Abstract
Many aspects of metabolism exhibit daily rhythmicity under the control of endogenous circadian clocks, and disruptions in circadian timing result in dysfunctions associated with the metabolic syndrome. Nocturnin (Noc) is a robustly rhythmic gene that encodes a deadenylase thought to be involved in the removal of polyA tails from mRNAs. Mice lacking the Noc gene display resistance to diet-induced obesity and hepatic steatosis, due in part to reduced lipid trafficking in the small intestine. In addition, Noc appears to play important roles in other tissues and has been implicated in lipid metabolism, adipogenesis, glucose homeostasis, inflammation and osteogenesis. Therefore, Noc is a potential key post-transcriptional mediator in the circadian control of many metabolic processes.
Keywords: Nocturnin, Ccrn4l, Circadian, Deadenylase, Post-transcriptional, Metabolic syndrome
The circadian clock exerts central and peripheral effects that are vital for proper metabolic homeostasis
Obesity and the metabolic syndrome
A variety of conditions collectively grouped and diagnosed as the metabolic syndrome are on the rise world-wide and causing serious health concerns. Nearly 40% of men and women in the United States qualified for such diagnosis as of 2005 [1]. Among the conditions associated with the metabolic syndrome are obesity and dysregulation of glucose levels [1]. Indeed, excess weight is the leading cause of poor health and is associated with development of cardiovascular disease and diabetes [2]. Interestingly, alterations in biological timing occurring through shift work and jet lag, result in major disruptions in physiology that manifest as symptoms of the metabolic syndrome [reviewed in 3]. Examining the regulation of rhythmic metabolic processes is therefore vital to our understanding and treatment of this disorder.
Research investigating the intimate relationship between daily “circadian” cycles and metabolic rhythms has benefitted from examining metabolic phenotypes in mutant mice with targeted disruptions to genes involved in both rhythm generation and rhythmic output. One such rhythmic output gene is Nocturnin (Noc), which codes for a circadian deadenylase and is the focus of this review. Deadenylases participate in post-transcriptional mRNA regulation through destabilization of target transcripts, and circadian control of deadenylase activity is one mechanism whereby the clock regulates rhythmic gene expression. Noc has been implicated in many aspects of lipid metabolism presumably through the post-transcriptional circadian regulation of genes involved in metabolizing fat [4]. More recently, its roles in metabolism have expanded to include lipogenesis, adipogenesis and osteogenesis [4–7]. Here we highlight the recent advances that place Nocturnin at a unique intersection between both circadian clocks and metabolism, and we emphasize how these two important processes interact to maintain proper balance in the face of metabolic challenges.
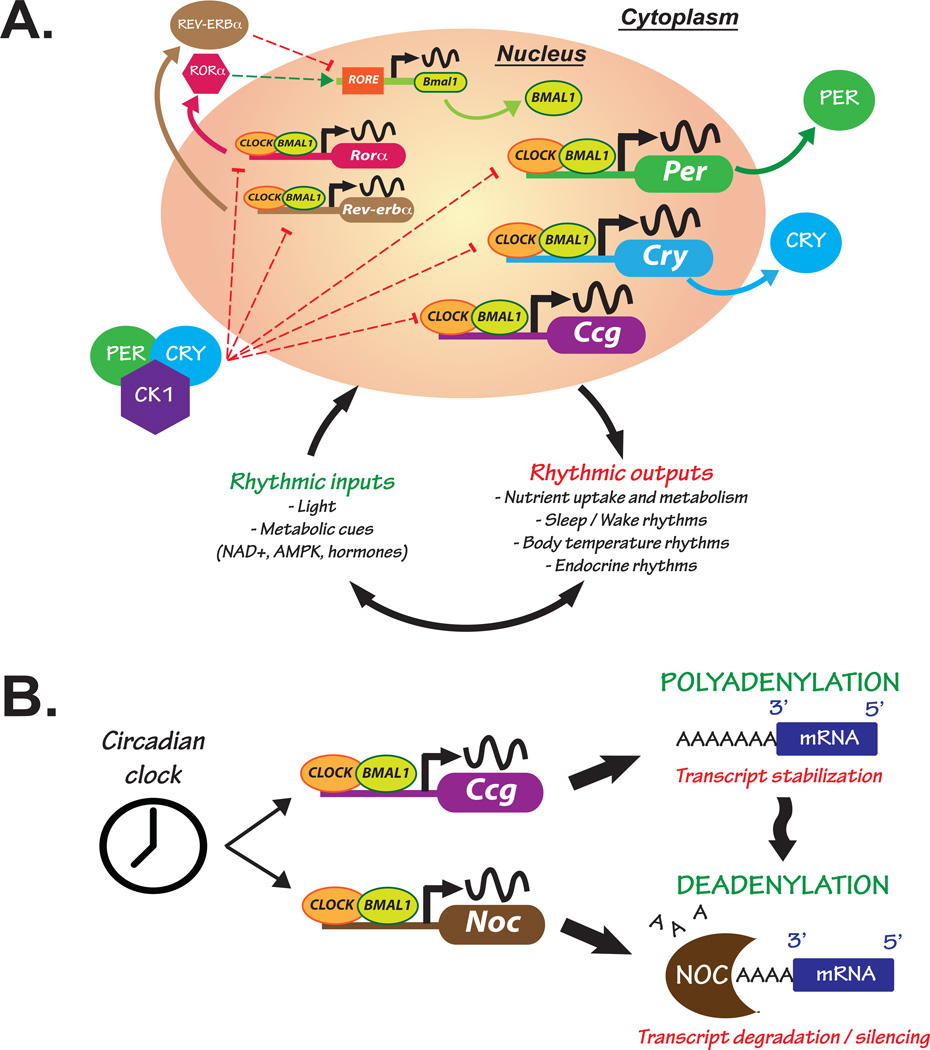
The circadian clock and metabolism
The circadian clock drives rhythmic processes in both physiology and behavior and synchronizes them to the environment. In mammals, clocks are found within cells throughout the body and are coordinated by a “master” pacemaker located within the hypothalamic suprachiasmatic nucleus(SCN) [reviewed in 8]. The circadian clock mechanism consists of auto-regulatory transcription/translation feedback loops resulting in rhythmic production and degradation of core “clock” genes/proteins (Figure 1A). In one loop, the proteins CLOCK and BMAL1 form a heterodimer that binds to E-box enhancer elements of target genes, including other core clock genes such as Period and Cryptochrome. The protein products PERIOD (PER) and CRYPTOCHROME (CRY) accumulate in the cytoplasm and form a complex with Casein Kinase 1 (CK1), followed by translocation back into the nucleus where they repress the CLOCK/BMAL1 activity and thus turn off their own transcription. In an interlocking loop of the clock, the nuclear receptors RORα and REV-ERBα direct alternating activation and repression, respectively, of Bmal1 expression (Figure 1A). Together, these loops drive a 24 h oscillation that coordinates the timing of processes, e.g. metabolic reactions, centrally in the brain and peripherally throughout the body. In turn, the core clock genes receive inputs from the periphery to ensure proper between-function synchronization. For example, NAD+, a major contributor of cell metabolism, shows 24-hour oscillations [9] and has been shown to modulate the circadian activity of different key metabolites such as SIRT1 [10], AMPK [11] or PGC1-α [12]. Overall, there is growing evidence that circadian rhythms and metabolic processes maintain intricate interactions in order to ensure energy homeostasis [13–16].
Figure 1. The molecular circadian clock is responsive to environmental and metabolic cues while maintaining tight control over gene expression.
(A) Cells from the suprachiasmatic nucleus (SCN) within the brain receive external environmental (e.g. light) and internal (e.g. nutrients and hormones) cues that influence gene expression of core “clock” genes. The CLOCK/BMAL1 heterodimer binds to E-box enhancer elements in the promoter of core clock genes, such as Period (Per) and Cryptochrome (Cry), and other clock-controlled genes (Ccg) responsible for clock output. PER and CRY proteins accumulate in the cytoplasm where they complex with Casein Kinase 1 (CK1) and translocate back into the nucleus inhibiting their own transcription. Clock output is responsible for synchronizing rhythms in peripheral clocks and influencing processes such as rhythmic nutrient metabolism and uptake, and the sleep/wake and body temperature cycles. Nutrient signals (e.g. NAD+, AMPK) participate in crosstalk with the core clock by feeding back and influencing nuclear receptors such as Rev-erbα and the retinoic acid-related orphan receptor α (RORα), thus contributing to molecular rhythm generation. Positive and negative regulatory mechanisms are represented with green and red broken lines, respectively. (B) The circadian clock generates rhythms in gene expression, but post-transcriptional mechanisms such as deadenylation can also alter rhythmic mRNA processing. mRNA stability is maintained in part through polyadenylation, a process involving the addition of 3’ adenosine residues creating a polyA tail on messenger transcripts. Conversely, polyA tail removal through deadenylation leads to transcript degradation or silencing. The clock and metabolic cues influence rhythmic expression of the gene Nocturnin (Noc), encoding a circadian deadenylase. This could be one mechanism whereby the clock exerts tight control over expression of genes involved in nutrient metabolism through regulating post-transcriptional modifications.
Peripheral tissues can respond directly to clock output, or local oscillators within the tissues can be entrained by systemic cues and affect tissue-specific gene expression [14]. Likewise, metabolic perturbations such as high fat diet (HFD) feeding produce nutrient signals that can feedback and influence clock gene expression [17]. This intimate relationship between clocks and metabolism was notably exhibited in mice harboring a mutation in the Clock gene sequence (ClockΔ19). These mice display many physiological disruptions associated with the metabolic syndrome including hyperlipidemia, hyperglycemia and hypoinsulinemia [18]. Disruptions in other core clock genes such as Bmal1 or Per2 lead to disruptions in metabolism as well [reviewed in 3, 19]. While the autoregulatory feedback loops comprising the core oscillator affect gene transcription, there is increasing evidence for circadian control over post-transcriptional modifications [20, 21]. Examples of these modifications include mRNA splicing, silencing and deadenylation [21] which allow for the precise temporal control of gene expression at times that are most beneficial to the energetics, and thus survival, of an organism. Study of the circadian deadenylase Nocturnin is therefore vital to understanding this link between the circadian clock and metabolism.
Nocturnin Discovery and Function
Nocturnin is a circadian deadenylase
At a time when little was known of the components comprising the vertebrate circadian clock, Noc was discovered using a differential display screen in a search for cycling transcripts in retinal photoreceptors of Xenopus laevis [22, 23]. Noc mRNA displays high amplitude, rhythmic expression in isolated Xenopus eye cups in cyclic light (light:dark) and constant (light:light or dark:dark) conditions [23]. Significant sequence similarity is found between Noc and the yeast transcription factor Carbon catabolite repressor 4 (yCCR4) and thus its official gene name is carbon-catabolite repressor 4-like (Ccrn4l) [23, 24]. yCCR4 is a multifunctional protein that is part of large transcriptional complexes in yeast, and has deadenylase activity confined to its C-terminal domain. Nocturnin lacks the N-terminal activation domains and leucine-rich repeat region necessary for yCCR4’s interaction with transcription complexes [23]. However, conserved in the Noc sequence is a C-terminal Mg2+-dependent endonuclease-like domain, which was found in yCCR4 to be required for proper polyA-specific mRNA degradation, implicating it as a functional deadenylase [25]. Deadenylation is one type of post-transcriptional modification occurring with mRNA transcripts whereby their 3’ polyA tails are removed, thus destabilizing the transcript and leading to its degradation or silencing (Figure 1B). The ability of Nocturnin to function as a deadenylase and remove 3’ adenosine residues from mRNA transcripts was confirmed in cell-based assays both with Xenopus (xNOC) and mouse (mNOC) Nocturnin protein [24, 26].
While first discovered in Xenopus, Noc homologues were found in other species with a high degree of similarity in coding sequence [27]. Noc mRNA levels are highly rhythmic with peak amplitude occurring in the early evening, both in Xenopus and mouse [23, 27]. In mouse, Noc is expressed rhythmically in almost every tissue but with a particularly high amplitude rhythm in the liver [27]. In Huh7 cells, a human hepatoma cell line, luciferase assays demonstrated that Noc is transcriptionally regulated by the CLOCK/BMAL1 heterodimer [28]. Li and colleagues also found that Noc has E-boxes upstream of its transcription start site and chromatin immunoprecipitation (ChIP) revealed direct binding of the CLOCK/BMAL1 heterodimer to these sequences [28]. The rhythmic profile of Noc is regulated by additional mechanisms beyond CLOCK/BMAL1 transcription. For example, recent evidence has demonstrated an important role for the liver-specific microRNA (miRNA) miR-122 in the proper shaping of rhythmic Noc expression [29].
Noc is not critical for rhythm generation by the core clock, as mice deficient for Nocturnin (Noc−/−) do not have overt circadian phenotypes [30]. More importantly, Nocturnin's circadian profile remains rhythmic in ClockΔ19 animals, though with damped amplitude, suggesting that other signals or factors also exert control over Noc's expression [31]. Similarly, Kornmann and colleagues utilized transgenic mice in which the liver clock could be conditionally inactivated while the remainder of the clocks throughout the body remained unaffected. In these animals lacking a local liver clock, Nocturnin was among a small group of genes that remained rhythmic in the liver [32]. This further supports a role for systemic cues, potentially those arising from feeding and nutrient metabolism, in effecting Noc expression. Although NOC is not directly involved in regulating core clock gene expression, it does have functions in mediating its rhythmic output. Because many of the rate-limiting enzymes in metabolic reactions are under circadian control, gene expression must be tightly controlled and post-transcriptional mechanisms such as deadenylation play an important part in this process [20, 33]. Nocturnin, as a circadian deadenylase, is poised to play an important role in post-transcriptional regulation of metabolic genes under circadian control [20].
Metabolic challenges reveal a role for Nocturnin in lipid metabolism
Noc−/− mice are resistant to diet-induced obesity (DIO) and hepatic steatosis
Noc is expressed rhythmically throughout the body, including many tissues vital for metabolism, such as intestine and liver [27, 34] (Figure 2). Noc−/− mice exhibit several metabolic phenotypes when challenged with a HFD [30], with the pronounced phenotype being that Noc−/− mice remain lean compared to their wildtype (WT) littermates. This “leanness” manifests itself peripherally, and a reduction in epididymal fat pad weight and hepatic lipid accumulation is observed [30]. However, these mice are not more active and do not consume less food than WT littermates. Additionally, their oxygen and carbon dioxide production/consumption is not altered and the Noc−/− mice exhibit a lower body temperature. Interestingly, their respiratory exchange ratio is slightly elevated, indicative of an altered utilization of lipid as an energy source. Hepatic expression levels of several genes associated with lipid metabolism is significantly lower in Noc−/− mice, including Srebp-1c, Scd1 and L-Fabp. If NOC acts on these lipid-associated transcripts directly, one expectation would be that these transcripts are more stable in the absence of deadenylation, potentially leading to higher levels. However, the opposite is observed suggesting that NOC may be acting indirectly on other targets that in turn mediate the expression of these genes, or that it may have deadenylase-independent functions. This is a major mechanistic question that when answered will help in understanding the lean phenotype of Noc−/− mice and the role NOC plays in lipid metabolism. The ubiquitous expression of Nocturnin also confounds the interpretation of where NOC is acting and whether it has tissue-specific functions. Recent work has elucidated the role of NOC in several tissues responsible for the processing of fat and that will be the focus of the rest of this review.
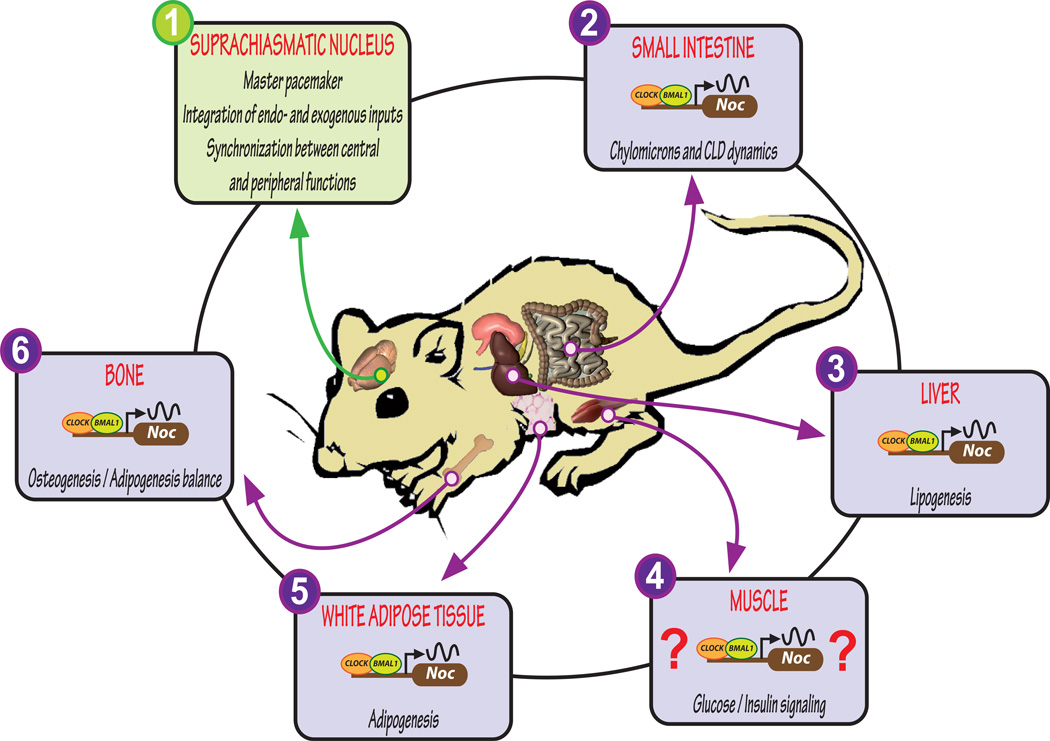
Figure 2. Nocturnin is rhythmically expressed throughout the body and is intimately linked with metabolism.
The master pacemaker in the hypothalamic suprachiasmatic nucleus (SCN) of the mammalian brain drives rhythms in gene/protein expression that synchronizes rhythmic behaviors and processes such as feeding and nutrient uptake and metabolism to the environment (1). Nutrients, especially lipid, are taken up by the body primarily in the small intestine. The deadenylase Nocturnin (Noc) is rhythmically expressed in the proximal small intestine and is involved in lipid processing into chylomicrons and the formation of cytoplasmic lipid droplets (CLD) (2). The highest amplitude expression of Noc is seen in the liver and it is here that NOC participates in lipogenesis (3). Noc expression in muscle has not been studied to date, but muscle is a key tissue in glucose metabolism and Noc−/− mice show altered glucose/insulin processing, suggesting a role for NOC in this tissue (4). Adipose tissue does not exhibit rhythmic Noc expression under basal conditions. However, placing mice under a restricted feeding paradigm induces Noc rhythms. NOC is involved in adipogenesis through its interactions with PPARγ (5). Noc is also rhythmic in bone and it participates in lineage determination of mesenchymal stromal cells towards osteogenesis or adipogenesis (6). Crosstalk exists between the core clock in the brain and the periphery through metabolic signals resulting from nutrient metabolism.
Noc responds acutely to external stimuli
Noc is unique among deadenylases in that it is an immediate early gene and responds to various stimuli such as the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) and serum in cell culture [24]. Given that TPA and serum are known factors capable of synchronizing clocks in cultured fibroblasts [35, 36], these results show that Nocturnin can be acutely regulated by physiological cues, thus suggesting that it plays a role in regulating responses to external signals in addition to its circadian role. In normal ad libitum (ad lib) feeding conditions, Noc expression peaks in the early evening in the small intestine [4], consistent with other tissues examined. This peak occurs concomitantly with peak food intake by mice. However, a daytime bolus of lipid given at a time when Noc expression is normally low and only modest quantities of food are consumed, acutely induces Noc in the small intestine [4]. This may reflect a role for NOC in the processing of lipid at this abnormal time of consumption. Alignment of Noc expression with food intake, as well as its acute responsiveness to various stimuli including lipid, supports the idea that NOC might function to regulate nutrient metabolism, i.e. ingestion, trafficking and distribution of nutrients, with possible specificity for lipids.
Nutrient excess is linked with obesity through the development of increasing intracellular and systemic lipid products such as free fatty acids (FFA) and diacylglycerol (DAG) [37]. This results in a need to process and store these extra metabolites, and expansion of adipose tissue is one mechanism facilitating lipid storage. Kawai and colleagues found that Noc is highly upregulated in 3T3-L1 cells undergoing adipogenesis [38, 39], and similarly, mice chronically exposed to a HFD exhibit increased expression of Noc in epididymal white adipose tissue (eWAT) [40]. Under ad lib feeding conditions on a standard chow diet, Noc expression is not rhythmic in eWAT [5]. However, when access to food is restricted to a narrow window during the daytime, Noc expression in eWAT becomes rhythmic, and fasting animals following this restricted feeding paradigm significantly induced Noc expression in eWAT [5]. Coordination of Noc expression with the time of food intake implies a critical role for NOC in the regulation of metabolism.
Animals maintain metabolic homeostasis when fasted, in part, through upregulation of gluconeogenesis and mobilization of glucose, both of which help preserve proper blood glucose levels and prevent hypoglycemia. One of the key players involved in activation of gluconeogenic genes during fasting is the forkhead box (FoxO) family of transcription factors [41]. Paik et al. found that Nocturnin is one of the most highly downregulated genes in liver endothelial cells from a FoxO 1/3/4 conditional knockout mouse model [42]. The Noc gene sequence was found to have conserved FoxO binding elements and FoxO binding to these sites has been confirmed by ChIP [42]. When animals end their fast, they must likewise shut down gluconeogenesis and peripheral tissues such as liver and muscle take up glucose for utilization and storage [43]. Kubota and colleagues found that impairment of insulin signaling in endothelial cells is associated with reduced glucose uptake in skeletal muscle of mice [44]. With loss of FoxO in hepatic endothelial cells affecting Noc expression levels, NOC may play a role in regulating the metabolic response to the fed/fasting states. Furthermore, it is not known whether regulation of Noc by FoxO also occurs in other cell types, such as the hepatocytes, where FoxO’s regulation of gluconeogenesis is best known. Regardless, Noc−/− mice given a bolus of glucose following an overnight fast, show significantly higher blood glucose values compared to WT littermates and they take longer to return to baseline levels [30]. This suggests a deficit in the regulation of glucose levels in Noc−/− mice. Taken together, these findings imply a role for NOC in glucose/insulin signaling, an area that needs further study.
A role for Nocturnin in lipid metabolism in the small intestine
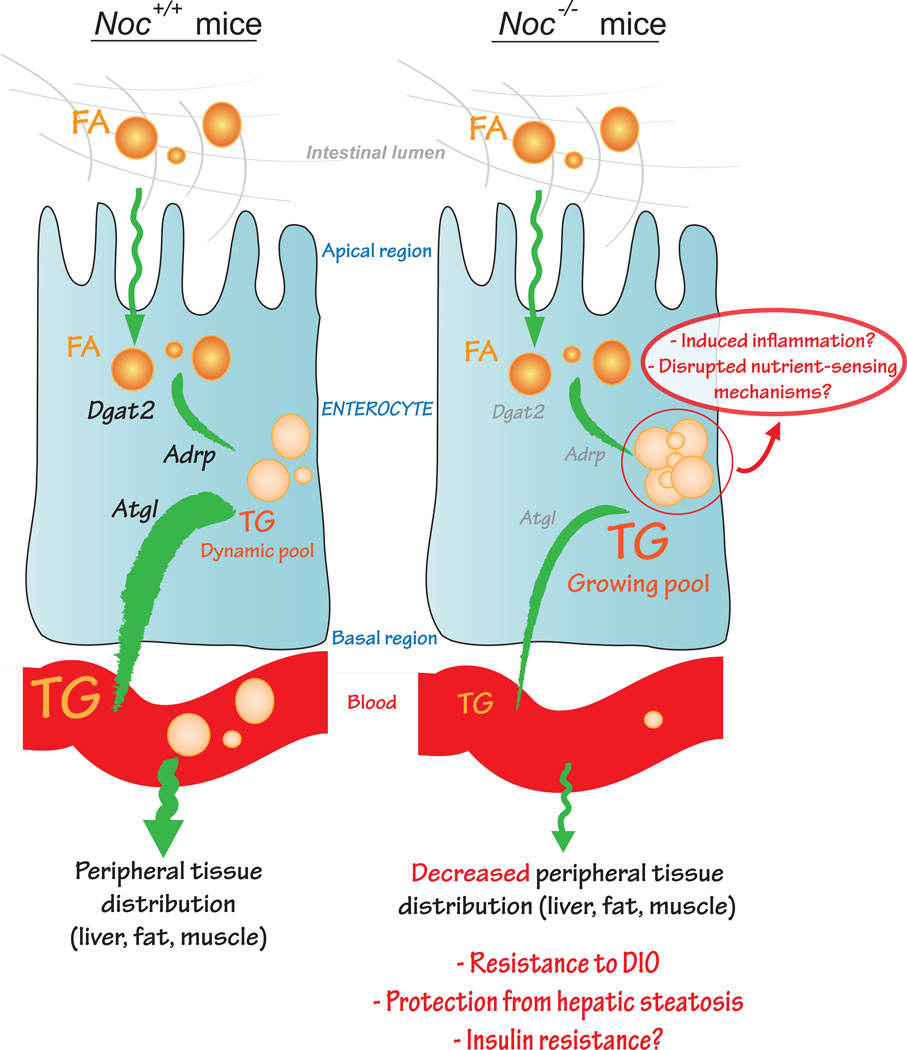
The small intestine functions to break down and absorb dietary nutrients from food with the proximal region acting as the main site of lipid uptake [45]. Enterocytes, specialized epithelial cells lining the lumen of the intestine, take up lipid as fatty acids (FA) and convert it to neutral triglyceride (TG) stored as cytoplasmic lipid droplets (CLDs) or packaged into chylomicrons (CMs) for subsequent release into the blood [45]. Noc−/− mice might have a deficit in CM synthesis or release because they show greater sequestration of TG in CLDs in enterocytes after olive oil gavage, compared with WT animals [4]. This phenotype is accompanied by decreased expression levels of Adipophilin (Adrp/Plin2), adipocyte triglyceride lipase (Atgl/Pnpla2) and diacylglycerol acetyltransferase 2 (Dgat2), genes essential for regulating the flux of TG between storage in CLDs and packaging into CMs [reviewed in 46, 47, 48] (Figure 3). Disrupted lipid trafficking is also observed in isolated enterocytes from Noc−/− animals exposed to radio-labeled FA, and Noc−/− enterocytes show higher intracellular accumulation and decreased release of lipoprotein particles into the medium [4]. Surprisingly, Noc−/− enterocytes display increased activity of microsomal triglyceride transfer protein (MTP), a protein responsible for the formation of CM particles [4], most likely a compensatory response to the increased accumulation of TG within the enterocyte. Although a precise mechanism of action for NOC in intestinal lipid trafficking has not yet been elucidated, these findings suggest that at least part of the lean phenotype observed in Noc−/− mice exposed to HFD is due to decreased release of intestinal lipid into the circulation. In addition to the importance of timing in chylomicron dynamics [49, 50], the observed sequestration of lipids in Noc−/− enterocytes emphasizes the fact that enterocytes, although not intuitively considered as a lipid storage organ, undergo active lipid dynamics [51]. Interestingly, Dgat1 KO mice exhibit similar intestinal lipid sequestration, thus manifesting a major defect in TG synthesis in enterocytes [48]. Given the absence of alteration of Dgat1 expression levels in the Noc−/− model [4], it is likely that NOC’s effect on intestinal lipid processing is through an independent mechanism and not the result of a direct effect on Dgat1 mRNA levels. [50, 52].
Figure 3. Proposed working hypothesis for the development of resistance to DIO in Noc−/− under high-fat diet feeding.
In Noc+/+ mice, fatty acids (FA) are processed from the lumen of the intestine to the peripheral blood circulation after undergoing chylomicron formation. This step requires the activity of diacylglycerol O-acyltransferase 2 (DGAT2) to transform FA into triglycerides (TG), adipose differentiation-related protein (ADRP) to transiently constitute a poolof TG, and adipose triglyceride lipase (ATGL) to release TG to the blood. In Noc−/− mice, Dgat2, Adrp and Atgl mRNA levels are decreased, thus reducing chylomicron formation in the enterocytes. Lipid sequestration results in decreased TG release and distribution to the periphery. These alterations contribute to the development of resistance to DIO in Noc−/− mice but could also lead to the development of insulin resistance via inflammatory mechanisms or disrupted nutrient sensing.
While trapped lipid in the enterocytes of Noc−/− mice may be beneficial to preventing DIO, intestinal lipid accumulation might have several detrimental consequences. Lipid metabolites can interfere directly with insulin signaling or produce an inflammatory response that likewise disrupts proper responses to insulin [43, 53–59]. Interestingly, Nocturnin has recently been implicated in the inflammatory response and its maintenance [60]. NOC is acutely induced by LPS in mouse embryonic fibroblasts (MEFs) [60] and MEFs harvested from Noc−/− mice showed drastically decreased levels of the pro-inflammatory transcript iNOS 24 hr after LPS treatment. This effect is not observed with another pro-inflammatory gene, TNFα. Data on mRNA stability also demonstrated that Noc deficiency decreased the half-life of iNOS, but not of TNFα mRNA. In addition, Noc−/− mice show increased survivability to LPS. The stabilization, rather than destabilization, of the iNOS message by NOC is surprising since NOC’s known activity is to deadenylate targets. Perhaps iNOS is an indirect target of NOC, or perhaps NOC has other deadenylase-independent functions. Regardless, these data indicate that NOC may be playing a role in the link between clocks and the inflammatory response [61, 62]. It has been proposed that type-2 diabetes might also be an inflammatory disease, as many immunological changes at the cytokine and chemokine levels occur along with the development of insulin-resistance phenotypes [63]. Since Noc function might be linked to glucose and insulin signaling [30], studies examining the existence or extent of gut inflammation in Noc−/− mice on a HFD will help assess if these animals are insulin resistant.
Another potential consequence of the increased intestinal lipid accumulation in Noc−/− mice could be disruption of the gut/brain axis, i.e. the central homeostatic regulation that is mediated through nutrient-sensing. There is increasing evidence to support hypothalamic control of energy and glucose homeostasis via nutrient-sensing mechanisms and disruption of these processes might contribute to metabolic diseases [64, 65]. Therefore, the observed lipid sequestration in Noc−/− enterocytes could trigger inappropriate sympathetic signaling to the brain in order to regulate energy metabolism and lipid trafficking. Further experiments are needed to determine whether NOC disruption impairs the intestine-to-brain communication and therefore alters the nutrient-sensing properties of the hypothalamus.
Nocturnin is a key regulator of the adipogenesis / osteogenesis balance
As discussed above, Noc expression is induced in fasted animals and after lipid administration [4, 5]. Nuclear receptors that bind ligands, which include steroid hormones and dietary lipids, also play critical role in nutrient sensing [reviewed in 66]. One such receptor is Peroxisome proliferator-activated receptor γ (PPARγ), a master regulator of adipogenesis. PPARγ is also involved in other metabolic processes such as fatty acid storage, glucose metabolism and osteogenesis [reviewed in 6]. Recent evidence from Noc−/− mice suggested a mechanistic link between NOC and PPARγ, as these mice exhibit altered hepatic Pparg rhythms [30]. Noc is also significantly induced in mesenchymal stromal cells (MSCs) transfected with Pparg2 (one of two major forms of the PPARγ protein expressed mainly in adipogenic cells) and treated with the PPARγ agonist rosiglitazone [4]. Together, these data suggest that Nocturnin is a transcriptional target of PPARγ [38]. As previously mentioned, a pro-adipogenic function has been associated with Noc as its expression is highly upregulated in cells undergoing adipogenesis and Noc expression is gradually increased in eWAT of mice fed a HFD over 4 generations [39, 40]. Expansion of adipose tissue occurs through development of preadipocyte progenitors both locally from the adipose tissue itself, and remotely through recruitment of progenitors from tissues such as bone [reviewed in 67]. Lineage determination in these cells is therefore important as they can differentiate into multiple cell types including adipocytes or osteoblasts. NOC has a vital role in promoting adipogenesis rather than osteoblastogenesis and this could occur through its interactions with PPARγ [39]. Cellular studies in HEK293 cells revealed that Noc and Pparg overexpression results in PPARγ nuclear, rather than cytoplasmic accumulation [39]. The mechanism behind this finding is not known, but is likely to be distinct from a deadenylase function, since this effect was observed using either NOC constructs with functional deadenylase activity or catalytically-dead mutant NOC. The ability of NOC to influence adipogenesis vs. osteogenesis could therefore occur through its mediation of PPARγ nuclear translocation.
Supporting this, Noc−/− mice exhibit an increased bone mass phenotype and decreased marrow adiposity [39]. Bone mass and structure correlates with Igf1 expression levels, themselves depending on PPARγ activity. Igf1 displays rhythmic expression in the murine femur antiphase to Noc [68]. Indeed, immunoprecipitation experiments revealed that a protein-RNA complex exists between NOC and Igf1 [68]. These findings reveal a unique place for NOC in the adipocyte program of both adipose and bone tissue. Profound changes in bone mass and body composition are associated not only with metabolic disturbances, but also with aging and Noc is proposed to play a role in both of these processes [7]. Evidence demonstrating that NOC’s effects on PPARγ occur independently of its deadenylase function lends further evidence for a new mechanism whereby NOC can contribute to the regulation of gene expression.
Concluding remarks and perspectives
There is increasing evidence supporting a pivotal role for NOC in lipid metabolism but many questions remain (Box 1). Rhythmic expression of Noc is seen in virtually all tissues and phenotypes have emerged implicating involvement of Nocturnin in lipogenesis, adipogenesis and osteogenesis. Noc expression is also responsive to a variety of metabolic signals including FoxO, PPARγ and dietary lipid. Expression levels and rhythms of several key genes necessary for maintaining proper lipid metabolism are altered in both intestine and liver of Noc−/− mice. Nocturnin may be regulating these transcripts directly or indirectly through its deadenylase activity and perhaps by deadenylase-independent functions as well. Future research will focus on using Noc conditional knockout mice to determine the tissue-specific functions of Nocturnin and how these contribute to lipid processing in the whole body. Furthermore, since Noc is widely expressed, it is likely that additional tissue-specific phenotypes in the Noc−/− mice will emerge with further study. Already, the observation that the Noc−/− mouse is resistant to DIO, yet has altered glucose/insulin signaling suggests that a number of different tissues and cell types may contribute to this phenotype. This will be an exciting area of study as Nocturnin may provide a point of dissociation between “beneficial” lipid processing deficits and detrimental effects of the metabolic syndrome such as obesity and diabetes.
Box 1. Outstanding Questions.
Does NOC contribute to the regulation of gene expression via mechanisms independent of its mRNA deadenylase activity?
Is the altered gene expression of lipid processing genes in Noc−/− mice due to a direct or indirect regulation by NOC?
What is the fate of excess lipid in Noc−/− intestines?
Does the increased sequestration of lipid in enterocytes of Noc−/− mice cause gut inflammation?
Are Noc−/− mice insulin resistant on a standard or HFD?
What role does NOC play in glucose/insulin signaling in muscle and liver?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberti KG, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 2.Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 3.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douris N, et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Current biology : CB. 2011;21:1347–1355. doi: 10.1016/j.cub.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert MR, et al. Nocturnin expression is induced by fasting in the white adipose tissue of restricted fed mice. PloS one. 2011;6:e17051–e17051. doi: 10.1371/journal.pone.0017051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai M, Rosen CJ. PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nature reviews. Endocrinology. 2010;6:629–636. doi: 10.1038/nrendo.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecka-Czernik B, et al. Skeletal aging and the adipocyte program: New insights from an "old" molecule. Cell cycle (Georgetown, Tex.) 2010;9:3648–3654. doi: 10.4161/cc.9.18.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Human Molecular Genetics. 2006;15(Spec No):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassone-Corsi P. Minireview: NAD+, a circadian metabolite with an epigenetic twist. Endocrinology. 2012;153:1–5. doi: 10.1210/en.2011-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Um JH, et al. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PloS one. 2011;6:e18450. doi: 10.1371/journal.pone.0018450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naimi M, et al. Energetic cell sensors: a key to metabolic homeostasis. Trends Endocrinol Metab. 2010;21:75–82. doi: 10.1016/j.tem.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Lin JD, et al. Integration of energy metabolism and the mammalian clock. Cell Cycle. 2008;7:453–457. doi: 10.4161/cc.7.4.5442. [DOI] [PubMed] [Google Scholar]
- 14.Asher G, Schibler U. Crosstalk between Components of Circadian and Metabolic Cycles in Mammals. Cell Metabolism. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Froy O. Metabolism and Circadian Rhythms-Implications for Obesity. Endocrine Reviews. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- 16.Peek CB, et al. Nutrient sensing and the circadian clock. Trends Endocrinol Metab. 2012 doi: 10.1016/j.tem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green CB, et al. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harbor Symposia on Quantitative Biology. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- 21.Kojima S, et al. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green CB, et al. Tryptophan hydroxylase mRNA levels are regulated by the circadian clock, temperature, and cAMP in chick pineal cells. Brain Res. 1996;738:1–7. doi: 10.1016/0006-8993(96)00743-3. [DOI] [PubMed] [Google Scholar]
- 23.Green CB, Besharse JC. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14884–14888. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garbarino-Pico E, et al. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupressoir A, et al. Identification of four families of yCCR4− and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Current Biology. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, et al. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Developmental Biology. 2001;1:9–9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, et al. CLOCK/BMAL1 regulates human nocturnin transcription through binding to the E-box of nocturnin promoter. Molecular and Cellular Biochemistry. 2008;317:169–177. doi: 10.1007/s11010-008-9846-x. [DOI] [PubMed] [Google Scholar]
- 29.Kojima S, et al. MicroRNA-122 Modulates the Rhythmic Expression Profile of the Circadian Deadenylase Nocturnin in Mouse Liver. Plos One. 2010;5 doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green CB, et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oishi K, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. Journal of Biological Chemistry. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 32.Kornmann B, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahar S, Sassone-Corsi P. Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab. 2012;23:1–8. doi: 10.1016/j.tem.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupressoir A, et al. Characterization of a mammalian gene related to the yeast CCR4 general transcription factor and revealed by transposon insertion. J Biol Chem. 1999;274:31068–31075. doi: 10.1074/jbc.274.43.31068. [DOI] [PubMed] [Google Scholar]
- 35.Balsalobre A, et al. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 36.Tsuchiya Y, et al. Temperature compensation and temperature resetting of circadian rhythms in mammalian cultured fibroblasts. Genes to cells : devoted to molecular & cellular mechanisms. 2003;8:713–720. doi: 10.1046/j.1365-2443.2003.00669.x. [DOI] [PubMed] [Google Scholar]
- 37.Prentki M, Madiraju SR. Glycerolipid metabolism and signaling in health and disease. Endocr Rev. 2008;29:647–676. doi: 10.1210/er.2008-0007. [DOI] [PubMed] [Google Scholar]
- 38.Kawai M, et al. Nocturnin: a circadian target of Pparg-induced adipogenesis. Annals of the New York Academy of Sciences. 2010;1192:131–138. doi: 10.1111/j.1749-6632.2009.05221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawai M, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massiera F, et al. A Western-like fat diet is sufficient to induce a gradual enhancement in fat mass over generations. Journal of Lipid Research. 2010 doi: 10.1194/jlr.M006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature reviews. Molecular cell biology. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuel VT, et al. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubota T, et al. Impaired insulin signaling in endothelial cells reduces insulin-induced glucose uptake by skeletal muscle. Cell Metab. 2011;13:294–307. doi: 10.1016/j.cmet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 45.Iqbal J, Hussain MM. Intestinal lipid absorption. American Journal of Physiology-Endocrinology and Metabolism. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bickel PE, et al. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochimica et Biophysica Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lass A, et al. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yen CLE, et al. DGAT enzymes and triacylglycerol biosynthesis. Journal of Lipid Research. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hussain MM, Pan X. Clock genes, intestinal transport and plasma lipid homeostasis. Trends in Endocrinology and Metabolism. 2009;20:177–185. doi: 10.1016/j.tem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. Journal of Lipid Research. 2009;50:1800–1813. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J, et al. A dynamic, cytoplasmic triacylglycerol pool in enterocytes revealed by ex vivo and in vivo coherent anti-Stokes Raman scattering imaging. J Lipid Res. 2009;50:1080–1089. doi: 10.1194/jlr.M800555-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gimble JM, Floyd ZE. Metabolism: What Causes the Gut's Circadian Instincts? Current biology : CB. 2011;21:R624–R626. doi: 10.1016/j.cub.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Ding S, Lund PK. Role of intestinal inflammation as an early event in obesity and insulin resistance. Curr Opin Clin Nutr Metab Care. 2011;14:328–333. doi: 10.1097/MCO.0b013e3283478727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bradley WD, et al. The emerging role of the intestine in metabolic diseases. Arch Physiol Biochem. 2011;117:165–176. doi: 10.3109/13813455.2011.578651. [DOI] [PubMed] [Google Scholar]
- 55.Hsieh J, et al. Postprandial dyslipidemia in insulin resistance: mechanisms and role of intestinal insulin sensitivity. Atheroscler Suppl. 2008;9:7–13. doi: 10.1016/j.atherosclerosissup.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Ji Y, et al. Nutrient-induced inflammation in the intestine. Curr Opin Clin Nutr Metab Care. 2011;14:315–321. doi: 10.1097/MCO.0b013e3283476e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serino M, et al. Lipid-induced peroxidation in the intestine is involved in glucose homeostasis imbalance in mice. PLoS One. 2011;6:e21184. doi: 10.1371/journal.pone.0021184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schenk S, et al. Insulin sensitivity: modulation by nutrients and inflammation. The Journal of clinical investigation. 2008;118:2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kahn SE, et al. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 60.Niu S, et al. The circadian deadenylase Nocturnin is necessary for stabilization of the iNOS mRNA in mice. PloS one. 2011;6:e26954. doi: 10.1371/journal.pone.0026954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibbs JE, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castanon-Cervantes O, et al. Dysregulation of inflammatory responses by chronic circadian disruption. Journal of Immunology. 2010;185:5796–5805. doi: 10.4049/jimmunol.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11:98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- 64.Lam TKT. Neuronal regulation of homeostasis by nutrient sensing. Nature medicine. 2010;16:392–395. doi: 10.1038/nm0410-392. [DOI] [PubMed] [Google Scholar]
- 65.Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metab. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang X, et al. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- 67.Hausman GJ, Hausman DB. Search for the preadipocyte progenitor cell. J Clin Invest. 2006;116:3103–3106. doi: 10.1172/JCI30666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawai M, et al. Nocturnin Suppresses Igf1 Expression in Bone by Targeting the 3' Untranslated Region of Igf1 mRNA. Endocrinology. 2010 doi: 10.1210/en.2010-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]