Abstract
Pairing limited proteolysis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) to probe clostridial collagenase collagen binding domain (CBD) reveals the solution dynamics and stability of the protein, as these factors are crucial to CBD effectiveness as a drug-delivery vehicle. MS analysis of proteolytic digests indicates initial cleavage sites, thereby specifying the less stable and highly accessible regions of CBD. Modulation of protein structure and stability upon metal binding is shown through MS analysis of calcium-bound and cobalt-bound CBD proteolytic digests. Previously determined X-ray crystal structures illustrate that calcium binding induces secondary structure transformation in the highly mobile N-terminal arm and increases protein stability. MS-based detection of exposed residues confirms protein flexibility, accentuates N-terminal dynamics, and demonstrates increased global protein stability exported by calcium binding. Additionally, apo- and calcium-bound CBD proteolysis sites correlate well with crystallographic B-factors, accessibility, and enzyme specificity. MS-observed cleavage sites with no clear correlations are explained either by crystal contacts of the X-ray crystal structures or by observed differences between Molecules A and B in the X-ray crystal structures. The study newly reveals the absence of the βA strand and thus the very dynamic N-terminal linker, as corroborated by the solution X-ray scattering results. Cobalt binding has a regional effect on the solution phase stability of CBD, as limited proteolysis data implies the capture of an intermediate-CBD solution structure when cobalt is bound.
Introduction
Activation of clostridial collagenases by a physiological calcium concentration is a contributing agent for gas gangrene [1]; however, the collagenolytic activity is also beneficial for treatment of Dupuytren’s disease and removal of dead tissue from burns and ulcers [2,3]. Mature collagenases from Clostridium histolyticum are divided into two classes, class I (ColG) and class II (ColH), and are characterized by a differing segmental domain structure consisting of a large N-terminal gluzincin catalytic domain, one or two polycystic kidney disease (PKD) domains, and one or two C-terminal collagen binding domains (CBD) [1,4]. A single CBD is the minimal unit responsible for targeting collagenase to the C-terminus of a mini-collagen in a unidirectional fashion [5]. Without CBD, the enzyme acts as a gelatinase that only cleaves soluble, non-triple helical collagen.
High resolution X-ray crystal structures of the 13.8kDa CBD (apo- and calcium-bound) [6] reveal that the protein crystallizes as two molecules (Molecules A and B) per asymmetric unit and that each molecule consists of a ten β-sheet core region. The core region is flanked with a 14 amino acid N-terminal arm region that is a calcium sensor thought to alter the thermodynamic stability of CBD, cause domain rearrangement, and operate as an on/off switch for collagenolysis [7]. This on/off switch allows CBD to serve as a drug-delivery system regulated by the presence of a physiological calcium concentration. One molecule of the apo-CBD crystal structure (Molecule B) shows that the N-terminal arm possesses α-helical structure and is highly mobile compared to calcium-bound CBD (Fig. 1a,b), in which the arm region in both molecules is a fixed parallel β-sheet that wraps around the face of the protein, hugging the protein core and coordinating with the two calcium ions present per molecule. The binding of calcium to the protein dramatically increases the stability of the already very stable apo- protein due to the stabilizing effect of the secondary structural transition of the N-terminal arm from a highly mobile region to an inflexible β-sheet.
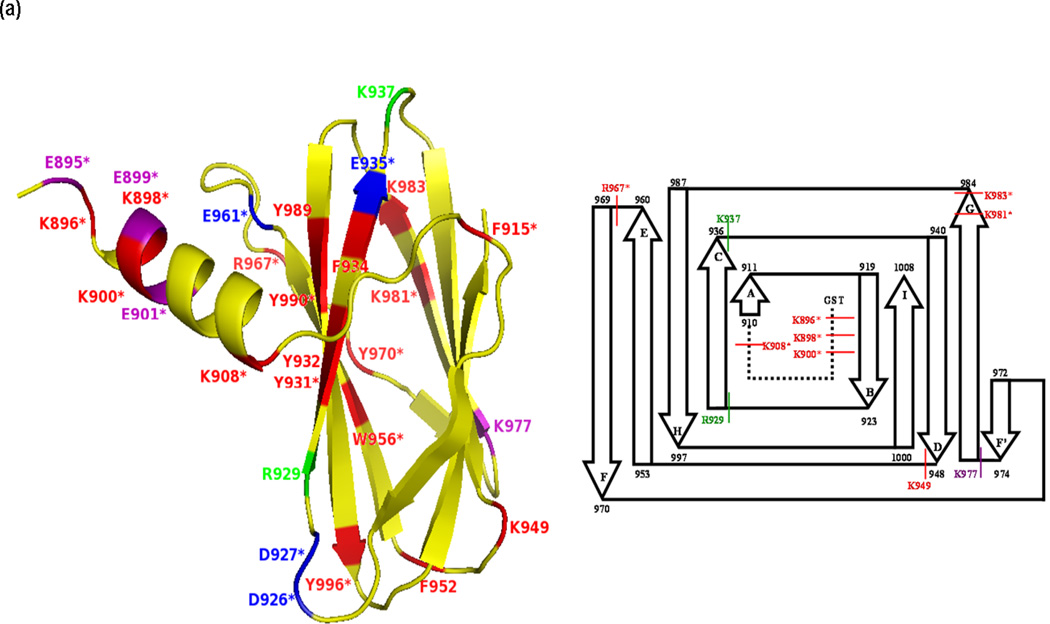
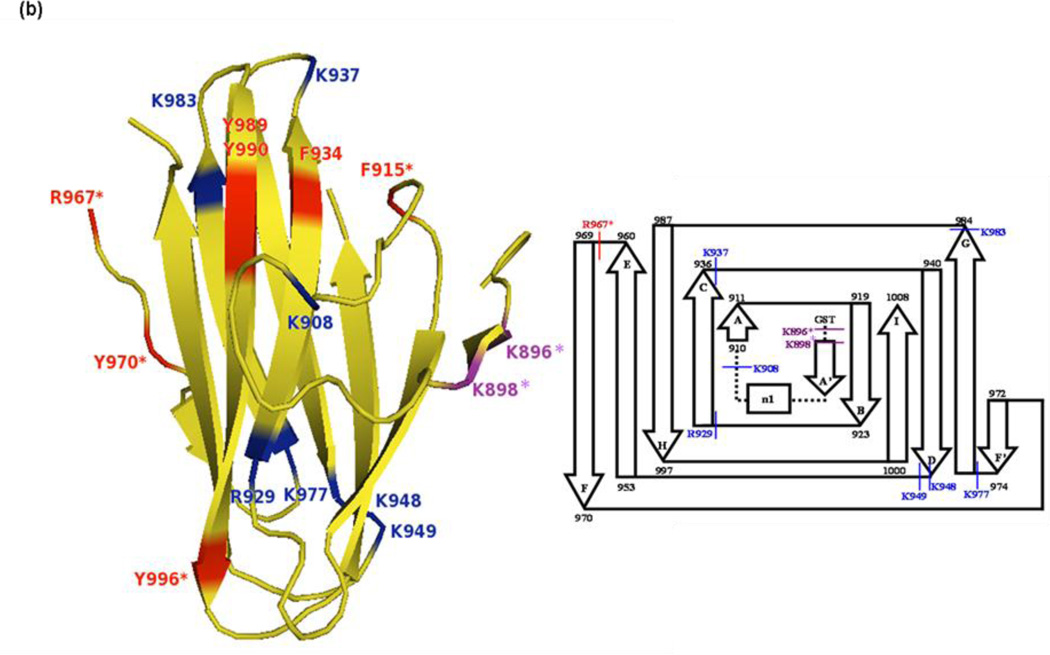
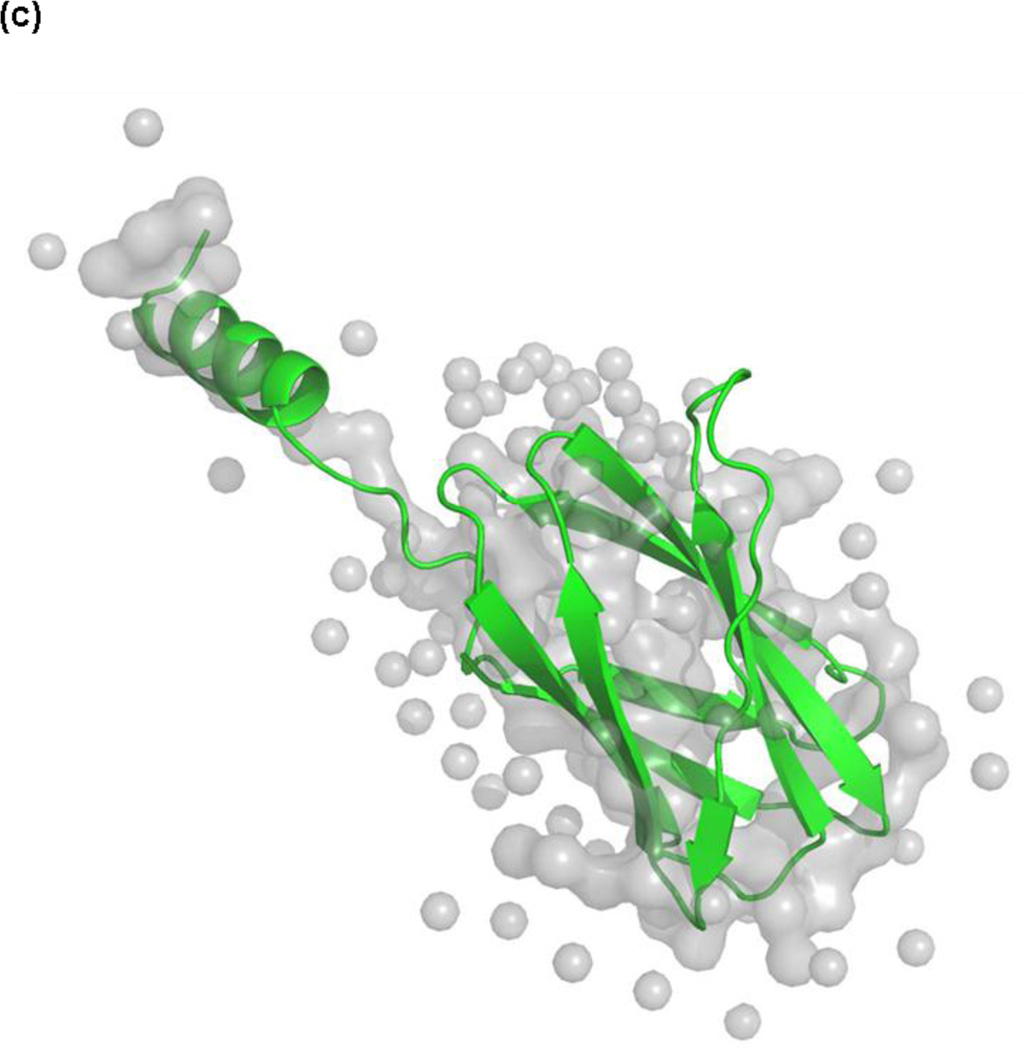
Fig. 1.
X-ray crystal structures of Molecule B for (a) apo-CBD and (b) calcium-bound CBD and (c) the X-ray scattering solution phase structure and the best fit for Molecule B of apo-CBD. Proteolytic cleavages determined by MALDI-TOF MS primarily occurred in loops and unstructured regions of the protein, as seen in the labeled crystal structures and in the corresponding inserts showing a comparison of cleavage sites for 1:100 trypsin digests of (a) apo- and (b) calcium-bound CBD. Labeled residues in (a) and (b) show points of cleavage for trypsin, chymotrypsin, and Glu-C digests at t=0 (red), t=10min (green), t=20–30min (purple), and t=50min and beyond (blue), where starred (*) residues denote primary cleavage sites determined by MS analysis. Structures were rendered using The PyMOL Molecular Graphics System, Version 1.4, Schrödinger, LLC
The in vitro stability of CBD has been proposed to be therapeutically beneficial. Clostridial CBD is being tested as a novel drug-delivery vehicle for various growth factors [8]. Cytokines and growth factors are often easily diffused and typically have short half-lives in vivo; therefore, linking these molecules to a CBD renders them non-diffusible and increases their half-lives [6,8–11]. Consequently, these fusion proteins are potentially excellent therapeutic agents for drug-delivery systems [8,12,13]. Understanding the behavior of CBD with respect to calcium binding and collagen interaction is essential to the development of therapeutic agents by rational drug design.
The pairing of matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis with limited proteolysis studies to map the surface of proteins with unknown three-dimensional structures is becoming an invaluable technique in the rapidly growing proteomics field [14–17]. Even if the X-ray crystal structure of a protein is known, there is an ambiguity in the correlation of the X-ray structure to the solution structure. While X-ray structures provide detailed information, crystal packing interactions can impact the three-dimensional structure. Methods such as NMR, H/D exchange mass spectrometry (HXMS) [18],[19,20] and chemical modifications followed by MS [21] can be designed to obtain similar information; however, NMR data is difficult to interpret for regions with correlated mobility. Compared to NMR, HXMS and chemical modification mass spectrometry do not require large amounts of protein. Alternatively, limited proteolysis coupled with mass spectrometry, particularly MALDI-TOF MS, is easy to execute while maintaining high sensitivity in monitoring proteolytic cleavage of proteins in solution[17,22,23]. Despite that accepted methods of limited proteolysis have been established, unanswered questions remain as to why a protease preferentially cleaves at one site versus another site of a protein [24]. For this reason, increased attention has been given to the properties of proteases, including their cleavage patterns and specificities. Previous limited proteolysis studies have concluded that proteases disfavor cleavage in more stable areas of proteins exhibiting secondary structure, such as helical chains and β-sheets, and favor cleavage of native proteins in unstructured regions, such as flexible loops [25] [26–28]. Cleavage in preferred locations has been associated with the mobility of the target protein’s side-chains as indicated by crystallographic B-factors (or temperature factors [29]), exposure, accessibility, and protrusion [30,31]; however, even these factors do not reportedly account for all proteolytic events [14,32]. Despite discrepancies in correlating proteolytic cleavage sites with protein and/or protease characteristics, limited proteolysis, often coupled with other spectroscopic and biophysical techniques, has been proven effective to investigate the structure-function relationships, conformational features, topology, and stability of biomolecules in their native or partially unfolded states [26,27,33–38] in the solution phase.
To probe the solution structure of CBD and the dynamics of the N-terminal arm in the presence and absence of calcium, limited proteolytic fragments of apo- and calcium-bound CBD, obtained using trypsin, chymotrypsin, or endopeptidase Glu-C, were analyzed using MALDI-TOF MS. Cleavage sites were mapped to the respective previously determined X-ray crystal structures (Fig. 1a, b) and were correlated with the B-factors and the surface accessibility obtained from the crystal structures, as well as enzyme specificity. Crystallographic B-factors, or Debye-Waller temperature factors, provide information about the motional disorder of a protein, including the static or dynamic mobility of each backbone and side-chain atom, and serve as a measure of uncertainty of atomic position (or scattering) in the crystal structure[39]. The more an atom, or group of atoms, vibrates and oscillates, the higher the B-factor will be for the mobile area; therefore, a protein’s solvent exposed side-chain atoms typically exhibit greater mobility than the main-chain atoms, thus resulting in higher B-factors. Additionally, in light of recent growing interest in the application of X-ray scattering techniques to biomolecules in solution, small angle X-ray scattering (SAXS) analysis of CBD was employed, despite the limited resolution afforded by SAXS, to further educe solution phase structural and dynamic information, thereby affording a comparison of MS limited proteolysis results of the apo- protein to the corresponding SAXS-derived structure of apo-CBD (Fig. 1c). CBD proteolytic digests in the presence of cobalt, a known modulator [40–44], and in the presence of both cobalt and calcium were also analyzed using MALDI-TOF MS and were compared to apo- and calcium-bound CBD proteolysis results.
Experimental
Protein Expression and Purification
Native C. histolyticum class I ColG CBD (residues N894-K1008, numbering from mature collagenase) was expressed as a glutathione S-transferase (GST)-fusion protein and purified as previously reported [4]. Purification of the protein revealed that seven amino acids (GSPGIPG) of the GST-fusion tag remained at the N-terminus of the protein after thrombin cleavage. The CBD was prepared in a 50mM Tris-HCl pH 7.5 buffer containing 100mM NaCl.
Limited Proteolysis
Apo-CBD and holo-CBD forms (calcium-bound, cobalt-bound, and calcium- and cobalt-bound CBD) were digested using proteases with varying substrate specificities. Trypsin (Sigma-Aldrich), immobilized chymotrypsin (Princeton Separations), and endopeptidase Glu-C (Calbiochem) were individually used to digest CBD. All proteases were prepared using 50mM Tris-HCl pH 7.5 buffer containing 100mM NaCl. Proteolytic digestions were performed at 25°C with enzyme:protein ratios of 1:100 and 1:1000 for trypsin digests, 1:100, 1:500, and 1:1000 for immobilized chymotrypsin digests, and 1:10 for Glu-C digests. For holo-CBD studies, CBD was incubated with 0.005M CaCl2 and/or CoCl2 for 30 minutes prior to introduction of protease to ensure calcium and/or cobalt binding. All CaCl2 and CoCl2 solutions were prepared using the same protein and protease buffer. Each digest lasted at least four hours (t=4hr), and data were collected every ten minutes for the first hour and every hour for the next three hours. Additional data were collected for time-points beyond t=4hr for some digest experiments. Data for each digest were also collected for the time-point t=0, indicating that once the protease had been added to the protein, a sample of the proteolytic digest reaction was immediately removed and quenched by rapid, thorough mixing with an equal volume of 1M 2,5-dihydroxybenzoic acid (DHB) matrix prepared in 90:10 MeOH/Water 0.1% TFA. Minor differences in the procedure for digestions using immobilized chymotrypsin, particularly with respect to quenching the protease, were necessary due to the solid bead support covalently bonded to the protease; therefore, the procedure recommended in the technical notes and procedure guide (Princeton Separations EN-261) provided with the protease was used in conjunction with the quenching procedure used for the other proteases.
Two forms of control samples were introduced for each digest as necessary references to differentiate between background peaks and peaks generated from cleaved fragments in the MS spectra. One control included data for each digest collected for the native protein sample before adding the protease. A comparison of peaks from the intact, undigested CBD protein (control samples) to peaks from digested CBD allowed for the identification of background peaks and noise, thus resulting in a more accurate analysis of peaks representing fragments produced during CBD digestion. The second control involved only the individual proteases. Proteases are capable of auto-digestion during the cleavage of other proteins. To discriminate CBD data from protease auto-digestion data, parallel protease auto-digestion experiments were concurrently conducted with the corresponding CBD experiments. Each protease was allowed to undergo autolysis (in conditions of apo- and in the presence of calcium, cobalt, or calcium and cobalt) using the same protease concentration, buffer, and proteolysis methods as used in the previously described digestions of CBD, and data were collected at the same time-points as for the corresponding protein digests. Comparison of MS peaks due to protease auto-digestion to peaks from the CBD cleavage allowed for discernment of peaks representing true fragments that evolved from CBD digestion.
MALDI-TOF Mass Spectrometry
Each quenched CBD digest sample was spotted onto a Bruker MTP 384 ground stainless steel MALDI target plate and allowed to air dry. Samples from each digestion reaction were analyzed using a Bruker Reflex III (Bruker Daltonic GMbH, Bremen, Germany) MALDI-TOF mass spectrometer. The theoretical m/z values corresponding to in silico proteolytic digest products were obtained using the Protein Prospector server (Baker, P.R. and Clauser, K.R. http://prospector.ucsf.edu) and the Bruker Daltonics sequence editor 3.1 software using the CBD sequence. These m/z values were then searched both manually and automatically in the acquired MALDI-TOF MS spectra using the Bruker BioTools 3.2 software.
Calculations for Cleavage Probability Determined by Solvent Accessibility, Side-chain Mobility, and Protease Specificity
The Protein Data Bank (PDB) structure coordinates for apo- and calcium-bound CBD (1NQJ and 1NQD, respectively, as identified in PDB) were submitted to the Dictionary of Secondary Structure of Proteins (DSSP) server [45] to calculate the solvent accessibility for each residue in both molecules (Molecules A and B) of the apo- and calcium-bound X-ray crystal structures (NCBI Accession/gi numbers: 1NQJ_A/157879415, 1NQJ _B/157879416, 1NQD_A/30749689, 1NQD_B/30749690 for apo- Molecule A, apo- Molecule B, calcium-bound Molecule A, and calcium-bound Molecule B, respectively). The DSSP values, given in terms of solvent exposure for the entire residue, were then used to calculate the accessibility of each residue’s specific side-chain based on values for the accessible surface areas of the residue’s total atoms and its side-chain atoms as previously determined for a Gly-X-Gly tripeptide [46]. The coordinate files for CBD X-ray crystal structures also provided B-factors (measure of the flexibility and mobility) for all atoms in each molecule of apo- and calcium-bound CBD. The reported B-factors were averaged for the side-chain for each residue in each molecule for both the apo- and calcium-bound structures. Effective B-factor and side-chain accessibility values in the highly mobile N-terminal arm region (Fig. 1a) were most likely underestimated due to additional rotational motion relative to the core of the protein as described in more detail in the Results and Discussion section of the manuscript.
The specificities (%) for trypsin and chymotrypsin were determined by entering the CBD sequence into the Expasy Peptide Cutter server (http://us.expasy.org/tools/peptidecutter). This server generated a list showing the percent probability of either tryptic or chymotryptic cleavage for each residue in CBD, assuming a linear protein chain exhibiting no secondary structure, based solely on protease specificity. No probabilities were obtained based on Glu-C specificity because the server did not provide probability values based on Glu-C cleavage.
The relative probability of proteolytic cleavage at specific CBD residues was calculated for each residue in each molecule (Molecules A and B) by multiplying the values of the relative (percent) accessibility, the averaged B-factor, and the protease specificity (for trypsin or chymotrypsin) for each residue’s side-chain, giving equal weight to all factors, as described in the formula: Relative Cleavage Probability Per Residue = %Side-Chain Accessibility × Side-Chain B-factor × %Protease Specificity. The calculated cleavage probability for each residue was then normalized using calculated relative probabilities for all CBD residues to determine an overall value (on a scale of 0–1, where 0 reflected no cleavage is expected to occur and 1 reflected the residue most susceptible to proteolytic cleavage) indicating the probability of primary cleavage (defined in the following section) for each CBD residue. The normalized cleavage probabilities were graphically plotted for each residue of both molecules of apo- and calcium-bound CBD.
Small Angle X-ray Scattering (SAXS)
The SAXS data were collected for samples of apo-CBD in 10mM Tris-HCl pH 7.5 buffer containing 100mM NaCl at the Advanced Photon Source (APS) XOR beamline of sector 12-ID at the Advanced Photon Source (APS) in Argonne National Laboratory. All data collection and analysis, calculations, and modeling used in the determination of the apo-CBD X-ray scattering solution phase structure were conducted as previously reported [5].
Results and Discussion
MS Determination of Cleavage Sites from CBD Digest Fragments
Each peak in mass spectrum corresponding to a particular proteolytic digest time-point for either apo-, calcium-bound, or cobalt-bound CBD provided data for the determination of primary cleavage sites and subsequent cleavage sites. Primary cleavage sites were determined to be the initial points of protease attack from which all subsequent digest fragments were generated. Primary cleavage sites were typically detected as single cleavage sites. Single cleavage sites were indicated by two peaks on the same time-point spectrum corresponding to two fragments that add up (both in mass and corresponding sequence) to the intact, parent CBD protein (see Fig. 1a–e in Supplemental Data). However, not all primary cleavage sites were indicated by single cleavages and two peaks. In some instances, primary cleavage sites were determined by one or more peaks corresponding to masses of digest fragments that indicated specific points of cleavage but that did not add (in mass or sequence) to total the intact protein (see Fig. 1a–e in Supplemental Data). Because of the differences in the protein affinity of the two initial fragments, their ions in the mass spectra were not of equal intensity. Also, the initial fragments could rapidly disappear because of facile secondary cleavage. Compilation of all fragments detected in all time-points for the same digest allowed for the creation of maps (not shown) indicating the primary cleavage sites of the protein and the most probable lineage of each peptide fragment detected during the overall digest. MS determination of the primary cleavage sites and the possible subsequent cleavage sites from which other fragments descend allowed for the prediction of the most accessible cleavage sites in the protein.
The following sections in the manuscript are divided into subsections to provide as much detailed information as possible from the observed MS data. The protein scheme for CBD shown as insets in Fig. 1a,b (also in Supplemental Data Fig. 2a,b) is first used to compare MS results for trypsin (Fig. 2), Glu-C (Fig. 3), and chymotrypsin (Fig. 4) to the obvious secondary structure features seen in the X-ray crystal structures of apo- and calcium-bound CBD. Except for Glu-C, this leads to the discussion of MS data using more detailed correlation maps created using flexibility/mobility (B-factor) and solvent accessibility for CBD residues in Molecules A and B and enzyme specificity for each protease. It is believed that these results demonstrate that there are clear differences between the solution phase and crystal structures. The fact that there are structural differences between Molecules A and B in the X-ray crystal structure provides a benchmark for the differences to expect in the solution phase CBD structure. This is also evident from the SAXS-derived structure (Fig. 1c) and is further used to correlate MS data.
Fig. 2.
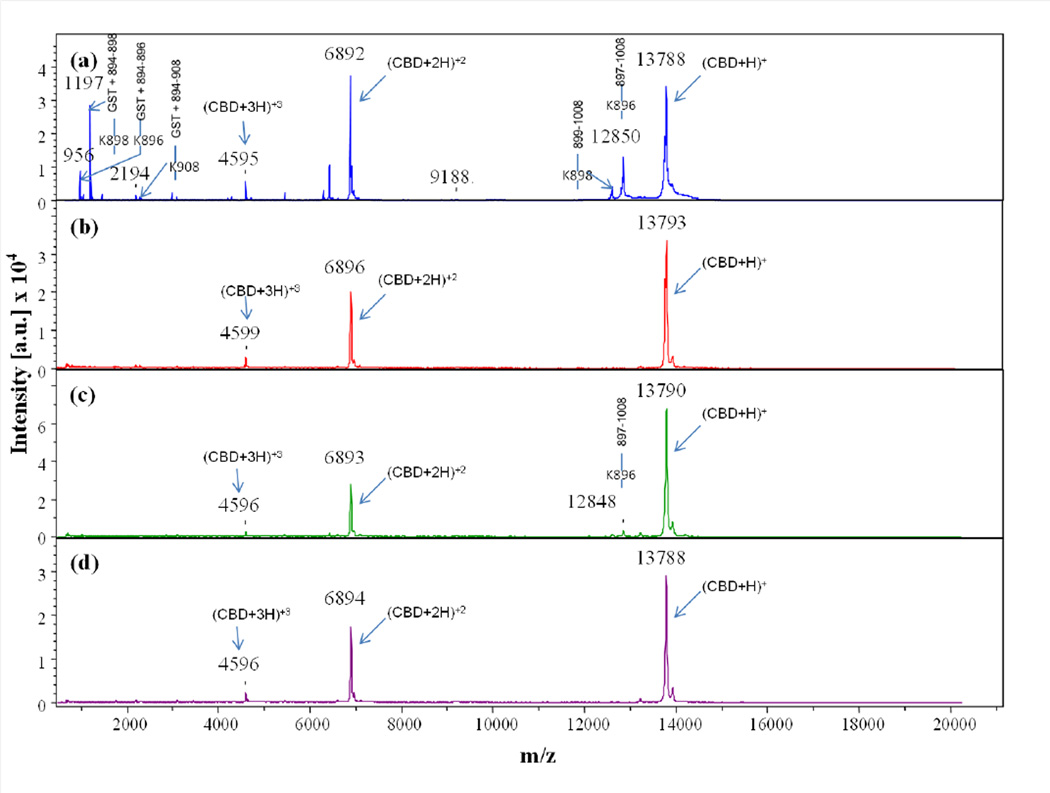
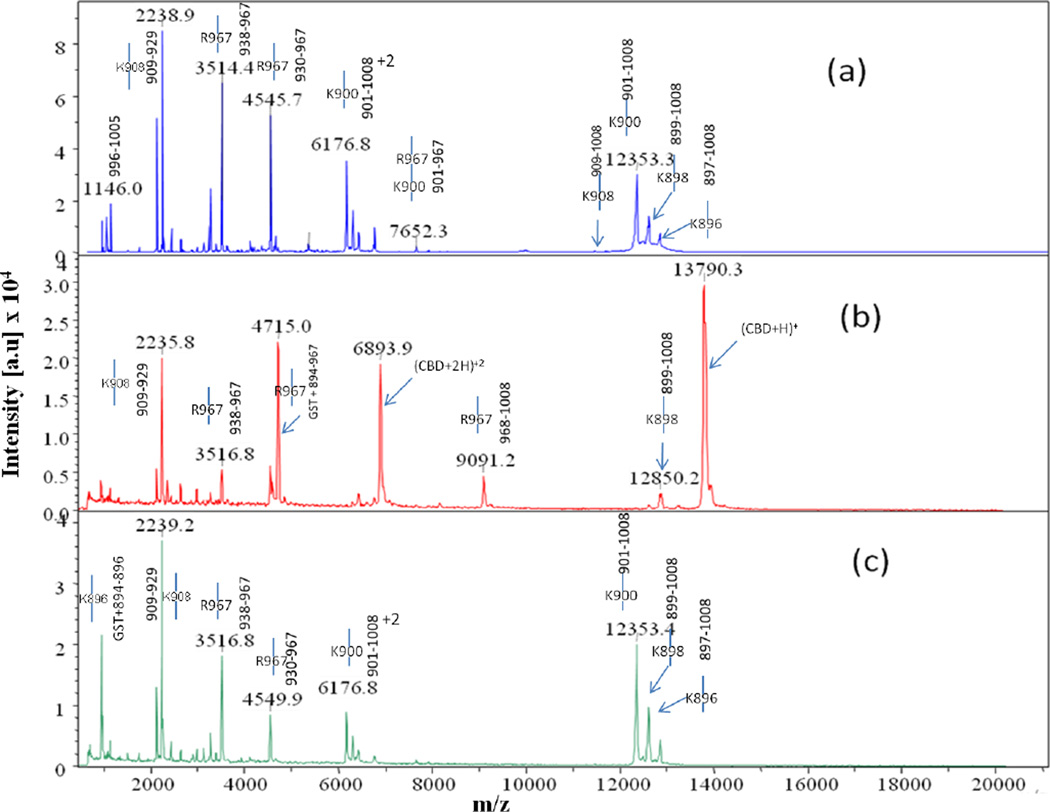
Comparison of MALDI-TOF MS spectra collected at time point t = 0 for trypsin digests of (a) apo-, (b) calcium-bound, (c) cobalt-bound, and (d) calcium- and cobalt-bound CBD. Full-length CBD construct is GST tag + 894–1008, where the GST tag is the seven residue peptide (GSPGIPG) remaining after thrombin cleavage of the GST-CBD fusion protein
Fig. 3.
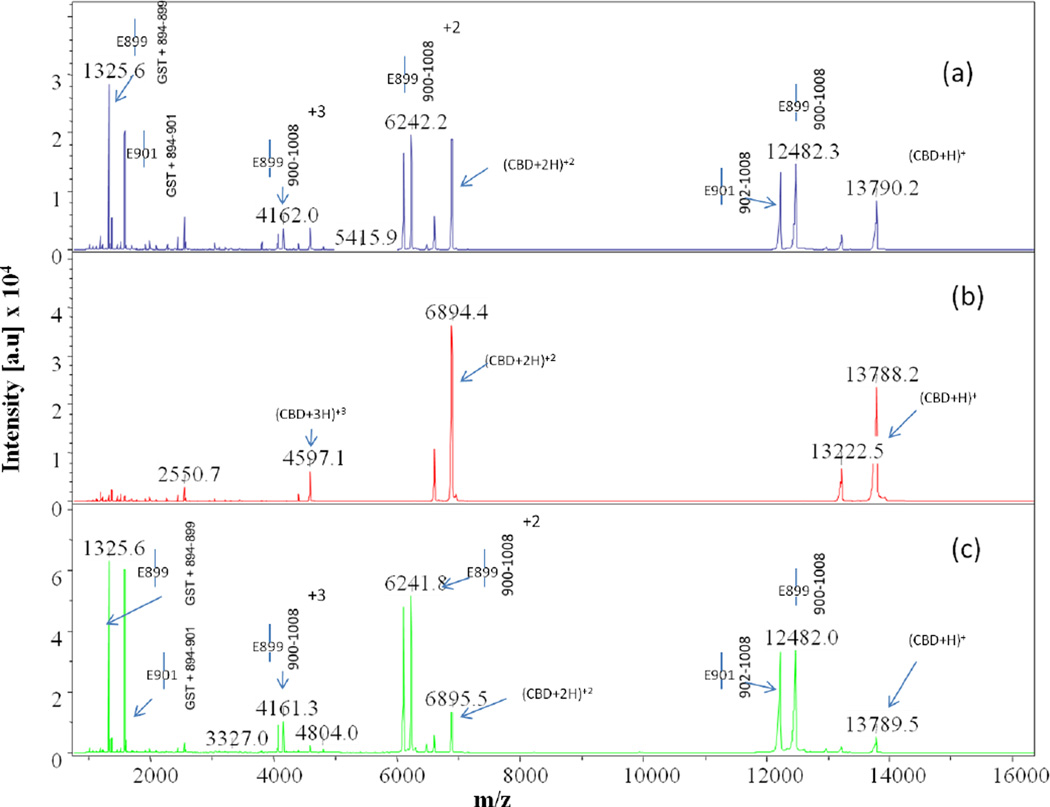
Comparison of MALDI-TOF MS spectra collected at time point t = 4hr for trypsin digests of (a) apo-, (b) calcium-bound, and (c) cobalt-bound CBD. Full-length CBD construct is GST tag + 894–1008, where the GST tag is the seven residue peptide (GSPGIPG) remaining after thrombin cleavage of the GST-CBD fusion protein
Fig. 4.
Comparison of MALDI-TOF MS spectra collected at time point t = 3hr for Glu-C digests of (a) apo-, (b) calcium-bound, and (c) cobalt-bound CBD. Spectrum (b) reveals intense m/z peaks at 13788, 6894, and 4597 representing the intact parent peak and the corresponding doubly and triply charged peaks, respectively, clearly showing that no digestion of the protein occurs when calcium is bound. The peaks next to these labeled intact peaks represent the protein minus the GST tag
Time-course Analysis
Comparison of the time-course data for each protease digestion provided insight into the stability of CBD. Analysis of the digests also afforded significant information about regional structural changes and dynamics of the protein in solution (in apo- and holo- forms).
Analysis of Trypsin Digests
Time-course cleavage for the trypsin digests of apo-CBD revealed that the protease targets the protruding regions of the protein first (Fig. 1a). As predicted from the apo- crystal structure, the highly mobile N-terminal arm region of the protein is projected to be initially attacked by the protease as determined by intense MS peaks corresponding to single, primary cleavage sites at K896 (m/z 956, 12850), K898 (m/z 1197, 12609), and K900 (m/z 1454, 12352) occurring immediately in the t=0 1:100 (Fig. 2a). Cleavage at residue K908, located before the beginning of the βA strand connecting the N-terminal arm to the protein core (Fig. 1a inset scheme; Supplemental Data Fig. 2a,c), was also quickly targeted by the protease. Although two corresponding t=0 MS peaks indicating K908 as a single cleavage site were not observed, the intensity of the peak representing the N-terminal fragment GST tag through residue K908 (m/z 2344) suggests that primary cleavage occurs at this last residue in the N-terminal arm region connecting it to the core of the protein. A less intense peak corresponding to the cleavage at K908 compared to peak intensities corresponding to other cleavages at lysine arm residues can possibly be explained by the position of K908 in the X-ray crystal structure of the apo- protein.
Other single, primary cleavages occurring in the apo-CBD t=0 trypsin digest include R967 (m/z 9090, 4713), located in a loop region just before the βF strand, and K983 (m/z 10827, 2982), located at the very end of the βG strand. Due to the β-sandwich “jelly-roll” structure of the protein core, these two β-strands are located on each side of the protein (Fig. 1a). Despite that analysis of the apo- crystal structure shows K983 hydrogen bonding with a residue in a neighboring β-sheet, this residue is clearly exposed and susceptible for cleavage in the solution structure of the protein as seen in the MS spectrum for the t=0 digest. Cleavage of the apo- protein also occurred at K981 (m/z 3209) in the t=0 digest. This residue located within the βG strand, but still closer to the end of the βG strand, is most likely a primary cleavage site based on the MS analysis. Since this represents the C-terminal fragment (V982-K1008) of the cleaved protein, it is not a possible product of successive lineage resulting from cleavage at K983. Similar to K981, cleavage at K949 would be likely since it is located at the beginning of a loop region following the βD strand; however, lineage of enzymatic fragments observed in MS digest data suggests that this location is not a primary cleavage site and that cleavage at this residue cannot occur without initial cleavage at R967 and K983.
Time-course cleavage patterns of the cobalt-bound and the calcium-bound CBD digests indicated that metal binding modulates the stability of the CBD structure. The overall MS spectra for the cobalt-bound CBD trypsin digests closely resembled those for the apo-CBD digests (Fig. 2a,c and 3a,c). For example, in the cobalt-bound t=0 trypsin digest, cleavage sites similar to apo- results were observed for residues K896 (m/z 1408, 12848), K898 (m/z 12610; C-terminal fragment), K900 (m/z 1452; N-terminal fragment), and K908 (m/z 2344; N-terminal fragment) in the N-terminal arm of the protein and for K983 (m/z 2983; C-terminal fragment) located at the end of the βG strand. As discussed for the t=0 apo-CBD data, cleavage of the residues in the N-terminal arm emphasizes the free-range of mobility in this region of the protein, thus accentuating its susceptibility to proteolysis. MS data revealing cleavage at residues in the arm region further shows that cobalt does not bind to this area of the protein. However, unlike apo-CBD, residue R967 (m/z 9092, 4714) is not cleaved until t=10min. This delay in cleavage is explained by the nearby cobalt binding site at H959 as seen in the cobalt-bound CBD crystal structure (unpublished data). MS observation of changes in the fragmentation patterns of CBD in the presence of cobalt probably suggests that cobalt binds to stabilize omega loop 960–968 enough to see a delay in the observation of peaks corresponding to cleavage at R967. The stability of the CBD protein due to cobalt binding is also evident by the comparison of the MS data for the t=0 digestion of apo-CBD to the 1:1000 trypsin digest of cobalt-bound CBD (results not shown).
The MS data for the calcium-bound CBD trypsin digests drastically differ from the apo- CBD and from the cobalt-bound CBD digests (Fig. 2 and 3). The only cleavage fragments observed from t=0 to t=40min for calcium-bound digests correspond to cleavage sites at R967 (m/z 9087, 4712; t=0), K896 (m/z 12851, 956; t=30min), and K898 (m/z 12608, 1197; t=30min). All of these cleavage sites were observed at low intensities compared to apo- or cobalt-bound CBD (Fig. 2 and 3). The R967 cleavage site, located in a highly flexible loop region, is extremely consistent between the digests of all forms of the CBD protein. This residue is clearly exposed and very vulnerable to proteolytic cleavage despite metal binding. Residue K896 is clearly located outside the newly formed βA’ strand, which is constructed from the highly mobile arm in apo-CBD during the transition from apo- to calcium-bound CBD. The intensity associated with cleavage at K898 is even lower than the corresponding intensity for K896. This is consistent with the fact that K898 is located at the end of the βA’ strand (Fig. 1b). No other cleavage sites are apparent for the calcium-bound CBD digest until t=50min to t=1hr. At these time-points, new cleavage sites at R929 (m/z 2234, 4547), K937 (m/z 3267, 3514), K948 (m/z 6611), K949 (m/z 2128, 3636), K983 (m/z 1746, 1510, 2637, and 2978), and K977 (m/z 3639) result from the previous cleavage at R967. This indicates that when calcium is bound, the protein core remains stable and intact much longer than the apo- or cobalt-bound CBD core. The successive cleavage pattern is similar to that of apo- and cobalt-bound digests; however, the digestion time is significantly delayed due to an increase in stability felt throughout the protein due to calcium binding.
The increased stability exported throughout CBD upon calcium binding is evident even after four hours of trypsin digestion (Fig. 3b). In the MS data for the t=4hr digest of apo-CBD, there is an absence of the parent peak representing native CBD, thus indicating that all CBD molecules have been cleaved at least once, leaving no intact protein (first observed at t=30min). By comparison, the corresponding calcium-bound CBD t=4hr spectrum clearly shows an intense peak for the intact protein (m/z 13793), indicating increased stability upon calcium binding.
MS data from the calcium- and cobalt-bound CBD digest resemble data from the calcium-bound digest (Fig. 2b,d). This clearly demonstrates the global influence of calcium binding over the regional effect of cobalt binding. Analysis of the trypsin limited proteolysis experiments involving cobalt establishes the coordination of cobalt with CBD, particularly with respect to a manner different from calcium binding, and suggests that an intermediate CBD structure between apo- and calcium-bound CBD is captured in solution when cobalt is bound to the protein.
Analysis of Glu-C Digests
MS data for the Glu-C digests of CBD did not reveal the same detailed information regarding time-course cleavage patterns as did the analysis of spectra for the trypsin digests. The first Glu-C digest peaks for apo-CBD were not observed until t=20min (data not shown) of the 1:10 digest. The peaks representing cleavage at N-terminal arm glutamic acid residues in the t=20min digest were very small, and it was not until t=3hr that these peaks were seen with significant intensity (Fig. 4a). This notable difference when compared to intense initial digest peaks observed at t=0 for trypsin digests of apo-CBD at significantly lower enzyme:protein digest ratios is easily explained by the enzyme activity of the Glu-C protease at neutral pH. Initial cleavage sites for apo-CBD digested by Glu-C, however, did correspond well with regions initially cleaved by trypsin. As predicted from the apo- crystal structure, the highly mobile N-terminal arm region of the protein was initially attacked. In addition to lysine residues, the N-terminal arm is also rich with glutamic acid residues. Similar to the N-terminal arm lysine residues concluded to be primary cleavage sites by trypsin limited proteolysis experiments, all glutamic acid residues (E895, E899, and E901) in the N-terminal arm were also determined to be single, primary cleavage sites by analysis of Glu-C digests (m/z 827, 12979; m/z 1326, 12480; and m/z 1583, 12223, respectively). Because Glu-C digestion of the protein, potentially retarded by the suppressed activity of the enzyme, was not observed until a significantly later time-point as compared to other enzyme digestions, the digest was allowed to continue and data were collected at various time-points past the scheduled t=4hr. Further digestion of the apo- protein was eventually observed at cleavage sites D926 (m/z 4206, N-terminal fragment), D927 (m/z 1182, 1538, 2415), E935 (m/z 5368, N-terminal fragment) located at the end of the βC sheet, and E961 (m/z 5414, C-terminal fragment) located in the flexible loop connecting βE and βF (mass spectra not shown). D926, D927, and E961 cleavages are expected since these are located on loops connecting β-sheets. D926 and D927 are located between βB and βC, as seen in Fig. 1a. E961 is located between βE and βF, similar to R967 which was observed in the above trypsin studies. Even though E935 is located at the end of the βC sheet, this residue is still a possible cleavage site because β-sheets could be ill-defined at the end of the strands.
As seen in the trypsin digestion, the Glu-C calcium-bound digest clearly reveals that metal binding modulates the stability of the CBD structure. A comparison of calcium-bound MS spectra to the corresponding apo- spectra reveals that a significant decrease in cleavage occurs when calcium is bound. This further suggests a global stability imparted throughout the protein when the metal is present and indicates that the difference in stability can easily be detected by limited proteolysis. Throughout the entire digest, no peaks representing cleavage of the three N-terminal glutamic acid residues were ever observed (Fig. 4b), thereby indicating that the arm region is no longer mobile as indicated in the calcium-bound X-ray structure. In the crystal structure, the arm forms a β-sheet as it wraps around and hugs the protein core. It is likely that the arm region adopts the same secondary structure in solution when calcium is bound. The X-ray crystal structure of calcium-bound CBD shows the coordination of arm residues E899 and E901 and residue D927 with calcium. No digestion at these sites in the presence of calcium, as observed in the MS data, emphasizes calcium binding in this region of the protein and corroborates the X-ray crystal structure.
Analysis of the MS data for the Glu-C digestion of cobalt-bound CBD revealed that the digest data of the cobalt-bound CBD more closely resembled apo-CBD (Fig. 4a,c) and that the digest spectra of the calcium- and cobalt-bound CBD (not shown) very closely resembled that of calcium-bound CBD, thus reinforcing the structural data and the trypsin proteolysis data by indicating that in solution cobalt may not have high affinity for the calcium binding site. Trypsin data revealed that the binding of cobalt provided localized stability as observed in the delay of cleavage at R967 when comparing apo- and calcium-bound digests. The absence of cleavage in the cobalt-bound Glu-C data at the corresponding E961residue, located on the loop connecting βE and βF in the apo- structure (Fig. 1a), further revealed localized stability in this loop region upon cobalt binding. Additionally, the Glu-C results emphasized that the binding of cobalt does not afford the same global stability to the protein as does the binding of calcium, as was seen by comparing the t=28hr Glu-C spectra for all digestions involving calcium and cobalt (not shown). The comparison clearly indicated the retention of the parent peak (m/z 13788) when calcium is present versus little or no retention of the intact protein during the apo- or cobalt-bound digests, thus reflecting much stronger binding interactions between the protein and calcium.
Analysis of Chymotrypsin Digests
Unlike trypsin and Glu-C, chymotrypsin targets large, hydrophobic residues that are not typically mobile and are generally located in the highly inaccessible hydrophobic core of the protein; therefore, widespread proteolytic digestion of CBD was not expected from the chymotrypsin experiments. While the MS data for CBD chymotrypsin digests supported this proteolytic tendency, the t=0 chymotrypsin digest spectrum for apo-CBD revealed significant information about the relative flexibility of the core region of the protein.
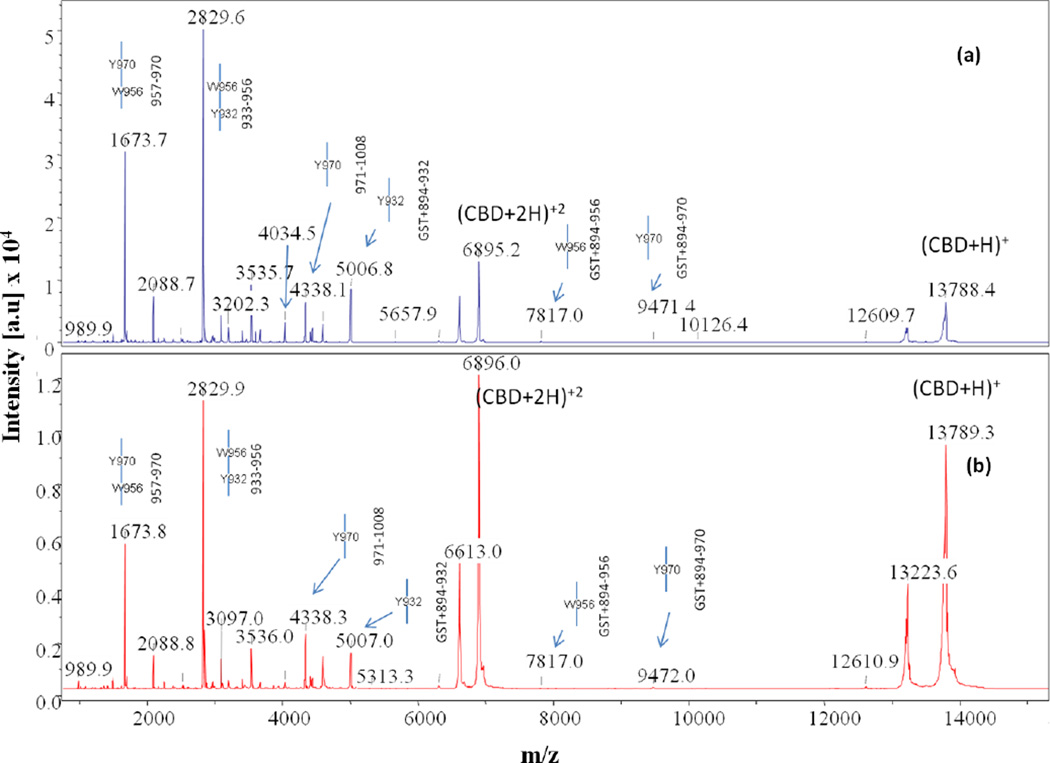
MS data for the t=0 chymotrypsin digest of apo-CBD clearly identified residues Y970 and W956 as a single, primary cleavage sites. According to the apo- crystal structure, Y970 is an exposed residue located at the base of a very short loop between the βF and βF’ sheets and faces the outside of the protein (Fig. 1a). Cleavage at Y970 results in two protein fragments corresponding to intense MS peaks (m/z 9468, 4336) at t=0 (Fig. 5a). The cleaved portion represented by the peak at m/z 4336 is a highly stable fragment, as witnessed in corresponding peaks in spectra throughout the apo- digest from t=0 to t=4hr. Once cleavage at Y970 occurs, the protein is opened, allowing chymotrypsin to digest core residues. The intriguing single, primary cleavage at W956 was also undeniably observed, as witnessed by MS peaks (m/z 7814, 5976) in the t=0 apo-CBD digest. Located on sheet βE, residue W956 is positioned directly behind Y970 and is buried with its side-chain facing the protein core. It is surprising that both fragments indicating single cleavage at W956 were detected in the spectrum since the residue is strongly situated in the assumed impenetrable, hydrophobic core of the protein. According to NMR data [47], the region of residue W956 is the least mobile/least accessible area of the protein in solution. Furthermore, the environment around W956 changes very little despite the presence or absence of calcium as monitored by fluorescence spectrometry [7]. MS data representing single cleavage at W956 is likely explained by the dynamic internal cavities of the apo- protein [7].
Fig. 5.
Comparison of MALDI-TOF MS spectra collected at time point t = 4hr for chymotrypsin digests of (a) apo- and (b) calcium-bound CBD. Full-length CBD construct is GST tag + 894–1008, where the GST tag is the seven residue peptide (GSPGIPG) remaining after thrombin cleavage of the GST-CBD fusion protein
Other primary cleavage sites determined from the t=0 apo-CBD chymotrypsin digest spectrum include residues Y996 (m/z 1562), Y990 (m/z 2251), Y931 (m/z 4841, N-terminal fragment), and F915 (m/z 3087, N-terminal fragment; 2168). Residue Y996 is located at the end of the βH sheet in the same tyrosine-rich region as Y970 is located (Fig. 1a). According to the apo-CBD structure, the side-chain of Y996 is solvent exposed and located in close proximity to Y970, thereby indicating that cleavage is likely to occur quickly at Y996. As with cleavage at Y970, cleavage at Y996 is instrumental in opening CBD and allowing successive cleavage of core residues, such as cleavage at F952 (m/z 6435) located on the βE sheet as indicated by t=0 spectrum. The cleavage at F952 is clearly determined to be a secondary cleavage site by MS, even though it has an MS peak with a reasonably high intensity. According to Wilson et al. [6], mutation analysis reveals that F952 is essential to CBD core packing. Therefore, cleavage at F952 results in disturbance of the core arrangement, thus rendering the core more vulnerable to chymotrypsin proteolytic attack.
Residues Y990 and Y931 are also significant t=0 chymotrypsin primary cleavage sites in apo-CBD that potentially open the CBD core for successive cleavage. Initial cleavages at residues Y990, positioned only a few residues away from Y996 on the βH sheet, and Y931, located on the βC sheet, likely open CBD and create pathways for further proteolytic cleavage at core residues, such as Y989 and Y932. These were determined to be secondary cleavage sites by lineage maps created from MS data. According to the apo- crystal structure, residue F915 is situated at the top of the first loop region after the βA strand and the N-terminal arm and is also determined to be a clear primary cleavage site based on the MS analysis.
As witnessed in the trypsin CBD digests, the binding of calcium to CBD decreases the proteolytic effect of chymotrypsin in the digestion of CBD. In the t=0 MS spectrum for calcium-bound CBD, only three primary cleavage sites are observed: Y970, Y996, and F915. The peaks representing cleavage at Y970 (m/z 4337, C-terminal fragment), Y996 (m/z 1563), and F915 (m/z 3087, N-terminal fragment) at t=0 are also clearly less intense compared to the corresponding apo- spectrum, suggesting that when calcium is bound there is a decrease in the rate of proteolytic digestion and an increase in global stabilization of the protein. For instance, primary cleavage at Y970 occurs at a significantly lower rate when calcium is bound, and the cleavage at the core residue W956 is not observed. The decrease in the proteolytic effect of chymotrypsin due to calcium binding to CBD is further evident by comparing the peak intensities of the intact protein for apo- and calcium-bound CBD (Fig. 5).
An overall increased global stability of the protein upon calcium binding was also confirmed in an MS spectral comparison of apo- versus calcium-bound CBD time-wise chymotrypsin proteolysis digests. No cobalt-bound experiments were conducted using chymotrypsin.
Correlation of Limited Proteolysis with Accessibility and Mobility
Data for plots of residue side-chain accessibility and mobility for each molecule (Molecules A and B) of either the apo- or calcium-bound CBD crystal structure were individually calculated and then compared to the limited proteolysis results as determined by MS (see Supplemental Data, Tables I and II). As expected, the general trend for apo- and calcium-bound CBD showed that when the accessibility of the side-chain for a residue increased, the mobility of the side-chain also increased. After comparing each parameter individually for both molecules in each structure, the plots were then analyzed with respect to residues predicted by MS data to be easily cleaved sites. The predicted sites from the limited proteolysis experiments correlated well with the plots representing accessibility and mobility per residue side-chain (plots not shown; see Supplemental Data, Tables I and II). Any errors in correlating cleavage with the accessibility and mobility were easily explained by either crystal contacts or properties of the protease. For example, based on accessibility and mobility, residue R985 should have been a primary cleavage site for the apo-CBD trypsin digests; however, because a proline (P986) follows this residue, trypsin did not cleave at this site due to the specificity of trypsin.
Probabilities for primary cleavage sites of the CBD protein for the various protease digests were therefore determined using the criteria of relative solvent accessibility, mobility (crystallographic B-factors), and protease specificity (trypsin and chymotrypsin only) for the side-chain of each residue, as explained in the Experimental section. Fig. 6a,b indicates the probability of proteolytic cleavage as a function of the amino acid residues for apo- and calcium-bound CBD. It is important to note that these calculated probabilities are only effective in predicting initial primary cleavage sites because these values are based on the intact protein. Once the protease initially attacks, the protein adopts new conformations and the criteria (i.e. accessibility and B-factors) change for the residues, thereby not affording the prediction of secondary cleavage sites and successive cleavages.
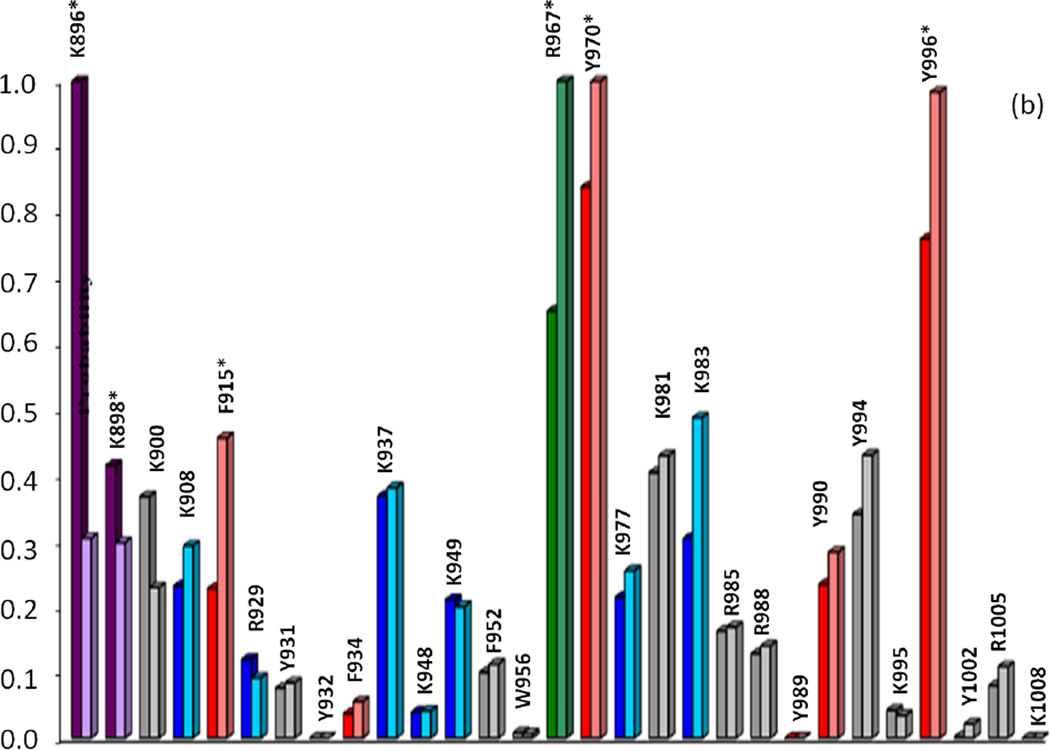
Fig. 6.
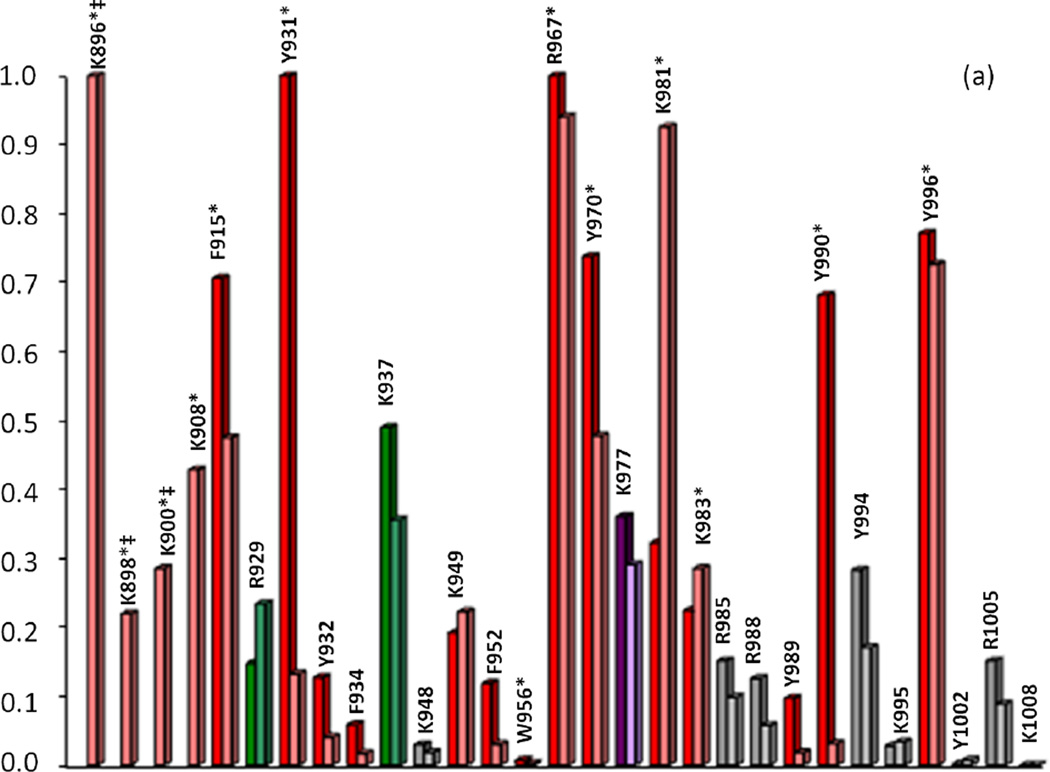
Correlation of calculated primary cleavage probability of CBD residues in Molecules A (left) and B (right) with MS trypsin and chymotrypsin (a, apo-; b, calcium-bound) digest results. Colored bars indicate cleavage at t=0 (red), t=10min (green), t=20–30min (purple), t=50min-1hr (blue), or no observed cleavage (gray). Primary cleavage sites determined by MS are indicated by starred (*) residues. ‡Note: Probability values for residues K896, K988, K900 (a, apo-, trypsin) should be significantly higher than all other residues due to high mobility of the N-terminal arm region, yet the values as shown reflect the underestimated crystallographic B-factors used to calculate probability for these residues. See Tables I and II in Supplemental Data for raw data values
Calculated cleavage probabilities for the trypsin digests of apo- and calcium-bound CBD correlated well with MS analysis of the corresponding digests. As previously discussed in the analysis of the trypsin time-course studies, MS data for the apo-CBD indicated that primary cleavage occurs at three lysine residues (K896, K898, and K900) on the flexible N-terminal arm and at residues R967 and K983. Calculated probabilities for the corresponding digest suggest that R967 and arm residue K896 are the two residues most likely to be the initial primary tryptic cleavage sites in the apo-CBD protein (Fig. 6a). However, when the high mobility of the N-terminal arm region is taken into account, the calculated probabilities of the arm residues should be dramatically higher compared to residues in other regions of the protein. Existence of high mobility in the arm region is evident from comparing Molecule A (where the arm region is unresolved) in the crystal structure to Molecule B (where the arm itself exhibits high mobility). Because the calculated probabilities are based on crystallographic B-factors, the values for K896, K898, and K900 unfortunately misrepresent the highly dynamic N-terminal arm as a static region (see note in Fig. 6 caption). In the crystal structure, the helix that whips around in solution is locked in structure by symmetry related neighboring molecules, thereby radically suppressing the B-factors of the residues located in the arm. Another factor to consider concerning the suppressed B-factors for the N-terminal arm residues is the distance of the arm residue from the core of the protein. Therefore, an extrapolation of decreased stability (thereby increased mobility and higher B-factor) proportional to an increase in distance of residues on the N-terminal arm away from the protein core could be purported. For example, the cleavage at K908 can be explained by a comparison of differences in detail between the X-ray crystal structure and the SAXS-derived structure for apo-CBD. In the crystal structure, residue K908 is not highly exposed and the βA strand bridging K908 to the protein core is structured (Fig. 1a). However, in the SAXS apo- structure, the βA strand appears to be unwound (Fig. 1c), thus increasing the susceptibility of K908 to be cleaved and confirming the vulnerability of this region to proteolytic cleavage as shown in the MS data. MS analysis of the time-course CBD proteolytic fragments revealed the unprecedented absence of the βA strand and confirmed its flexibility in solution. Although there is no proper way to estimate what the B-factors should be for the N-terminal arm residues, these residues are clearly expected to have the highest B-factors, thereby, the highest cleavage probabilities. The apo-CBD t=0 trypsin data (Fig. 2a) clearly suggests that MS high peak intensities corresponding to K896 (m/z 956, 12850), K898 (m/z 1197, 12609), K900 (m/z 1454, 12352), K908 (m/z 2433, 11461), and R967 (m/z 4713, 9091) correlate well with the calculated high cleavage probabilities (Fig. 6a).
Another correlation regarding the crystal structure and the solution behavior of apo-CBD can be detected by comparing the calculated cleavage probabilities for residues K981 and K983. While MS data revealed that primary cleavages occur at K983 and K981, the calculated probabilities indicate that primary cleavage is more probable at K981 than at K983 (Fig. 6a). The specificity value for these residues is equal, yet the overall calculated probability for initial cleavage at K983 versus K981 varies more in Molecule B (which includes the arm region of the protein) than in Molecule A (where the arm region is absent). The most noticeable distinction between residues K983 and K981 is the difference in crystallographic B-factors (see Supplemental Data, Table I). The B-factor for K981 in Molecule A is slightly higher than K983, but is significantly higher for Molecule B, thus explaining the dramatic difference in probabilities calculated for K981 and K983. Low calculated probability for K983 can be attributed to hydrogen bonding based on the X-ray crystal structure. This further clarifies the discrepancy between the calculated probabilities for cleavage at K981/K983. The results witnessed in the MS cleavage data can only be attributed to structural differences between solution and crystal adaptations, thus proving how MS analysis of proteolytic cleavages can be instrumental in detecting or predicting even slight residue variations in orientation when a protein is in solution versus its crystal structures, particularly the residues and regions pertaining to the surface of the protein.
A comparison of calculated probabilities with MS time-course data for the trypsin digest of calcium-bound CBD further highlights how MS analysis successfully explains the likelihood of residues being proteolytically cleaved. As seen in Fig. 6b, residue K896 is predicted to have a higher probability of cleavage than R967 in Molecule A, whereas R967 is predicted to be the initial cleavage site in Molecule B. If probability values for Molecules A and B are averaged for each residue in the protein, residues R967 and K896 are respectively the first and second highest predicted sites for proteolytic cleavage. This is consistent with the MS data obtained for all time-points from t=0 to t=4hr of the calcium-bound trypsin digestion. As seen in Fig. 4, peaks corresponding to cleavage at R967 (m/z 4713, 9091) and K896 (m/z 956, 12850) in the t=4hr digestion of calcium-bound CBD are the most intense. While there is little discrepancy between MS data and calculated probabilities for the predicted initial tryptic cleavage sites of calcium-bound CBD, the MS analysis affords a more resolute order of cleavage, thereby offering a more specific insight into the solution and surface dynamics of the protein.
Similar correlations for calculated cleavage probabilities were also noted for chymotrypsin digestions. Overall, the apo-CBD residues with the highest calculated probabilities for initial cleavage by chymotrypsin are consistent with MS results for the corresponding digest. MS analysis revealed that residues F915, Y931, Y970, Y990, and Y996 are candidates for initial primary cleavage by chymotrypsin, all of which are among the residues with values of the highest calculated probabilities (Fig. 6a). The MS data for primary cleavage at F915 is also supported by the SAXS-derived apo- structure. As discussed in the trypsin results, the SAXS apo-CBD structure suggests that the βA strand is unwound, thereby making this unstructured region a likely target susceptible to proteolytic cleavage. There are, however, some calculated probability differences between Molecules A and B in the crystal structure, as previously discussed. For example, calculated probabilities for cleavage of the apo- protein at residues Y931 and Y990 (Fig. 6a) are higher in Molecule A than Molecule B indicating more solvent accessibility/mobility in Molecule A than in Molecule B of the crystal structure. The difference in probability between molecules was not expected to be reflected in the MS results because the solution phase represents an average of Molecules A and B, as evident from the trypsin studies. MS analysis deemed both residues significant primary cleavage sites in the apoprotein, as previously described.
As seen in Fig. 6b, the residues predicted to be the initial primary cleavage site for calcium-bound CBD during chymotrypsin digestion are Y970, F915, and Y996 for both Molecules A and B. MS data plainly revealed these residues as the definitive initial primary chymotrypsin cleavage sites when calcium is bound.
Conclusions
In this study, the application of MALDI-TOF MS analysis combined with limited proteolysis was demonstrated as a technique to probe and identify dynamic regions in a protein. Results from this method distinguished the structural differences that the presence of calcium or cobalt induce on CBD and identified possible structural variations between crystal and solution structures. Some of these small structural variations were elucidated by comparing MS proteolysis results with the SAXS-derived apo-CBD structure. Cleavage sites predicted by analysis of MS data correlated well with side-chain accessibility, X-ray B-factors, and protease specificity. The calculated cleavage probabilities for residues in Molecules A and B of the X-ray crystal structures are good indications of the uncertainty in flexibility and accessibility to be observed in the corresponding solution phase structure, as was demonstrated by a better correlation of the average probability values to the MS data. Correlation of calculated probabilities with MS data and X-ray crystal structures signified that crystal structures cannot be used alone in determining potential protease cleavage and revealed that the structural dynamics of the protein residues may dictate protease cleavage. Determination of primary cleavage sites by MS was also beneficial in monitoring the solution stability of CBD and the dynamics of the N-terminal arm region of the protein in its transition from apo- to various holo- forms. The SAXS-derived structure of the apo- protein, although much lower in resolution than the corresponding crystal structure, confirmed the absence of the βA strand and the dynamic nature of the N-terminal linker as shown by the MS data. Furthermore, the very flexible collagenase structure adopted inside the clostridial cell could be flexible enough to be able to be secreted. Once secreted in the extracellular matrix of animals, the enzyme may adopt a very rigid structure to proceed in collagenolysis.
Acknowledgments
The authors gratefully acknowledge support, in whole or in part, by the National Institutes of Health Center for Protein Structure and Function Grants NCRR COBRE 1 P20RR15569 and INBRE P20RR16460. This work was also supported by the AR Biosciences Institute (ABI) and a grant-in-aid for scientific research (C) from the Japan Society for the Promotion of Science and Kagawa University Project Research Fund 2005–2006. We thank Dr. Soenke Seifert at the X-ray Science Division, Advanced Photon Source at Argonne National Laboratory for SAXS assistance.
References
- 1.Matsushita O, Jung CM, Katayama S, Minami J, Takahashi Y, Okabe A. Gene duplication and multiplicity of collagenases in Clostridium histolyticum. J. Bacteriol. 1999;181:923–933. doi: 10.1128/jb.181.3.923-933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starkweather KD, Lattuga S, Hurst LC, Badalamente MA, Guilak F, Sampson SP, Dowd A, Wisch D. Collagenase in the treatment of Dupuytren's disease: an in vitro study. J. Hand Surg. [Am] 1996;21:490–495. doi: 10.1016/S0363-5023(96)80368-6. [DOI] [PubMed] [Google Scholar]
- 3.Badalamente MA, Hurst LC. Enzyme injection as nonsurgical treatment of Dupuytren's disease. J. Hand Surg. [Am] 2000;25:629–636. doi: 10.1053/jhsu.2000.6918. [DOI] [PubMed] [Google Scholar]
- 4.Matsushita O, Koide T, Kobayashi R, Nagata K, Okabe A. Substrate recognition by the collagen-binding domain of Clostridium histolyticum class I collagenase. J. Biol. Chem. 2001;276:8761–8770. doi: 10.1074/jbc.M003450200. [DOI] [PubMed] [Google Scholar]
- 5.Philominathan ST, Koide T, Hamada K, Yasui H, Seifert S, Matsushita O, Sakon J. Unidirectional binding of clostridial collagenase to triple helical substrates. J. Biol. Chem. 2009;16:10868–10876. doi: 10.1074/jbc.M807684200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson JJ, Matsushita O, Okabe A, Sakon J. A bacterial collagen-binding domain with novel calcium-binding motif controls domain orientation. EMBO J. 2003;22:1743–1752. doi: 10.1093/emboj/cdg172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Philominathan ST, Matsushita O, Gensure R, Sakon J. Ca2+-induced linker transformation leads to a compact and rigid collagen-binding domain of Clostridium histolyticum collagenase. FEBS J. 2009;276:3589–3601. doi: 10.1111/j.1742-4658.2009.07078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishi N, Matsushita O, Yuube K, Miyanaka H, Okabe A, Wada F. Collagen-binding growth factors: production and characterization of functional fusion proteins having a collagen-binding domain. Proc. Natl. Acad. Sci. U S A. 1998;95:7018–7023. doi: 10.1073/pnas.95.12.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyoshima T, Matsushita O, Minami J, Nishi N, Okabe A, Itano T. Collagen-binding domain of a Clostridium histolyticum collagenase exhibits a broad substrate spectrum both in vitro and in vivo. Connect. Tissue Res. 2001;42:281–290. doi: 10.3109/03008200109016842. [DOI] [PubMed] [Google Scholar]
- 10.Vogel WF. Collagen-receptor signaling in health and disease. Eur. J. Dermatol. 2001;11:506–514. [PubMed] [Google Scholar]
- 11.Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 2002;277:4223–4231. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 12.Ponnapakkam T, Katikaneni R, Miller E, Ponnapakkam A, Hirofumi S, Miyata S, Suva LJ, Sakon J, Matsushita O, Gensure RC. Monthly administration of a novel PTH-collagen binding domain fusion protein is anabolic in mice. Calcif. Tissue Int. 2011;88:511–520. doi: 10.1007/s00223-011-9485-1. [DOI] [PubMed] [Google Scholar]
- 13.Ponnapakkam T, Katikaneni R, Nichols T, Tobin G, Sakon J, Matsushita O, Gensure RC. Prevention of Chemotherapy-Induced Osteoporosis by Cyclophosphamide with a long-acting form of Parathyroid Hormone. J. Endocrinol. Invest. 2011 doi: 10.3275/7864. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard SJ, Beynon RJ, Thornton JM. Assessment of conformational parameters as predictors of limited proteolytic sites in native protein structures. Protein Eng. 1998;11:349–359. doi: 10.1093/protein/11.5.349. [DOI] [PubMed] [Google Scholar]
- 15.Cohen SL, Ferre-D'Amare AR, Burley SK, Chait BT. Probing the solution structure of the DNA-binding protein Max by a combination of proteolysis and mass spectrometry. Protein Sci. 1995;4:1088–1099. doi: 10.1002/pro.5560040607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winston RL, Fitzgerald MC. Mass spectrometry as a readout of protein structure and function. Mass Spectrom. Rev. 1997;16:165–179. doi: 10.1002/(SICI)1098-2787(1997)16:4<165::AID-MAS1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 17.Gheyi T, Rodgers L, Romero R, Sauder JM, Burley SK. Mass spectrometry guided in situ proteolysis to obtain crystals for X-ray structure determination. J. Am. Soc. Mass Spectrom. 2010;21:1795–1801. doi: 10.1016/j.jasms.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liyanage R, Devarapalli N, Puckett LM, Phan NH, Gidden J, Stites WE, Lay JO. Comparison of two ESI-MS based H/D exchange methods for extracting protein folding energies. International Journal of Mass Spectrometry. 2009;287:96–104. doi: 10.1016/j.ijms.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Englander SW, Sosnick TR, Englander JJ, Mayne L. Mechanisms and uses of hydrogen exchange. Curr. Opin. Struct. Biol. 1996;6:18–23. doi: 10.1016/s0959-440x(96)80090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J. Am. Soc. Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams JG, Tomer KB, Hioe CE, Zolla-Pazner S, Norris PJ. The antigenic determinants on HIV p24 for CD4+ T cell inhibiting antibodies as determined by limited proteolysis, chemical modification, and mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:1560–1569. doi: 10.1016/j.jasms.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, Bain K, Bonanno JB, Buchanan M, Henderson D, Lorimer D, Marsh C, Reynes JA, Sauder JM, Schwinn K, Thai C, Burley SK. High-throughput limited proteolysis/mass spectrometry for protein domain elucidation. J. Struct. Funct. Genomics. 2005;6:129–134. doi: 10.1007/s10969-005-1918-5. [DOI] [PubMed] [Google Scholar]
- 23.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 24.Tao L, Kiefer SE, Xie D, Bryson JW, Hefta SA, Doyle ML. Time-resolved limited proteolysis of mitogen-activated protein kinase-activated protein kinase-2 determined by LC/MS only. J. Am. Soc. Mass Spectrom. 2008;19:841–854. doi: 10.1016/j.jasms.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Kazanov MD, Igarashi Y, Eroshkin AM, Cieplak P, Ratnikov B, Zhang Y, Li Z, Godzik A, Osterman AL, Smith JW. Structural determinants of limited proteolysis. J. Proteome Res. 2011;10:3642–3651. doi: 10.1021/pr200271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta. 1998;1382:191–206. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- 27.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta. Biochim. Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- 28.Fontana A, Zambonin M, Polverino de Laureto P, De Filippis V, Clementi A, Scaramella E. Probing the conformational state of apomyoglobin by limited proteolysis. J. Mol. Biol. 1997;266:223–230. doi: 10.1006/jmbi.1996.0787. [DOI] [PubMed] [Google Scholar]
- 29.Fontana A, Fassina G, Vita C, Dalzoppo D, Zamai M, Zambonin M. Correlation between sites of limited proteolysis and segmental mobility in thermolysin. Biochemistry. 1986;25:1847–1851. doi: 10.1021/bi00356a001. [DOI] [PubMed] [Google Scholar]
- 30.Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- 31.Novotny J, Bruccoleri RE. Correlation among sites of limited proteolysis, enzyme accessibility and segmental mobility. FEBS Lett. 1987;211:185–189. doi: 10.1016/0014-5793(87)81433-3. [DOI] [PubMed] [Google Scholar]
- 32.Hubbard SJ, Eisenmenger F, Thornton JM. Modeling studies of the change in conformation required for cleavage of limited proteolytic sites. Protein Sci. 1994;3:757–768. doi: 10.1002/pro.5560030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suh MJ, Pourshahian S, Limbach PA. Developing limited proteolysis and mass spectrometry for the characterization of ribosome topography. J. Am. Soc. Mass Spectrom. 2007;18:1304–1317. doi: 10.1016/j.jasms.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinkse MW, Heck AJ, Rumpel K, Pullen F. Probing noncovalent protein-ligand interactions of the cGMP-dependent protein kinase using electrospray ionization time of flight mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:1392–1399. doi: 10.1016/j.jasms.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Leite JF, Amoscato AA, Cascio M. Coupled proteolytic and mass spectrometry studies indicate a novel topology for the glycine receptor. J. Biol. Chem. 2000;275:13683–13689. doi: 10.1074/jbc.275.18.13683. [DOI] [PubMed] [Google Scholar]
- 36.Palaiomylitou M, Tartas A, Vlachakis D, Tzamarias D, Vlassi M. Investigating the structural stability of the Tup1-interaction domain of Ssn6: evidence for a conformational change on the complex. Proteins. 2008;70:72–82. doi: 10.1002/prot.21489. [DOI] [PubMed] [Google Scholar]
- 37.Huang S, Zou X, Guo P, Zhong L, Peng J, Jing G. Probing the subtle conformational state of N138ND2-Q106O hydrogen bonding deletion mutant (Asn138Asp) of staphylococcal nuclease using time of flight mass spectrometry with limited proteolysis. Arch. Biochem. Biophys. 2005;434:86–92. doi: 10.1016/j.abb.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Esteban B, Carrascal M, Abian J, Lamparter T. Light-induced conformational changes of cyanobacterial phytochrome Cph1 probed by limited proteolysis and autophosphorylation. Biochemistry. 2005;44:450–461. doi: 10.1021/bi0484365. [DOI] [PubMed] [Google Scholar]
- 39.Li DW, Bruschweiler R. All-atom contact model for understanding protein dynamics from crystallographic B-factors. Biophys. J. 2009;96:3074–3081. doi: 10.1016/j.bpj.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guncar G, Wang CI, Forwood JK, Teh T, Catanzariti AM, Ellis JG, Dodds PN, Kobe B. The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:209–213. doi: 10.1107/S1744309107004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glusker JP. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 42.Harding MM. The architecture of metal coordination groups in proteins. Acta Crystallogr. D Biol. Crystallogr. 2004;60:849–859. doi: 10.1107/S0907444904004081. [DOI] [PubMed] [Google Scholar]
- 43.Maret W, Vallee BL. Cobalt as probe and label of proteins. Methods Enzymol. 1993;226:52–71. doi: 10.1016/0076-6879(93)26005-t. [DOI] [PubMed] [Google Scholar]
- 44.Ulmer DD, Vallee BL. Advances in Chemistry. Anonymous American Chemical Society; 1991. Structure and Function of Metalloenzymes; pp. 187–218. [Google Scholar]
- 45.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 46.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 47.Philominathan ST, Matsushita O, Jordan JB, Sakon J. 1H, 13C and 15N resonance assignments of Ca2+ bound collagen-binding domain derived from a clostridial collagenase. Biomol. NMR Assign. 2008;2:127–129. doi: 10.1007/s12104-008-9102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]