, Isolation and Structural Determination. 2. Colegate SM, Molyneux RJ, editors. Chapter 4. CRC Press; Boca Raton: 2008. pp. 113–142. [Google Scholar]
- 49.Farrant RD, Hollerton JC, Lynn SM, Provera S, Sidebottom PJ, Upton RJ. Magn Reson Chem. 2010;48:753–762. doi: 10.1002/mrc.2647. [DOI] [PubMed] [Google Scholar]
- 50.Walker GS, Ryder TF, Sharma R, Smith EB, Freund A. Drug Metab Dispos. 2011;39:433–440. doi: 10.1124/dmd.110.036343. [DOI] [PubMed] [Google Scholar]
- 51.Wider G, Dreier L. J Am Chem Soc. 2006;128:2571–2576. doi: 10.1021/ja055336t. [DOI] [PubMed] [Google Scholar]
- 52.Mocak J, Bond AM, Mitchell S, Scollary G. Pure Appl Chem. 1997;69:297–328. [Google Scholar]
- 53.Dietary Supplements / Ginkgo. The United States Pharmacopeia and The National Formulary (USP34-NF29) Vol. 1. The United States Pharmacopeial Convention; Rockville: 2011. pp. 1148–1153. [Google Scholar]
- 54.Liang YZ, Xie P, Chan K. J Chromatogr B. 2004;812:53–70. doi: 10.1016/j.jchromb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 55.Mustafa O, Brendan M, Pei C. J Food Drug Anal. 2007;15:55–62. [Google Scholar]
- 56.Rasmussen B, Cloarec O, Tang H, Stærk D, Jaroszewski JW. Planta Med. 2006;72:556–563. doi: 10.1055/s-2006-931567. [DOI] [PubMed] [Google Scholar]
- 57.van der Kooy F, Maltese F, Choi YH, Kim HK, Verpoorte R. Planta Med. 2009;75:763–775. doi: 10.1055/s-0029-1185450. [DOI] [PubMed] [Google Scholar]
- 58.Liang YZ, Xie P, Chan K. Planta Med. 2010;76:1997–2003. doi: 10.1055/s-0030-1250541. [DOI] [PubMed] [Google Scholar]
- 59.Trygg J, Lundstedt T. In: Handbook of Metabonomics and Metabolomics. 1. Lindon JC, Nicholson JK, Holmes E, editors. Chapter 6. Elsevier; Amsterdam: 2007. pp. 171–199. [Google Scholar]
- 60.Zhao J, Muhammad I, Dunbar DC, Khan IA, Fischer NH, Fronczek FR. Acta Cryst. 2002;C58:o195–o198. doi: 10.1107/s0108270102000689. [DOI] [PubMed] [Google Scholar]
- 61.Wojdyr M. J Appl Cryst. 2010;43:1126–1128. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.