Abstract
Interspecific hybridization and allopolyploidization contribute to the origin of many important crops. Synthetic Brassica is a widely used model for the study of genetic recombination and “fixed heterosis” in allopolyploids. To investigate the effects of the cytoplasm and genome combinations on meiotic recombination, we produced digenomic diploid and triploid hybrids and trigenomic triploid hybrids from the reciprocal crosses of three Brassica diploids (B. rapa, AA; B. nigra, BB; B. oleracea, CC). The chromosomes in the resultant hybrids were doubled to obtain three allotetraploids (B. juncea, AA.BB; B. napus, AA.CC; B. carinata, BB.CC). Intra- and intergenomic chromosome pairings in these hybrids were quantified using genomic in situ hybridization and BAC-FISH. The level of intra- and intergenomic pairings varied significantly, depending on the genome combinations and the cytoplasmic background and/or their interaction. The extent of intragenomic pairing was less than that of intergenomic pairing within each genome. The extent of pairing variations within the B genome was less than that within the A and C genomes, each of which had a similar extent of pairing. Synthetic allotetraploids exhibited nondiploidized meiotic behavior, and their chromosomal instabilities were correlated with the relationship of the genomes and cytoplasmic background. Our results highlight the specific roles of the cytoplasm and genome to the chromosomal behaviors of hybrids and allopolyploids.
Keywords: synthetic Brassica, chromosome pairing, fluorescence in situ hybridization, interspecific hybridization, allopolyploid, cytoplasmic effect
POLYPLOIDY has played a crucial role in the evolutionary history of higher plants. Up to 80% of flowering plant species have been estimated to have undergone one or more polyploidization events in their ancestry (Masterson 1994; Ramsey and Schemske 1998; Otto 2007; Wood et al. 2009). Interspecific hybridization and allopolyploidization contribute to the origin of many important crops, including canola (Brassica), cotton (Gossypium), tobacco (Nicotiana), and wheat (Triticum). Among the six Brassica crops in the U-triangle (Nagaharu 1935), Brassica carinata Braun (2n = 34, BBCC), B. juncea (L.) Czern. (2n = 36, AABB), and B. napus L. (2n = 38, AACC) are allotetraploids, which originated naturally through convergent alloploid evolution between any two of the three diploid species B. nigra (L.) Koch (2n = 16, BB), B. oleracea L. (2n = 18, CC), and B. rapa L. (syn. B. campestris, 2n = 20, AA). This complex of diploids and allopolyploids is now considered as a model system for studying polyploidization in crop species (Lukens et al. 2006; Pires et al. 2006). Synthetic Brassica, especially B. napus, has become one of the most widely used models to study the genetic and epigenetic alterations caused by meiosis-driven genome reshuffling in allopolyploids (Gaeta et al. 2007; Nicolas et al. 2007, 2009; Szadkowski et al. 2010, 2011; Xiong et al. 2011) since the seminal work of Song et al. (1995).
Previous studies suggested that the genomes from two ancestral diploids in natural Brassica allotetraploids have different stabilities and that cytoplasm has exerted considerable influence on the evolution of nuclear genomes of alloploids (Prakash et al. 2009). It has been confirmed that B. nigra and B. rapa have contributed the cytoplasm to B. carinata and B. juncea, respectively. However, it is still uncertain about the cytoplasm donor of B. napus. B. rapa has been suggested as a potential plastid genome donor to B. napus (Flannery et al. 2006; Allender and King 2010). When the parental diploid species of allopolyploid has highly differentiated cytoplasm, as in B. juncea and B. carinata, the nuclear genomes contributed by the male parents are considerably altered compared to the nuclear genomes of female parents (Song et al. 1988, 1995). The A genome in B. juncea has remained mostly intact while the B genome has changed considerably; the B genome in B. carinata has unchanged but the C genome has considerably altered. In B. napus, both A and C genomes have undergone a similar extent of changes. Nevertheless, recent comparative sequence analysis between homeologous genome segments of B. napus and its two progenitor species showed that the C-genome segments were expanded in size relative to their A-genome counterparts in the majority of the genomic regions studied and revealed that the C genome is more vulnerable to undergoing changes than the A genome after the formation of B. napus (Cheung et al. 2009). The cytoplasm background of resynthesized B. napus from its progenitors significantly affects the transmission frequency of the meiotic-driven genetic changes to the progenies (Szadkowski et al. 2010).
In spite of sharing considerable homeology between the partaking genomes, these Brassica allopolyploid species exclusively exhibit diploid-like meiosis. It has been proposed that diploid-like meiosis is genetically regulated in Brassica and its related genera (Prakash et al. 2009). However, so far only the major gene PrBn (Pairing regulator in Brassica napus) in B. napus was demonstrated to be responsible for inhibiting the homeologous pairing in its haploids (Jenczewski et al. 2003), which has been mapped on linkage group C9, and to display incomplete penetrance (Liu et al. 2006). It was further shown that the variation in crossover frequency among B. napus accessions representing a range of genetic and geographic origins roughly correlates with the multiple origins of B. napus and PrBn diversity (Cifuentes et al. 2010). The findings highlight the diverse nature of homeologous recombination regulation in the wild. The diploid-like meiotic behavior of allopolyploids is also thought to result from the divergence between homeologous chromosomes, which may already exist and/or be accentuated at the onset of polyploid formation (Le Comber et al. 2010), and involve the rearrangement of large chromosome fragments (reviewed in Jenczewski and Alix 2004).
Genome structure and genomic relationships of the six Brassica species have been extensively investigated in the diversely originated natural types and artificially synthesized Brassica crop species (for review see Prakash et al. 2009), which, however, makes it difficult to compare the results from different reports because the extent of homeologous recombination in resynthesized B. napus is influenced by progenitor genotype and/or combination (Prakash and Hinata 1980; Attia et al. 1986) and cytoplasm backgrounds (Szadkowski et al. 2010). In the present investigation, the di- and trigenomic hybrids were produced from the reciprocal pair crosses of three Brassica diploids to study and compare the precise effects of genome combinations and cytoplasm on the meiotic recombination of each genome through genomic in situ hybridization (GISH) and BAC-FISH. The chromosome pairing in the synthetic Brassica allotetraploids showed nondiploidized behavior and was related to the genome affinity and cytoplasmic background. Our results provide new insight into the effects of the cytoplasm background and genome combinations on the chromosomal recombination in Brassica hybrids and allopolyploids and, more importantly, on the chromosome stability and diploidization in synthetic allopolyploids.
Materials and Methods
Plant material and crosses
Each genotype or cultivar of three cultivated Brassica diploids was used as parents in reciprocal crosses to produce interspecific hybrids: B. rapa (2n = 20, AA genome, genotype 3H120), B. nigra (L.) Koch cv. Giebra (2n = 16, BB), and B. oleracea var. alboglabra L. (2n = 18, CC, genotype Chi Jie Lan) (Figure 1). These materials were grown in the experimental fields on the campus of Huazhong Agricultural University, Wuhan, China. One plant in each genotype was selected for the reciprocal crosses performed by hand emasculation and pollination. After ∼20 days of pollination, the immature embryos were cultured on MS medium (Murashige and Skoog 1962). To ensure the homogenous identity of the hybrid plants from the same combination, the plantlet from one embryo of each cross was successively subcultured on MS medium with 1.5 mg/liter−1 6-benzyl aminopurine (6-BA) and 0.25 mg/liter−1 α-naphthalenacetic acid (NAA) to generate enough cloned plantlets for study. The cloned plantlets were cultured on MS agar medium with 1.5 mg/liter−1 6-BA, 0.25 mg/liter−1 NAA, and 100 mg/liter−1 colchicine for ∼10 days to double the number of chromosomes for synthesizing the allotetroploids and subsequently transferred to MS medium without colchicine until plantlets were regenerated from callus. Three allotetraploids (AA.BB, AA.CC, and BB.CC) were synthesized by doubling the chromosome numbers of the respective digenomic hybrids (see below), and CC.AA was directly obtained from the cultured embryo-plantlet, probably by the spontaneous chromosome doubling in vitro.
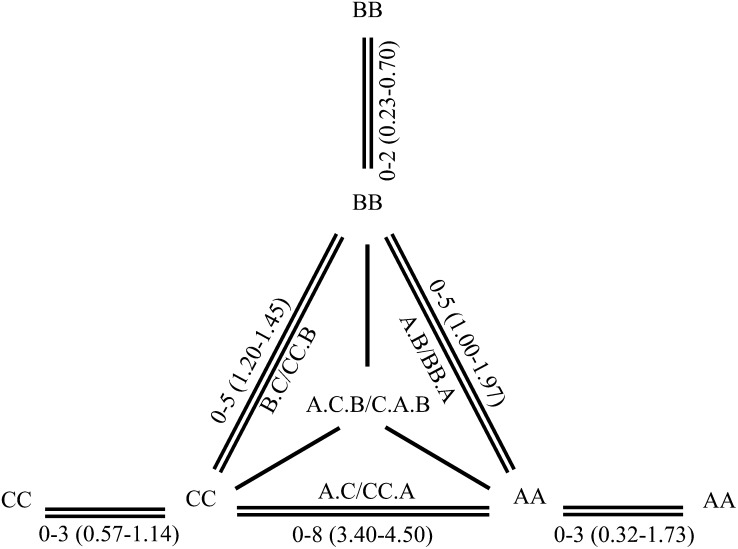
Figure 1 .
Schematic illustration showing the production of hybrids from pair crosses of three Brassica diploids and their chromosome pairing. AA, BB, and CC designate B. rapa (AA genome), B. nigra (BB genome), and B. oleracea (CC genome). The digenomic diploid or triploid hybrids obtained are indicated between two parents, and the trigenomic triploids are at the center of the triangle. The ranges and averages of allosyndetic bivalents between three pairs of genomes in digenomic and trigenomic hybrids are shown on the double lines connecting two parents, and those of autosyndetic bivalents are shown on the double lines connecting the same genome at the three joints.
Cytological investigation and pollen fertility analysis
The ovaries from young flower buds were collected and treated with 8-hydroxyquinoline for 3–4 hr at room temperature before fixed in Carnoy’s solution I (3:1 ethanol:glacial acetic acid, v/v) and stored at –20° for further chromosome counting in somatic cells. The young flower buds were fixed directly in Carnoy’s solution I and stored at −20° for meiosis study. Cytogenetic observation was carried out according to the methods of Li et al. (1995). More than 300 pollen grains from three flowers of each plant were stained with acetocarmine (1%), and the percentage of stainable pollen grains was calculated to measure pollen fertility.
Probes and chromosome preparations
The probes used for GISH and BAC-FISH were the following: (1) total genomic DNA of B. nigra cv. Giebra labeled with biotin-11-dUTP (Fermentas) by nick translation; (2) the plasmid DNA of BAC BoB014O06 (provided by Susan J. Armstrong, University of Birmingham, Birmingham, UK) labeled with biotin-11-dCTP (for A.C and CC.A hybrids) by random priming using the BioPrime DNA Labeling System kit (Invitrogen, Life Technologies) or with digoxigenin-11-dUTP (Roche, Basel, Switzerland) (for A.C.B combination) by random priming using the BioPrime Array CGH Genomic Labeling System kit according to the manufacturer’s protocol (Invitrogen, Life Technologies).
Total genomic DNA of B. rapa (3H120) and 45S rDNA was boiled for 15 min twice to obtain DNA fragments of 100–500 bp and used as blocks. The 45S rDNA was used to replace the intergenic spacer of the B. oleracea 45S rDNA as the block agent (Howell et al. 2008) to reduce the intensity of strong signals and to produce a more even distribution of hybridization signal intensity. The C0t-1 DNA was prepared from B. oleracea A12DHd genomic DNA (Zwick et al. 1997). Chromosome preparation for FISH was carried out according to Zhong et al. (1996) with minor modifications (Ge and Li 2007); an enzyme mixture containing 0.6% cellulase Onozuka RS (Yakult, Tokyo), 0.2% pectinase (Merck, Darmstadt, Germany), and 0.5% snailase (Beijing Baitai Biochem, Beijing) was used for digestion.
GISH and BAC-FISH
Slides were pretreated with 0.01% pepsin in 10 mM HCl for 20 min and with RNase in 2× SSC (DNase-free, 100 µg/ml, 1 hr at 37°), fixed in 4% paraformaldehyde for 10 min, dehydrated in 75 and 100% ethanol for 3 min each, and air dried. Hybridization mixture contained 50% deionized formamide, 2× SSC, 10% dextran sulfate, 0.5% SDS, and 100-ng probes for each slide, while ∼1000 ng blocking A-genome DNA, 1 µg C0t-1 DNA to block repeated sequence, and 100 ng 45S rDNA were added for blocking in A.C, CC.A, and A.C.B hybrids. The mixture was denatured at 70° for 10 min. The probes and chromosomal DNA on the slides were co-denatured at 80° for 5 min in a thermal cycler and hybridized at 37° overnight in a humid chamber. Slides were washed stringently for 10 min in 0.1× SSC with 20% deionized formamide at 42°. The immunodetection of biotinylated and digoxigenated DNA probe was carried out by using Cy3-labeled streptavidin (KPL, St. Louis) and anti-digoxigenin conjugate-FITC (Roch, Basel, Switzerland), respectively. Finally, preparations were counterstained with 4′-6-diamidino-2-phenylindole (DAPI) solution (Roche, Basel, Switzerland) (1 mg/ml), mounted in antifade solution (Vector Laboratories, Peterborough, UK).
Image capturing, processing, and statistical analysis
All images were captured with a CCD camera attached to a fluorescence microscope (Nikon Eclipse 80i). Images were processed by Adobe Photoshop (Adobe Systems, San Jose, CA) to adjust contrast and brightness. Two-by-two chi-square contingency tests were used to test the difference in pairing configuration.
Results
Morphology, cytology, and pollen fertility in hybrids
From reciprocal pair crosses of three Brassica diploids, the following digenomic diploid and triploid hybrids were produced: A.B/BB.A, A.C/CC.A, and B.C/CC.B (Figure 1). The production of hybrids (BB.A, CC.A, and CC.B) likely resulted from the fusion of unreduced gametes by the female parent and reduced gametes by the male parent in the three crosses, although the possibility of chromosome duplication for one genome during the mitotic divisions of the zygote after fertilization could not be excluded. Two trigenomic hybrids (A.C.B and C.A.B) were obtained from the crosses between the synthesized B. napus (AA.CC/CC.AA) and B. nigra, which contained the cytoplasm of B. rapa and B. oleracea, respectively. No hybrids were obtained from the crosses synthetic B. juncea (AA.BB) × B. oleracea and synthetic B. carinata (BB.CC) × B. rapa, although many pollinations were carried out. The crossability between the synthesized allotetraploids and the diploids was quite low.
These hybrids generally showed an intermediate morphology, while the reciprocal hybrids inclined more to the maternal parents, especially BB.A, CC.A, and CC.B, probably due to the one more copy of the maternal genome (Supporting Information, Figure S1). The three digenomic hybrids (A.B, A.C, B.C) had very low pollen fertility, and the two trigenomic hybrids (A.C.B, C.A.B) were male sterile. Among the three digenomic triploids (BB.A, CC.A, CC.B), BB.A and CC.B had much higher pollen fertility than CC.A and produced some seeds after open pollination. The pairing frequency in A.C was obviously higher than in A.B and B.C (Table S1), indicating the close homology between A and C genomes. The average of bivalents in C.A.B was significantly lower than that in A.C.B (χ2 = 927.48, P < 0.01), indicating the cytoplasmic effect on the chromosome pairing. The pairing frequency in BB.A and CC.B was similar, but much lower than that in CC.A. The pairing differences in these hybrids were further analyzed using FISH to examine the intra- and intergenomic pairing.
Variable chromosome pairing in different genome combinations
Parental chromosomes in the A.B, B.C, BB.A, and CC.B hybrids were easily distinguished by using the labeled B. nigra genomic DNA probe and those in the A.C and CC.A hybrids by using the B. oleracea BoB014O06 probe. The chromosomes from A, B, and C genomes in the A.C.B hybrid were identified by dual-color FISH with the labeled B. nigra DNA and BoB014O06 probes. The BoB014O06 hybridized strongly to the centromeric region; therefore, 45S rDNA and C0t-1 DNA blocking in hybridization mixture could reduce the intensity of the strong signals and produce an even hybridization signal intensity distribution. Additionally, the chromosome pairings within each genome (autosyndesis) and between different genomes (allosyndesis) were quantified and compared (Table 1 and Table 2).
Table 1 . Chromosome associations in PMCs of hybrids at diakinesis and metaphase I as revealed by FISH.
| Chromosome associations (ranges) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | |||||||||||||
| Hybrids | 2n | Total | IA | IB | IC | Total | IIA-A | IIB-B | IIC-C | IIA-B | IIA-C | IIB-C | Total | Total | PMCs with multivalent (%) | Total PMCs |
| A.B | 18 | 8.26 (3–16) | 4.74a (2–9) | 3.53b (1–7) | — | 2.13 (0–5) | 0.53b (0–3) | 0.23c (0–2) | — | 1.38c (0–5) | — | — | 0.75 (0–3) | 0.70 (0–2) | 86.89 | 61 |
| A.C | 19 | 4.86 (1–13) | 2.64b (0–8) | — | 2.23a (0–5) | 5.77 (3–9) | 1.18a (0–3) | — | 1.14b (0–3) | — | 3.45c (0–7) | — | 0.86 (0–3) | — | 83.05 | 22 |
| B.C | 17 | 9.43 (3–13) | — | 4.72a (1–7) | 4.72d (1–9) | 2.10 (0–5) | — | 0.30b,c (0–2) | 0.57c (0–2) | — | — | 1.23c (0–5) | 0.65 (0–3) | 0.30 (0–1) | 74.17 | 120 |
| A.C.B | 27 | 5.27 (2–8) | 0.64c (0–2) | 4.32a (2–7) | 0.32c (0–2) | 10.32 (2–12) | 1.73d (0–3) | 0.45b,a (0–1) | 1.14b (0–3) | 1.00c (0–3) | 4.50b (1–8) | 1.50c (0–5) | 0.41 (0–1) | — | 40.91 | 22 |
| BB.A | 26 | 7.21 (3–10) | 5.65d(1–9) | 1.56c (0–6) | — | 8.12 (6–10) | 0.65b (0–2) | 5.35d (2–7) | — | 2.12b (0–5) | — | — | 0.85 (0–2) | — | 67.65 | 34 |
| CC.A | 28 | 5.91 (3–10) | 5.18a,d (2–9) | — | 0.74c (0–4) | 10.38 (8–12) | 0.32c (0–2) | — | 6.29a (2–9) | — | 3.76c (0–8) | — | 0.44 (0–1) | — | 44.12 | 34 |
| CC.B | 26 | 6.40 (4–14) | — | 4.80a (3–7) | 1.60b (0–7) | 8.70 (6–10) | — | 0.70a (0–1) | 6.70a (5–8) | — | — | 1.20c (0–3) | 0.80 (0–2) | — | 70 | 10 |
I, univalent; II, bivalent; III, trivalent; IV, quadrivalent. IA, IB, and IC indicate univalents belonging to the A, B, and C genomes, respectively; IIA-A, IIB-B, and IIC-C indicate autosyndetic bivalents formed between two chromosomes of A, B, or C genomes in A.B, A.C, B.C, and A.C.B hybrids, but IIB-B in BB.A and IIC-C in CC.A and CC.B indicate homologous bivalents; IIA-B, IIA-C, and IIB-C indicate allosyndetic bivalents formed between A and B chromosomes, A and C chromosomes, and B and C chromosomes, respectively.
Groups significantly different by χ2 -test, p < 0.05.
Table 2 . Means of chromosome numbers and ratios for no pairing, autosyndesis, and allosyndesis within each genome in hybrids.
| A genome | B genome | C genome | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Hybrids | No pairing | Autosyndesis | Allosyndesis | No pairing | Autosyndesis | Allosyndesis | No pairing | Autosyndesis | Allosyndesis |
| A.B | 4.74 (0.47a) | 1.93b (0.19b,*) | 3.33b (0.33c,**) | 3.53 (0.44b) | 1.15c (0.14c,*) | 3.33b (0.42b**) | — | — | — |
| A.C | 2.64 (0.26b) | 3.50a (0.35a,*) | 3.86b (0.39c,**) | — | — | — | 2.23 (0.25b) | 2.91d (0.32b,*) | 3.86b (0.43b,**) |
| B.C | — | — | — | 4.72 (0.59a) | 1.23c (0.15c,*) | 2.06c (0.26c,**) | 4.72 (0.53a) | 2.23a (0.25b,c,*) | 2.06c (0.23c,**) |
| A.C.B | 0.64 (0.06c) | 3.68a (0.37a,*) | 5.68a (0.57b,**) | 4.32 (0.54a,b) | 1.00c (0.13c) | 2.68c, b (0.34c,b) | 0.32 (0.04c) | 2.36a,d (0.26b,c,*) | 6.32a (0.70a,**) |
| BB.A | 5.65 (0.57a) | 1.68b (0.17b,c,*) | 2.68c (0.27c,**) | 1.56 (0.20c) | 1.06c (0.13c,*) | 2.68b,c (0.34b,c,**) | — | — | — |
| CC.A | 5.18 (0.52a) | 0.74c (0.07c,*) | 4.09b (0.41c,**) | — | — | — | 0.74 (0.08c) | 0.59c (0.07c,*) | 4.09b (0.45b,**) |
| CC.B | — | — | — | 4.80 (0.60a) | 1.40c (0.18c,*) | 1.80c (0.23c,**) | 1.60 (0.18b,c) | 1.20b (0.13b,c,*) | 1.80c (0.20c,**) |
| Average | 4.21 (0.42) | 2.07 (0.21*) | 3.72 (0.37**) | 4.34 (0.54) | 1.19 (0.15*) | 2.47 (0.31**) | 3.79 (0.42) | 2.34 (0.26*) | 2.87 (0.32**) |
For the calculation of the total chromosome numbers involved in autosyndesis within each genome in hybrids (A.B, A.C, B.C, A.C.B), the bivalent, trivalent, and quadrivalent of the same genome mean two, three, and four chromosomes, while the trivalent of two genomes, such as A-A-C, is taken as 1.5 A-genome chromosomes for autosyndesis and 0.5 A-genome chromosome and 1 C-genome chromosome for allosyndesis, and the quadrivalent (A-A-A-C) is divided into 2.5 A-genome chromosomes for autosyndesis and 0.5 A-genome chromosome and 1 C-genome chromosome for allosyndesis. For the calculation of the total chromosome number for allosyndesis within each genome, the bivalent means one chromosome for one of two genomes, the chromosome shared by two associations in trivalent or quadrivalent is taken as one-half for one genome and another half for the other genome. The ratios of autosyndesis, allosyndesis, and no pairing within each genome are the observed chromosome numbers/the total chromosome numbers of each genome. In digenomic triploid hybrids (BB.A, CC.A, and CC.B), the homologous bivalents in Table 1 are excluded from the chromosomes for autosyndesis. For the division of pairing in trivalents, the B-B pairing in A-B-B of BB.A is assumed homologous pairing, and A-B pairing is allosyndetic pairing; for B-B-B, both homologous and autosyndetic pairings are assumed. The same calculation is made for CC.A and CC.B. Single (*) and double (**) asterisks indicate that the averages of autosyndesis or allosyndesis, respectively, from different genomes in each hybrid are insignificantly different (the chi-square test: α = 0.05).
Groups significantly different by χ2 -test, p < 0.05.
In A.B, the average of univalents for A genome (4.74) was higher than that for B genome (3.53); the difference might result from the presence of two more chromosomes in the A genome than in the B genome because their rates of the univalents to the chromosome number of the genome were nearly the same. The maximum autosyndetic bivalent of the A genome was 3, one more than that of the B genome, and the average of the A genome (0.53) was higher than that of the B genome (0.23). The average and maximum of allosyndetic bivalents were 1.38 and 5, respectively—much higher than those of autosyndesis (Table 1). The trivalents A-A-B and A-B-B occurred in 37.7 and 26.23% of pollen mother cells (PMCs), respectively (Table S2). The total average chromosomes per cell and the rates for autosyndesis within the A or B genome were significantly lower than those for allosyndesis (Table 2). The average of the autosyndetic chromosomes in A genome (1.93) was higher than that in B genome (1.15), but the rates were similar. The same 3.33 chromosomes showed allosyndesis in the two genomes at similar rates (0.33 and 0.42). In A.C, the average and the rate of univalents for A (2.64/0.26) and C genomes (2.23/0.25) were comparable; the autosyndetic bivalents within A and C genomes were 1.18 and 1.14, with a maximum of 3.0. The average (3.45) and maximum (7.0) of allosyndetic bivalents were much higher than those of autosyndesis (Table 1). The trivalents A-A-C and A-C-C were observed in 27.27 and 22.73% of PMCs, respectively (Table S2). Total numbers of chromosomes and rates for autosyndesis were 3.50 and 0.35 in the A genome and 2.91 and 0.32 in the C genome, which are similar numbers to those for allosyndesis, the same 3.86 chromosomes at the similar rates (0.39 and 0.43) (Table 2). In B.C, the univalent frequency for both B and C genomes was 4.72; the autosyndetic bivalents for B genome were 0.30, lower than 0.57 for C genome. The average and maximum of allosyndetic bivalents were 1.23 and 5.0, respectively—much higher than those of autosyndesis (Table 1, Figure 2C). The trivalents B-B-C and B-C-C appeared in 23.23% of PMCs (Table S2). The chromosomes for allosyndesis in B genome (2.06) were significantly higher than those for autosyndesis (1.23), but not for the rates (0.26, 0.15). The C genome had similar chromosomes for auto- and allosyndesis. The B and C genomes had different chromosomes (1.23 and 2.23) for autosyndesis, respectively, but the rates were similar (0.15 vs. 0.25). They happened to have the same 2.06 chromosomes for allosyndesis (Table 2). In A.C.B, the univalent frequencies for A and C genomes (0.64, 0.32) were much lower than for the B genome (4.32). Inversely, the autosyndetic bivalents within A and C genomes (1.73, 1.14) were much higher than within the B genome (0.45). The average and maximum of allosyndetic bivalents were 4.50 and 8.0 for A.C, >1.00 and 3.0 for A.B, and 1.50 and 5.0 for B.C, respectively, (Table 1). Trivalents A-B-C and A-A-A were observed in 18.18 and 13.64% of PMCs (Table S2). As a whole, the chromosome frequencies for autosyndesis were significantly lower than those for allosyndesis in all three genomes. The chromosomes and rates for autosyndesis in A and C genomes (3.68/0.37 and 2.36/0.26) were similar but higher than those in B genome (1.00/0.13), and the values for allosyndesis in A and C genomes (5.68/0.57 and 6.32/0.70) were also similar but higher than those in B genome (2.68/0.34) (Table 2).
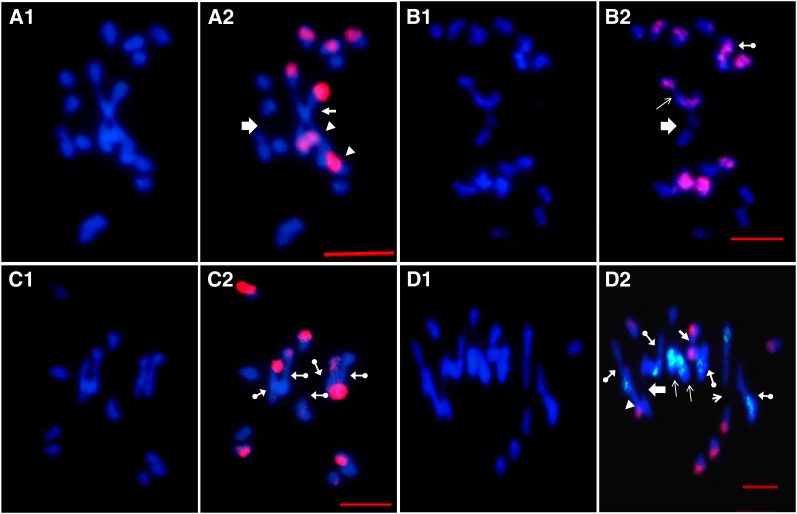
Figure 2 .
GISH/BAC-FISH analyses of meiotic chromosome pairings in PMCs of Brassica hybrids. DAPI (blue) and merged images are given for each cell. (A1 and A2) One diakinesis PMC from A.B with the following pairing: 6 IA + 4 IB + 1 IIA-A (big arrow) + 1 IIB-B (solid arrow) + 2 IIA-B (solid arrowhead). Red signals are from labeled B. nigra probe. (B1 and B2) One diakinesis PMC from A.C with the following pairing: 7 IA + 6 IC +1 IIA-A (open arrow) + 1 IIC-C (arrow) + 1IIA-C (solid arrows with ball tail). Red signals are from C genome specific BAC BoB014O06. (C1 and C2) One Metaphase I (MI) PMC from B.C with the following pairing: 3 IB + 4 IC + 5 IIB-C (solid arrow with ball tail). Red signals are from B. nigra probe. (D1 and D2) One MI PMC from A.C.B with the following pairing: 3 IA +4 IB + 1 IIA-A (big arrow) + 1 IIB-B (solid arrow) + 2 IIC-C (arrow) + 1 IIA-B (solid arrowhead) + 4 IIA-C (solid arrow with ball tail) + 1 IIB-C (arrowhead). Red signals are from B. nigra probe, and green signals are from the C-genome-specific BAC BoB014O06. All bars, 5μm.
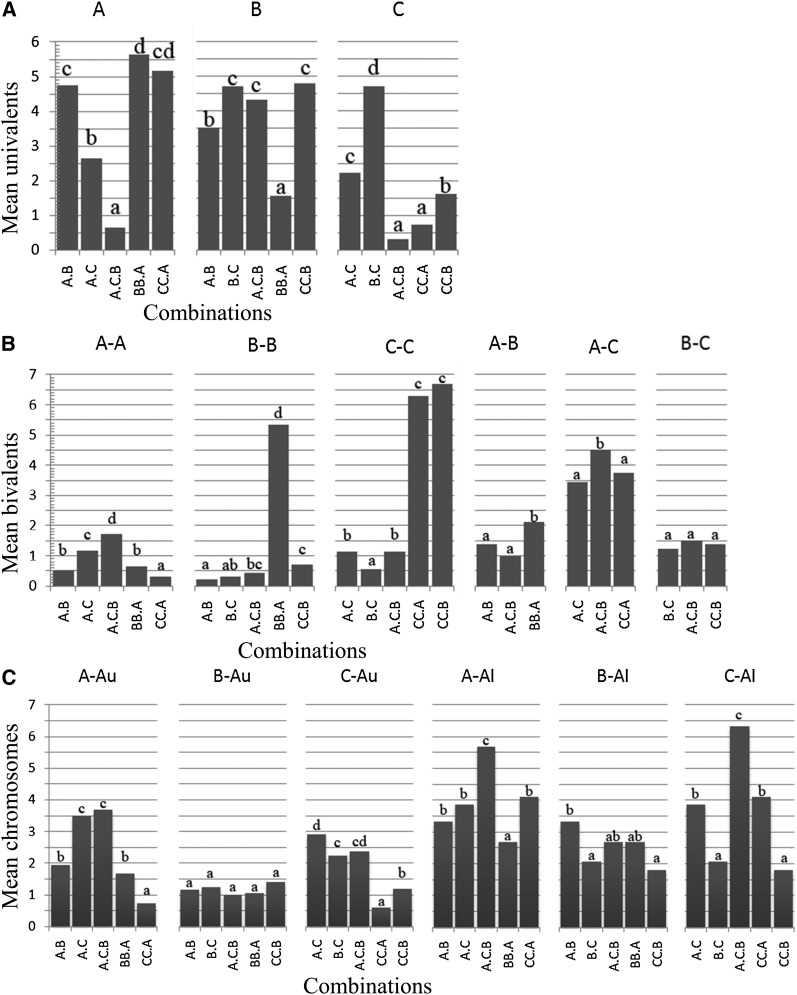
Among the three digenomic diploid hybrids A.B, A.C, and B.C, the average of bivalents in A.C was higher than that in A.B and B.C. Such difference resulted mainly from higher allosyndetic bivalents for A-C (3.45) than for A-B (1.38) and B-C (1.50) (Table 1, Figure 3B). Moreover, the bivalents and chromosomes of autosyndesis within A and C genomes in A.C was much higher than that for A genome in A.B and for C genome in B.C, respectively (Table 1 and Table 2). The average of bivalents in A.B and B.C was similar. In triploid A.C.B, the univalents were mainly from B genome (Tables 1 and 2; Figure 3A). The autosyndetic bivalents and chromosomes within A and C genomes were similar to those in A.C, but autosyndesis for B genome was similar to A.B and B.C (Table 1 and Table 2; Figure 3, B and C). The average of allosyndetic bivalents for A-B and B-C was comparable to A.B and B.C hybrids, respectively, and that for A-C was higher than in A.C. The total chromosomes involving in allosyndesis in A genome were significantly higher in A.C.B than in A.C and A.B, while those in A.C and A.B were similar. The values in C genome were also higher in A.C.B than in A.C and B.C, and those in A.C were higher than in B.C. The values of B genome in A.C.B were similar to those in A.B or B.C, but the values in A.B were higher than in B.C (Table 2; Figure 3C). These pairing results not only showed that A and C genomes were more closely related than A and B genomes or B and C genomes, but also that the auto- and allosyndesis was suppressed or enhanced simultaneously. The pairing involving B genome was less affected, while that involving A or C genome was prone to more alterations.
Figure 3 .
Comparisons of the averages of univalents within each genome (A), autosyndetic and allosyndetic bivalents in hybrids (B), and the averages of chromosomes involving autosyndesis and allosyndesis (Au, Al) within each genome across hybrids (C). Shared letters within each type denote that the values are not significantly different (contingent chi-square tests: α = 0.05).
In three digenomic triploid hybrids (BB.A, CC.A, CC.B), the chromosomes from one parent existed as homozygous pairs from chromosome duplication and others were in haploidy state. In BB.A, the average and maximum of autosyndetic bivalents within A genome were 0.65 and 2.0, respectively—fewer than those of A-B allosyndetic bivalents (2.12 and 5.0). The average and maximum of homologous bivalents within B genome were 5.35 and 7.0, which means that allosyndesis occurred in all cells. Trivalents A-B-B and B-B-B occurred in 38.24 and 17.65% of PMCs, respectively. The 1.68 chromosomes for autosyndesis in A genome were fewer than the 2.68 chromosomes for allosyndesis. Of 16 chromosomes of B genome, 10.70 showed homologous pairing, 1.06 autosyndetic pairing, 2.68 allosyndetic pairing, and 1.56 nonpairing (Table 2). In CC.A, the average and maximum of autosyndetic bivalents within A genome were 0.32 and 2.0, respectively, which is fewer than those of A-C allosyndetic bivalents (3.76 and 8.0). The average and maximum of homologous bivalents within C genome were 6.29 and 9.0, respectively. Trivalents A-C-C and C-C-C occurred in 26.47 and 11.76% of PMCs, respectively. The 0.74 chromosomes for autosyndesis in A genome were much fewer than 4.09 for allosyndesis. Among 18 chromosomes of C genome, 12.58 formed homologous bivalents, 0.60 and 4.09 gave auto- and allosyndesis, and 0.74 unpaired (Table 2). The autosyndetic bivalents within A genome in BB.A (0.65) was significantly higher than that in CC.A (0.32) and also for the chromosomes for autosyndesis (Table 2 and Table 3). However, the frequency of allosyndetic bivalents for A-B (2.12) was lower than that for A-C (3.76), and the chromosomes for allosyndesis in A genome in BB.A (2.68) were also fewer than 4.09 in CC.A (Table 2). In CC.B, the average and maximum of autosyndetic bivalents within B genome were 0.70 and 1.0, respectively, which were lower than those of B-C allosyndetic bivalents (1.20 and 3.0). The average and maximum of homologous pairing within C genome were 6.70 and 8.0, respectively. Trivalents B-C-C and C-C-C were observed in 50 and 10% of PMCs, respectively. The chromosomes for auto- and allosyndesis in B genome were similar (1.40 and 1.80, respectively). The associations of 18 chromosomes of C genome occurred as 13.40 for homologous pairing, 1.20 for autosyndesis, 1.80 for allosyndesis, and 1.60 for no pairing (Table 2). The number of chromosomes in C genome for allosyndesis (4.09) in CC.A were much higher than those in CC.B (1.80), while the chromosomes for autosyndesis in CC.A (0.59) were fewer than those in CC.B (1.20).
Table 3 . Meiotic pairings and pollen fertility in synthetic allotetraploids.
| I | II | III | IV | |||||
|---|---|---|---|---|---|---|---|---|
| Allotetraploids | 2n | Means (ranges) | Means (ranges) | Means (ranges) | Means (ranges) | PMCs with multivalent (%) | Total PMCs | Pollen fertility (%) |
| AA.BB | 36 | 0.04 (0–2) | 17.85 (16–18) | — | 0.08 (0–1) | 7.09 | 141 | 88.20 |
| AA.CC | 38 | 0.45a (0–10) | 18.64a (14–19) | — | 0.07a (0–2) | 4.49 | 89 | 83.89 |
| CC.AA | 38 | 0.36a (0–6) | 18.64a (12–19) | 0.01 (0–2) | 0.10a (0–2) | 11.68 | 182 | 33.33 |
| BB.CC | 34 | 0.73 (0–6) | 16.62 (14–17) | — | 0.01 (0–1) | 0.73 | 137 | 60.52 |
I, univalent; II, bivalent; III, trivalent; IV, quadrivalent.
Shared letters within each association type denote that the values are insignificantly different (the chi-square test: α= 0.05)
The autosyndetic bivalents and total chromosomes within A genome in A.B and BB.A had no significant difference, but the chromosomes of A genome for allosyndesis in BB.A were significantly lower than those in A.B (Table 2). This suggested that the haploid or diploid state of B genome affected mainly the allosyndetic pairing of A genome. The autosyndesis within B genome and B-C allosyndesis in the two-hybrids B.C and CC.B were similar (Table 1 and Table 2), suggesting that the haploid or diploid state of C genome has no obvious effect on the pairing of B genome. The fewer C-genome univalents in CC.A than in CC.B suggested that the C-genome pairing was enhanced by A genome but reduced by B genome.
As A, B, and C genomes in these hybrids were from the same three diploids (Figure 1), the chromosome pairing of each genome could be compared, which should reveal the effects of different genome combinations and different types of cytoplasm on the chromosome pairing (Figure 3; Table 1, Table 2, and Table 3). The total chromosomes per cell involved in auto- and allosyndesis within B genome in these hybrids were generally lower than those in A and C genomes and varied in narrower ranges (Table 2, Figure 3C), but the rates of the total chromosome number of the three genomes were similar in most hybrids; only in A.C.B were the rates for auto- and allosyndesis within B genome significantly lower than those within A and C genomes (Table 2, Figure 3C). The averages and rates for autosyndesis in each hybrid were significantly lower than those for allosyndesis, but not for those of A and C genomes in A.C, of C genome in B.C, and of B genome in CC.B. The gross averages and rates for autosyndesis within each genome across all hybrids were also significantly lower than those of allosyndesis (Table 2). Although the average number of chromosomes for auto- and allosyndesis in B genome (1.19, 2.47) were fewer than those in A (2.07, 3.72) and C (2.33, 2.87) genomes, their rates were similar, e.g., 0.21 and 0.37, 0.15 and 0.31, and 0.26 and 0.32 for A, B, and C genomes, respectively. Among the hybrids with haploid A genome (A.B, A.C, A.C.B, BB.A, and CC.A), the chromosomes and rates of autosyndesis within A genome in A.C.B and A.C were much higher than in BB.A and A.B and higher than in CC.A, except the rates between A.B and BB.A, and BB.A and CC.A. In A.B, A.C, and A.C.B with donor cytoplasm from B. rapa, those values in A.C.B and A.C with similar frequencies were significantly higher than that in A.B (Table 2). The difference was insignificant between A.B and BB.A, but significant between A.C and CC.A. This further showed that the haploid or diploid of B or C genome had different effects on the pairing of A genome. However, the chromosomes and rates for autosyndesis in B genome showed insignificant differences among the hybrids with haploid B genome (A.B, B.C, A.C.B, and CC.B), which were also similar to those in BB.A, after excluding homologous pairing (Table 2). This showed that the haploidy or diploidy of C genome had no or limited effects on the pairing of B genome. Among the hybrids with haploid C genome (A.C, B.C, A.C.B), the number of chromosomes for autosyndesis within C genome in A.C were higher than those in B.C, but not than those in A.C.B (Table 2), suggesting that the A and B genomes had different effects on influencing the homologous pairing of C genome. The autosyndetic chromosomes in C genome in these hybrids were much higher than those in CC.A and CC.B with dominant homologous pairing. The higher frequency in CC.B (1.20) than in CC.A (0.59) was possibly caused by the lower allosyndesis for the more distant relationship between B and C genomes than between A and C genomes.
For the allosyndesis within A genome, the chromosomes in A.C.B were higher than those in CC.A, A.C, and A.B and higher than in BB.A, and the rate in A.C.B was higher than that in the other four with no significant differences (Figure 1 and Figure 3C; Table 1). This result showed that the introduction of B genome could enhance the A-C association and also that a close homeology existed between A and C genomes because the haploid or diploid C genome did not change its homeologous pairing. For allosyndesis within B genome, the chromosomes in A.B were significantly higher than those in BB.A, B.C, and CC.B, but not in A.C.B; those in A.C.B and BB.A were higher than in B.C and CC.B, and the latter two had similar values. The rates in A.B, A.C.B, and BB.A were higher than those in B.C and CC.B. The A-B associations and the chromosomes for allosyndesis in A and B genomes in BB.A were significantly higher than in A.B, suggesting that the haploid or diploid B genome had obvious effect on the homeologous pairing of the two related genomes. The B-genome chromosomes for allosyndesis in B.C and CC.B were fewer than those in A.B. Once again, the similar frequencies in B.C and CC.B proposed that the haploid or diploid C genome had no obvious effect on the homeologous pairing of the two related genomes. For the allosyndesis within C genome, the chromosomes and rates in A.C.B were higher than those in CC.A and A.C and higher than B.C and CC.B, while those between CC.A and A.C or B.C and CC.B were similar. Notably, the allosyndesis frequency in A and C genomes was highest in A.C.B, while the frequency in B genome was somehow similar to those in other hybrids.
Chromosome pairing in synthetic allotetraploids
The chromosome pairings in the synthetic allotetraploids (AA.BB, AA.CC/CC.AA, and BB.CC) were not fully diploidized, and the univalents and multivalents appeared frequently (Table 3). The data from conventional and GISH observations (Table 3 and Table 4) showed that the average of univalents in AA.BB was significantly lower than in the other three, and that in AA.CC and CC.AA was lower than that in BB.CC, while the difference between AA.CC and CC.AA was insignificant (χ2 = 0.95, P > 0.05). The trivalents were observed only in CC.AA with low frequency. The frequencies of quadrivalents in AA.BB, AA.CC, and CC.AA were similar, but significantly higher than that in BB.CC. The differences among the bivalents were expected, considering their different chromosome numbers. The pollen fertility in these synthetics seemed to be negatively correlated with the frequency of univalents, while the different pollen fertilities between the reciprocal AA.CC and CC.AA (83.89/33.33%) were possibly attributable to the cytoplasmic effect, since their chromosome pairing behaviors were identical except for the different rates of PMCs with multivalents (4.49/11.68%) (Table 3).
Table 4 . FISH studies of chromosome pairing in synthetic allotetraploids.
| Chromosome associations (ranges) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | ||||||||||||
| Allotetraploids | 2n | Total | IA | IB | IC | Total | IIA-A | IIB-B | IIC-C | IIA-C | Total | Total | A-A-A-A | Others | Total PMCs |
| AA.BB | 36 | — | — | — | — | 17.74 (16–18) | 9.80a (8–10) | 7.94a (7–8) | — | — | — | 0.13 (0–1) | 0.07a (0–1) | A-A-B-B 0.06 (0–1) | 69 |
| AA.CC | 38 | 0.52 (0–10) | 0.35a (0–6) | — | 0.17a (0–4) | 18.22 (14–19) | 9.52b (6–10) | — | 8.61b (6–9) | 0.09 (0–2) | — | 0.26 (0–2) | — | A-A-C-C 0.26b (0–2) | 23 |
| CC.AA | 38 | 0.11 (0–8) | 0.08b (0–6) | — | 0.03b (0–2) | 18.41 (12–19) | 9.66a, b (5–10) | — | 8.75b (7–9) | — | 0.03 (0–2) | 0.25 (0–2) | 0.03b (0–1) | A-A-C-C 0.22b (0–2) | 73 |
| BB.CC | 34 | 0.67 (0–4) | — | 0.46 (0–4) | 0.21a (0–2) | 16.62 (15–17) | — | 7.74b (6–8) | 8.87b (8–9) | — | — | 0.03 (0–1) | — | B-B-C-C 0.03 (0–1) | 39 |
I, univalent; II, bivalent; III, trivalent; IV, quadrivalent. IA, IB, and IC indicate univalents belonging to the A, B, and C genomes, respectively; IIA-A, IIB-B, and IIC-C indicate homologous bivalents formed between a pair of A or a pair of B or a pair of C chromosomes, respectively.
Groups significantly different by χ2 -test, p < 0.05.
With GISH/BAC-FISH analyses, the genome-specific homologous and homeologous pairings were distinguished (Table 4). The genome-specific univalents occurred in these allotetraploids except AA.BB. In both AA.CC and CC.AA, A-genome univalents were more frequent than those in C genome. The mean of A-genome univalents in AA.CC was significantly higher than that in CC.AA. In BB.CC, B-genome univalents were more frequent than C-genome ones (χ2 = 4.36, P < 0.05). The C-genome univalents in CC.AA were significantly lower than in AA.CC and BB.CC, but the differences in AA.CC and BB.CC were insignificant. The averages of A-genome homologous bivalents in AA.CC and CC.AA were similar, but only the value in AA.CC was significantly lower than in AA.BB. The B-genome homologous bivalents in BB.CC were significantly lower than in AA.BB, which resulted from the occurrence of B-genome univalents in BB.CC, but not in AA.BB. The means of C-genome homologous bivalents were similar among AA.CC, CC.AA, and BB.CC. The homeologous bivalents and trivalents were observed only in AA.CC and CC.AA, respectively. The quadrivalents involving the chromosomes of one or two genomes were formed in these allotetraploids, their means varied (0.03–0.26), and the maximum was 1–2, being higher in AA.CC/CC.AA. The A-A-A-A quadrivalents occurred with the similar rates in AA.BB and CC.AA. The A-A-B-B quadrivalents appeared in AA.BB. The A-A-C-C quadrivalents occurred in AA.CC and CC.AA at relatively high rates (0.26 and 0.22), and two such pairings per cell were observed (Table 4, Figure 4B), but their difference was insignificant (χ2 = 0.01, P > 0.05). The B-B-C-C quadrivalents also occurred in BB.CC at a low rate. The lowest rates of C-genome-specific univalents within these synthetics and the similar rate of C-genome bivalents among these synthetics suggested that the C-genome chromosomes generally showed more normal pairing than those of A and B genomes (Table 4).
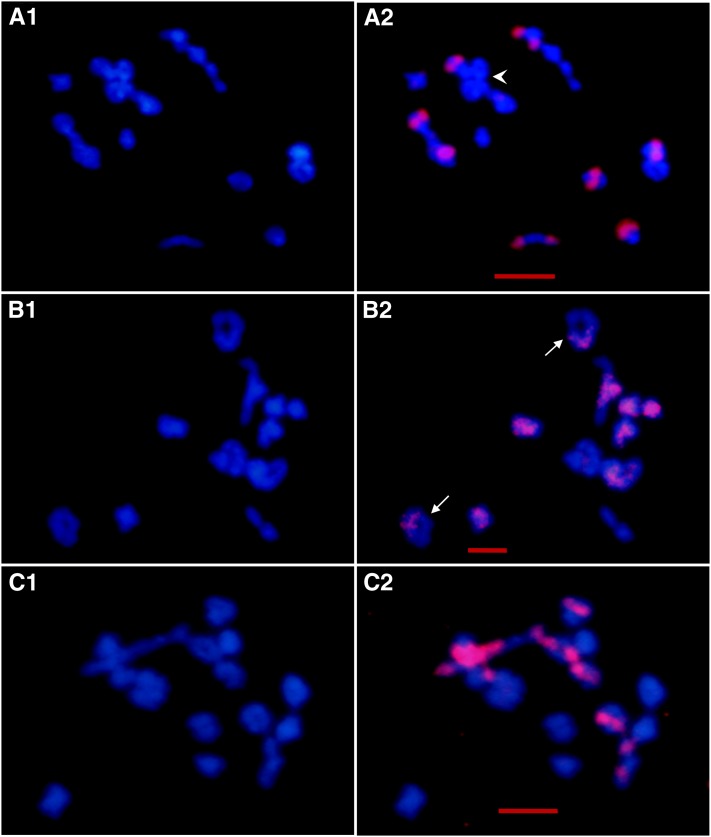
Figure 4 .
GISH/BAC-FISH analyses of meiotic chromosome pairings in synthetic Brassica allotetraploids. A1–C1 are DAPI (blue) images, and A2–C2 are merged images. (A1 and A2) One diakinesis of AA.BB with the following pairing: 10 IIA-A + 8 IIB-B. Arrowhead in A2 shows association of bivalents. Red signals are from B. nigra. (B1 and B2) One diakinesis of AA.CC with the following pairing: 8 IIA-A + 7 IIC-C + 2 IVA-A-C-C (solid arrow). Red signals are from the C-genome-specific BAC BoB014O06. Note that the two C-genome chromosomes labeled by the BAC in the two quadrivalents are homogenously and deeply stained by DAPI. (C1 and C2) One diakinesis of BB.CC with the pairing: 8 IIB-B + 9IIC-C. Red signals are from B. nigra. All bars, 5μm.
For the prevalence of homologous pairing in these allotetraploids, the averages of chromosomes involved in auto- and allosyndesis within each genome were low (0–0.35), but allosyndetic chromosomes in AA.CC and CC.AA were much higher (Table 5). In AA.BB, both pairings within A genome were observed with higher frequency for autosyndesis, but only allosyndesis within B genome at the same frequency as A genome was observed. In AA.CC, the two genomes showed only allosyndesis at the same frequency (0.35). In CC.AA, the two genomes showed auto- and allosyndesis, with the latter at much higher frequency. In BB.CC, only allosyndesis within two genomes occurred at low frequency (0.03). For the comparison of pairing within A genome, AA.BB and CC.AA had similar frequencies for autosyndesis, but AA.CC did not have this pairing; AA.CC and CC.AA showed similar frequencies for allosyndesis but higher than that in AA.BB. For the pairing within B genome, AA.BB and BB.CC had no autosyndesis but had similar low frequencies for allosyndesis. For the pairing with C genome, AA.CC, CC.AA, and BB.CC exhibited no autosyndesis, and AA.CC and CC.AA presented similar frequencies for allosyndesis but much higher than that in BB.CC. The higher frequency of allosyndesis in AA.CC and CC.AA than in AA.BB and BB.CC was attributable to the closer relationships between A and C genomes than with B genome.
Table 5 . Means of chromosome numbers for auto- and allosyndesis within each genome in synthetic allotetraploids.
| A genome | B genome | C genome | ||||
|---|---|---|---|---|---|---|
| Allotetraploids | Autosyndesis | Allosyndesis | Autosyndesis | Allosyndesis | Autosyndesis | Allosyndesis |
| AA.BB | 0.14a | 0.06a | 0 | 0.06a | — | — |
| AA.CC | 0 | 0.35b | — | — | 0 | 0.35b |
| CC.AA | 0.08a | 0.23b | — | — | 0 | 0.23b |
| BB.CC | — | — | 0 | 0.03a | 0 | 0.03a |
| Average | 0.10 | 0.18 | 0 | 0.05 | 0 | 0.19 |
For the calculation of the total chromosome numbers involved in autosyndesis and allosyndesis within each genome in synthetic allotetraploids, A-A-C is taken as 1.5 A-genome chromosomes for autosyndesis and 0.5 A-genome chromosome and 1 C-genome chromosome for allosyndesis; A-A-A is taken as 1.5 for autosyndesis and 1.5 for allosyndesis; the homologous and homeologous ring quadrivalent (A-A-C-C) is divided into 1 A-genome and 1 C-genome chromosome for autosyndesis and 1 A-genome and 1 C-genome chromosome for allosyndesis; the homologous and homeologous ring quadrivalent (A-A-A-A) is divided into 2 equal A-genome chromosomes for homologous pairing and autosyndesis.
Groups significantly different by χ2 -test, p < 0.05.
Discussion
In this study, the chromosome pairings in the synthetic Brassica hybrids and allopolyploids with different genomic composition and cytoplasm were characterized by GISH and BAC-FISH, and the impacts of the hybrid genomic structure on the rates of auto- and allosyndesis and on the stability of the genomes at diploid stage were revealed. The level and frequency of auto- and allosyndesis for each genome varied significantly across hybrids, and those for A and C genomes were in wider ranges of variations than for B genome. The level of autosyndesis was lower than that of allosyndesis (Table 1 and Table 2; Figure 1 and Figure 3). The meiotic pairing in the synthesized allotetraploids was nondiploidized and was affected by the genome and cytoplasmic types (Table 3). The use of the same three parents in pair crosses eliminates the genotypic effect and makes the results in these hybrids comparable.
Genome structure of Brassica diploids
There has been a continuous debate and conflicting views on the origin and evolution of basic karyotypes in Brassica. The pachytene chromosome analysis of the basic genomes (Röbbelen 1960) provided compelling evidence in support of x = 6 as the constitution of the basic archetype. On the basis of marker arrangement conservation, Truco et al. (1996) also proposed a model of genome evolution that described these basic genomes as derived from six ancestral chromosomes that underwent several duplications and rearrangements. Recently, an ancestral Brassiceae karyotype (ABK) with six haploid chromosomes was proposed as the progenitor of tribe Brassiceae, which resulted from a reduction in chromosome number in ancestral crucifer karyotype (ACK, n = 8) or Proto-Calepineae karyotype (n = 7) (Lysak et al. 2006; Mandáková and Lysak 2008). This prototype (ABK) subsequently has diverged into nigra, rapa, and oleracea lineages 7.3–4 MYA (Wroblewski et al. 2000) or ∼7.9 MYA (Lysak et al. 2005). x = 6 is most likely the basic chromosome number in the tribe Brassiceae and the genus Brassica (Prakash et al. 2011). Meiotic chromosome pairing in the haploids of B. campestris (syn. B. rapa, 2n = 10, 3I + 2II + 1III) (Armstrong and Keller 1981), B. oleracea (2n = 9, 4I + 1II + 1III) (Thompson 1956; Armstrong and Keller 1982), B. nigra (2n = 8, 4 I + 2II) (Prakash 1973), and a related species, B. tournefortii (2n =10, 3I + 2II + 1III) (Prakash 1974) also favored this proposal. Two A-A autosyndetic pairs and two C-C autosyndetic pairs were observed in haploids of B. napus cv. Darmor-bzh (Nicolas et al. 2007). In trigenomic interspecific hybrids (AABC, BBAC, and CCAB) from the crosses of B. napus (AACC), B. juncea (AABB), and B. carinata (BBCC), a maximum of three A-A pairs, two B-B pairs, or two C-C pairs were observed across all hybrid types (Mason et al. 2010), although these genomes from natural allotetraploids have experienced the evolutionary process. A maximum of two B-B pairs appeared in trigenomic hybrids from crosses between natural and synthetic B. napus and B. nigra (AC.B, A.C.B) and between B. carinata and B. rapa (BC.A) (Ge and Li 2007), while the B genome was from allotetraploid B. carinata or diploid B. nigra. Our results showed that the maximum of two or three autosyndetic bivalents could occur in the A or C genomes in the haploid genomes of hybrids, but only two could occur in the B genome. The similar extents of autosyndesis within each genome from diploids and allotetraploids showed that the genome structure was largely maintained during allopolyploidization, but B genome was more stable than A or C genome. The consistent formation of two autosyndetic bivalents within B genome of diverse sources provided the chromosomal evidence for the proposal that the present Brassica genomes were derived from the basic karyotype with x = 6. Although the pairing is dependent on the structure of hybrids and on the presence or not of genetic control, no genetic factor for suppression of pairing was found in the B genome in trigenomic Brassica hybrids (Busso et al. 1987). The autosyndesis within one genome reflects the segmental homology between chromosomes caused by the rearrangements of the blocks or by the common origin of the chromosomes involved (Truco et al. 1996). Our present result also showed that the pairing in B genome was less affected by the genome and cytoplasm types. The recent draft genome sequence of B. rapa provided new data for the genome structure of Brassica diploids and revealed the almost complete triplication of the B. rapa genome relative to Arabidopsis thaliana and to ACK (n = 8) (Wang et al. 2011). But the triplication theory still fails to explain the origin of extant chromosome numbers.
Cytoplasmic and genomic effects on meiotic pairing in hybrids
Nuclear-cytoplasmic interactions are predicted to be important in allopolyploid and hybrid evolution (Gill 1991; Wendel 2000; Levin 2003) because the presence of a foreign nuclear genome in the cytoplasm from the female parent can result in nuclear-cytoplasmic incompatibilities. Chromosomal rearrangements in hybrids and allopolyploids could potentially occur in response to changes in nuclear-cytoplasmic interactions. Two types of cytoplasm exist in three Brassica diploids: the B type in B. nigra and the A/C type in B. rapa and B. oleracea. The A and B types are quite distinct although they retain homology to a large extent (Palmer et al. 1983; Yanagino et al. 1987; Warwick and Black 1991; Pradhan et al. 1992). The reciprocal synthetics of B. juncea (AABB/BBAA) showed directional genomic changes, with the significant alterations of the paternal genome. The reciprocal synthetic B. napus (AACC/CCAA) did not show different genomic changes. Song et al. (1995) suggested that this was attributable to the more closely related A and C cytoplasmic genomes and to the more compatible nuclear-cytoplasmic genomes in the AC and CA polyploids. Chromosome pairing studied in the large number of allohaploids produced from a wide range of B. napus accessions revealed two main clear-cut meiotic phenotypes that showed a twofold difference in the number of univalents at metaphase I and correlated with the only two plastid haplotypes identified in these accessions (Cifuentes et al. 2010). The segregation of two alleles at PrBn might explain a large part of the variation in meiotic behavior found among B. napus allohaploids. The results indicated that variation in crossover frequency among allohaploid genotypes generally correlates with the multiple origins of B. napus and PrBn diversity and also suggested the cytoplasmic and genetic effects on the meiotic crossover in allohaploids (Cifuentes et al. 2010). The significant reduction in crossover in all B. napus allohaploids compared with synthesized B. oleracea × B. rapa hybrids could reflect principally chromosome rearrangements that accentuated the divergence between B. napus homeologous chromosomes after the inception of this species. But the possibility that new nuclear-cytoplasmic interactions in new hybrids promote the crossover cannot be excluded.
The significant difference in chromosome pairing in the two hybrids A.C.B and C.A.B with the same nuclear genomes but a different cytoplasmic genome also showed the role of the cytoplasm in crossover frequency, although the A and C cytoplasmic genomes were closely related. The complex effects of the cytoplasm and nuclear genome interaction obviously resulted in the variation in crossover frequency of each genome among different hybrids (Table 1 and Table 2; Figure 3).
The trigenomic hybrids (AABC, BBAC, and CCAB) likely had the A/C type cytoplasm because they were produced from the crosses B. juncea × B. napus, B. juncea × B. carinata and B. napus × B. carinata (Mason et al. 2010). The comparison between the results from these hybrids and ours should reveal the effects of the cytoplasm and genome structure because both studies used the dual-color GISH and the same BAC clone (BoB014O06) from B. oleracea. Hybrids in the present study showed higher levels of autosyndetic pairing within haploid A and C genomes compared to the haploid B genome. Interestingly, the frequency of B-C pairs in AABC hybrids was much lower than in our B.C hybrid, but the frequency of A-C pairs in BBAC was higher than in our A.C hybrid, and also the frequency of A-B pairs in CCAB was lower than in our A.B hybrid. The frequency of A-B pairs in BBAC was a little lower than those in our BB.A, and A-C pairs in CCAB were nearly the same as those in our CC.A, but B-C pairs in CCAB were lower than in our CC.B. The different results from the two studies might be due to the presence of diversity in the genome structure in three diploids and allotetraploids of different origins or could be the effects of cytoplasmic genome. The allosyndetic bivalents in BB.A, CC.A, and CC.B were a little higher than in A.B, A.C, and B.C, respectively, although only the difference between BB.A and A.B was significant (Table 1); the chromosome numbers for allosyndesis in CC.A and A.C showed only such a trend (Table 2). The information that the presence of a complete diploid genome enhanced homeologous pairing in trigenomic hybrids in comparison to allodiploids was also revealed in other Brassica hybrids (Nagpal et al. 1996), probably because digenomic triploids had more potential partners for homeologous pairing than allodiploids. Another reason might be related to the different ploidy levels, as meiotic recombination increases in Arabidopsis auto- and allopolyploids relative to diploids (Pecinka et al. 2011).
The genetic control of chromosome pairing in Brassica allopolyploids may be very complex. Even the major genetic factor PrBn that controls homeologous pairing at the haploid stage was detected (Jenczewski et al. 2003) and localized to linkage group C9 (Liu et al. 2006). But triploid AAC hybrids of the two B. napus genotypes for mapping PrBn as male with the same B. rapa variety displayed a similar meiotic behavior, which showed that PrBn had no effect on the meiotic behavior of triploid hybrids (Leflon et al. 2006) and that the gene was ineffective at hemizygous stage (Jenczewski and Alix 2004). But two AAC hybrids showed significant genotypic variation in crossover rates along a pair of A-genome chromosomes observed (Nicolas et al. 2009). The hybrids with Darmor-bzh of high pairing presented a reduction of autosyndesis within C genome, particularly of A-C allosyndesis compared with its haploid (Nicolas et al. 2007), showing that PrBn on the C genome failed to enhance or affect the pairing of its own chromosome in the AAC background. However, cytogenetic estimation of class I crossovers (interfering crossovers) in the entire genome by immunolocalization of a key protein, MutL Homolog1, showed that crossover rates were significantly higher in the allotetraploid AACC hybrid than in the diploid AA hybrid and were highest in the allotriploid AAC hybrid (Leflon et al. 2010).
Genome relatedness and chromosome behavior in synthetic allotetraploids
These synthetic Brassica allotetraploids showed substantial differences in their meiotic behaviors (Table 3 and Table 4). The associations between or among bivalents occurred in these allotetraploids with high frequencies (Figure 4), as observed in Brassica species previously (Maćkowiak and Heneen 1999). In the present study, AA.CC and CC.AA presented relatively high rates of univalents and the highest rate of homeologous pairing, as in other synthetic B. napus observed with the same BAC clone BoB014O06 from B. oleracea (Leflon et al. 2010). But AA.CC had high pollen fertility while CC.AA had much lower fertility, which was possibly caused by the cytoplasmic effects. The differences in cytological behaviors in our synthetics could be explained partly by their genome relatedness as revealed by the chromosome pairing in respective dihaploid hybrids from which they were derived (Tables 1–4) and as shown by other Brassica allopolyploids (Yao et al. 2012). The A-C bivalents in A.C were much higher than the A-B bivalents in A.B and the B-C bivalents in B.C. Hence, AA.CC formed A-C bivalents and A-A-C-C quadrivalents at a high rate, while AA.BB and BB.CC produced no homeologous bivalents except some homeologous quadrivalents (A-A-B-B or B-B-C-C) at low rates. Subsequently, AA.CC and CC.AA showed much higher frequencies for allosyndesis than AA.BB and BB.CC (Table 5). It was difficult to explain the much higher rate of univalents, particularly those of B genome in BB.CC than in AA.BB, because the cytoplasm from the B-genome donor in BB.CC should assist in stabilizing its chromosomes, while the B-genome chromosomes in the cytoplasm from the A-genome donor B. rapa in AA.BB was expected to show more aberrations.
The genetic changes caused by homeologous chromosome rearrangement were found to be common in newly resynthesized B. napus allotetraploids (Gaeta et al. 2007; Szadkowski et al. 2010, 2011). In the very first meiosis of synthetic B. napus, the frequent occurrence of A-C bivalents and/or multivalents and univalents was detected, which resulted in the production of gametes with unbalanced chromosomal composition and/or carrying chromosomal rearrangements (Szadkowski et al. 2010). The frequency of the meiotic-driven genetic changes depends significantly on the cytoplasm background inherited from the progenitors because the progenies of the synthetics with the B. rapa cytoplasm showed an excess of plants without rearrangements and a lower frequency of plants carrying A1 marker loss than the one with B. oleracea cytoplasm. By contrast, no difference was found between C1 marker loss frequencies in the progenies of reciprocal synthetics. Conversely, the genetic backgrounds on B. oleracea cytoplasm did not influence the frequency of rearrangements. Furthermore, homeolog pairing and chromosome rearrangements, aneuploidy, and homeologous chromosome compensation were identified in 50 resynthesized B. napus lines across generations S0:1–S5:6 and in the S10:11 generation by using a newly developed cytogenetic method to distinguish all 38 chromosomes in B. napus (Xiong et al. 2011). The data demonstrated that chromosome changes (aneuploidy and translocations) occurred most frequently on homeologous chromosome pairs that display the most extensive stretches of syntenic marker loci (Parkin et al. 2005; Udall et al. 2005; Gaeta et al. 2007; Nicolas et al. 2009). The two most unstable homeologous sets were A1/C1 and A2/C2, and their changes occurred in >50% of lines, including nullisomy, monosomy, trisomy, and tetrasomy because homeologous linkage groups A1/C1 and A2/C2 are each syntenic along their entire chromosome length (Parkin et al. 2005). Coincidently, two homeologous A-A-C-C quadrivalents formed in our synthetic B. napus (Figure 4B), which possibly involved the two groups A1/C1 and A2/C2. Accordingly, fewer chromosome changes were expected in synthetic B. juncea and B. carinata with more divergent genomes because their chromosomes showed a lower frequency of homeolog pairing (Table 4). Furthermore, the A, B, and C genomes showed different chromosomal stabilities in synthesized Brassica allohexaploids (Ge et al. 2009). Thus, it is worthwhile to trace the chromosomal rearrangements and stability across several generations in our synthesized allotetraploids in the future, especially in B. juncea and B. carinata.
Supplementary Material
Acknowledgments
We are grateful to Susan J. Armstrong and Steve L. Price (The University of Birmingham, Birmingham, UK) for providing the clone BoB014O06; to Genlou Sun (Saint Mary's University, Halifax, NS, Canada) for revising the manuscript; and to the two anonymous reviewers for their constructive comments. The study was supported by a special grant (Department of Science and Technology, Peoples' Republic of China) to National Key Laboratory of Crop Genetic Improvement.
Footnotes
Communicating editor: J. C. Schimenti
Literature Cited
- Allender C. J., King G. J., 2010. Origins of the amphiploid species Brassica napus L. investigeated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong K. C., Keller W. A., 1981. Chromosome pairing in haploids of Brassica campestris. Theor. Appl. Genet. 59: 49–52 [DOI] [PubMed] [Google Scholar]
- Armstrong K. C., Keller W. A., 1982. Chromosome pairing in haploids of Brassica oleracea. Can. J. Genet. Cytol. 24: 735–739 [DOI] [PubMed] [Google Scholar]
- Attia T., Busso C., Röbbelen G., 1986. Cytogenetic relationship within cultivated Brassica analyzed in amphihaploids from the three diploid ancestors. Can. J. Genet. Cytol. 28: 323–329 [Google Scholar]
- Busso C., Attia T., Röbbelen G., 1987. Trigenomic combination for the analysis of meiotic control in cultivated diploid Brassica species. Genome 29: 331–333 [Google Scholar]
- Cheung F., Trick M., Drou N., Lim Y. P., Park J. Y., et al. , 2009. Comparative analysis between Homeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21: 1912–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M., Eber F., Lucas M. O., Lode M., Chèvre A. M., et al. , 2010. Repeated polyploidy drove different levels of crossover suppression between homeologous chromosomes in Brassica napus allohaploids. Plant Cell 22: 2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery M. L., Mitchell F. J. G., Coyne S., Kavanagh T. A., Burke J. I., et al. , 2006. Plastid genome characterization in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 113: 1221–1231 [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Federico I. L., Leon E., Osborn T. C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. H., Li Z. Y., 2007. Intra- and intergenomic homology of B-genome chromosomes in trigenomic combinations of the cultivated Brassica species revealed by GISH analysis. Chromosome Res. 15: 849–861 [DOI] [PubMed] [Google Scholar]
- Ge X. H., Wang J., Li Z. Y., 2009. Different genome-specific chromosome stabilities in synthetic Brassica allohexaploids revealed by wide crosses with Orychophragmus. Ann. Bot. 104: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, B. S., 1991 Nucleocytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants, pp. 48–53 in Proceedings of the Kihara Memorial International Symposium on Cytoplasmic Engineering in Wheat, edited by T. Sasakurma. Yokohama, Japan.
- Howell E. C., Kearsey M. J., Jones G. H., King G. J., Armstrong S. J., 2008. A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics 180: 1849–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenczewski E., Alix K., 2004. From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23: 21–45 [Google Scholar]
- Jenczewski E., Eber F., Grimaud A., Huet S., Lucas M. O., et al. , 2003. PrBn, a major gene controlling homeologous chromosome pairing in oilseed rape (Brassica napus) haploids. Genetics 164: 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Comber S. C., Ainouche M. L., Kovarik A., Leitch A. R., 2010. Making a functional diploid: from polysomic to disomic inheritance. New Phytol. 186: 113–122 [DOI] [PubMed] [Google Scholar]
- Leflon M., Eber F., Letanneur J. C., 2006. Pairing and recombination at meiosis of Brassica rapa (AA) × Brassica napus (AACC) hybrids. Theor. Appl. Genet. 113: 1467–1480 [DOI] [PubMed] [Google Scholar]
- Leflon M., Grandont L., Eber F., Huteau V., Coriton O., et al. , 2010. Crossovers get a boost in Brassica allotriploid and allotetraploid hybrids. Plant Cell 22: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28: 5–11 [Google Scholar]
- Li Z. Y., Liu H. L., Luo P., 1995. Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus. Theor. Appl. Genet. 91: 131–136 [DOI] [PubMed] [Google Scholar]
- Liu Z. Q., Adamczyk K., Manzanares-Dauleux M., Eber F., Lucas M. O., 2006. Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.). Genetics 174: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens L. N., Pires J. C., Leon E., Vogelzang R., Oslach L., 2006. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140: 336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M. A., Koch M. A., Pecinka A., Schubert I., 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M., Berr A., Pecinka A., Schmidt R., Mcbreen K., et al. , 2006. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103: 5224–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maćkowiak M., Heneen W. K., 1999. Meiotic karyotype of the B genomes of B. nigra and B. carinata. Hereditas 130: 131–135 [Google Scholar]
- Mandáková T., Lysak M. A., 2008. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason A. S., Huteau V., Eber F., Coriton O., Yan G. J., et al. , 2010. Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosome Res. 18: 655–666 [DOI] [PubMed] [Google Scholar]
- Masterson J., 1994. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264: 421–424 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F., 1962. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant. 15: 473–479 [Google Scholar]
- Nagaharu U., 1935. Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452 [Google Scholar]
- Nagpal R., Raina S. N., Sodhi Y. S., Mukhopadhyay A., Arumugam N., et al. , 1996. Transfer of Brassica tournefortii (TT) genes to allotetraploid oilseed Brassica species (B. juncea AABB, B. napus AACC, B. carinata BBCC): homeologous pairing is more pronounced in the three-genome hybrids (TACC, TBAA, TCAA, TCBB) as compared to allodiploids (TA, TB, TC). Theor. Appl. Genet. 92: 566–571 [DOI] [PubMed] [Google Scholar]
- Nicolas S. D., Le Mignon G., Eber F., Coriton O., Momod H., et al. , 2007. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175: 487–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas S. D., Leflon M., Momod H., Eber F., Coriton O., et al. , 2009. Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. Plant Cell 21: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., 2007. The evolutionary consequences of polyploidy. Cell 131: 452–462 [DOI] [PubMed] [Google Scholar]
- Palmer J. D., Shields C. R., Cohen D. B., Orton T. J., 1983. Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor. Appl. Genet. 65: 181–189 [DOI] [PubMed] [Google Scholar]
- Parkin I. A. P., Gulden S. M., Sharpe A. G., Lukens L., Trick M., et al. , 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Fang W., Rehmsmeier M., Levy A. A., Scheid O. M., 2011. Polyploidization increases meiotic recombination frequency in Arabidopsis. BMC Biol. 9: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires J. C., Maureira I. J., Givnish T. J., Sytsma K. J., Seberg O., et al. , 2006. Phylogeny, genome size, and chromosome evolution of Asparagales, pp. 287–304 in Monocots: Comparative Biology and Evolution Poales, edited by Columbus J. T., Friar E. A., Porter J. M., Prince L. M., Simpson M. G. Rancho Santa Ana Botanic Garden, Claremont, CA [Google Scholar]
- Pradhan A. K., Prakash S., Mukhopadhyay A., Pental D., 1992. Phytogeny of Brassica and allied genera based on variation in chloroplast and mitochondrial DNA patterns: molecular and taxonomic classifications are incongruous. Theor. Appl. Genet. 85: 331–340 [DOI] [PubMed] [Google Scholar]
- Prakash S., 1973. Haploidy in Brassica nigra Koch. Euphytica 22: 613–614 [Google Scholar]
- Prakash S., 1974. Haploid meiosis and origin of Brassica tournefortii Gouan. Euphytica 23: 591–595 [Google Scholar]
- Prakash S., Hinata K., 1980. Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Bot. 55: 1–57 [Google Scholar]
- Prakash S., Bhat S. R., Quiros C. F., Kirti P. B., Chopra V. L., 2009. Brassica and its close allies: cytogenetics and evolution. Plant Breed. Rev. 31: 21–187 [Google Scholar]
- Prakash S., Wu X. M., Bhat S. R., 2011. History, evolution, and domestion of Brassica crops. Plant Breed. Rev. 35: 19–84 [Google Scholar]
- Ramsey J., Schemske D. W., 1998. Pathways, mechanisms and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501 [Google Scholar]
- Röbbelen G., 1960. Observations on the analysis of Brassica genomes. Chromosoma 11: 205–228 (in German) [DOI] [PubMed] [Google Scholar]
- Song K. M., Osborn T. C., Williams P. H., 1988. Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs) 1. Genome evolution of diploid and amphidiploid species. Theor. Appl. Genet. 75: 784–794 [Google Scholar]
- Song K., Lu P., Tang K., Osborn T. C., 1995. Rapid genome changes in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szadkowski E., Eber F., Huteau V., Lodé M., Huneau C., et al. , 2010. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186: 102–112 [DOI] [PubMed] [Google Scholar]
- Szadkowski E., Eber F., Huteau V., Lodé M., Coriton O., et al. , 2011. Polyploid formation pathways have an impact on genetic rearrangements in resynthesized Brassica napus. New Phytol. 191: 884–894 [DOI] [PubMed] [Google Scholar]
- Thompson K. F., 1956. Production of haploid plants of marrow stem kale. Nature 178: 74813369530 [Google Scholar]
- Truco M. J., Hu H., Sadowski J., Quiros C. F., 1996. Inter- and intra-genomic homology of the Brassica genomes: implications for their origin and evolution. Theor. Appl. Genet. 93: 1225–1233 [DOI] [PubMed] [Google Scholar]
- Udall J. A., Quijada P. A., Osborn T. C., 2005. Detection of chromosomal rearrangements derived from homologous recombination in four mapping populations of Brassica napus L. Genetics 169: 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. W., Wang H. Z., Wang J., Sun R. F., Wu J., et al. , 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1040 [DOI] [PubMed] [Google Scholar]
- Warwick S. I., Black L. D., 1991. Molecular systematics of Brassica and allied genera (subtribe Brassicinae, Brassiceae): chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82: 81–92 [DOI] [PubMed] [Google Scholar]
- Wendel J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249 [PubMed] [Google Scholar]
- Wood T. E., Takebayashi N., Barker M. S., Mayrose I., Greenspoon P. B., et al. , 2009. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T., Coulibaly S., Sadowski J., Quiros C. F., 2000. Variation and phylogenetic utility of the Arabidopsis thaliana Rps2 homolog in various species of the tribe Brassiceae. Mol. Phylogenet. Evol. 16: 440–448 [DOI] [PubMed] [Google Scholar]
- Xiong Z. Y., Gaeta R. T., Pires J. C., 2011. Homeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagino T., Takahata Y., Hinata K., 1987. Chloroplast DNA variation among diploid species in Brassica and allied genera. Jpn. J. Genet. 82: 119–125 [Google Scholar]
- Yao X. C., Ge X. H., Li Z. Y., 2012. Different fertility and meiotic regularity in allohexaploids derived from trigenomic hybrids between three cultivated Brassica allotetraploids and B. maurorum. Plant Cell Rep. 31: 781–788 [DOI] [PubMed] [Google Scholar]
- Zhong X. B., de Jong J. H., Zabel P., 1996. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res. 4: 24–28 [DOI] [PubMed] [Google Scholar]
- Zwick M. S., Hanson R. E., McKnight T. D., Islam-Faridi M. H., Stelly D. M., et al. , 1997. A rapid procedure for the isolation of C0t-1 DNA from plants. Genome 40: 138–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.