Abstract
Objective
Carotid plaque is a marker of subclinical atherosclerosis with a genetic component. The aim of this follow-up fine mapping study was to identify candidate genes for carotid plaque within four linkage regions.
Methods
We successfully genotyped 3,712 single nucleotide polymorphisms (SNPs) under the four linkage regions that were previously identified in 100 extended Dominican families. Family-based association tests were performed to investigate their associations with carotid plaque. Promising SNPs were evaluated in an independent population-based subcohort (N=941, 384 Dominicans) from the Northern Manhattan Study (NOMAS).
Results
In the family study, evidence for association (p<0.0005) was found regarding several genes (NAV2, EFCAB11/TDP1, AGBL1, PTPN9, LINGO1 and LOC730118), with the strongest association at rs4143999 near EFCAB11/TDP1 (p=0.00001 for carotid presence and 0.00003 for plaque area, multiple testing corrected p≤0.02). The association in AGBL1 and PTPN9 was mainly driven by the families with linkage evidence (p=0.00008~0.00001 and 0.76~0.32, respectively, in the families with and without linkage evidence). However, these associations explained only a small portion of the observed linkage. In NOMAS, replication (p<0.05 in the whole NOMAS subcohort and p<0.10 in the smaller Dominican subcohort) was found for SNPs within/near EFCAB11, NAV2, AGBL1 and other genes.
Conclusion
This follow-up study has identified multiple candidate genes for carotid plaque in the Dominican population. Many of these genes have been implicated in neurodegenerative and cardiovascular diseases. Further studies with in-depth re-sequencing are needed to uncover both rare and common functional variants that contribute to the susceptibility to atherosclerosis.
Keywords: Carotid plaque, Linkage, Fine mapping, Atherosclerosis, Hispanics
INTRODUCTION
Carotid plaque is an established measure of subclinical atherosclerosis and a better predictor of future vascular events than carotid intima media thickness (IMT) [1, 2]. Although several family studies have shown that atherosclerotic plaque has a considerable genetic component, few genome-wide linkage and association studies have been reported, with substantial variability in the findings [3, 4]. To date, only two genetic loci (chromosome 4q31 and 7q22) have been associated with the presence of plaque with genome-wide significance (p<5×10−8) in a meta-analysis among 31,211 individuals of European ancestry from the Cohort for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium [5]. As with other common diseases, the variability and lack of replication among studies is in part due to phenotypic and genetic complexity, small effects for individual susceptibility genes, and multiple study differences, including the research protocols, ascertainment methods, choice of genetic model, marker density, and the population under study.
One strategy to rectify some of these issues is to follow-up with association in the same pedigrees used to identify linkage, utilizing increased marker density in the candidate regions. Family-based designs have several favorable features for gene discovery as summarized by Laird and Lange [6]: “They are robust to confounding and variance inflation, which can arise in standard designs in the presence of population substructure; they test for both linkage and association; and they offer a natural solution to the multiple comparison problem.”. In a previous 10- centimorgan (cM) genome scan of 100 multi-generation Dominican families with a median family size of 14, we found 4 chromosomal regions on 7q, 11p, 14q and 15q with suggestive linkage (logarithm of odds (LOD) ≥2.0) to carotid plaque presence or area [3]. In the present study, we report a follow-up fine mapping study on carotid plaque presence and area by surveying these four regions with 3,712 densely mapped single nucleotide polymorphisms (SNP) (average 1 SNP per 17.5 kb) in the same Dominican family data set and in an independent population-based subcohort.
MATERIALS AND METHODS
Subjects
The family study was derived from the Northern Manhattan Study (NOMAS) and has been described in detail previously [7]. In brief, probands for the family study were selected from Caribbean Hispanic members of NOMAS at high risk for cardiovascular disease based on the following criteria: (1) having a sibling with a history of myocardial infarction or stroke; or (2) having 2 of 3 quantitative risk phenotypes (maximal carotid plaque thickness, left ventricular mass, or homocysteine level above the 75th percentiles in the NOMAS cohort). Families were then enrolled if the proband was able to provide a family history, obtain the family members' permission to contact them, and had at least 3 first-degree relatives able to participate. The probands were initially identified in Northern Manhattan and family members were enrolled in New York at Columbia University as well as in the Dominican Republic (DR) at the Clinicas Corazones Unidos in Santo Domingo. In addition, a subset of 941 (384 Dominicans) subjects in the NOMAS cohort who had carotid measurements and were not members of the Family Study served as an independent validation sample.
Demographic, socioeconomic and risk factor data were collected for all the subjects through direct interview based on the NOMAS instruments [8]. Written informed consent was provided by the participants. The study was approved by the Institutional Review Boards of Columbia University, University of Miami, the National Bioethics Committee, and the Independent Ethics Committee of Instituto Oncologico Regional del Cibao in the DR.
Carotid Plaque Phenotypes
High-resolution B-mode 2-dimensional ultrasound was performed for the examination of carotid plaque according to the standard scanning and reading protocols [9]. Carotid bifurcation and internal and common carotid arteries were examined for plaque defined as an area of focal wall thickening >50% greater than surrounding wall thickness in millimeters. Once plaques were detected, in-depth imaging of plaques was performed in long axes and multiple angles. The optimized and normalized images were analyzed offline by automated computerized edge detection system M'Ath (Intelligence in Medical Technologies, Inc, Paris, France) and area of each plaque was measured.. The sum of all plaque areas (mm2) within each subject was calculated and expressed as a total carotid plaque area.
SNP Selection and Genotyping
In a subcohort of nearly 1200 NOMAS subjects, we had genome-wide data available from the Human SNP Array 6.0 chip (AffyMetrix). Among the genotyped individuals, there were 384 DR subjects, thus providing a unique database of SNP allele frequency and linkage disequilibrium (LD) structure in the DR population. Using this database, we selected SNPs with minor allele frequency >0.05 and pairwise LD (r2) <0.8 to cover the one-LOD-unit down critical regions on chromosomes 7q, 11p, 14q, and 15q. Assays for selected SNPs were manufactured on Infinium iSelectHD Custom BeadChips (Illumina Inc). Genotyping results were analyzed using the Illumina GenomeStudio® Genotyping module. To ensure high genotyping quality, samples with a GenTrain score <0.15 were dropped from further analysis. Genotypes on SNPs with a GenCall score > 0.8 were automatically called by the GenomeStudio® Genotyping module, whereas SNPs with lower GenCall scores were manually called. SNPs with <95% call rate and DNA samples with genotyping efficiency <95% were removed from the final statistical analysis in both the family data set and population-based NOMAS DR subset. No SNP was found to be severely deviated from Hardy-Weinberg equilibrium (p>10−3).
Statistical Analysis
Using SOLAR[10], polygenic models for carotid plaque presence and total carotid plaque area were employed to screen for the significant covariates (p<0.10) that would be adjusted for in all final association analyses. Covariates that were tested included age, sex, waist hip ratio (WHR), body mass index (BMI), hypertension, hypercholesterolemia, diabetes, and smoking (pack years). Hypertension was defined as having a history of high blood pressure, anti-hypertensive medication usage, systolic blood pressure ≥140 mmHg, or diastolic blood pressure ≥90 mmHg. Diabetes was defined by having a history of diabetes, anti-diabetic medication usage, or fasting blood sugar ≥126 mg/dL. Hypercholesterolemia was defined by having a history of high cholesterol, lipid-lowering medication usage, or total cholesterol ≥240 mg/dL. Smoking was defined as pack years of smoking and calculated as the number of cigarette packs per day*years smoked.
For the association analysis in the family data set, the Quantitative Transmission-Disequilibrium test (QTDT) was used as the primary analytical tool because it is designed to evaluate association in the presence of linkage without inflating type I error, is robust against population stratification, and is able to use data from all available relatives in a pedigree, which makes it more powerful in analyzing data sets with missing parental data [11]. In addition, family-based association analysis based on generalized estimating equations (GEE) was performed with PBAT to assess consistency of results[12]. QTDT analysis was then performed for each peak in each of two subsets defined by the family-specific LOD score on the trait of interest: one consisted of the families with a positive LOD score and another included the families with a zero or negative LOD score at the peak loci of each of our 4 regions. To correct for the multiple testing in the peak-wide association mapping, we used SOLAR to estimate the effective number of tests based on the dependence (i.e. LD) between SNPs. The peak-wide significance threshold was defined as 0.05/effective number of tests under each linkage peak.
To validate the SNPs showing an association with a plaque phenotype in the family study (p<0.05 for QTDT), we carried out logistic regression for plaque presence and linear regression for plaque area in an independent population-based sample from the NOMAS cohort, using an additive genetic model and adjusting for the same covariates as in the family data set, as well as for the top 2 principal components (PC) of ancestry estimated with EIGENSTRAT [13]. We used the first top 2 PCs for the adjustment because among the 10 top PCs, none of other 8 top PCs showed a plaque area-associated p <0.1 and majority of genetic variation can be explained by the first and second principal components [14]. We also explored the potential function of the associated SNPs using FuncPred, one of a set of web-based tools for SNP information (SNPinfo Web Server at http://snpinfo.niehs.nih.gov/index.html).
RESULTS
Association Mapping in the Whole Family Dataset
Among the four regions identified in our linkage study, the one-LOD-unit-down region extends from 152 to 156 megabases (Mb) on chromosome 7q (4Mb total), from 8 to 25 Mb on chromosome 11p (17Mb), from 82 to 101 Mb on chromosome 14q (19 Mb), and from 68 to 93 Mb on chromosome 15q (25Mb). To fine map these linkage regions, 4048 highly informative tagSNPs were selected for genotyping. 306 SNPs failed manufacturing and 30 SNPs did not pass genotyping quality controls. As a result, 3712 SNPs (100, 1190, 1304, and 1118 on chromosomes 7q, 11p, 14q, and 15p respectively) were available for the family-based association analysis. The number of effective independent tests is 80, 558, 598, and 536 for each linkage region, respectively. Overall, 1306 subjects from the 100 DR families had complete genotype and phenotype data for analysis.
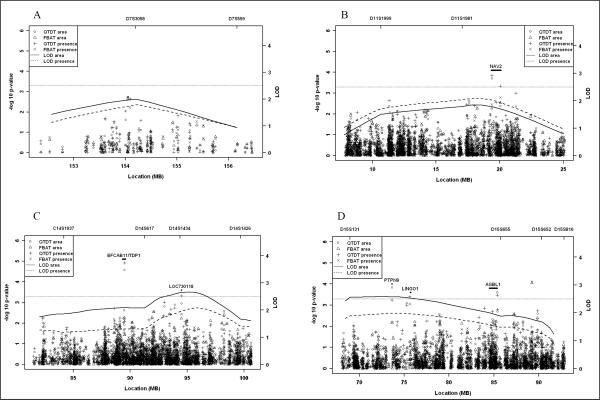
As all four regions had a LOD score >2.00 for at least one plaque phenotype (plaque area or plaque presence) and LOD scores >1.50 for both plaque phenotypes, we evaluated the associations to both plaque phenotypes within each region (Figure 1). Table 1 displays the top associated SNPs (p <0.0005 for QTDT for either plaque phenotype) in the fine mapping family study. On chromosome 11p, two SNPs (rs2702663, intronic; rs1442710, coding-synonymous) in neuron navigator 2 (NAV2) gene were associated with both plaque presence and area. On chromosome 14q, the most significant association with both plaque presence (p=0.00001, multiple testing corrected p=0.006) and plaque area (p=0.00003, multiple testing corrected p=0.02) was found for an intergenic SNP (rs4143999) between tyrosyl-DNA phosphodiesterase 1 (TDP1) and EF-hand calcium binding domain 11 (EFCAB11) genes. Another association was seen for rs10144811 near a hypothetical gene LOC730118 to plaque presence. On chromosome 15q, the top three associations were found for rs16939720 in ATP/GTP binding protein-like 1 (AGBL1), rs11635996 in protein tyrosine phosphatase, non-receptor type 9 (PTPN9), and rs12902898 near leucine rich repeat and Ig domain containing 1(LINGO1). The results from PBAT also supported these associations (Table 1).
Figure 1.
Maximum multipoint linkage and fine mapping association results for carotid plaque on chromosome 7q (A), 11p (B), 14q (C) and 15 (D). Multipoint LOD score curves over the one-LOD unit down region are displayed as a solid line for carotid plaque area and a dash line for presence of carotid plaque. Fine mapping association results of quantitative transmission-disequilibrium test (QTDT) and family-based association test (FBAT) are represented as a circle symbol for plaque area QTDT, a triangle symbol for plaque area FBAT, a plus symbol for plaque area, and a times symbol for plaque area FBAT.
Table 1.
SNPs Associated With Plaque Presence or Area With a QTDT Test P Value < 0.0005
| Region | SNP | Position | Nearby Gene† | SNP Function | Minor Allele | Allele Frequency | Plaque Presence P Values* |
Plaque Area P Values* |
||
|---|---|---|---|---|---|---|---|---|---|---|
| QTDT | PBAT | QTDT | PBAT | |||||||
| 11p | rs2702663 | 19371742 | NAV2 | intronic | G | 0.12 | 0.00014 | 0.00348 | 0.00019 | 0.00538 |
| rs1442710 | 20046520 | NAV2 | cds-syn | C | 0.29 | 0.00046 | 0.00163 | 0.00417 | 0.00292 | |
| 14q | rs4143999 | 89491246 | EFCAB11/TDP1 | intergenic | T | 0.23 | 0.00001 ‡ | 0.00743 | 0.00003 ‡ | 0.00533 |
| rs10144811 | 94499493 | LOC730118 | intergenic | A | 0.40 | 0.00047 | 0.00356 | 0.00109 | 0.00589 | |
| 15q | rs11635996 | 73641787 | PTPN9 | intronic | A | 0.08 | 0.00058 | 0.00655 | 0.00014 | 0.03210 |
| rs12902898 | 75636245 | LINGO1 | intergenic | G | 0.20 | 0.00040 | 0.05350 | 0.00089 | 0.05260 | |
| rs16939720 | 85358908 | AGBL1 | intronic | A | 0.09 | 0.00034 | 0.04160 | 0.00025 | 0.00652 | |
SNP: single nucleotide polymorphism; QTDT: quantitative transmission disequilibrium test; PBAT: pedigree-based association test.
Bold for a QTDT p value < 0.0005.
The distance from the gene <100 kb.
Bonferroni-corrected p value < 0.05 based on the effective test number.
On chromosome 7q, although no association was detected with QTDT p<0.0005 (Figure 1), six SNPs in dipeptidyl-peptidase 6 (DPP6) showed an association with QTDT p<0.05 (supplemental table), including rs3807218 (QTDT p=0.002 for plaque presence and 0.006 for plaque area) at a splicing site in a coding region.
Association Tests in the Families with and without Linkage Evidence
In our original linkage analysis, not all families showed evidence for linkage to these regions, suggesting that some families may carry genetic variants outside these regions that contribute to the inter-individual plaque variations. To further reduce genetic heterogeneity, we performed a subset analysis using families with and without evidence for linkage based on family-specific LOD scores at each locus. For each of the loci, roughly half of the 1306 individuals were in families contributing LOD≥0 (Table 2). In the subset of families with evidence for linkage, despite a smaller sample size (about half of the overall analysis), the associations became more significant between rs2702663 in NAV2 and plaque presence (p=0.00005) and between the two SNPs in AGBL1 and PTPN9 and both plaque phenotypes (p=0.00001~0.00008). In the subset of families without evidence for linkage, the association for these SNPs were not significant (p=0.17~0.76) (Table 2). Therefore, the evidence for association to these SNPs was mainly driven by the families with evidence for linkage. In contrast, rs1442710 in NAV2, rs4143999 near EFCAB11/TDP1, rs10144811 near LOC7311, and rs12902898 near LINGO1 showed an association (p ≤ 0.05) in both subsets. A stronger association was found in the subset of families with linkage evidence, suggesting that variants at or linked to these SNPs may contribute to carotid plaque variations in broader populations.
Table 2.
Subset Association Analysis of the Top SNPs in Dominican Families With and Without Evidence for Linkage
| Plaque Presence QTDT P Values* |
Plaque Area QTDT P Values* |
|||||
|---|---|---|---|---|---|---|
| Region | SNP | Nearby Gene | Families With Linkage Evidence† | Families Without Linkage Evidence‡ | Families With Linkage Evidence† | Families Without Linkage Evidence‡ |
| N=614 | N=691 | N=607 | N=699 | |||
| 11p | rs2702663 | NAV2 | 0.00005 § | 0.17026 | 0.00065 | 0.09430 |
| rs1442710 | NAV2 | 0.00974 | 0.01647 | 0.05616 | 0.02645 | |
| N=698 | N=608 | N=761 | N=545 | |||
| 14q | rs4143999 | EFCAB11/TDP1 | 0.00155 | 0.00305 | 0.00240 | 0.03080 |
| rs10144811 | LOC730118 | 0.00296 | 0.05338 | 0.00474 | 0.01934 | |
| N=690 | N=616 | N=672 | N=634 | |||
| 15q | rs11635996 | PTPN9 | 0.00003 § | 0.59487 | 0.00008 § | 0.32063 |
| rs12902898 | LINGO1 | 0.00422 | 0.02608 | 0.00307 | 0.06903 | |
| rs16939720 | AGBL1 | 0.00006 § | 0.76357 | 0.00001 § | 0.59568 | |
SNP: single nucleotide polymorphism; QTDT: quantitative transmission disequilibrium test, N: number of individuals.
Bold for a QTDT p value < 0.0005.
Family-specific LOD score > 0.
Family-specific LOD score ≤ 0.
Bonferroni-corrected p value < 0.05 based on the effective test number.
Replication in the NOMAS Subcohort
In the QTDT analysis of 3,712 SNPs in the family data set, a total of 263 SNPs showed an association with carotid plaque or presence (Supplemental Table) with a p value <0.05. We further evaluated the association of these SNPs with carotid plaque in our independent NOMAS subcohort. Table 3 presents the replicating SNPs for a plaque phenotype with p < 0.05 in the NOMAS subcohort (n=941) and ≤0.10 in the NOMAS DR subset (n=384). Although no replication was found for the top associated SNPs listed in Table 1, multiple replications in the independent sample were found for SNPs within/near the same three genes (NAV2, EFCAB11 and AGBL1) listed in Table 1 and DPP6 on 7q. In addition, replications were found for SNPs within/near other genes, including 4 genes showing a replication with a p <0.005 in the whole NOMAS subcohort: GALNTL4, (UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4), LOC730217, EFTUD1(elongation factor Tu GTP binding domain containing 1) and C15orf38-AP3S2 (Read-through transcription between the neighboring chromosome 15 open reading frame 38 and adaptor-related protein complex 3, sigma 2 subunit).
Table 3.
Replication of SNPs With QTDT P Value <0.05 in NOMAS (P <0.05) and NOMAS DR Subset (P ≤0.10)
| Region | SNP | Position | Nearby Gene | SNP Function | Carotid Plaque | P Values |
||
|---|---|---|---|---|---|---|---|---|
| DR Families (QTDT) | NOMAS Subcohort |
|||||||
| All | DR Subset | |||||||
| 7q | rs12537575 | 153608357 | DPP6 | intronic | area | 0.023 | 0.018 | 0.035 |
| rs12537575 | 153608357 | DPP6 | intronic | presence | 0.020 | 0.021 | 0.098 | |
| 11p | rs7935223 | 11552909 | GALNTL4 | intronic | presence | 0.038 | 0.001 | 0.003 |
| rs12275977 | 11589215 | GALNTL4 | intronic | presence | 0.010 | 0.046 | 0.051 | |
| rs884272 | 12412257 | PARVA | intronic | presence | 0.029 | 0.018 | 0.086 | |
| rs11605946 | 19503602 | NAV2 | intronic | area | 0.041 | 0.025 | 0.031 | |
| rs7109727 | 20823009 | NELL1 | intronic | area | 0.040 | 0.047 | 0.051 | |
| 14q | rs1667540 | 84742976 | intergenic | presence | 0.030 | 0.025 | 0.054 | |
| rs7148298 | 88914604 | FOXN3 | intronic | presence | 0.033 | 0.011 | 0.036 | |
| rs765031 | 89463274 | EFCAB11 | intronic | area | 0.022 | 0.043 | 0.107 | |
| rs1384981 | 97030127 | LOC730217 | intronic | presence | 0.013 | 0.004 | 0.018 | |
| 15q | rs7162859 | 79147092 | MESDC1 | flanking | presence | 0.028 | 0.035 | 0.032 |
| rs11638444 | 79321874 | IL16 | intronic | presence | 0.025 | 0.020 | 0.044 | |
| rs8039027 | 79343326 | IL16 | intronic | presence | 0.017 | 0.023 | 0.013 | |
| rs1969090 | 80175005 | EFTUD1 | flanking | area | 0.028 | 0.001 | 0.026 | |
| rs1969090 | 80175005 | EFTUD1 | flanking | presence | 0.031 | 0.002 | 0.062 | |
| rs1444298 | 84398616 | AGBL1 | flanking | area | 0.042 | 0.013 | 0.026 | |
| rs3784752 | 87254791 | MFGE8 | intronic | area | 0.025 | 0.040 | 0.037 | |
| rs3784752 | 87254791 | MFGE8 | intronic | presence | 0.012 | 0.047 | 0.052 | |
| rs3853638 | 88244143 | C15ORF38-AP3S2 | intronic | area | 0.009 | 0.040 | 0.055 | |
| rs3853638 | 88244143 | C15ORF38-AP3S2 | intronic | presence | 0.015 | 0.006 | 0.033 | |
| rs3853640 | 88244723 | C15ORF38-AP3S2 | intronic | area | 0.007 | 0.027 | 0.061 | |
| rs3853640 | 88244723 | C15ORF38-AP3S2 | intronic | presence | 0.006 | 0.001 | 0.019 | |
DR: Dominican Republic; SNP: single nucleotide polymorphism; QTDT: quantitative transmission disequilibrium test.
DISCUSSION
In this follow up study of our previously identified linkage peaks, several peak-wide significant associations were found between four SNPs in or near five genes (NAV2, EFCAB11/TDP1, AGBL1, PTPN9) and carotid plaque in the family-based association analysis of overall 100 Dominican families or the subset of the families with evidence for linkage. In addition, multiple replications were observed for the SNPs in/near NAV2, EFCAB11, AGBL1, and multiple other genes in an independent community-based subcohort that is mainly composed of Caribbean Hispanics. To our knowledge, the current study presents the first effort to follow up the linkage regions and fine map the genes that influence carotid plaque burden in Caribbean Hispanics.
Among the four regions, the most significant association was found for rs4143999 with both plaque area and presence on 14q, a region reported to be linked to coronary artery calcification by two other family studies [15, 16]. SNP rs4143999 is located within a regulatory element (as suggested by histone acetylation marker) between the 5' end of EFCAB11 and TDP1. EFCAB11 contains 3 EF-hand domains and is probably involved in muscle relaxation via its calcium-binding activity [17]. TDP1 is involved in repairing stalled topoisomerase I-DNA complexes by catalyzing the hydrolysis of the phosphodiester bond between the tyrosine residue of topoisomerase I and the 3-prime phosphate of DNA. TDP1 may also remove glycolate from single-stranded DNA containing 3-prime phosphoglycolate, suggesting a role in repair of free-radical mediated DNA double-strand breaks [18]. Mutations in/near TDP1 and EFCAB11 are associated with neurodegenerative diseases [19, 20] and type II diabetes [21]. Interestingly, rs10144811, with the second strongest association on 14q, is also located in a transcription factor binding site. These observations suggest a potential role of these associated SNPs in transcriptional regulation of the nearby genes by altering the regulatory element. Moreover, in this region, replication was found for an EFCAB11 SNP rs765031 and rs7148298 in forkhead box N3 (FOXN3) gene (Table 3), a member of the forkhead/winged helix transcription factor family [22]. Genetic variations in FOXN3 have been complicated with serum cholesterol levels [23], type II diabetes mellitus [24], and Alzheimer's disease [25].
On chromosome 11p, the two strongest associations fell at rs2702663 and rs1442710 in NAV2. The association (rs11605946) for NAV2 was also extended to our replication sample of a more generalized population (Table 3). Functionally, NAV2 encodes a member of the neuron navigator gene family, which may involve in cellular growth and migration. Animal models indicated that NAV2 functions in mammalian nervous system development, and is necessary for normal cranial nerve development and blood pressure regulation in the adult [26]. NAV2 polymorphisms may cause abnormalities in the baroreceptor reflex leading to poor response to acute changes in blood pressure and development of atherosclerosis.
Evidence for association with carotid plaque on chromosome 15q was marked by three SNPs in AGBL,1, LINGO1, and PTPN9. Among them, AGBL1 and LINGO1 have been reported to be associated with neurodegenerative diseases. Proteins encoded by AGBL1 are expected to have multiple molecular functions (binding, metal ion binding, metallopeptidase activity, metallocarboxypeptidase activity, zinc ion binding) and the variations in AGBL1 are associated with migraine and schizophrenia [27, 28]. The protein encoded by LINGO1 is an NGR1 binding partner and a component of the NGR1/NGFR signaling complex [29]. Variations in LINGO1 are associated with neurodegenerative diseases such as Parkinson disease and essential tremor [30, 31]. The protein encoded by PTPN9 is a member of the protein tyrosine phosphatase (PTP) family, which are signaling molecules that regulate a variety of cellular processes, including cell growth, differentiation, mitotic cycle, neuronal activation, platelet activation, and insulin signaling [32]. The variation near PTPN9 was found to be associated with adult height [33] and Alzheimer's disease [25].
On chromosome 7q, the most consistent evidence for association with carotid plaque was found for the SNPs in DPP6. Variants in this gene have been implicated in amyotrophic lateral sclerosis (ALS) [34], narcolepsy [20], Alzheimer's disease [25], familial idiopathic ventricular fibrillation [35], coronary artery disease (CAD) [21], and type I diabetes [36] in several GWAS. Expression studies have also showed a 20-fold increase of DPP6 mRNA levels in the myocardium of risk-haplotype carriers as compared to controls [35], suggesting that DPP6 is an attractive candidate for neurodegenerative and vascular disorders.
Some genes that were less significant in the family data set (0.0005<p<0.05) but replicated in the NOMAS, may be interesting because of their biological functions and association with neurodegenerative, vascular and metabolic diseases found in GWAS of large populations. For example, the protein encoded by GALNTL4 is a member of the large subfamily of glycosyltransferases, participates in metabolic pathways, and plays a role in insulin stimulated glucose transport in muscle. The variations in GALNTL4 have been implicated in narcolepsy [20], Alzheimer's disease [25], diabetes [37] and subclinical brain infract [38]. PARVA (parvin, alpha) is highly expressed in many tissues and has a role in cell adhesion, motility and survival. Variations in PARVA have been associated with CAD [21], heart failure mortality [39], sleep and circadian phenotypes [40], and narcolepsy [20]. The protein encoded by IL16 (interleukin 16) is a modulator of T cell activation, and its variations have been implicated in CAD [41] and fibrinogen levels [23].
There are several implications in the present study. First, the majority of the genetic variants associated with carotid plaque reside in regions harboring genes implicated in vascular and neurodegenerative diseases. Although atherosclerosis is an established mechanism underlying cardiovascular diseases and stroke, the nature of the connection between atherosclerosis and neurodegenerative disorders remains less clear. One explanation is that neurodegenerative diseases could be related to atherosclerosis as they may share the same vascular risk factors. Another explanation is that atherosclerosis and neurodegeneration are independent but share convergent disease processes [42], given their shared epidemiology and pathophysiological elements with aging. Understanding of the genetic basis for these diseases will provide new promise toward the research for their prevention and treatment in the elderly individuals with vascular risk factors. Second, none of our top SNPs in QTDT analysis largely explained the observed linkage peaks although the LOD score for plaque presence on 14q dropped by 0.39 in the linkage analysis conditional on rs4143999 and rs10144811 (p=0.04 for likelihood test, data not shown), suggesting that there are multiple genetic variants, each with a small effect, contributing to the linkage peak. In addition, with denser coverage of 18,904 SNPs in these four regions, we previously found other associations in/near LOC730217, EFTUD1 and AP3S2 [3]. Our tagSNP approach may also miss both rare and common variants responsible for the detected linkage. Therefore, future studies via in-depth re-sequencing of these regions using the extremes of unexplained variation of plaque area as the phenotype, an approach described by Lanktree et al[43], are needed to uncover both rare and common functional variants underlying the variations in carotid plaque burden. Third, some top associations were mainly driven by the families with evidence for linkage, highlighting the need for prioritizing the informative families for deeper sequencing to maximize the probability to capture both common and rare causative mutations.
CONCLUSION
Our follow-up study has mapped multiple candidate genes for carotid plaque in Dominican families, showing some genetic effects within a specific population and others in a more generalized population. Many of these genes have been implicated in cardiovascular and neurodegenerative diseases suggesting overlapping pathways. Future studies on various atherosclerotic phenotypes (IMT versus plaque area) via in-depth re-sequencing of these regions are needed to uncover both rare and common functional variants that influence the susceptibility to atherosclerosis.
Supplementary Material
Acknowledgements
The authors are grateful to the study participants for their collaboration and to all staff of the Northern Manhattan Study and Family Study for their energetic efforts to this study and in particular Edison Sabala and Janet DeRosa.
This research was supported by grants from the National Institute of Neurological Disorders and Stroke (R01 NS40807, RO1 NS047655, K24 NS 062737) and Evelyn F. McKnight Brain Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have no conflicts of interest to disclose regarding this work.
REFERENCES
- 1.Inaba Y, Chen JA, Bergmann SR. Carotid plaque, compared with carotid intima-media thickness, more accurately predicts coronary artery disease events: A meta-analysis. Atherosclerosis. 2011 doi: 10.1016/j.atherosclerosis.2011.06.044. 2011 Jun 2030. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Spence JD, Rundek T. Toward Clinical Applications of Carotid Ultrasound: Intima-Media Thickness, Plaque Area, and Three-Dimensional Phenotypes. Springer-Verlag London Ltd; London: 2012. [Google Scholar]
- 3.Dong C, Beecham A, Slifer S, Wang L, Blanton SH, Wright CB, Rundek T, Sacco RL. Genomewide linkage and peakwide association analyses of carotid plaque in Caribbean Hispanics. Stroke. 2010;41(12):2750–2756. doi: 10.1161/STROKEAHA.110.596981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt KJ, Duggirala R, Goring HH, Williams JT, Almasy L, Blangero J, O'Leary DH, Stern MP. Genetic basis of variation in carotid artery plaque in the San Antonio Family Heart Study. Stroke. 2002;33(12):2775–2780. doi: 10.1161/01.str.0000043827.03966.ef. [DOI] [PubMed] [Google Scholar]
- 5.Bis JC, Kavousi M, Franceschini N, Isaacs A, Abecasis GR, Schminke U, Post WS, Smith AV, Cupples LA, Markus HS, et al. Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nature genetics. 2011;43(10):940–947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laird NM, Lange C. Family-based methods for linkage and association analysis. Adv Genet. 2008;60:219–252. doi: 10.1016/S0065-2660(07)00410-5. [DOI] [PubMed] [Google Scholar]
- 7.Sacco RL, Sabala EA, Rundek T, Juo SH, Huang JS, DiTullio M, Homma S, Almonte K, Lithgow CG, Boden-Albala B. Design of a family study among high-risk Caribbean Hispanics: the Northern Manhattan Family Study. Ethn Dis. 2007;17(2):351–357. [PMC free article] [PubMed] [Google Scholar]
- 8.Sacco RL, Khatri M, Rundek T, Xu Q, Gardener H, Boden-Albala B, Di Tullio MR, Homma S, Elkind MS, Paik MC. Improving global vascular risk prediction with behavioral and anthropometric factors. The multiethnic NOMAS (Northern Manhattan Cohort Study) J Am Coll Cardiol. 2009;54(24):2303–2311. doi: 10.1016/j.jacc.2009.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rundek T, Arif H, Boden-Albala B, Elkind MS, Paik MC, Sacco RL. Carotid plaque, a subclinical precursor of vascular events: the Northern Manhattan Study. Neurology. 2008;70(14):1200–1207. doi: 10.1212/01.wnl.0000303969.63165.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. American journal of human genetics. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. American journal of human genetics. 2000;66(1):279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. American journal of human genetics. 2004;74(2):367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 14.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science (New York, NY. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 15.Lange LA, Lange EM, Bielak LF, Langefeld CD, Kardia SL, Royston P, Turner ST, Sheedy PF, 2nd, Boerwinkle E, Peyser PA. Autosomal genome-wide scan for coronary artery calcification loci in sibships at high risk for hypertension. Arterioscler Thromb Vasc Biol. 2002;22(3):418–423. doi: 10.1161/hq0302.105721. [DOI] [PubMed] [Google Scholar]
- 16.O'Donnell CJ, Cupples LA, D'Agostino RB, Fox CS, Hoffmann U, Hwang SJ, Ingellson E, Liu C, Murabito JM, Polak JF, et al. Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI's Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S4. doi: 10.1186/1471-2350-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nature genetics. 2004;36(1):40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 18.Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, Shiloh Y, Pommier Y. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009;28(23):3667–3680. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434(7029):108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 20.Miyagawa T, Kawashima M, Nishida N, Ohashi J, Kimura R, Fujimoto A, Shimada M, Morishita S, Shigeta T, Lin L, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nature genetics. 2008;40(11):1324–1328. doi: 10.1038/ng.231. [DOI] [PubMed] [Google Scholar]
- 21.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pati D, Keller C, Groudine M, Plon SE. Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol Cell Biol. 1997;17(6):3037–3046. doi: 10.1128/mcb.17.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strachan DP, Rudnicka AR, Power C, Shepherd P, Fuller E, Davis A, Gibb I, Kumari M, Rumley A, Macfarlane GJ, et al. Lifecourse influences on health among British adults: effects of region of residence in childhood and adulthood. Int J Epidemiol. 2007;36(3):522–531. doi: 10.1093/ije/dyl309. [DOI] [PubMed] [Google Scholar]
- 24.Florez JC, Manning AK, Dupuis J, McAteer J, Irenze K, Gianniny L, Mirel DB, Fox CS, Cupples LA, Meigs JB. A 100K genome-wide association scan for diabetes and related traits in the Framingham Heart Study: replication and integration with other genome-wide datasets. Diabetes. 2007;56(12):3063–3074. doi: 10.2337/db07-0451. [DOI] [PubMed] [Google Scholar]
- 25.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature genetics. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNeill EM, Roos KP, Moechars D, Clagett-Dame M. Nav2 is necessary for cranial nerve development and blood pressure regulation. Neural Dev. 2010;5:6. doi: 10.1186/1749-8104-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ligthart L, de Vries B, Smith AV, Ikram MA, Amin N, Hottenga JJ, Koelewijn SC, Kattenberg VM, de Moor MH, Janssens AC, et al. Meta-analysis of genome-wide association for migraine in six population-based European cohorts. Eur J Hum Genet. 2011;19(8):901–907. doi: 10.1038/ejhg.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13(6):570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, Levesque M, Allaire N, Perrin S, Sands B, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7(3):221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 30.Inoue H, Lin L, Lee X, Shao Z, Mendes S, Snodgrass-Belt P, Sweigard H, Engber T, Pepinsky B, Yang L, et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson's disease models. Proc Natl Acad Sci U S A. 2007;104(36):14430–14435. doi: 10.1073/pnas.0700901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefansson H, Steinberg S, Petursson H, Gustafsson O, Gudjonsdottir IH, Jonsdottir GA, Palsson ST, Jonsson T, Saemundsdottir J, Bjornsdottir G, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nature genetics. 2009;41(3):277–279. doi: 10.1038/ng.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu M, Warshawsky I, Majerus PW. Cloning and expression of a cytosolic megakaryocyte protein-tyrosine-phosphatase with sequence homology to retinaldehyde-binding protein and yeast SEC14p. Proc Natl Acad Sci U S A. 1992;89(7):2980–2984. doi: 10.1073/pnas.89.7.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, Ohmiya H, Daigo Y, Yamamoto K, Kubo M, Nakamura Y, et al. A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum Mol Genet. 2010;19(11):2303–2312. doi: 10.1093/hmg/ddq091. [DOI] [PubMed] [Google Scholar]
- 34.van Es MA, van Vught PW, Blauw HM, Franke L, Saris CG, Van den Bosch L, de Jong SW, de Jong V, Baas F, van't Slot R, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nature genetics. 2008;40(1):29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- 35.Alders M, Koopmann TT, Christiaans I, Postema PG, Beekman L, Tanck MW, Zeppenfeld K, Loh P, Koch KT, Demolombe S, et al. Haplotype-sharing analysis implicates chromosome 7q36 harboring DPP6 in familial idiopathic ventricular fibrillation. American journal of human genetics. 2009;84(4):468–476. doi: 10.1016/j.ajhg.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature genetics. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer ND, McDonough CW, Hicks PJ, Roh BH, Wing MR, An SS, Hester JM, Cooke JN, Bostrom MA, Rudock ME, et al. A genome-wide association search for type 2 diabetes genes in african americans. PLoS One. 2010;7(1):e29202. doi: 10.1371/journal.pone.0029202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debette S, Bis JC, Fornage M, Schmidt H, Ikram MA, Sigurdsson S, Heiss G, Struchalin M, Smith AV, van der Lugt A, et al. Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke. 2010;41(2):210–217. doi: 10.1161/STROKEAHA.109.569194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison AC, Felix JF, Cupples LA, Glazer NL, Loehr LR, Dehghan A, Demissie S, Bis JC, Rosamond WD, Aulchenko YS, et al. Genomic variation associated with mortality among adults of European and African ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2010;3(3):248–255. doi: 10.1161/CIRCGENETICS.109.895995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottlieb DJ, O'Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Wang Y, Zhang Y, Li L. Association between interleukin-16 polymorphisms and risk of coronary artery disease. DNA Cell Biol. 2011;30(5):305–308. doi: 10.1089/dna.2010.1145. [DOI] [PubMed] [Google Scholar]
- 42.Casserly I, Topol E. Convergence of atherosclerosis and Alzheimer's disease: inflammation, cholesterol, and misfolded proteins. Lancet. 2004;363(9415):1139–1146. doi: 10.1016/S0140-6736(04)15900-X. [DOI] [PubMed] [Google Scholar]
- 43.Lanktree MB, Hegele RA, Schork NJ, Spence JD. Extremes of unexplained variation as a phenotype: an efficient approach for genome-wide association studies of cardiovascular disease. Circ Cardiovasc Genet. 2010;3(2):215–221. doi: 10.1161/CIRCGENETICS.109.934505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.