Abstract
6-Hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK), a key enzyme in the folate biosynthesis pathway catalyzing the pyrophosphoryl transfer from ATP to 6-hydroxymethyl-7,8-dihydropterin, is an attractive target for developing novel antimicrobial agents. Previously, we studied the mechanism of HPPK action, synthesized bisubstrate analogue inhibitors by linking 6-hydroxymethylpterin to adenosine through phosphate groups, and developed a new generation of bisubstrate inhibitors by replacing the phosphate bridge with a piperidine-containing linkage. To further improve linker properties, we have synthesized a new compound, characterized its protein binding/inhibiting properties, and determined its structure in complex with HPPK. Surprisingly, this inhibitor exhibits a new binding mode in that the adenine base is flipped when compared to previously reported structures. Furthermore, the side chain of amino acid residue E77 is involved in protein-inhibitor interaction, forming hydrogen bonds with both 2' and 3' hydroxyl groups of the ribose moiety. Residue E77 is conserved among HPPK sequences, but interacts only indirectly with the bound MgATP via water molecules. Never observed before, the E77-ribose interaction is compatible only with the new inhibitor-binding mode. Therefore, this compound represents a new direction for further development.
Keywords: Antibacterial, Bisubstrate, Folate, HPPK, Pterin
1. Introduction
Folate cofactors are essential for all forms of life. With an active transport system, mammals obtain folates from their diet; lacking an active transport system, most microorganisms must synthesize folates de novo.1 Hence, the folate biosynthesis pathway is one of the major targets for the development of antimicrobial agents.2–6 For example, inhibitors of dihydropteroate synthase and dihydrofolate reductase, two enzymes in the pathway, are currently used in clinic as antibiotics.7–9 6-Hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK) is another enzyme in the folate pathway and is absent from mammals. Therefore, HPPK is an attractive target for developing novel antimicrobial agents; attempts have been made for decades.10–15
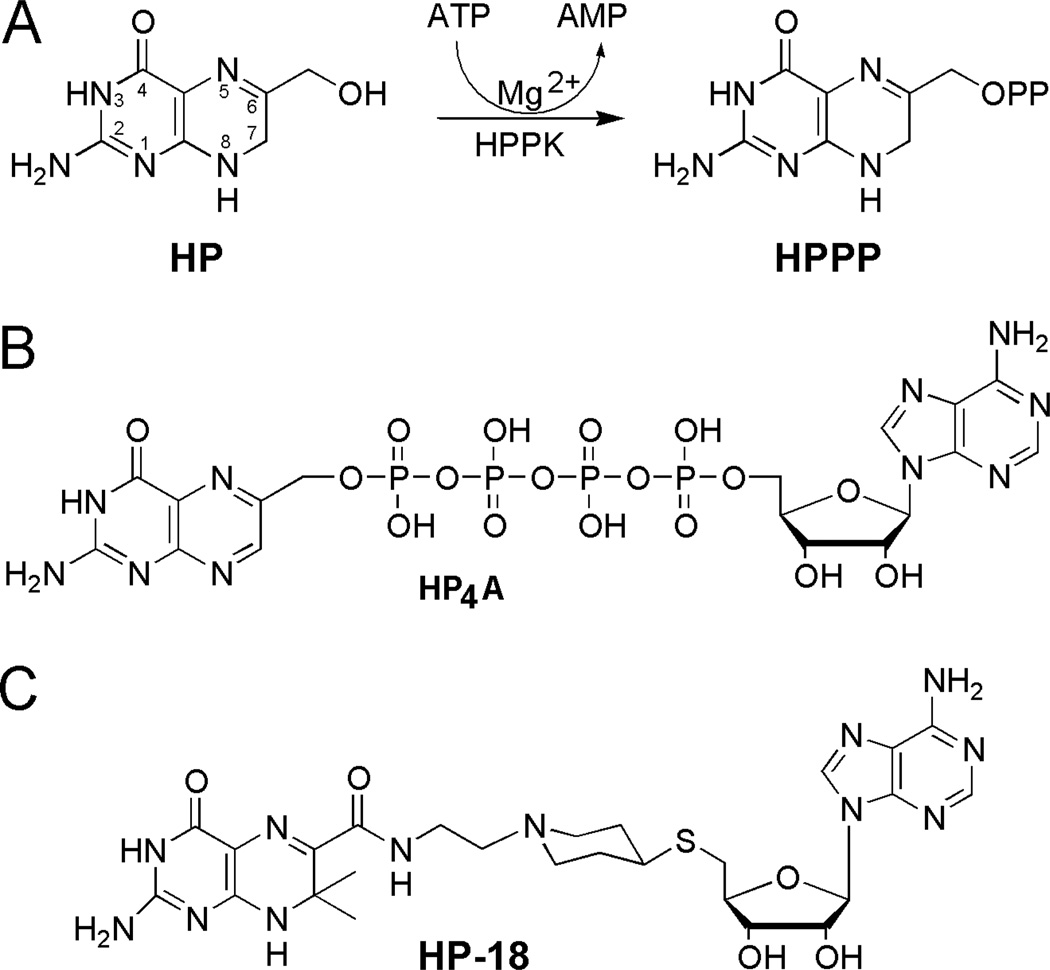
HPPK is currently the best understood pyrophosphokinase.16,17 As shown in Fig. 1A, HPPK catalyzes the transfer of pyrophosphate from ATP to 6-hydroxymethyl-7,8-dihydropterin (HP); the products of the reaction are AMP and 6-hydroxymethyl-7,8-dihydropterin pyrophosphate (HPPP).18 The reaction follows an apparently ordered kinetic mechanism with MgATP binding to the enzyme first (Kd = 2.6–4.5 µM) followed by rapid addition of HP (Kd in the sub-µM range).19–21 The catalytically competent active center is assembled only after both MgATP and HP are bound.22,17
Figure 1.
HPPK-catalyzed reaction and bisubstrate analogue inhibitors of the enzyme. (A) The pyrophosphoryl transfer catalyzed by HPPK and chemical structures of the substrate and product of the reaction. (B) Chemical structure of bisubstrate analogue inhibitor HP4A.14 (C) Chemical structure of bisubstrate analogue inhibitor HP-18.15
We have been developing bisubstrate analogue inhibitors of HPPK, among which P1-(6-hydroxymethylpterin)-P4-(5'-adenosyl)tetraphosphate (HP4A, Fig. 1B)14 and 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine (HP-18, Fig. 1C)15 show promising enzyme binding and inhibition properties. The Kd value for the binding of HP4A to HPPK is 0.47 µM with IC50 = 0.44 µM.14 The Kd value for the binding of HP-18 to HPPK is 2.55 µM with IC50 = 3.16 µM.14 As aforementioned, the Kd value for the binding of MgATP to HPPK is 2.6–4.5 µM. Therefore, both HP4A and HP-18 are good inhibitors of the enzyme.
For optimized ligand binding, the adenine base and phosphate tail of ATP each forms four hydrogen bonds with HPPK, while HP forms six hydrogen bonds with the enzyme.22,13,23 The crystal structures of HP4A and HP-18 each in complex with HPPK [Protein Data Bank (PDB) entry 1EX8 for HPPK•HP4A and 3UDV for HPPK•HP-18] show that (1) the adenine base in each inhibitor forms four hydrogen bonds, (2) the phosphate bridge of HP4A forms nine, whereas the linker in HP-18 forms three hydrogen bonds, and (3) the pterin moiety in HP4A forms five, whereas the pterin moiety in HP-18 forms six hydrogen bonds with the enzyme.14,15 Although HP-18 appears to bind to the enzyme not as tight as HP4A does, it has two advantages. First, HP-18 optimizes the binding of both the pterin and adenine base moieties to the enzyme. Second, HP-18 has a linker that is more drug-like than the phosphate bridge that carries many negative charges and thereby leads to poor bioavailability. Furthermore, the structure of HPPK•HP-18 (PDB entry 3UDV) indicates that the oxidation of the sulfide in HP-18 to sulfone could enable the formation of more hydrogen bonds between the linker of the inhibitor and the enzyme. Here, we report a new bisubstrate analogue inhibitor of HPPK, which contains such a sulfone group but exhibits an unexpected binding mode when it is bound to HPPK. As an effort to maximize the interaction between the linker and the enzyme, the aminoethylpiperidine spacer in HP-18 (Fig. 1C) has been replaced with a glycyl aminoethyl spacer (Fig. 2) in the new compound.
Figure 2.
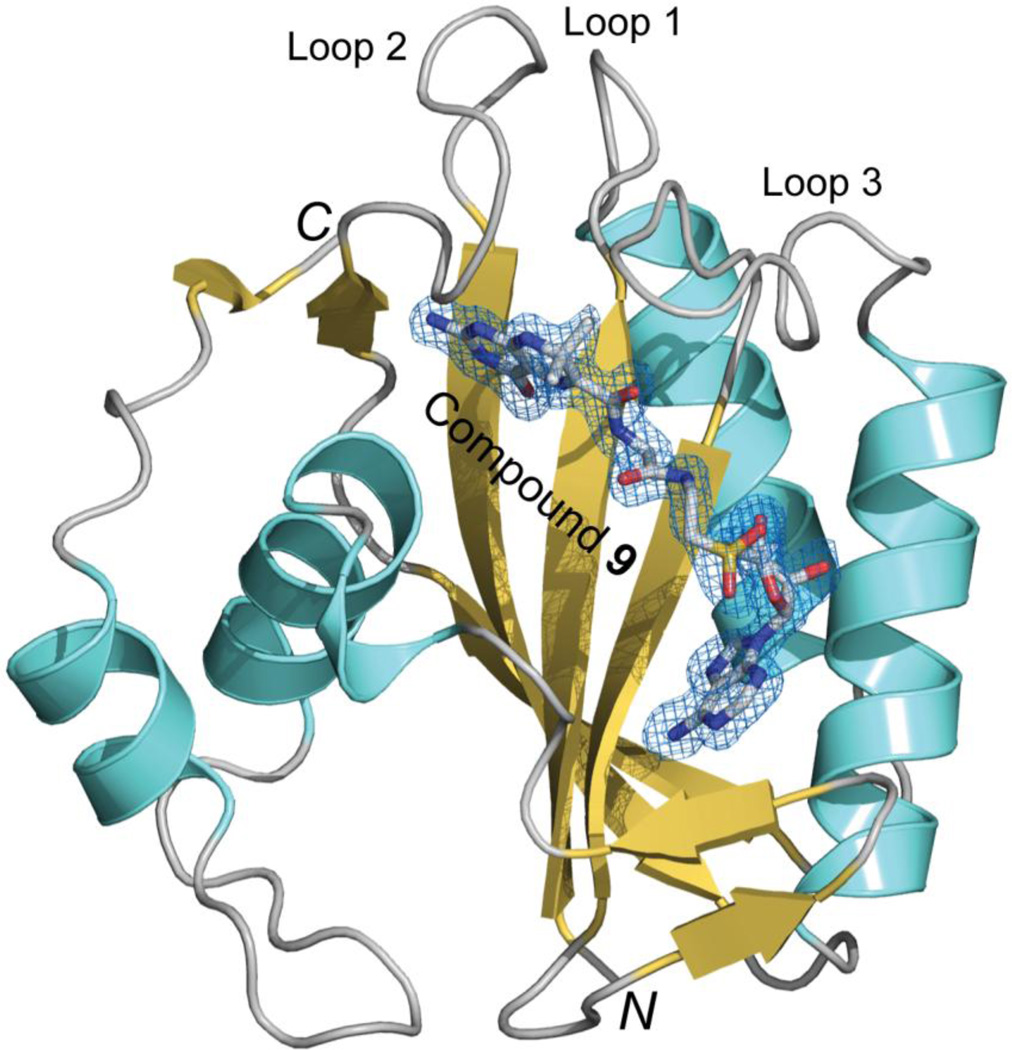
Schematic illustration of the HPPK•9 structure. Polypeptide chains are shown as a ribbon diagrams with helices (spirals) in cyan, strands (arrows) in orange, and loops (tubes) in grey. Compound 9 is shown as a stick model in atomic color scheme (C in grey, N in blue, O in red, and S in yellow) with electron density map (2Fo - Fc; contoured at 1.0 σ) as a blue net.
2. Results and discussion
2.1. Synthesis
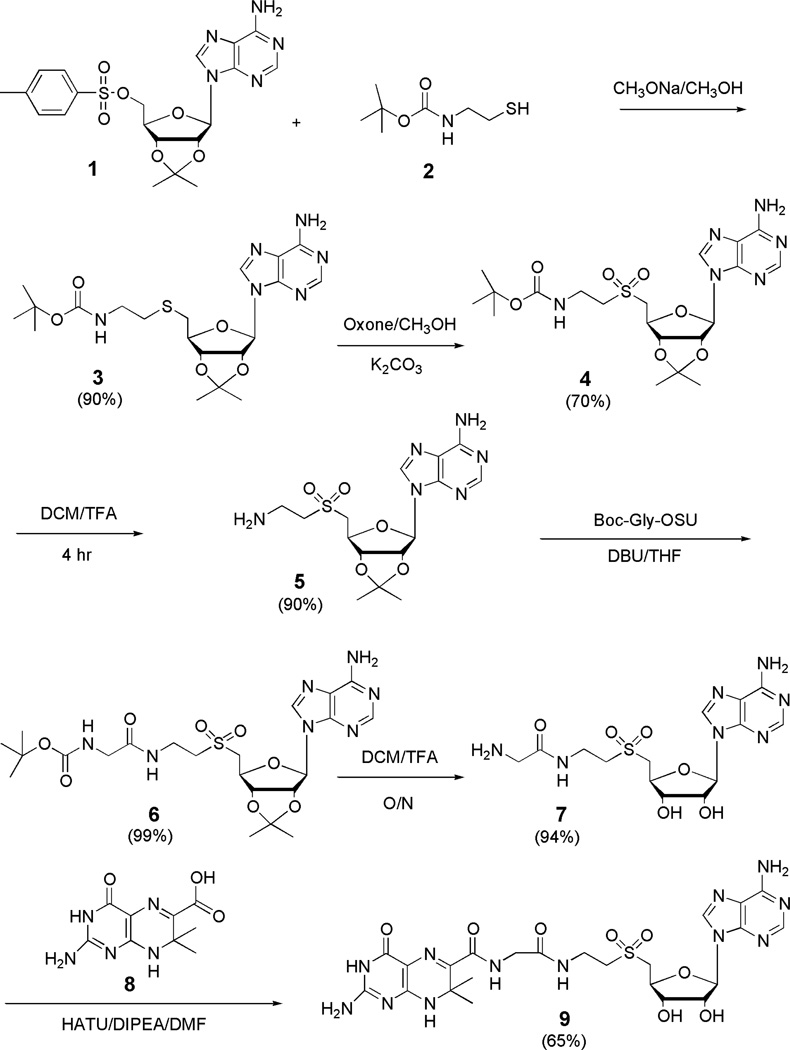
As illustrated in Scheme 1, under basic condition the sulfur nucleophile of (2-mercapto-ethyl)-carbamic acid tert-butyl ester (2) attacked 2′,3′-O-isopropylidene-5′-O-toluene-p-sulfonyl adenosine (1) and gave compound 2-[6-(6-Amino-purin-9-yl)-2,2-dimethyl-tetrahydro-furo[3,4-d][1,3]dioxol-4-ylmethylsulfanyl]-ethyl}-carbamic acid tert-butyl ester (3). The sulfide in 3 was then oxidized to the corresponding sulfone in {2-[6-(6-Amino-purin-9-yl)-2,2-dimethyltetrahydro-furo[3,4-d][1,3]dioxol-4-ylmethanesulfonyl]-ethyl}-carbamic acid tert-butyl ester (4) using potassium hydrogen persulfate (oxone®) at 0 °C in methanol and sodium bicarbonate (pH 5). Under the TFA/DCM condition, cleavage of the BOC protection group yielded 9-[6-(2-Amino-ethanesulfonylmethyl)-2,2-dimethyl-tetrahydro-furo[3,4-d][1,3]dioxol-4-yl]-9H-purin-6-ylamine (5). Amidation of 5 with Boc-glycine N-hydroxysuccinimide ester gave (2-{2-[6-(6-amino-purin-9-yl)-2,2-dimethyl-tetrahydro-furo[3,4-d][1,3]dioxol-4-ylmethanesulfonyl]-ethylcarbamoyl}-ethyl)-carbamic acid tert-butyl ester (6). Under the TFA/DCM condition, cleavage of the BOC protection group 1,3-dioxolanes gave compound 3-amino-N-{2-[5-(6-amino-purin-9-yl)-3,4-dihydroxy-tetrahydro-furan-2-ylmethanesulfonyl]-ethyl}-propionamide (7). Amidation of 7 with 2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydro-pteridine-6-carboxylic acid (8)24,15 gave the final product 2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydro-pteridine-6-carboxylic acid (2-{2-[5-(6-amino-purin-9-yl)-3,4-dihydroxy-tetrahydro-furan-2-ylmethanesulfonyl]-ethylcarbamoyl}-ethyl)-amide (9).
Scheme 1.
2.2. HPPK binding and inhibition
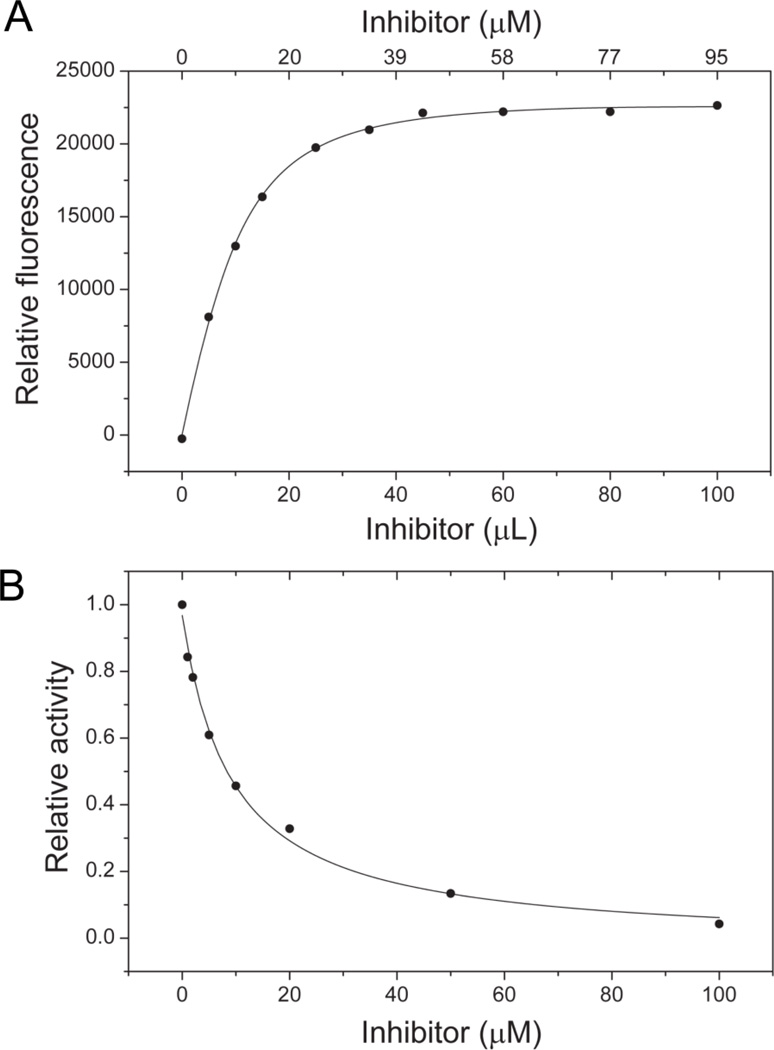
The Kd and IC50 measurements were carried out as described25,14 with sufficient details presented in the experimental methods (4.2 and 4.3). The Kd value is 4.16 ± 0.25 µM, and the IC50 value is 9.53 ± 0.96 µM for compound 9.
2.3. Crystal structure of compound 9 in complex with HPPK
The HPPK•9 structure (Fig. 2) contains 1 HPPK (residues 1–158), 1 compound 9, and 171 water molecules in the asymmetric unit. The statistics of X-ray diffraction data and structural refinement is summarized in Table 1.
Table 1.
Crystal data, X-ray diffraction, and structure
| Crystal | ||
| Space group | P21212 | |
| Unit cell parameters: | a (Å) | 53.12 |
| b (Å) | 70.58 | |
| c (Å) | 36.26 | |
| Data | Overall (last shell) | |
| Resolution (Å) | 30.00-1.67 (1.73-1.67) | |
| Unique reflections | 14746 (802) | |
| Redundancy | 4.9 (2.5) | |
| Completeness (%) | 89.0 (49.1) | |
| Rmergea | 0.086 (0.411) | |
| I/σ | 15.3 (1.8) | |
| Refinement | Overall (last shell) | |
| Resolution (Å) | 27.57-1.73 (1.82-1.73) | |
| Unique reflections | 13829 (1419) | |
| Completeness (%) | 93.6 (70.0) | |
| Data in the test set | 940 (97) | |
| R-work | 0.172 (0.277) | |
| R-free | 0.222 (0.349) | |
| Structure | ||
| Protein non-H atoms / B (Å2) | 1290 / 19.6 | |
| Ligand atoms / B (Å2) | 44 / 19.0 | |
| Water oxygen atoms / B (Å2) | 171 / 27.3 | |
| RMSD | ||
| Bond lengths (Å) | 0.012 | |
| Bond angles (°) | 1.379 | |
| Coordinate error (Å) | 0.51 | |
| Ramachandran plot b | ||
| Favored regions (%) | 96.8 | |
| Disallowed regions (%) | 0.0 | |
Rmerge = Σ|(I-<I>)| / Σ(I), where I is the observed intensity.
Obtained using Ramachandran data by Lovell and coworkers.36
2.4. Distinct binding mode of compound 9 to HPPK
2.4.1. Loop 3 of HPPK in the HPPK•9 structure is ordered
HPPK has three flexible loops (Loop 1, residues 8–15; Loop 2, residues 43–53; Loop 3, residues 82–92. Fig. 2), among which Loop 3 undergoes dramatic conformational changes during catalysis.26,27 The catalytic trajectory of HPPK can be described by six consecutive states: apo-HPPK, HPPK•MgATP, HPPK•MgATP•HP, HPPK•MgAMP-PP-HP (the transition state), HPPK•AMP•HPPP, and HPPK•HPPP.21,28 In the HPPK•MgATP•HP, HPPK•MgAMP-PP-HP, and HPPK•MgAMP•HPPP states, Loop 3 is closed.28,22
Although Loop 3 in the HPPK•HP-18 structure (PDB entry 3UDV) appears to assume the closed conformation, about 40% of the loop (residues 83–86) is disordered (Fig. 3A), which we believe is due to insufficient interaction between Loop 3 and the linker connecting the pterin and adenine moieties of the inhibitor.15 In contrast, Loop 3 in the HPPK•9 structure, which assumes the closed conformation, is completely ordered (Fig. 2, 3A).
Figure 3.
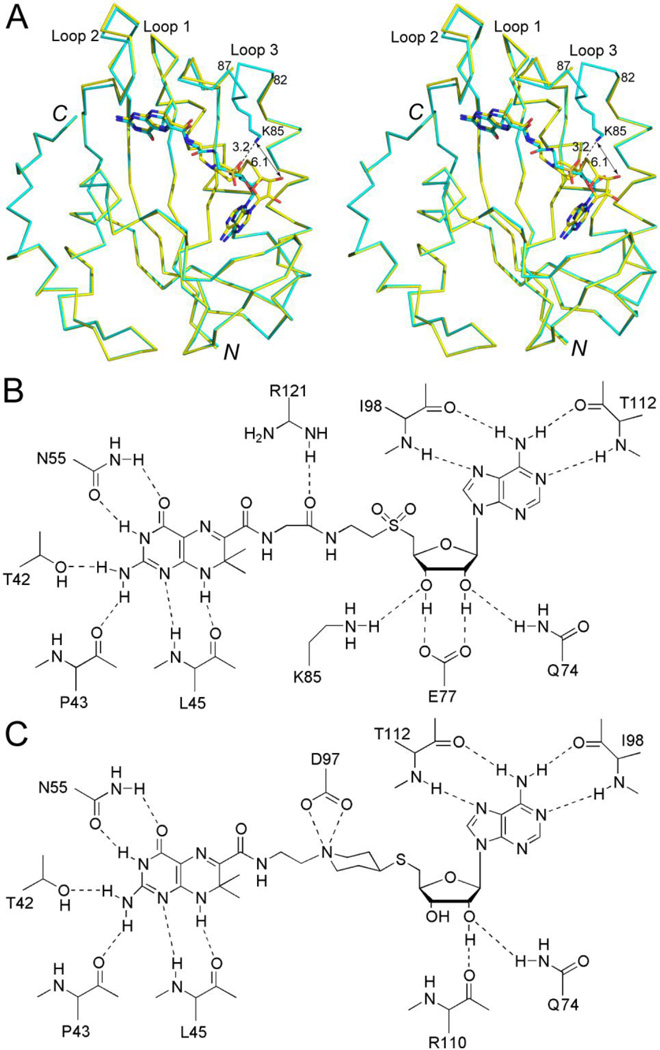
Structural comparison. (A) Stereoview showing the superimposed HPPK•9 (in cyan, this work) and HPPK•HP-18 (in yellow, PDB entry 3UDV) structures. Proteins are shown as Cα traces and ligands as sticks. The side chain of K85 in the HPPK•9 structure is also shown as a stick model. The dotted line in black indicates the hydrogen bond between the K85 side chain and the compound 9 ribose moiety. The dashed line indicates a hydrogen bond (3.2 Å) between the K85 amino group of HPPK and the ribose 2' hydroxyl group of compound 9. The double-headed arrow indicates a distance of 6.1 Å between the K85 amino group of HPPK and the ribose 2' hydroxyl group of HP-18. (B) Protein-inhibitor interactions as observed in the HPPK•9 structure. (C) Protein-inhibitor interactions as observed in the HPPK•HP-18 structure.
The structure shows that a hydrogen bond is formed between the amino group of K85 and the ribose 2' hydroxyl group of compound 9 (Fig. 3A). No other favored interactions are observed between Loop 3 and compound 9. The HPPK•9 structure shares the same crystal form with HPPK•HP-18 (PDB entry 3UDV) and the two structures superimpose very well with a root-mean-square deviation (RMSD) value of 0.133 Å for 518 out of 609 pairs of backbone atom (N, Cα, C, and O) positions. Therefore, it is this hydrogen bond between Loop 3 (side chain amino group of K85) and compound 9 (2' hydroxyl group of ribose) that leads to the completely ordered Loop 3.
2.4.2. Adenine base of compound 9 is flipped
In the HPPK•HP-18 (PDB entry 3UDV) and HPPK•9 structures, the interactions between the protein and the pterin moiety of inhibitors are identical (Fig. 3B, 3C). In contrast, the binding mode of the adenine base is distinct. The binding mode of the adenine base in HPPK•HP-18 is the catalytic binding model as observed in the catalytic center assembly of HPPK (PDB entry 1Q0N).22 The adenine base in HPPK•9 is, however, flipped with respect to the catalytic binding mode (Fig. 3A). Interestingly, the pattern of protein-adenine base interaction remains (Fig. 3B, 3C). In the superimposed HPPK•HP-18 and HPPK•9 structures (Fig. 3A), the distance between the amino group of K85 (in HPPK•9) and the 2' hydroxyl group of HP-18 (in HPPK•HP-18) is 6.1 Å, whereas the distance between the amino group of K85 (in HPPK•9) and the 2' hydroxyl group of compound 9 (in HPPK•9) is 3.2 Å, indicating that the base flipping repositions the ribose moiety and thereby enables the formation of the hydrogen bond between Loop 3 and compound 9. More importantly, such a gain in inhibitor binding is not at the expense of weakening protein-adenine base interaction.
2.4.3. Conserved E77 side chain is involved in the binding of compound 9
Residue E77 is conserved among HPPK sequences. In the catalytic center assembly (PDB entry 1Q0N), it interacts with Mg2+ via a water molecule and with -phosphate via another water molecule.22 In HPPK•HP-18 (PDB entry 3UDV), E77 has a van der Waals interaction (3.5 Å) with the inhibitor.15 In the HPPK•9 complex, however, the E77 side chain forms two hydrogen bonds with the ribose (Fig. 3B), which is only possible for the repositioned ribose moiety. Taking into account the hydrogen bond between Loop 3 and ribose, the new binding mode of the adenosine moiety enhances the protein-adenosine interaction by two more hydrogen bonds than HP-18 (Fig. 3B, 3C).
2.4.4. The sulfone group of compound 9 is not in the position to interact with HPPK
The sulfone group of compound 9 is not positioned to interact with the protein due to the novel binding mode, but the structure suggests moderate modifications of compound 9 by moving the sulfone group away from the ribose may enhance protein-inhibitor interactions.
2.4.5. The new spacer in compound 9 does not form more hydrogen bonds with HPPK
As aforementioned, the aminoethylpiperidine spacer in HP-18 (Fig. 1C) was replaced with the glycyl aminoethyl spacer in compound 9 (Fig. 2). The new spacer forms only one hydrogen bond with HPPK (Fig. 3B), no better than the aminoethylpiperidine spacer (Fig. 3C). Nevertheless, the flexibility of the new spacer may be necessary for the new binding mode of the adenosine moiety of compound 9.
3. Conclusions
We have been developing bisubstrate analogue inhibitors for HPPK, which contains the pterin moiety, the adenine base moiety, and the linker connecting the two moieties. Here, we report a new lead molecule of such inhibitors, which was designed for improved protein-linker interactions. As aforementioned, the Kd value for the binding of HP-18 to HPPK is 2.55 µM with an IC50 value of 3.16 µM.14 With a Kd value of 4.16 ± 0.25 µM and an IC50 value of 9.53 ± 0.96 µM, compound 9 is not better than HP-18. Nevertheless, the new compound exhibits an unexpected, distinct binding mode to the protein. With respect to the catalytic binding mode, which has been observed in the catalytic center assembly, the new inhibitor (compound 9) exhibits a flipped adenine base and repositioned ribose. This rearrangement enables the hydrogen bond formation between the hydroxyl groups of ribose with the side chains of E77 and K85. The K85-ribose interaction stabilizes Loop 3 and the E77-ribose interaction suggests how to use another conserved residue in the development of HPPK inhibitors. Compound 9, therefore, represents a new lead for further improvement. Relocation of the sulfone group closer to the pterin moiety may results in protein-sulfone interactions that will certainly improve the affinity of bisubstrate analogue inhibitors. To facilitate the sulfone relocation, the new glycyl aminoethyl spacer in compound 9 has certain advantage over the aminoethylpiperidine spacer in HP-18.
4. Experimental methods
4.1. Chemistry
4.1.1. General methods
All chemicals were purchased from Sigma-Aldrich except that compound 1 was purchased from TCI America. Starting materials and solvents were used without further purification. Anhydrous reactions were conducted under a positive pressure of dry N2. Reactions were monitored by TLC, on Baker-flex Silica Gel IB-F (J. T. Baker). All compounds and intermediates were purified by flash chromatography performed on Teledyne ISCO Combiflash Rf system using RediSep Rf columns. Ion exchange chromatography was performed using strata Scx (50 µm particle size, 70 Å pore) resin cartridges. Preparative high pressure liquid chromatography (HPLC) was conducted using a Waters 600E system with a Waters 2487 dual λ absorbance detector and Phenomenex C18 columns (250 mm × 21.2 mm, 5 µm particle size, 110 Å pore) at a flow rate of 10 mL/min. A binary solvent systems consisting of A = 0.1% aqueous TFA and B = 0.1% TFA in acetonitrile was employed with the gradients as indicated. 1H and 13C NMR data were obtained on a Varian 400 MHz spectrometer and are reported in ppm relative to TMS (tetramethylsilane). Mass spectra were measured with Agilent 1100 series LC/Mass Selective Detector, Agilent 1200 LC/MSD-SL system and Thermoquest Surveyor Finnigan LCQ deca. IR spectra were measured with a JASCO FT/IR 4100 spectrometer. Chemical purity was determined by HPLC analysis with a Zorbax Eclipse plus C18 column (Narrow Bore RR 2.1 mm × 50 mm, 3.5 micron; flow rate of 0.3 mL/min; solvent, methanol:H2O gradient, 0.1% acetic acid; detection at 260 nm), confirming > 95% purity.
4.1.2. 2-[6-(6-Amino-purin-9-yl)-2,2-dimethyl-tetrahydro-furo[3,4-d][1,3]dioxol-4-ylmethylsulfanyl]-ethyl}-carbamic acid tert-butyl ester (3)
A solution of compound 2 (1.77 g, 10.0 mmol) in NaOCH3 (0.5 M in methanol, 20.2 mL, 10.1 mmol) was stirred under a nitrogen atmosphere for 15 min. Compound 1 (3.23 g, 7.0 mmol) was then added and the mixture was reflux for 5 h. After the reaction was finished, the solvent was removed under vacuum; the crude material was dissolved in ethyl acetate. The ethyl acetate solution was washed with saturated NaHCO3, 5% HCl, and brine. The ethyl acetate solution was then dried with MgSO4, filtered and evaporated to give the crude product. The title compound was obtained in 90% yield (2.94 g) after purification by flash chromatography. IR, vmax (neat, cm−1) 3332, 2980, 2934, 1696, 1640, 1598, 1366, 1248, 1208, 1161, 1086, 868. NMR δ H (400 MHz; CD3OD), 1.38 (3H, s), 1.41 (9H, s), 1.58 (3H, s), 2.60 (2H, t), 2.83 (2H, m), 3.17 (2H, m), 4.34 (1H, m), 5.06 (1H, m), 5.52 (1H, m), 6.18 (1H, d), 8.25 (1H, s), 8.28 (1H, s); δ13C (100 MHz; CD3OD), 158.03 (1C), 157.23 (1C), 153.95 (1C), 150.08 (1C), 141.66 (1C), 120.48 (1C), 115.36 (1C), 91.52 (1C), 87.73 (1C), 85.06 (1C), 84.93 (1C), 79.97 (1C), 41.08 (1C), 34.90 (1C), 33.03 (1C), 28.74 (3C), 27.39 (1C), 25.55 (1C); HRMS (ESI-MS) calculated for C20H30N6O5S (MH+): 467.2071; found: 467.2082.
4.1.3. {2-[6-(6-Amino-purin-9-yl)-2,2-dimethyl-tetrahydro-furo[3,4-d][1,3]dioxol-4-ylmethanesulfonyl]-ethyl}-carbamic acid tert-butyl ester (4)
Potassium carbonate powder (0.28g, 2.0 mmol) was carefully added to a solution of potassium hydrogen persulfate (1.84 g, 3.0 mmol), water (15 mL), until the final pH of the aqueous solution was 5. The oxone solution was added dropwise to a solution of compound 3 (0.47g 1.0 mmol) in methanol (15 mL) which was cooled to 0 °C while stirring, and the reaction mixture was stirred at room temperature overnight. The solution was extracted three times between ethyl acetate and water, and the organic layer was dried over MgSO4. The organic layer was concentrated and the title compound was obtained in 70% yield (0.35 g) after purification by flash chromatography. IR, vmax (neat, cm−1) 3345, 2979, 2921, 1694, 1651, 1236, 1165, 1090, 1022, 894, 871. NMR δ H (400 MHz; CD3OD), 1.37 (12H, s), 1.59 (3H, s), 3.09–3.21 (2H, m), 3.27–3.39 (2H, m), 3.61 (1H, q), 3.82 (1H, q), 4.71 (1H, m), 5.23 (1H, q), 5.47 (1H, q), 6.26 (1H, d), 8.25 (1H, s), 8.27 (1H, s); δ13C (100 MHz; CD3OD), 157.66 (1C), 157.30 (1C), 154.04 (1C), 149.86 (1C), 142.14 (1C), 120.71 (1C), 115.78 (1C), 90.90 (1C), 85.26 (1C), 85.06 (1C), 82.91 (1C), 80.41 (1C), 57.12 (1C), 54.69 (1C), 34.82 (1C), 28.68 (3C), 27.40 (1C), 25.56 (1C); HRMS (ESI-MS) calculated for C20H30N6O7S (MH+): 499.1969; found: 499.1976.
4.1.4. 2-Amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydro-pteridine-6-carboxylic acid (2-{2-[5-(6-amino-purin-9-yl)-3,4-dihydroxy-tetrahydro-furan-2-ylmethanesulfonyl]-ethylcarbamoyl}-ethyl)-amide (9)
To a solution of compound 4 (0.50 g, 1.0 mmol) in 5 mL DCM, 1 mL TFA was added dropwise at −20 °C; then the reaction mixture was stirred at room temperature for a few hours. After the reaction was finished, the solvent was removed under vacuum, and the residue was purified by silica gel chromatography, eluting with 15% DCM/methanol, to afford 0.36 g (90%) of the compound 5 as white foam. A solution of Boc-l-Gly-OSu (0.27 g, 1.0 mmol) in dry THF (10 mL) was treated with the product, and the resulting suspension was cooled at 0 °C under nitrogen. A solution of DIPEA (0.2 mL, 1.1 mmol) in dry THF (4.0mL) was added. Stirring was continued overnight, and then the solvent was evaporated. The crude product was dissolved in ethyl acetate and purified by flash chromatography. The product, compound 6 (0.5 g, 99%), was dissolved in 5 mL DCM, and 2 mL TFA was added dropwise at −20 °C. Then, the reaction mixture was stirred at room temperature overnight. After the reaction was finished, the solvent was removed under vacuum, and the residue was purified by silica gel chromatography, eluting with 15% DCM/methanol, to afford 0.35 g (94%) of compound 7. The product was added to a solution of compound 8 (0.20 g, 0.84 mmol), O-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) (0.33g, 0.88 mmol), and DIPEA (411 µL, 2.4 mmol)) in anhydrous DMF (50 mL). After 18 h at room temperature, the solvent was evaporated under high vacuum, and the reaction residue was purified by HPLC (H2O:Methanol) to give the final product, compound 9 (0.35g, 0.55 mmol, 65%). IR, vmax (neat, cm−1) 3101, 2796, 2360, 1655, 1508, 1432, 1292, 1196, 1130, 834, 798, 721. NMR δ H (400 MHz; CD3OD), 1.57 (6H, s), 3.23–2.28 (2H, m), 3.30–3.33 (1H, m), 3.43–3.52 (3H, m) 3.54–3.88 (3H, m), 4..43–4.45 (1H,t), 4.49–4.53 (1H, m), 6.07 (1H, d,), 8.45 (1H, s), 8.48 (1H, s); δ13C (100 MHz; DMSO-d6), 169.15 (1C), 163.72 (1C), 158.84 (1C), 155.39 (1C), 149.03 (1C), 143.80 (1C), 142.48 (1C), 119.67 (1C), 117.78 (1C), 114.86(1C), 110.00 (1C), 100.80 (1C), 88.62 (1C), 79.36 (1C), 73.37 (1C), 72.93 (1C), 56.38 (1C), 53.97 (1C), 53.18 (1C), 42.04 (1C), 32.58 (1C), 28.49 (2C); HRMS (ESI-MS) calculated for C23H30N12O8S (MH+): 635.2103; found: 635.2116.
4.2. Fluorometric titration
The dissociation constants of the inhibitors were measured as described25 for the fluorometric measurement of the dissociation constant of Ant-ATP with modifications. Briefly, the inhibitors were dissolved in dimethylsulfoxide. Dimethylsulfoxide concentrations were kept within 1.7% during the titration experiments and control experiments showed that dimethylsulfoxide at these concentrations had no effects on activity (substrate binding and catalysis) of the enzyme. E. coli HPPK was dissolved in 100 mM Tris, pH 8.3. The titration was performed at 23 °C by adding aliquots of a 2 mM compound 9 stock solution to the HPPK solution. The initial HPPK concentration and volume were 10 µM and 2 mL, respectively. The excitation and emission wavelengths were 450 and 480 nm, respectively. The excitation and emission slits were 1 and 4 nm, respectively. The Kd values were obtained by nonlinear least-squares regression of the data to equation 1 as described25
| (Eq. 1) |
where ΔFobs and ΔFmol are observed and molar fluorescence changes caused by binding, Et is the total concentration of HPPK, and Lt is the total concentration of the inhibitor. As an example, the fluorometric titration of HPPK with compound 9 is shown in Fig. 4A.
Figure 4.
Kd and IC50 measurements. (A) Fluorometric titration of HPPK with compound 9. (B) Inhibition of HPPK by compound 9.
4.3. Enzyme inhibition assay
IC50 measurements were carried out essentially as described.14 Briefly, the initial reaction mixtures in 30 µL contained 1 nM E. coli HPPK, 2 µM ATP, 1 µM HP, 5 mM MgCl2, 25 mM DTT, and a trace amount of [α-32P]-ATP (~1 Ci) in 100 mM Tris, pH 8.3. The experiments were conducted at 23 °C. The reaction was initiated by the addition of the enzyme and stopped 30 min later by the addition of 6 µL of 0.5 M EDTA. The radioactive reactant and product were separated by thin-layer chromatography, using a PEI-cellulose plastic plate (EMD) with 0.3 M KH2PO4 as the mobile phase, and quantified by a Phosphor-Imager system (Amersham Typhoon 9200). The IC50 values were obtained by fitting the data to a logistic equation by nonlinear least-squares regression of the data to equation 2 as described29
| (Eq. 2) |
where v is the reaction rate, vmin the minimum reaction rate, vmax the maximum reaction rate, and [I] the concentration of the inhibitor. The inhibition of HPPK by compound 9 is shown in Fig. 4B.
4.4. Crystallization, X-ray diffraction, structure solution, and refinement
A Hydra II-Plus-One crystallization robot (Matrix Technologies, Hudson, New Hampshire, USA) and Crystal Screen kits from Hampton Research (Laguna Niguel, California, USA) were used. Crystals of HPPK•9 were grown at 19±1 °C in sitting drops containing 0.3 µl protein solution (10 mg/mL HPPK with saturated compound 9 in 20 mM Tris-HCl pH 8.0) and 0.3 µl well solution (25% PEG-3350 and 0.2 M NaCl in 0.1 M HEPES, pH 7.5). A crystal of HPPK•9 was soaked in a cryoprotectant solution containing 75% (v/v) well solution and 25% (v/v) ethylene glycol, and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at 100K with an MARCCD detector mounted at the synchrotron Beamline 22 at the Advanced Photon Source, Argonne National Laboratory. Data processing was carried out with the HKL2000 program suite.30 The structure was solved by Fourier synthesis starting with the HPPK•HP-18 structure (PDB entry 3UDV).15 Multiple conformations of amino acid residues, ligands, and solvent molecules were removed from the starting model. Structure solution and refinement were done with PHENIX.31 All graphics work, including model building and rebuilding, was performed with COOT.32 The structures were verified with annealed omit maps and the geometry was assessed using PROCHECK33 and WHAT IF.34 The statistics of X-ray diffraction data and the HPPK•9 structure are summarized in Table 1. Illustrations were prepared with PyMOL.35
Supplementary Material
Acknowledgements
This research was supported by NIH grant R01GM084402 (H.Y.), NIAID Trans NIH/FDA Intramural Biodefense Program Y3-RC-8007-01 (X.J.), and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Mass spectrometry experiments were conducted on an Agilent 1100 series LC/Mass Selective Detector maintained by the Biophysics Resource in the Structural Biophysics Laboratory, an Agilent 1200 LC/MSD-SL system in the Chemical Biology Laboratory, and a Thermoquest Surveyor Finnigan LCQ deca maintained by the Comparative Carcinogenesis Laboratory of National Cancer Institute. IR spectrometry experiments were conducted on a JASCO FT/IR 4100 spectrometer in the Chemical Biology Laboratory of National Cancer Institute. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID and 22-BM beamlines at the Advanced Photon Source (APS), Argonne National Laboratory (ANL).
The coordinates and structure factors for the HPPK•9 complex have been deposited in the PDB under entry code 4F7V.
Abbreviations
- HP4A
P1-(6-hydroxymethylpterin)-P4-(5'-adenosyl)tetraphosphate
- HPPK
6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase
- HPPP
6-hydroxymethyl-7,8-dihydropterin pyrophosphate
- HP-18
5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine
- PDB
Protein Data Bank
- RMSD
root-mean-square deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data (the IR spectra and the 1H and 13C NMR spectra of compounds 3, 4, and 9) associated with this article can be found, in the online version, at …
References
- 1.Hitchings GH, Burchall JJ. Adv. Enzymol. Relat. Areas Mol. Biol. 1965;27:417–468. doi: 10.1002/9780470122723.ch9. [DOI] [PubMed] [Google Scholar]
- 2.Cohen ML. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 3.Neu HC. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 4.Murray BE. Adv. Intern. Med. 1997;42:339–367. [PubMed] [Google Scholar]
- 5.Bermingham A, Derrick JP. Bioessays. 2002;24:637–648. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- 6.Walsh C. Nat Rev Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 7.Hughes DTD. Sulphonamides. Antibiotic and Chemotherapy. 7th ed. New York: Churchill Livingstone; 1997. [Google Scholar]
- 8.Hughes DTD. In: Antibiotics and Chemotherapy. O'Grady F, Lambert HP, Finch RG, Greenwood D, editors. New York: Churchill Livingstone; 1997. pp. 346–356. [Google Scholar]
- 9.Zinner SH, Mayer KH. In: Principles and Practice of Infectious Diseases. Mandell GL, Bennett JE, Dolin R, editors. Philadelphia: Churchill Livingstone; 2005. pp. 440–451. [Google Scholar]
- 10.Wood HCS. In: Chemistry and Biology of Pteridines. Pfleiderer W, editor. Berlin-New York: Walter de Gruyter; 1975. [Google Scholar]
- 11.Al-Hassan SS, Cameron RJ, Curran AWC, Lyall WJS, Nicholson SH, Robinson DR, Stuart A, Suckling CJ, Stirling I, Wood HCS. J. Chem. Soc. Perkin Trans. 1985;1:1645–1659. [Google Scholar]
- 12.Hennig M, Dale GE, D'Arcy A, Danel F, Fischer S, Gray CP, Jolidon S, Muller F, Page MG, Pattison P, Oefner C. J. Mol. Biol. 1999;287:211–219. doi: 10.1006/jmbi.1999.2623. [DOI] [PubMed] [Google Scholar]
- 13.Stammers DK, Achari A, Somers DO, Bryant PK, Rosemond J, Scott DL, Champness JN. FEBS Lett. 1999;456:49–53. doi: 10.1016/s0014-5793(99)00860-1. [DOI] [PubMed] [Google Scholar]
- 14.Shi G, Blaszczyk J, Ji X, Yan H. J. Med. Chem. 2001;44:1364–1371. doi: 10.1021/jm0004493. [DOI] [PubMed] [Google Scholar]
- 15.Shi G, Shaw G, Liang YH, Subburaman P, Li Y, Wu Y, Yan H, Ji X. Bioorg Med Chem. 2012;20:47–57. doi: 10.1016/j.bmc.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derrick JP. In: Folic Acid and Folates. Litwack G, editor. Oxford, UK: Academic Press; 2008. pp. 411–433. [Google Scholar]
- 17.Yan H, Ji X. Protein Pept. Lett. 2011;18:328–335. doi: 10.2174/092986611794654003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiota T. In: Chemistry and Biochemistry of Folates. Blakley RT, Benkovic SJ, editors. New York: John Wiley & Sons; 1984. pp. 121–134. [Google Scholar]
- 19.Shi G, Gong Y, Savchenko A, Zeikus JG, Xiao B, Ji X, Yan H. Biochim. Biophys. Acta. 2000;1478:289–299. doi: 10.1016/s0167-4838(00)00043-1. [DOI] [PubMed] [Google Scholar]
- 20.Bermingham A, Bottomley JR, Primrose WU, Derrick JP. J. Biol. Chem. 2000;275:17962–17967. doi: 10.1074/jbc.M000331200. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Gong Y, Shi G, Blaszczyk J, Ji X, Yan H. Biochemistry. 2002;41:8777–8783. doi: 10.1021/bi025968h. [DOI] [PubMed] [Google Scholar]
- 22.Blaszczyk J, Shi G, Yan H, Ji X. Structure. 2000;8:1049–1058. doi: 10.1016/s0969-2126(00)00502-5. [DOI] [PubMed] [Google Scholar]
- 23.Blaszczyk J, Li Y, Shi G, Yan H, Ji X. Biochemistry. 2003;42:1573–1580. doi: 10.1021/bi0267994. [DOI] [PubMed] [Google Scholar]
- 24.Shi G, Ji X. Tetrahedron Lett. 2011;52:6174–6176. doi: 10.1016/j.tetlet.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Wu Y, Blaszczyk J, Ji X, Yan H. Biochemistry. 2003;42:1581–1588. doi: 10.1021/bi026800z. [DOI] [PubMed] [Google Scholar]
- 26.Xiao B, Shi G, Chen X, Yan H, Ji X. Structure. 1999;7:489–496. doi: 10.1016/s0969-2126(99)80065-3. [DOI] [PubMed] [Google Scholar]
- 27.Xiao B, Shi G, Gao J, Blaszczyk J, Liu Q, Ji X, Yan H. J. Biol. Chem. 2001;276:40274–40281. doi: 10.1074/jbc.M103837200. [DOI] [PubMed] [Google Scholar]
- 28.Blaszczyk J, Shi G, Li Y, Yan H, Ji X. Structure (Camb) 2004;12:467–475. doi: 10.1016/j.str.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Graeser D, Neubig RR. In: Signal transduction. Milligan G, editor. Oxford: IRL Press; 1992. pp. 1–30. [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.Vriend G. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: Delano Scientific; 2002. [Google Scholar]
- 36.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins: Struct. Funct. Genet. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.