Abstract
Background
Treating postoperative pain remains a significant challenge for perioperative medicine. Recent studies have shown that nerve growth factor is upregulated and contributes to incisional pain. To date, few studies have examined expression of other neurotrophin-related mediators that may contribute to the development and/or maintenance of incisional pain.
Methods
Male, Sprague-Dawley rats underwent a plantar incision and pain behaviors were examined (n = 6). In a separate group of rats, expression of neurotrophic factors were studied. At various times after incision (n = 4) or sham surgery (n = 4), the skin, muscle and dorsal root ganglia were harvested and total RNA isolated. Real-time reverse transcription polymerase chain reaction was performed and the fold change in gene expression was analyzed using significance analysis of microarrays.
Results
Several genes were changed (p < 0.05) as early as one hour after incision. Expression of artemin and nerve growth factor were increased in both incised skin and muscle. Brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-5 were all downregulated in the skin but upregulated in the muscle 48 h after incision. Few genes changed in the dorsal root ganglion. Most changes in expression occurred in the first 48 h after incision, a timeframe when pain behavior was the greatest.
Conclusion
Surgical incision is associated with pain-related gene expression changes in skin, muscle, and to a lesser extent, dorsal root ganglion. The gene expression profile provides clues as to mediators that are involved in peripheral sensitization and pain transmission after surgical incision and also suggest mechanisms for resolution of postoperative pain when more persistent pain syndromes like neuropathic pain continue.
Introduction
New analgesic therapies devoid of serious side effects are needed to advance perioperative pain management. In effort to develop new therapies, we must discover factors potentially contributing to incisional pain, evaluate their time course in relation to pain and understand how these substances affect nociception. Several mediators such as nerve growth factor (NGF),1,2 complement,3,4 prostaglandins,5 and lactic acid6–8 have been evaluated and likely contribute to nociception after incision in animal models.
Evaluation of pain mediators from surgical incisions in patients is restricted to serum and wound exudates. However, in animal models, gene expression profiles can be studied in injured tissues and dorsal root ganglion (DRG) innervating those tissues. Array technology or targeted gene expression profiles have been used to evaluate the patterns of nociceptive mediators in preclinical neuropathic pain models like sciatic nerve transection,9,10 nerve root ligation,11,12 and spinal nerve transection13 but not in incisional pain.
Global expression patterns can be ascertained using high-density oligonucleotide microarrays, but suffer from false-positives, and difficulty in determining the thresholds for biological relevance. Recent meta-analysis of microarray studies in neuropathic and inflammatory pain models suggest that immune-related genes are highly regulated across different pain models and have also identified new candidate genes.14 Because sensory neurons from different tissues respond uniquely to injury,15 the study of tissue specific expression of nociception-related genes will advance our understanding of postoperative pain. In the current study, we examined pain behaviors and gene expression after plantar incision of the rat hindpaw. We hypothesized that neurotrophin expression would be modulated by surgical incision in incised tissues. We determined the time course of expression of 84 genes from the neurotrophin and inflammatory cytokine families in skin, muscle and DRG using quantitative, real-time RT-PCR (reverse transcription polymerase chain reaction) using a commercially available, targeted array.
Materials and Methods
Animals
This study was approved by the Institutional Animal Care and Use Committee at the University of Iowa (Iowa City, Iowa). Male Sprague-Dawley adult rats (Harlan, Indianapolis, IN) weighing 275 to 300 g were housed 2 per cage with a 12-h light-dark cycle. Food and water were available ad libitum. All experiments, including incision, behavioral testing, tissue isolation, and processing were carried out by the same person (CMS) to reduce interindividual total variability. A total of 46 rats were used for this study; the behavior testing used 6 rats and the gene expression analysis used 8 rats per time point (4 rats per group). Data from all rats were included in the analysis.
Plantar Incision
The rat hindpaw plantar incision model of postoperative pain was used as previously described.16 Briefly, rats were anesthetized with 3% isoflurane delivered via nose cone. The plantar aspect of the right hindpaw was prepared with povidone iodine and a 1.0 cm longitudinal incision was made 0.5 cm from the end of the heel. The underlying flexor muscle was elevated and incised longitudinally allowing the muscle origin and insertion to remain intact. After hemostasis with gentle pressure, the skin was closed with two mattress sutures of 5-0 nylon on an FS-2 needle. Antibiotic ointment was applied to the incision immediately after surgery. Sutures were removed on postoperative day 2. Another group of rats served as controls and underwent sham surgery in which all procedures were performed except they did not undergo incision.
Tissue Harvest
Gene expression was examined 1 h, 4 h, 1 day, 2 days, and 10 days after incision or sham surgery. The times when pain behaviors are greatest after incision, postoperative days 0 through 2, were chosen to evaluate changes in gene expression. In addition, a time when pain behaviors have resolved, 10 days after incision, was chosen to determine if the gene expression had recovered. To collect tissues for real time RT-PCR, rats were deeply anesthetized with isoflurane (4–5%), and right hindpaw skin, muscle and the right sided L4 and L5 dorsal root ganglia from sham and incised rats were removed and immediately placed in RNA Later® solution (Ambion, Austin, TX). Tissues were stored at 4oC until further processing. Rats were then euthanized.
Behavioral Testing
In a separate group of rats, pain behaviors were tested through postoperative day (POD) 10. We elected to use another group of rats in order to minimize any changes in gene expression that might be affected by testing responses to noxious stimuli. Rats were acclimated to the testing environment for 3 days. Baseline pain scores were measured and rats underwent plantar incision.
Guarding pain
A cumulative pain score was used to assess nonevoked pain behaviors as described previously.17 Briefly, unrestrained rats were placed on a plastic mesh floor (8 × 8 mm). The incised and non-incised paws were viewed. Both paws of each animal were closely observed during a 1-min period repeated every 5 min for 1 h. Depending on the position in which each paw was observed during the majority of the 1-min scoring period, a 0, 1, or 2 was given. Full weightbearing of the paw (score = 0) was present if the wound was blanched or distorted by the mesh. If the paw was completely off the mesh, a score of 2 was recorded. If the area of the wound touched the mesh gently without any blanching or distorting, a 1 was given. The sum of the 12 scores (0–24) obtained during the 1-h session for each paw was obtained. The difference between the scores from the incised paw and nonincised paw is the cumulative pain score for that 1-h period. This test was performed before evoked tests were undertaken to minimize influence of evoked tests on guarding.
Withdrawal response to mechanical stimulation
Unrestrained rats were placed on mesh (grid 12 × 12 mm) and allowed to acclimate. Withdrawal responses to mechanical stimuli were determined using calibrated von Frey filaments applied from underneath the cage through openings in the plastic mesh to an area adjacent to the wound. Each filament was applied once, and increasing forces were applied until a withdrawal response was achieved or the 522 mN filament was reached. Each test was repeated after a 5-min test-free interval, for a total of three tests per rat. The lowest force from the three tests producing a response was considered the withdrawal threshold.18
Withdrawal latency to heat
Rats were placed individually on a glass floor (3 mm) covered with clear plastic cage and allowed to acclimate. Withdrawal latencies to radiant heat were assessed by applying a focused radiant heat source underneath a glass floor on the middle of incision. Before the experiments began, the intensity of the heat was adjusted to produce withdrawal latency in normal rats of 12 to 15 s. The latency time to evoke withdrawal was determined with a cut-off value of 20 s. Each rat was tested at least 3 times, at an interval of 10 min. The average of three trials was used to obtain paw withdrawal latency.
Real time quantitative RT-PCR
Total RNA was extracted with TRIzol® (Invitrogen, Carlsbad, CA) followed by DNAse treatment with Turbo Free DNAse® (Ambion) using the manufacturer’s recommended protocols. The samples underwent column purification using the RNeasy mini kit® (Qiagen, Valencia, CA). Total RNA underwent spectrophotometric analysis using a Nanodrop 1000 and samples with A260/A280 ratio greater than 1.7 were used. Random samples underwent further analysis with an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA) to assess RNA quality via confirmation of sharp peaks of 18S and 28S ribosomal RNA without “shoulders” or degradation. Reverse transcription of 500 ng of total RNA was carried out using First Strand Kit (SABiosciences, Frederick, MD) according to manufacturer’s directions. Complementary DNA was combined with a RT-PCR master mix containing SYBRR green (Life Technologies, Grand Island, NY) and ROX (Affymetrix, Santa Clara, CA) (loading control) and plated with primers for 84 different neurotrophins, neurotrophin receptors and related growth factor genes (SABiosciences). Controls wells were used to assess reverse transcription efficiency, presence of genomic DNA and positive RT-PCR controls. Those that failed control checks were removed from evaluation. Housekeeping genes included (1) hypoxanthine guanine phosphoribosyl transferase (2) ribosomal protein L13A (3) lactate dehydrogenase A (4) β actin and (5) ribosomal protein, large, P1. All 5 housekeeping genes were included in the calculation for relative gene expression as none changed with incision. Two step RT-PCR was performed on a 7900 Sequence Detection system (Applied Biosystems, Foster City, CA). Thermal cycling was initiated with an initial incubation at 95oC for 10 min, followed by 40 cycles with each cycle consisting of 95oC for 15 s and 60oC for 1 min. Dissociation curves were run for each gene and the presence of only one peak was confirmed for each gene of interest.
Behavior Data Analysis
All behavioral data were analyzed using Prism 5.0 software (GraphPad Software, Inc., San Diego, CA). Withdrawal threshold to mechanical stimuli are non-continuous and therefore were analyzed with nonparametric tests. The data were expressed as median and interquartile range. Differences were determined by Friedman’s ANOVA followed by Dunn’s post hoc test for comparing paw withdrawal threshold after incision to baseline. For guarding pain scores and withdrawal latencies to heat, data were expressed as mean ± SEM. One-way ANOVA followed by Dunnett’s post hoc test was used for comparisons between baseline and incision.
Gene Expression Data Analysis
All gene expression data were analyzed with the ABI SDS RQ Manager 1.2 software (Applied Biosystems). Each value is the average of tissue from 4 different animals. The formula used to calculate the relative gene expression level was the ΔΔ Ct method described in detail by Livak and Schmittgen.19 The mathematical formula is as follows: . The normalized data uses an average of 5 housekeeping genes.
Data are expressed as the fold change +/− standard deviation of the ΔΔCt. The equation for standard deviation is: , where σ = standard deviation of ΔCt and x and y are incision and sham groups, respectively. Differences between the incision group and sham group at each time point were determined using an uncorrected t-test.
In an effort to control for the relative change in gene expression with respect to the standard deviation of repeated measurements in a small gene array, we also performed the significance analysis of microarrays as described by Tusher et al.20 In this analysis, we increased the stringency for calling significant changes in gene expression by increasing the threshold value, delta (Δ). Using Δ = 1.2 allows for the most highly ranked genes to be further analyzed, and this value of delta has been previously shown to have the highest correlation with definitive measures of expression.20 The false discovery rate was defined as the percentage of falsely significant genes compared to the genes called significant. For reported genes, we maintained strict thresholds (Δ = 1.2) aiming for a false discovery rate close to zero. Data for all tissues and times after incision are contained within the supplemental digital content, (see supplemental digital content 1, tables 1–3). The supplemental content also has the additional significance analysis of microarray analyses with R values (R = fold change) set to 1.5 for those readers interested in the most robustly changed genes (see supplemental digital content 2, tables 1–3, which contain the false discovery rates and lists of significant genes).
Results
Pain behaviors
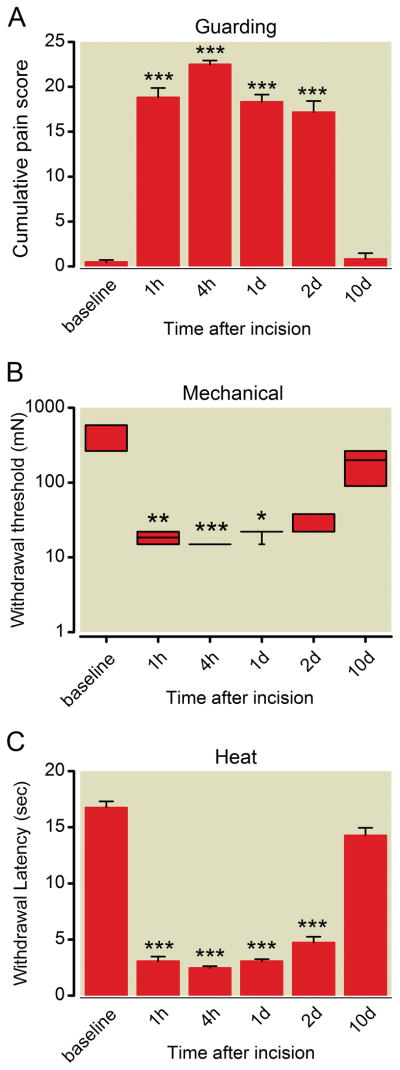
Similar to previous studies, the mean guarding pain score was increased after surgery through the first 2 postoperative days. The scores were 0.5 ± 0.5, 18.8 ± 2.6, 22.5 ± 1.0, 18.3 ± 2.0, and 17.2 ± 3.1 at baseline, POD 1 h, POD 4 h, POD 1, and POD 2, respectively (P < 0.001, n = 6, fig. 1A). The median withdrawal threshold decreased from 522 mN to 13 mN 1 hour after incision, (P < 0.01, n=6, fig. 1B) and recovered by day 10. The average heat withdrawal latencies were 16.8 ± 1.4, 3.1 ± 1.0, 2.5 ± 0.4, 3.1 ± 0.4, 4.7 ± 1.2, and 14.3 ± 1.7 s at baseline, POD 1 h, POD 4 h, POD 1, POD 2 respectively, (P < 0.05, n=6, fig. 1C). Pain related responses were greatest within 24 h after incision and all were not different than baseline by POD 10.
Figure 1.
Nonevoked guarding pain behavior (A), mechanical withdrawal threshold (B), and withdrawal latency to heat (C) after plantar incision. (A) Guarding data are expressed as mean ± SEM, n = 6. (B) Mechanical withdrawal threshold (milliNewtons) after plantar incision. Data are expressed as median (horizontal line) and box and whiskers with 1st and 3rd quartiles (box), and minimum and maximum (whiskers), n = 6. (C) Withdrawal latencies (seconds) to heat stimulation expressed as mean ± SEM, n = 6. * P < 0.05, ** P < 0.01, *** P < 0.001 versus baseline.
Gene expression trends
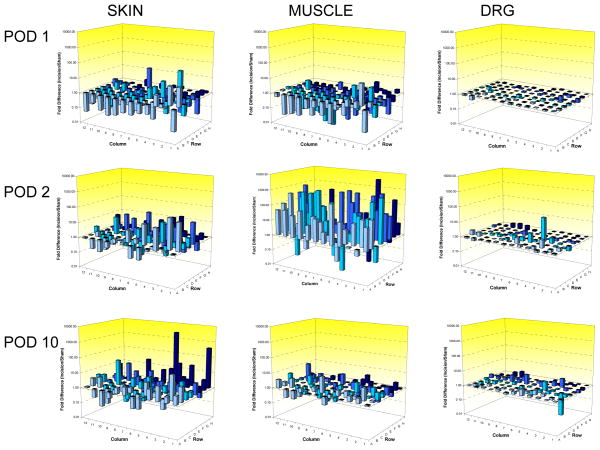
Compared to the sham group, plantar incision regulated 51 of 84 neurotrophin and cytokine profiles in the skin on postoperative day 1. Of these, 16 were increased by 1.5 fold or more; 9 were downregulated and 68 were unchanged (fig. 2). In muscle, 1 was upregulated, 11 were downregulated, and 72 were unchanged. In DRG, none were changed. On postoperative day 2, muscle was more reactive than skin; 10 factors were upregulated, 0 downregulated, and 74 were unchanged in skin, whereas in muscle 41 were upregulated, 4 were downregulated, and 39 were unchanged. There were no changes in DRG. The magnitude of regulation was 2 to 90 fold in the skin and 2 to 4,400 fold in the muscle. Altogether, more genes were upregulated in the skin compared to muscle; muscle regulation was mostly downregulation, except on postoperative day 2 when there was marked upregulation of many genes. The overall relative magnitude of effect was greatest in the muscle, modest in the skin and absent in the DRG after plantar incision.
Figure 2.
Three dimensional summary plate layouts depicting fold gene regulation for all genes studied. Data are expressed as fold difference of incision/sham. Values greater than 1.00 represent up regulation after incision compared to sham. Values less than 1.00 represent down regulation after incision compared to sham surgery. Each bar represents the average fold regulation from eight animals (four incision and four sham). POD = postoperative day. DRG = dorsal root ganglion
RT- PCR analysis of pronociceptive factors
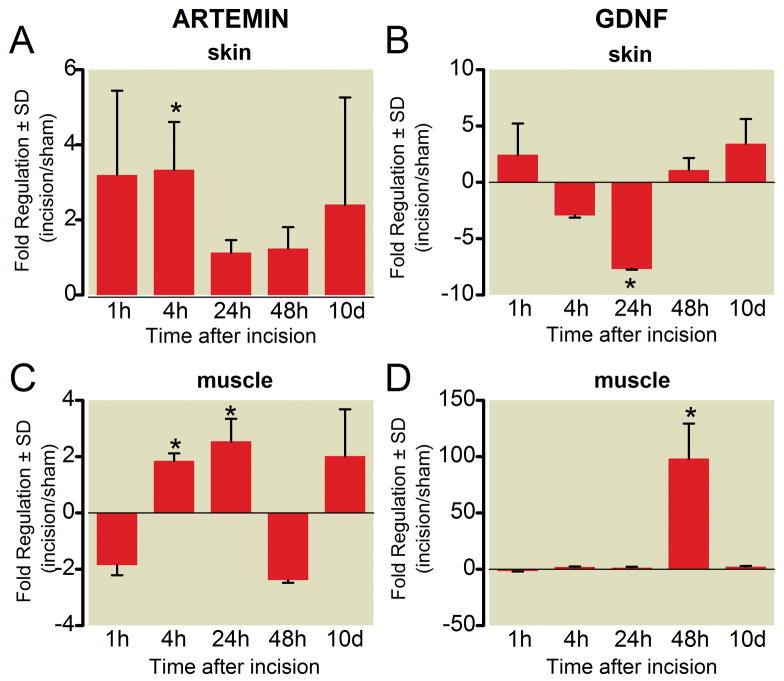
The GDNF family of neurotrophins, which includes GDNF and artemin, are each expressed in skin, muscle and DRG (fig. 3). In general, artemin expression was increased in both skin and muscle after incision. In skin, artemin messenger RNA (mRNA) was increased 4 h after incision (3.3 ± 1.3 fold, * p < 0.05, fig. 3A). In incised muscle, artemin was increased by 4 h (1.8 ± 0.3 at 4h, * p < 0.05, fig. 3B). GDNF expression in incised muscle was markedly increased on postoperative day 2 (97.9 ± 31.3, * p < 0.05, fig. 3D).
Figure 3.
Time course of gene expression for artemin and glial-derived neurotrophic factor (GDNF) messenger RNA in skin (A, B) and muscle (C, D) following plantar incision. Data are expressed as mean fold change (incision/sham) ± SD. * P < 0.05 versus sham, n = 4 per group.
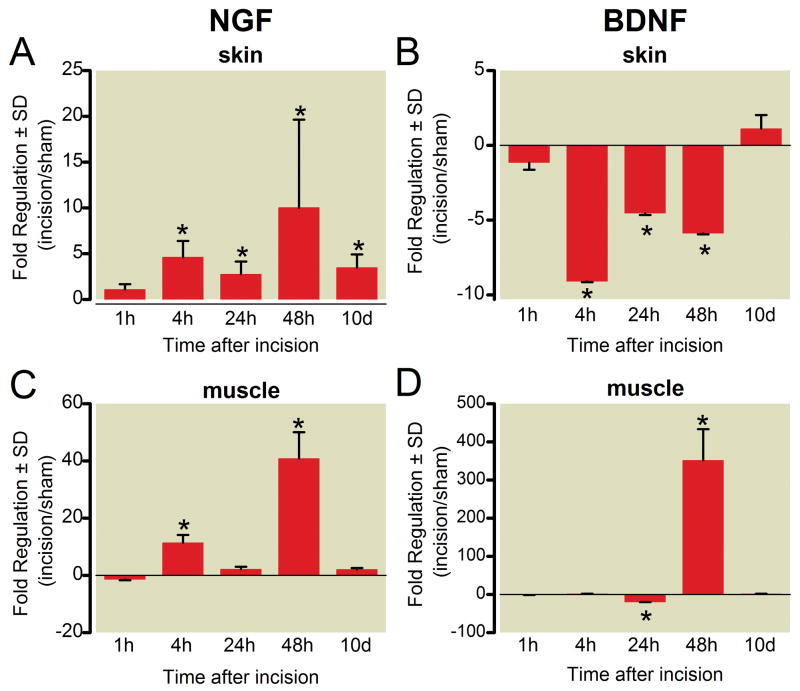
The NGF family of neurotrophins: NGF, brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), and neurotrophin-5 (NT-5) have been evaluated in several persistent pain models. These neurotrophins decreased in skin after incision (figs. 4 and 5), except NGF, which was increased, a finding supported by previous studies.1,2 In skin, the increase in NGF was apparent even 10 days after incision (3.47 ± 1.44 in skin,* p < 0.05, fig. 4A).
Figure 4.
Time course of gene expression for nerve growth factor (NGF) and brain derived neurotrophic factor (BDNF) messenger RNA in skin (A, B) and muscle (C, D) following plantar incision. Data are expressed as mean fold change (incision/sham) ± SD. * P < 0.05 versus sham, n = 4 per group.
Figure 5.
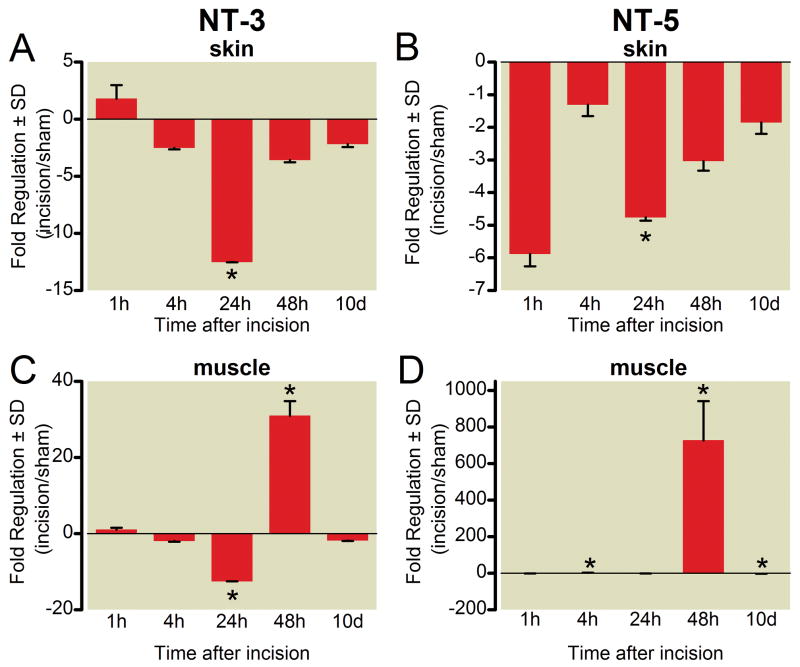
Time course of gene expression for neurotrophin-3 (NT-3) and neurotrophin-5 (NT-5) messenger RNA in skin (A, B) and muscle (C, D) following plantar incision. Data are expressed as mean fold change (incision/sham) ± SD. * P < 0.05 versus sham, n = 4 per group.
BDNF decreased after incision in the skin. The decrease in skin was apparent from 4 h through 48 h after incision, (fig. 4B). In muscle, BDNF was initially down regulated (20.0 ± 0.3 fold, * p < 0.05) followed by marked upregulation on postoperative day 2 (351.5 ± 82.1, * p < 0.05, fig. 4D). NT-3 and NT-5 were decreased in the skin at 24 h after incision, (figs. 5A and B). In muscle, NT-3 and NT-5 were increased at 48 h, (figs. 5C and D).
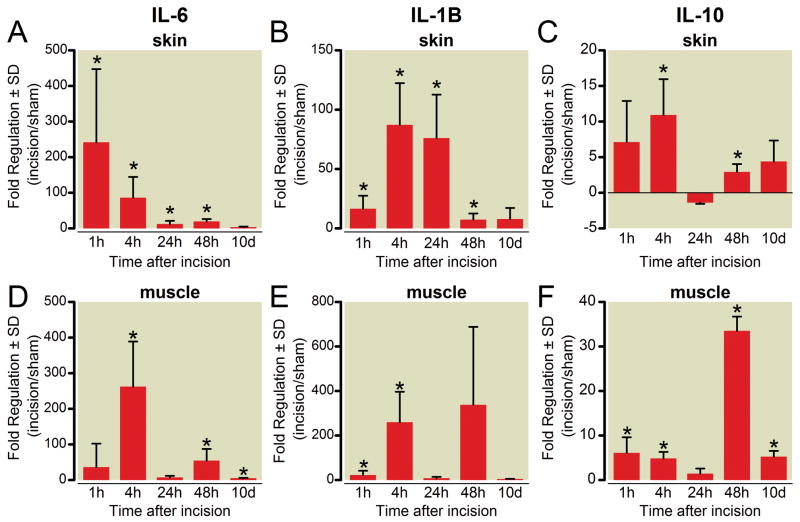
The pronociceptive interleukins (IL), IL-6, and IL-1β, were markedly increased in skin after plantar incision; the range of the peak increase was from 10 to 200 fold (fig. 6). In skin, IL-6 peaked immediately after incision (241.3 ± 41.7 fold, fig. 6A) and remained elevated through the first 2 days. IL-1β also remained elevated in skin through postoperative day 2, but the peak occurred later, at 4 h (87.0 ± 35.4 fold, fig. 6B). In incised plantar muscle, the gene expression of IL-6 and IL-1β were also increased, the magnitude of increases was similar to skin but the peak expression tended to occur later (figs. 6D and E).
Figure 6.
Time course of gene expression for interleukin (IL) messenger RNA in skin (A, B, C) and muscle (D, E, F) following plantar incision. Data are expressed as mean fold change (incision/sham) ± SD. * P < 0.05 versus sham, n = 4 per group.
The mRNAs for neurotrophin receptors were present in skin and muscle. Receptors for the GDNF family were generally downregulated in skin (table 1). For muscle, GFRα1 was downregulated for two days after incision, whereas, GFRα2 and GFRα3 were upregulated at 48 h. The receptors for the NGF family of neurotrophins also changed after incision. TrkA was downregulated in skin and muscle for the first 24 h, whereas p75 was increased in muscle (table 1). In the DRG, few genes changed after plantar incision, yet none were significant using significance analysis of microarrays set at Δ = 1.2 (supplemental digital content 2, table 3). Several genes were found to be significantly regulated in all three tissues at some point during the 10-day study period (table 2).
Table 1.
Fold change in gene expression for neurotrophin receptors in skin and muscle after hindpaw incision
| Skin | |||||
|---|---|---|---|---|---|
| Gene Name | 1 Hour | 4 hours | 24 hour | 48 Hour | 10 Days |
| GFRα1 | −1.94 ± 0.30 (0.330) | -2.38 ± 0.18 (0.096) | −4.98 ± 0.05 (0.001) | 2.18 ± 1.59 (0.325) | 3.03 ± 0.79 (0.005) |
| GFRα2 | 1.05 ± 0.42 (0.896) | 1.06 ± 0.16 (0.728) | 1.18 ± 0.14 (0.224) | 2.06 ± 0.59 (0.045) | 1.42 ± 0.69 (0.499) |
| GFRα3 | 1.06 ± 0.56 (0.897) | −2.23 ± 0.11 (0.020) | −5.86 ± 0.08 (0.009) | −4.19 ± 0.16 (0.083) | 1.17 ± 0.28 (0.527) |
| Trk A | 1.56 ± 2.28 (0.723) | −1.97 ± 0.16 (0.077) | −15.50 ± 0.05 (0.015) | −5.91 ± 0.14 (0.081) | −3.20 ± 0.25 (0.189) |
| Trk B | 1.07 ± 1.10 (0.938) | −1.70 ± 0.26 (0.279) | −3.22 ± 0.13 (0.036) | −3.48 ± 0.25 (0.206) | −1.36 ± 0.56 (0.700) |
| p75 | −1.38 ± 0.34 (0.514) | 2.54 ± 1.10 (0.075) | −1.70 ± 0.27 (0.297) | 2.38 ± 1.60 (0.244) | 3.24 ± 0.88 (0.005) |
| Muscle | |||||
|---|---|---|---|---|---|
| Gene Name | 1 Hour | 4 hours | 24 hour | 48 Hour | 10 Days |
| GFRα1 | 1.03 ± 0.34 (0.938) | −1.89 ± 0.13 (0.039) | −5.88 ± 0.08 (0.009) | −10.34 ± 0.07 (0.041) | 2.79 ± 1.33 (0.075) |
| GFRα2 | −1.47 ± 0.17 (0.204) | −1.00 ± 0.17 (0.991) | −1.14 ± 0.34 (0.744) | 42.52 ± 11.29 (0.0003) | 1.23 ± 0.50 (0.625) |
| GFRα3 | 1.58 ± 1.25 (0.621) | −1.42 ± 0.45 (0.603) | −3.26 ± 0.35 (0.342) | 50.81 ± 25.47 (0.002) | −1.26 ± 0.37 (0.638) |
| Trk A | −3.11 ± 0.35 (0.312) | −3.15 ± 0.11 (0.014) | −17.88 ± 0.07 (0.061) | 448.04 ± 142.05 (0.00007) | −3.60 ± 0.11 (0.016) |
| Trk B | −1.64 ± 0.50 (0.599) | −1.77 ± 0.23 (0.205) | −2.09 ± 0.33 (0.329) | 1.83 ± 0.92 (0.358) | 1.26 ± 0.54 (0.617) |
| p75 | −1.21 ± 0.34 (0.690) | 3.62 ± 1.84 (0.044) | −1.16 ± 0.27 (0.656) | 159.26 ± 16.53 (0.000002) | 2.83 ± 1.38 (0.077) |
Data are expressed as mean fold change (incision/sham) ± SD. Values in parenthesis are the P value. Boxes in red are significantly upregulated an those in blue are significantly downregulated
Table 2.
Genes regulated in all three tisues (skin, muscle and dorsal root ganglion) after plantar incision
| Pain Related Genes |
| Ciliary neurotrophic factor receptor |
| Fibroblast growth factor receptor 1 |
| Glial cell line derived neurotrophic factor family receptor alpha 2 |
| Interleukin 1 receptor, type I |
| Interleukin 6 |
| Leukemia inhibitory factor |
| Nerve growth factor receptor (tumor necrosis family receptor superfamily, member 16) |
| Neurotrophin 3 |
| Neurotrophic tyrosine kinase, receptor, type 1 |
| Fas (tumor necrosis family receptor superfamily, member 6) |
| Other Genes |
| Adenylate cyclase activating polypeptide 1 receptor 1 |
| FBJ murine osteosarcoma viral oncogene homolog |
| Fusion, derived from t(12;16) malignant liposarcoma (human) |
| Similar to cerebellin 1 precursor protein |
Discussion
Pronociceptive growth factors such as artemin, GDNF, and NGF are potential therapeutic targets for the treatment of postoperative pain. NGF monoclonal antibodies have been used in human clinical trials to treat various chronic pain conditions with moderate success, but have not been reported for clinical acute pain conditions such as postoperative pain.21,22 We found that expression of pronociceptive growth factors and immune-related genes were increased in incisions but, in general, upregulated to a greater extent and for a longer period of time in muscle compared to skin. Other neurotrophins, like NT-3, NT-5, and BDNF, could contribute to nociception after surgery as well, but the effect of these factors on sensory afferents, peripheral or central sensitization, and plasticity has not been as extensively studied. Importantly, these pronociceptive growth factors were concomitantly upregulated in incisions where they could interact to contribute to nociception. Combinations of pronociceptive factors have been measured in experimental human pain models as well as after surgical incision.23,24 The combination of NGF and artemin increases the duration of heat hyperalgesia compared to either alone.25 The observed increase in the expression of several neurotrophins within 48 h may contribute to sensitization and plasticity early after surgery, when pain behaviors and clinical postoperative pain are greatest.
Expression of glial-derived neurotrophic factor family ligands
Artemin is thought to be pronociceptive. It belongs to the GDNF family of neurotrophic factors which promote differentiation and survival of motor, sympathetic, and large and small sensory neurons during development.26–28 Artemin expression is increased after inflammation in skin25 and visceral tissues29 and sensitization of nociceptors occurs after acute artemin administration or with congenital overexpression.25,30,31 Pain related behaviors are also transiently increased after local artemin administration.25
After incision, artemin expression was immediately increased in skin. Muscle artemin expression was increased through 24 h and could contribute to deep tissue pain; however, artemin and muscle nociception have not been studied. In addition to pain, artemin could support restoration of sensory function after incision which includes tissue and nerve injury,32 or recovery of sympathetic innervation in blood vessels.33 These other roles for artemin could contribute to repair in incised tissues, a response that may be most pronounced in muscle tissue.
GDNF sensitizes nociceptors in a variety of pain models,25,31,34–36 however few studies have been able to evaluate the role of GDNF in pathophysiologic models by interfering with its action(s). Many pain studies have examined responses to intrathecally administered GDNF; however GDNF was not upregulated in the DRG and so spinal effects of GDNF are not likely germane to incisional nociception. GDNF may have a significant role in neuropathic pain models including partial sciatic nerve ligation or spinal nerve ligation37 brachial plexus avulsion38 or diabetes.39 After incision, GDNF mRNA increased in muscle, suggesting tissue specific regulatory pathways for expression after incision.
Expression of nerve growth factor family ligands and receptors
NGF is pronociceptive. In human volunteers, NGF produced myalgias and localized pain at the injection site40 and was not therapeutic in clinical trials for diabetic neuropathy.41,42 In animal models, NGF contributes to sensitization after inflammation, tissue damage and nerve injury.43,44 NGF is increased in skin and muscle following incision.1,2,45 Pain behaviors after incision can be reduced by NGF sequestration demonstrating a pronociceptive role.2,45,46 The current study demonstrated sustained NGF mRNA upregulation in both skin and muscle consistent with our previous protein measurements.1,2 This greater duration of increased mRNA expression for NGF in the present study may be a result of using five control, housekeeping genes.
The neurotrophin receptors include TrkA, TrkB, and p75 (also known as the low affinity NGF receptor). All the neurotrophins can bind to p75 and at least one of the Trk receptors. The TrkA receptor is the high affinity receptor for NGF, and signals nociceptive properties of NGF. p75 also is pronociceptive47. We found that TrkA and p75 mRNA were downregulated in skin. These changes may be due to reciprocal regulation such as ligand-receptor coupling downregulating the receptor.48 The receptors for NGF were upregulated in muscle at 48 h at a time when ligand and pain behaviors caused by muscle incision were also upregulated. The differences in muscle expression of NGF and its cognate receptors may contribute to greater activation of nociceptive sensory neurons by incision that includes muscle. These receptors may also contribute to muscle repair,49 apoptosis of sensory neurons,50 or regeneration of injured axons.47
BDNF, NT-3, and NT-5 influence nociceptor function. Most studies on the nociceptive actions of BDNF have largely been in the central nervous system, but the gene and protein are constitutively expressed in DRG neurons. BDNF expression can be upregulated by NGF.51 BDNF is downregulated in small DRG neurons after axotomy, but upregulated in large diameter DRG neurons.52 BDNF can sensitize c-fibers to noxious heat in vitro.53 The therapeutic efficacy of delivering BDNF to DRG in painful neuropathy has shown promising results in animals.54 In the current study, we find that incision decreased BDNF expression over a time course where pain behaviors were the greatest and future work could involve replacing BDNF in incised tissues as a potential analgesic treatment.
Neurotrophin-3 (NT-3) has antinociceptive properties in the peripheral nervous system. NT-3 reverses inflammatory hyperalgesia after complete Freund’s adjuvant,55 attenuates pain behaviors after nerve crush injury,56 and reverses mechanical hyperalgesia after intramuscular acid injection57. We demonstrate downregulation of NT-3 in skin and muscle 24 h after incision, with a rebound increase only in muscle at 48h after incision, patterns that may augment pain behavior after incision early and lead to recovery at later time periods. Neurotrophin-5 (NT-5) may have pronociceptive properties. Intraplantar injection of NT-5 induced thermal hyperalgesia in rats, an effect that was sustained for 24 h.53 Like NGF, NT-5 decreased heat threshold in c-fibers without altering mechanical responses.58 In our study, NT-5 was downregulated in skin, but upregulated in muscle, again peaking at 48 h after incision.
Expression of other neurotrophin receptors
Other receptors for the neurotrophins changed after incision (table 3). The receptor for artemin, GFRα-3, was downregulated after incision. GFRα-3 increased at 48 h when the ligand mRNA was decreased; thus, following a similar pattern to NGF and its receptors. The etiology of this biphasic regulation is unclear, but may represent negative feedback autoregulation similar to other growth factor receptors within the central nervous system.59
Table 3.
Fold change in gene expression from lumbar level 4 and 5 dorsal root ganglion after hindpaw incision
| Gene Name | 1 Hour | 4 hours | 24 hours | 48 Hours | 10 Days |
|---|---|---|---|---|---|
| Artemin | 1.14 ± 0.20 (0.45) | 1.03 ± 0.15 (0.87) | −1.19 ± 0.12 (0.26) | −1.15 ± 0.18 (0.59) | 1.02 ± 0.20 (0.94) |
| BDNF | 1.27 ± 0.28 (0.22) | −1.02 ± 0.15 (0.92) | −1.20 ± 0.14 (0.31) | 1.39 ± 0.56 (0.44) | 2.51 ± 1.39 (0.22) |
| GDNF | −1.05 ± −0.55 (0.91) | 1.27 ± 0.83 (0.73) | −1.23 ± 0.30 (0.59) | 2.02 ± 1.40 (0.36) | −1.69 ± 0.14 (0.08) |
| Gfra1 | 1.12 ± 0.25 (0.60) | 1.13 ± 0.13 (0.34) | −1.19 ± 0.07 (0.08) | 1.13 ± 0.24 (0.65) | 1.47 ± 0.57 (0.43) |
| Gfra2 | 1.01 ± 0.20 (0.97) | 1.02 ± 0.08 (0.86) | −1.20 ± 0.06 (0.04) | 1.12 ± 0.31 (0.72) | 1.25 ± 0.43 (0.59) |
| Gfra3 | −1.10 ± −0.28 (0.71) | 1.00 ± 0.07 (0.99) | −1.06 ± 0.13 (0.66) | −1.05 ± 0.22 (0.87) | 1.56 ± 0.78 (0.49) |
| IL-10 | 4.26 ± 3.49 (0.07) | −2.10 ± 0.20 (0.12) | −1.21 ± 0.46 (0.74) | −1.38 ± 0.47 (0.66) | −2.04 ± 0.34 (0.41) |
| IL-1β | 2.46 ± 1.54 (0.15) | −1.01 ± 0.28 (0.97) | 1.06 ± 0.30 (0.86) | 1.18 ± 0.42 (0.68) | −28.29 ± 0.05 (0.06) |
| IL-6 | 4.15 ± 6.04 (0.28) | 1.48 ± 0.58 (0.36) | −1.36 ± 0.15 (0.18) | 35.09 ± 41.54 (0.03) | 3.61 ± 1.46 (0.02) |
| NGF | 1.30 ± 0.29 (0.21) | −1.38 ± 0.22 (0.32) | −1.02 ± 0.44 (0.97) | 1.04 ± 0.18 (0.85) | 1.07 ± 0.18 (0.73) |
| p75 | 1.20 ± 0.33 (0.51) | 1.35 ± 0.08 (0.00) | 1.21 ± 0.19 (0.27) | −1.05 ± 0.28 (0.89) | 2.03 ± 1.21 (0.36) |
| NT-3 | −1.34 ± −0.36 (0.31) | −1.28 ± 0.28 (0.52) | −1.67 ± 0.12 (0.04) | −1.15 ± 0.09 (0.20) | 1.46 ± 0.69 (0.53) |
| NT-5 | −1.54 ± −0.45 (0.15) | 1.42 ± 0.70 (0.50) | 1.48 ± 0.52 (0.3) | −1.34 ± 0.29 (0.49) | 1.76 ± 0.92 (0.39) |
| trkA | −1.09 ± −0.63 (0.89) | 1.17 ± 0.27 (0.54) | −2.57 ± 0.09 (0.01) | 1.10 ± 0.43 (0.84) | −1.55 ± 0.13 (0.13) |
| trkB | 1.01 ± 0.28 (0.97) | 1.25 ± 0.14 (0.08) | −1.25 ± 0.10 (0.13) | −1.17 ± 0.16 (0.53) | −1.04 ± 0.19 (0.86) |
Data are expressed as mean fold change (incision/sham) ± SD. Values in parenthesis are the P value. Boxes in red are significantly upregulated an those in blue are significantly downregulated
BDNF=brain derived neurotrophic factor, GDNF = glial derived neurotrophic factor, IL = interleukin, NGF = nerve growth factor, NT = neurotrophin
RT-PCR analysis of interleukins
Inflammatory cytokines, including IL-1β and IL-6 are increased in inflammatory pain models and produce peripheral and central sensitization and hyperalgesia.60,61 IL-1 has been shown to be increased in the spinal cord dorsal horn in postoperative pain models,62 in peri-incisional tissues63,64 and serum23 of postoperative patients. IL-1 may cause hyperalgesia through the expression or release of pronociceptive compounds such as substance P,65 prostaglandins, and nerve growth factor.66 IL-1 may directly sensitize sensory neurons to heat.67 Although IL-1 is considered pronociceptive, the role for IL-1 in nociception after incision is not clear. Using IL-1 knockout mice, pain behavior was reduced in inflammatory and neuropathic models, but not after plantar incision.68 In contrast, using IL-1 receptor knockout mice and over-expression of the IL-1 receptor antagonists, mechanical hyperalgesia was reduced after incision.69
IL-6, a pronociceptive early marker of the inflammatory cascade, 70 can also enhance transient receptor vanilloid 1 activity, a key target in nociceptive heat responses.71 Similar to IL-1β, IL-6 is increased in inflammatory and neuropathic pain models.72–74 Increased levels of peri-incisional and plasma IL-6 are found after surgery in both humans64,75 and rodents76,77 The increases in IL-6 expression from skin and muscle parallel observations from surgical wound dialysates23 and muscle injury models.78,79 The therapeutic value of interfering with IL-6 function in incisional pain is not understood.
In contrast, IL-10 is an antiinflammatory cytokine that is increased in surgical wound exudates.23,80 In other models of pain, IL-10 can reduce pain behaviors and produce pronociceptive compounds such as and IL-1.81 The significance of modifying IL-10 expression in incisional pain models is also not known.
In summary, pronociceptive cytokines IL-1 beta and IL-6 were upregulated in both skin and muscle early after incision and return to near-baseline when pain behaviors resolve. The role of these cytokines in muscle and nociception is less clear compared to that of skin where pronociception is well studied. The interplay between antinociceptive and pronociceptive cytokines in surgical wounds is not yet understood; nonnociceptive functions like wound healing, reinnervation, neovascularization, reducing infection, and hemostasis82 must be considered when function of these cytokines is altered.
DRG gene changes in other pain models
There were few changes in gene expression in DRG after plantar incision. The time course and changes in neurotrophin mRNA expression in DRG after spinal nerve ligation are markedly different than incision. For example, BDNF increased 2–3 days after spinal nerve ligation, whereas NGF was increased 4-fold 3 weeks after nerve injury.11,83 After sciatic nerve axotomy, BDNF mRNA increased 1.6 fold and NT-3 decreased 1.6 fold three days after injury; no other changes in neurotrophins in the DRG were noted.9 Not all studies have found mRNA changes in the neurotrophin genes families. In two models of neuropathic pain, there were no changes in NGF, BDNF, NT-3, NT-5, GDNF, artemin, or their receptors.13 The lack of changes in gene expression in DRG is likely related to the small amount of nerve injury after plantar incision compared with a relatively large amount of nerve injury in other pain models. The time courses of changes in DRG and nerve injury models may be one of factors which contribute to persistent pain after nerve injury, but shorter lived pain after plantar incision.
Other models of peripheral inflammation result in short-lived changes in dorsal root ganglia neurotrophin expression. The mRNA for p75 and trkA were increased 8 and 24 h after injection of complete Freund’s adjuvant without change in NGF, BDNF, GDNF, artemin, NT-5 or trkB.84 In adult animals, BDNF mRNA a increased 6-fold in the L4/L5 DRG 1d after hindpaw complete Freund’s adjuvant administration.85
Statistical considerations
The expression data and subsequent analyses obtained in these experiments are reported in entirety in the supplemental digital content (see supplemental digital content, 1 & 2). The p-values are obtained from uncorrected t-tests, which may have inflated type-I error rates. The use of a post-hoc correction, such as the Bonferroni correction, is excessively conservative because the genes in any biological sample are not truly independent. Exact p-values are reported for all data so that interested readers may apply any correction. We used the significance analysis of microarrays to ascertain differentially regulated genes while allowing for some correction of multiple experiments.
Conclusion
Our data suggest that patterns of mRNA expression after incision are unique in magnitude and timing. Furthermore, specific factor expression in incisions is markedly different than other pain models. These factors may not only affect nociceptor activation, but also influence tissue remodeling, wound healing, reinnervation and the immune response depending on the tissue. The interaction among factors at different times after incision may contribute to pain after surgery and other processes in perioperative wound homeostasis. Our understanding of the interactions between the sensory, autonomic and the immune systems is incomplete, and further work is needed to determine how these mediators interact in injured tissues. Future studies will examine mechanisms for gene regulation in different tissues, the downstream signaling and subsequent impact of such changes, and a better understanding of how interference with these mediators can alter nociception after injury.
Supplementary Material
Final Boxed Summary Statement.
What we already know about this topic
Inflammatory product and neurotrophin genes are upregulatedin peripheral tissues, including peripheral nerves in animal models of chronic inflammation or nerve injury, but a survey of such genes after incisional injury has not been performed
What this article tells us that is new
Using a targeted array for 84 genes often altered in pain states, incisional injury was shown in rats to increase pronociceptive interleukins in skin as well as nerve growth factor and artemin in skin and muscle
The pattern of gene upregulation observed would be expected to result in and contribute to hypersensitivity and pain after incisional surgery
Acknowledgments
Funding: Supported by an educational grant from Endo Pharmaceuticals, Foundation for Anesthesia Education and Research Mentored Research Training Grant, Rochester, Minnesota, and National Institutes of Health T32 NS045549, Bethesda, Maryland, to CMS.
Footnotes
Work Attributed to: Department of Anesthesia, University of Iowa, Iowa City, IA
Contributor Information
Christina M. Spofford, Department of Anesthesia, University of Iowa.
Timothy J. Brennan, Department of Anesthesia, University of Iowa.
References
- 1.Wu C, Boustany L, Liang H, Brennan TJ. Nerve growth factor expression after plantar incision in the rat. Anesthesiology. 2007;107:128–35. doi: 10.1097/01.anes.0000267512.08619.bd. [DOI] [PubMed] [Google Scholar]
- 2.Wu C, Erickson MA, Xu J, Wild KD, Brennan TJ. Expression profile of nerve growth factor after muscle incision in the rat. Anesthesiology. 2009;110:140–9. doi: 10.1097/ALN.0b013e318190bc84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark JD, Qiao Y, Li X, Shi X, Angst MS, Yeomans DC. Blockade of the complement C5a receptor reduces incisional allodynia, edema, and cytokine expression. Anesthesiology. 2006;104:1274–82. doi: 10.1097/00000542-200606000-00024. [DOI] [PubMed] [Google Scholar]
- 4.Jang JH, Clark JD, Li X, Yorek MS, Usachev YM, Brennan TJ. Nociceptive sensitization by complement C5a and C3a in mouse. Pain. 2010;148:343–52. doi: 10.1016/j.pain.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kroin JS, Buvanendran A, Watts DE, Saha C, Tuman KJ. Upregulation of cerebrospinal fluid and peripheral prostaglandin E2 in a rat postoperative pain model. Anesth Analg. 2006;103:334–43. doi: 10.1213/01.ane.0000223674.52364.5c. [DOI] [PubMed] [Google Scholar]
- 6.Kim TJ, Freml L, Park SS, Brennan TJ. Lactate concentrations in incisions indicate ischemic-like conditions may contribute to postoperative pain. J Pain. 2007;8:59–66. doi: 10.1016/j.jpain.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Woo YC, Park SS, Subieta AR, Brennan TJ. Changes in tissue pH and temperature after incision indicate acidosis may contribute to postoperative pain. Anesthesiology. 2004;101:468–75. doi: 10.1097/00000542-200408000-00029. [DOI] [PubMed] [Google Scholar]
- 8.Deval E, Noël J, Gasull X, Delaunay A, Alloui A, Friend V, Eschalier A, Lazdunski M, Lingueglia E. Acid-sensing ion channels in postoperative pain. J Neurosci. 2011;31:6059–66. doi: 10.1523/JNEUROSCI.5266-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costigan M, Befort K, Karchewski L, Griffin RS, D'Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao HS, Huang QH, Zhang FX, Bao L, Lu YJ, Guo C, Yang L, Huang WJ, Fu G, Xu SH, Cheng XP, Yan Q, Zhu ZD, Zhang X, Chen Z, Han ZG, Zhang X. Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc Natl Acad Sci U S A. 2002;99:8360–5. doi: 10.1073/pnas.122231899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DS, Figueroa KW, Li KW, Boroujerdi A, Yolo T, Luo DZ. Profiling of dynamically changed gene expression in dorsal root ganglia post peripheral nerve injury and a critical role of injury-induced glial fibrillary acetic protein in maintenance of pain behaviors. Pain. 2009;143:114–22. doi: 10.1016/j.pain.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi H, Kawaguchi S, Mizuno S, Kirita T, Takebayashi T, Shimozawa K, Torigoe T, Sato N, Yamashita T. Gene expression profile of dorsal root ganglion in a lumbar radiculopathy model. Spine. 2008;33:2483–8. doi: 10.1097/BRS.0b013e318184acc3. [DOI] [PubMed] [Google Scholar]
- 13.LaCroix-Fralish ML, Tawfik VL, Tanga FY, Spratt KF, DeLeo JA. Differential spinal cord gene expression in rodent models of radicular and neuropathic pain. Anesthesiology. 2006;104:1283–92. doi: 10.1097/00000542-200606000-00025. [DOI] [PubMed] [Google Scholar]
- 14.LaCroix-Fralish ML, Austin J-S, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: Meta-analysis of microarray studies of pain. PAIN. 2011;152:1888–98. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Brennan TJ. Guarding pain and spontaneous activity of nociceptors after skin versus skin plus deep tissue incision. Anesthesiology. 2010;112:153–64. doi: 10.1097/ALN.0b013e3181c2952e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan TJ, Vandermeulen EP, Gebhart GF. Characterization of a rat model of incisional pain. Pain. 1996;64:493–501. doi: 10.1016/0304-3959(95)01441-1. [DOI] [PubMed] [Google Scholar]
- 17.Zahn PK, Gysbers D, Brennan TJ. Effect of systemic and intrathecal morphine in a rat model of postoperative pain. Anesthesiology. 1997;86:1066–77. doi: 10.1097/00000542-199705000-00010. [DOI] [PubMed] [Google Scholar]
- 18.Zahn PK, Brennan TJ. Primary and secondary hyperalgesia in a rat model for human postoperative pain. Anesthesiology. 1999;90:863–72. doi: 10.1097/00000542-199903000-00030. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Tusher V, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz N, Borenstein DG, Birbara C, Bramson C, Nemeth MA, Smith MD, Brown MT. Efficacy and safety of tanezumab in the treatment of chronic low back pain. Pain. 2011;152:2248–58. doi: 10.1016/j.pain.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Lane NE, Schnitzer TJ, Birbara CA, Mokhtarani M, Shelton DL, Smith MD, Brown MT. Tanezumab for the treatment of pain from osteoarthritis of the knee. N Engl J Med. 2010;363:1521–31. doi: 10.1056/NEJMoa0901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho B, Clark DJ, Angst MS. Local and systemic release of cytokines, nerve growth factor, prostaglandin E2, and substance P in incisional wounds and serum following cesarean delivery. J Pain. 2008;9:650–7. doi: 10.1016/j.jpain.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Carvalho B, Clark DJ, Yeomans DC, Angst MS. Continuous subcutaneous instillation of bupivacaine compared to saline reduces interleukin 10 and increases substance P in surgical wounds after cesarean delivery. Anesth Analg. 2010;111:1452–9. doi: 10.1213/ANE.0b013e3181f579de. [DOI] [PubMed] [Google Scholar]
- 25.Malin SA, Molliver DC, Koerber HR, Cornuet P, Frye R, Albers KM, Davis BM. Glial cell line-derived neurotrophic factor family members sensitize nociceptors in vitro and produce thermal hyperalgesia in vivo. J Neurosci. 2006;26:8588–99. doi: 10.1523/JNEUROSCI.1726-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buj-Bello A, Buchman VL, Horton A, Rosenthal A, Davies AM. GDNF is an age-specific survival factor for sensory and autonomic neurons. Neuron. 1995;15:821–8. doi: 10.1016/0896-6273(95)90173-6. [DOI] [PubMed] [Google Scholar]
- 27.Bespalov MM, Saarma M. GDNF family receptor complexes are emerging drug targets. Trends Pharmacol Sci. 2007;28:68–74. doi: 10.1016/j.tips.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Clarkson ED, Zawada WM, Freed CR. GDNF reduces apoptosis in dopaminergic neurons in vitro. Neuroreport. 1995;7:145–9. [PubMed] [Google Scholar]
- 29.Ceyhan GO, Bergmann F, Kadihasanoglu M, Erkan M, Park W, Hinz U, Giese T, Muller MW, Buchler MW, Giese NA, Friess H. The neurotrophic factor artemin influences the extent of neural damage and growth in chronic pancreatitis. Gut. 2007;56:534–44. doi: 10.1136/gut.2006.105528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elitt CM, McIlwrath SL, Lawson JJ, Malin SA, Molliver DC, Cornuet PK, Koerber HR, Davis BM, Albers KM. Artemin overexpression in skin enhances expression of TRPV1 and TRPA1 in cutaneous sensory neurons and leads to behavioral sensitivity to heat and cold. J Neurosci. 2006;26:8578–87. doi: 10.1523/JNEUROSCI.2185-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albers KM, Woodbury CJ, Ritter AM, Davis BM, Koerber HR. Glial cell-line-derived neurotrophic factor expression in skin alters the mechanical sensitivity of cutaneous nociceptors. J Neurosci. 2006;26:2981–90. doi: 10.1523/JNEUROSCI.4863-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, King T, Ossipov MH, Rossomando AJ, Vanderah TW, Harvey P, Cariani P, Frank E, Sah DW, Porreca F. Persistent restoration of sensory function by immediate or delayed systemic artemin after dorsal root injury. Nat Neurosci. 2008;11:488–96. doi: 10.1038/nn2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damon DH, teRiele JA, Marko SB. Vascular-derived artemin: A determinant of vascular sympathetic innervation? Am J Physiol Heart Circ Physiol. 2007;293:H266–73. doi: 10.1152/ajpheart.00859.2006. [DOI] [PubMed] [Google Scholar]
- 34.Bennett DL, Boucher TJ, Armanini MP, Poulsen KT, Michael GJ, Priestley JV, Phillips HS, McMahon SB, Shelton DL. The glial cell line-derived neurotrophic factor family receptor components are differentially regulated within sensory neurons after nerve injury. J Neurosci. 2000;20:427–37. doi: 10.1523/JNEUROSCI.20-01-00427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang M, Wang Y, He QH, Sun YX, Deng LB, Wang XM, Han JS. Glial cell line-derived neurotrophic factor contributes to delayed inflammatory hyperalgesia in adjuvant rat pain model. Neuroscience. 2003;117:503–12. doi: 10.1016/s0306-4522(02)00958-2. [DOI] [PubMed] [Google Scholar]
- 36.Amaya F, Shimosato G, Nagano M, Ueda M, Hashimoto S, Tanaka Y, Suzuki H, Tanaka M. NGF and GDNF differentially regulate TRPV1 expression that contributes to development of inflammatory thermal hyperalgesia. Eur J Neurosci. 2004;20:2303–10. doi: 10.1111/j.1460-9568.2004.03701.x. [DOI] [PubMed] [Google Scholar]
- 37.Boucher TJ, Okuse K, Bennett DL, Munson JB, Wood JN, McMahon SB. Potent analgesic effects of GDNF in neuropathic pain states. Science. 2000;290:124–7. doi: 10.1126/science.290.5489.124. [DOI] [PubMed] [Google Scholar]
- 38.Quintão NL, Santos AR, Campos MM, Calixto JB. The role of neurotrophic factors in genesis and maintenance of mechanical hypernociception after brachial plexus avulsion in mice. Pain. 2008;136:125–33. doi: 10.1016/j.pain.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 39.Christianson JA, Ryals JM, McCarson KE, Wright DE. Beneficial actions of neurotrophin treatment on diabetes-induced hypoalgesia in mice. J Pain. 2003;4:493–504. doi: 10.1016/j.jpain.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Dyck PJ, Peroutka S, Rask C, Burton E, Baker MK, Lehman KA, Gillen DA, Hokanson JL, O'Brien PC. Intradermal recombinant human nerve growth factor induces pressure allodynia and lowered heat-pain threshold in humans. Neurology. 1997;48:501–5. doi: 10.1212/wnl.48.2.501. [DOI] [PubMed] [Google Scholar]
- 41.Apfel SC, Schwartz S, Adornato BT, Freeman R, Biton V, Rendell M, Vinik A, Giuliani M, Stevens JC, Barbano R, Dyck PJ Group rCI. Efficacy and safety of recombinant human nerve growth factor in patients with diabetic polyneuropathy: A randomized controlled trial. JAMA. 2000;284:2215–21. doi: 10.1001/jama.284.17.2215. [DOI] [PubMed] [Google Scholar]
- 42.McArthur JC, Yiannoutsos C, Simpson DM, Adornato BT, Singer EJ, Hollander H, Marra C, Rubin M, Cohen BA, Tucker T, Navia BA, Schifitto G, Katzenstein D, Rask C, Zaborski L, Smith ME, Shriver S, Millar L, Clifford DB, Karalnik IJ. A phase II trial of nerve growth factor for sensory neuropathy associated with HIV infection. AIDS Clinical Trials Group Team 291. Neurology. 2000;54:1080–8. doi: 10.1212/wnl.54.5.1080. [DOI] [PubMed] [Google Scholar]
- 43.Diamond J, Foerster A, Holmes M, Coughlin M. Sensory nerves in adult rats regenerate and restore sensory function to the skin independently of endogenous NGF. J Neurosci. 1992;12:1467–76. doi: 10.1523/JNEUROSCI.12-04-01467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diamond J, Holmes M, Coughlin M. Endogenous NGF and nerve impulses regulate the collateral sprouting of sensory axons in the skin of the adult rat. J Neurosci. 1992;12:1454–66. doi: 10.1523/JNEUROSCI.12-04-01454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banik RK, Subieta AR, Wu C, Brennan TJ. Increased nerve growth factor after rat plantar incision contributes to guarding behavior and heat hyperalgesia. Pain. 2005;117:68–76. doi: 10.1016/j.pain.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 46.Zahn PK, Subieta A, Park SS, Brennan TJ. Effect of blockade of nerve growth factor and tumor necrosis factor on pain behaviors after plantar incision. J Pain. 2004;5:157–63. doi: 10.1016/j.jpain.2004.02.538. [DOI] [PubMed] [Google Scholar]
- 47.Nicol GD, Vasko MR. Unraveling the story of NGF-mediated sensitization of nociceptive sensory neurons: ON or OFF the Trks? Mol Interv. 2007;7:26–41. doi: 10.1124/mi.7.1.6. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Moheban DB, Conway BR, Bhattacharyya A, Segal RA. Cell surface Trk receptors mediate NGF-induced survival while internalized receptors regulate NGF-induced differentiation. J Neurosci. 2000;20:5671–8. doi: 10.1523/JNEUROSCI.20-15-05671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deponti D, Buono R, Catanzaro G, De Palma C, Longhi R, Meneveri R, Bresolin N, Bassi MT, Cossu G, Clementi E, Brunelli S. The low-affinity receptor for neurotrophins p75NTR plays a key role for satellite cell function in muscle repair acting via RhoA. Mol Biol Cell. 2009;20:3620–7. doi: 10.1091/mbc.E09-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukai J, Hachiya T, Shoji-Hoshino S, Kimura MT, Nadano D, Suvanto P, Hanaoka T, Li Y, Irie S, Greene LA, Sato TA. NADE, a p75NTR-associated cell death executor, is involved in signal transduction mediated by the common neurotrophin receptor p75NTR. J Biol Chem. 2000;275:17566–70. doi: 10.1074/jbc.C000140200. [DOI] [PubMed] [Google Scholar]
- 51.Pezet S, McMahon SB. Neurotrophins: mediators and modulators of pain. Annu Rev Neurosci. 2006;29:507–38. doi: 10.1146/annurev.neuro.29.051605.112929. [DOI] [PubMed] [Google Scholar]
- 52.Michael GJ, Averill S, Shortland PJ, Yan Q, Priestley JV. Axotomy results in major changes in BDNF expression by dorsal root ganglion cells: BDNF expression in large trkB and trkC cells, in pericellular baskets, and in projections to deep dorsal horn and dorsal column nuclei. Eur J Neurosci. 1999;11:3539–51. doi: 10.1046/j.1460-9568.1999.00767.x. [DOI] [PubMed] [Google Scholar]
- 53.Shu XQ, Llinas A, Mendell LM. Effects of trkB and trkC neurotrophin receptor agonists on thermal nociception: A behavioral and electrophysiological study. Pain. 1999;80:463–70. doi: 10.1016/S0304-3959(99)00042-1. [DOI] [PubMed] [Google Scholar]
- 54.Hao S, Mata M, Wolfe D, Huang S, Glorioso JC, Fink DJ. HSV-mediated gene transfer of the glial cell-derived neurotrophic factor provides an antiallodynic effect on neuropathic pain. Mol Ther. 2003;8:367–75. doi: 10.1016/s1525-0016(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe M, Endo Y, Kimoto K, Katoh-Semba R, Arakawa Y. Inhibition of adjuvant-induced inflammatory hyperalgesia in rats by local injection of neurotrophin-3. Neurosci Lett. 2000;282:61–4. doi: 10.1016/s0304-3940(00)00842-9. [DOI] [PubMed] [Google Scholar]
- 56.Wilson-Gerwing TD, Dmyterko MV, Zochodne DW, Johnston JM, Verge VM. Neurotrophin-3 suppresses thermal hyperalgesia associated with neuropathic pain and attenuates transient receptor potential vanilloid receptor-1 expression in adult sensory neurons. J Neurosci. 2005;25:758–67. doi: 10.1523/JNEUROSCI.3909-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gandhi R, Ryals JM, Wright DE. Neurotrophin-3 reverses chronic mechanical hyperalgesia induced by intramuscular acid injection. J Neurosci. 2004;24:9405–13. doi: 10.1523/JNEUROSCI.0899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rueff A, Mendell LM. Nerve growth factor NT-5 induce increased thermal sensitivity of cutaneous nociceptors in vitro. J Neurophysiol. 1996;76:3593–6. doi: 10.1152/jn.1996.76.5.3593. [DOI] [PubMed] [Google Scholar]
- 59.Fujikawa T, Soya H, Fukuoka H, Alam KSM, Yoshizato H, McEwen BS, Nakashima K. A biphasic regulation of receptor mRNA expressions for growth hormone, glucocorticoid and mineralocorticoid in the rat dentate gyrus during acute stress. Brain Res. 2000;874:186–93. doi: 10.1016/s0006-8993(00)02576-2. [DOI] [PubMed] [Google Scholar]
- 60.Watkins LR, Maier SF. Beyond neurons: evidence that immune and glial cells contribute to pathological pain states. Physiol Rev. 2002;82:981–1011. doi: 10.1152/physrev.00011.2002. [DOI] [PubMed] [Google Scholar]
- 61.Albertini R, Villaverde AB, Aimbire F, Bjordal J, Brugnera A, Mittmann J, Silva JA, Costa M. Cytokine mRNA expression is decreased in the subplantar muscle of rat paw subjected to carrageenan-induced inflammation after low-level laser therapy. Photomed Laser Surg. 2008;26:19–24. doi: 10.1089/pho.2007.2119. [DOI] [PubMed] [Google Scholar]
- 62.Fu D, Guo Q, Ai Y, Cai H, Yan J, Dai R. Glial activation and segmental upregulation of interleukin-1beta (IL-1β) in the rat spinal cord after surgical incision. Neurochem Res. 2006;31:333–40. doi: 10.1007/s11064-005-9032-4. [DOI] [PubMed] [Google Scholar]
- 63.Liang D, Shi X, Qiao Y, Angst MS, Yeomans DC, Clark JD. Chronic morphine administration enhances nociceptive sensitivity and local cytokine production after incision. Mol Pain. 2008;4:7. doi: 10.1186/1744-8069-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buvanendran A, Kroin JS, Berger RA, Hallab NJ, Saha C, Negrescu C, Moric M, Caicedo MS, Tuman KJ. Upregulation of prostaglandin E2 and interleukins in the central nervous system and peripheral tissue during and after surgery in humans. Anesthesiology. 2006;104:403–10. doi: 10.1097/00000542-200603000-00005. [DOI] [PubMed] [Google Scholar]
- 65.Skoff AM, Zhao C, Adler JE. Interleukin-1alpha regulates substance P expression and release in adult sensory neurons. Exp Neurol. 2009;217:395–400. doi: 10.1016/j.expneurol.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 66.Safieh-Garabedian B, Poole S, Allchorne A, Winter J, Woolf CJ. Contribution of interleukin-1 beta to the inflammation-induced increase in nerve growth factor levels and inflammatory hyperalgesia. Br J Pharmacol. 1995;115:1265–75. doi: 10.1111/j.1476-5381.1995.tb15035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Obreja O, Rathee P, Lips KS, Distler C, Kress M. IL-1 beta potentiates heat-activated currents in rat sensory neurons: Involvement of IL-1RI, tyrosine kinase, and protein kinase C. FASEB J. 2002;16:1497–503. doi: 10.1096/fj.02-0101com. [DOI] [PubMed] [Google Scholar]
- 68.Honore P, Wade CL, Zhong C, Harris RR, Wu C, Ghayur T, Iwakura Y, Decker MW, Faltynek C, Sullivan J, Jarvis MF. Interleukin-1alphabeta gene-deficient mice show reduced nociceptive sensitivity in models of inflammatory and neuropathic pain but not post-operative pain. Behav Brain Res. 2006;167:355–64. doi: 10.1016/j.bbr.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 69.Wolf G, Livshits D, Beilin B, Yirmiya R, Shavit Y. Interleukin-1 signaling is required for induction and maintenance of postoperative incisional pain: Genetic and pharmacological studies in mice. Brain Behav Immun. 2008;22:1072–7. doi: 10.1016/j.bbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 70.Sommer C, Kress M. Recent findings on how proinflammatory cytokines cause pain: Peripheral mechanisms in inflammatory and neuropathic hyperalgesia. Neurosci Lett. 2004;361:184–7. doi: 10.1016/j.neulet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 71.Andratsch M, Mair N, Constantin CE, Scherbakov N, Benetti C, Quarta S, Vogl C, Sailer CA, Uceyler N, Brockhaus J, Martini R, Sommer C, Zeilhofer HU, Muller W, Kuner R, Davis JB, Rose-John S, Kress M. A key role for gp130 expressed on peripheral sensory nerves in pathological pain. J Neurosci. 2009;29:13473–83. doi: 10.1523/JNEUROSCI.1822-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurek JB, Austin L, Cheema SS, Bartlett PF, Murphy M. Up-regulation of leukaemia inhibitory factor and interleukin-6 in transected sciatic nerve and muscle following denervation. Neuromuscul Disord. 1996;6:105–14. doi: 10.1016/0960-8966(95)00029-1. [DOI] [PubMed] [Google Scholar]
- 73.Okamoto H, Yamamura M, Morita Y, Harada S, Makino H, Ota Z. The synovial expression and serum levels of interleukin-6, interleukin-11, leukemia inhibitory factor, and oncostatin M in rheumatoid arthritis. Arthritis Rheum. 1997;40:1096–105. doi: 10.1002/art.1780400614. [DOI] [PubMed] [Google Scholar]
- 74.Wang XM, Hamza M, Wu TX, Dionne RA. Upregulation of IL-6, IL-8 and CCL2 gene expression after acute inflammation: Correlation to clinical pain. Pain. 2009;142:275–83. doi: 10.1016/j.pain.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahdy AM, Galley HF, Abdel-Wahed MA, El-Korny KF, Sheta SA, Webster NR. Differential modulation of interleukin-6 and interleukin-10 by diclofenac in patients undergoing major surgery. Br J Anaesth. 2002;88:797–802. doi: 10.1093/bja/88.6.797. [DOI] [PubMed] [Google Scholar]
- 76.Ishibashi S, Takeuchi H, Fujii K, Shiraishi N, Adachi Y, Kitano S. Length of laparotomy incision and surgical stress assessed by serum IL-6 level. Injury. 2006;37:247–51. doi: 10.1016/j.injury.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 77.Loram LC, Themistocleous AC, Fick LG, Kamerman PR. The time course of inflammatory cytokine secretion in a rat model of postoperative pain does not coincide with the onset of mechanical hyperalgesia. Can J Physiol Pharmacol. 2007;85:613–20. doi: 10.1139/y07-054. [DOI] [PubMed] [Google Scholar]
- 78.Ono T, Maekawa K, Watanabe S, Oka H, Kuboki T. Muscle contraction accelerates IL-6 mRNA expression in the rat masseter muscle. Arch Oral Biol. 2007;52:479–86. doi: 10.1016/j.archoralbio.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 79.Dina OA, Green PG, Levine JD. Role of interleukin-6 in chronic muscle hyperalgesic priming. Neuroscience. 2008;152:521–5. doi: 10.1016/j.neuroscience.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin F, Martinez V, Mazoit JX, Bouhassira D, Cherif K, Gentili ME, Piriou P, Chauvin M, Fletcher D. Antiinflammatory effect of peripheral nerve blocks after knee surgery: Clinical and biologic evaluation. Anesthesiology. 2008;109:484–90. doi: 10.1097/ALN.0b013e318182c2a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Plunkett JA, Yu CG, Easton JM, Bethea JR, Yezierski RP. Effects of interleukin-10 (IL-10) on pain behavior and gene expression following excitotoxic spinal cord injury in the rat. Exp Neurol. 2001;168:144–54. doi: 10.1006/exnr.2000.7604. [DOI] [PubMed] [Google Scholar]
- 82.Miller RJ, Jung H, Bhangoo SK, White FA. In: Cytokine and Chemokine Regulation of Sensory Neuron Function, Sensory Nerves. Canning BJ, Spina D, editors. Berlin: Springer-Verlag; 2009. pp. 417–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shen H, Chung JM, Chung K. Expression of neurotrophin mRNAs in the dorsal root ganglion after spinal nerve injury. Brain Res Mol Brain Res. 1999;64:186–92. doi: 10.1016/s0169-328x(98)00314-3. [DOI] [PubMed] [Google Scholar]
- 84.Chien CC, Fu WM, Huang HI, Lai YH, Tsai YF, Guo SL, Wu TJ, Ling QD. Expression of neurotrophic factors in neonatal rats after peripheral inflammation. J Pain. 2007;8:161–7. doi: 10.1016/j.jpain.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 85.Mannion RJ, Costigan M, Decosterd I, Amaya F, Ma QP, Holstege JC, Ji RR, Acheson A, Lindsay RM, Wilkinson GA, Woolf CJ. Neurotrophins: peripherally and centrally acting modulators of tactile stimulus-induced inflammatory pain hypersensitivity. Proc Natl Acad Sci U S A. 1999;96:9385–90. doi: 10.1073/pnas.96.16.9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.