Abstract
Heat-shock protein 90 (Hsp90) of Saccharomyces cerevisiae is an abundant essential eukaryotic molecular chaperone involved in the activation and stabilization of client proteins, including several transcription factors and oncogenic kinases. Hsp90 undergoes a complex series of conformational changes and interacts with partner co-chaperones such as Sba1, Cpr6, Cpr7, and Cns1 as it binds and hydrolyzes ATP. In the absence of nucleotide, Hsp90 is dimerized only at the carboxy-terminus. In the presence of ATP, Hsp90 also dimerizes at the amino-terminus, creating a binding site for Sba1. Truncation of a charged linker region of yeast Hsp90 (Hsp82Δlinker) was known to disrupt the ability of Hsp82 to undergo amino-terminal dimerization and bind Sba1. We found that yeast expressing Hsp82Δlinker constructs exhibited a specific synthetic lethal phenotype in cells lacking CPR7. The isolated tetratricopeptide repeat domain of Cpr7 was both necessary and sufficient for growth in those strains. Cpr6 and Cpr7 stably bound the carboxy-terminus of wild-type Hsp82 only in the presence of nonhydrolyzable ATP and formed an Hsp82–Cpr6–Cpr7 ternary complex. However, in cells expressing Hsp82Δlinker or lacking CPR7, Cpr6 was able to bind Hsp82 in the presence or absence of nucleotide. Overexpression of CNS1, but not of other co-chaperones, in cpr7 cells restored nucleotide-dependent Hsp82–Cpr6 interaction. Together, our results suggest that the in vivo functions of Cpr7 include modulating Hsp90 conformational changes, mediating proper signaling of the nucleotide-bound state to the carboxy-terminus of Hsp82, or regulating Hsp82–Cpr6 interaction.
Keywords: co-chaperones, heat-shock proteins, immunophilins, molecular chaperones
Hsp90 is an essential, abundant molecular chaperone critical for the folding and regulation of an estimated 5–10% of all cellular proteins, termed “client proteins” (Zhao et al. 2005; McClellan et al. 2007). The ability of Hsp90 to bind and hydrolyze ATP is essential for function, and many clients become destabilized or inactive in the presence of 17-AAG, a competitive inhibitor of ATP binding. Because a number of oncogenic signaling proteins, including Akt, Raf-1, Bcr-Abl, mutant p53, and HER-2/ErbB2, require Hsp90 for function, Hsp90 is a promising anti-cancer target, and Hsp90 inhibitors are currently in clinical trials (Pearl et al. 2008; Trepel et al. 2010).
Hsp90 contains three conserved domains: an amino-terminal ATP-binding domain, a middle domain, and a carboxy-terminal dimerization domain. Hsp90 binds clients that possess some secondary structure, and all three domains have been implicated in client binding (Jakob et al. 1995; Vaughan et al. 2006; Richter and Buchner 2011; Street et al. 2011). Hsp90 undergoes significant conformational changes upon nucleotide binding and hydrolysis (Ali et al. 2006; Shiau et al. 2006). Nucleotide-free Hsp90 dimerized at the carboxy-terminus is in an open conformation. ATP binding induces dimerization of amino-terminal domains. ATP hydrolysis requires contact between the amino-terminal domains and a flexible loop in the middle domains. Mutations in Hsp90 that disrupt the ability of Hsp90 to undergo conformational changes cause defects in ATPase and client activity. One such mutation, Hsp82Δlinker, is the deletion of a charged linker region (CLR) between the amino-terminal and middle domains. Increasingly larger deletions of the linker cause growth defects, ranging up to inviability, as well as defects in the ability of Hsp90 to undergo ATP-induced amino-terminal dimerization (Louvion et al. 1996; Hainzl et al. 2009).
Hsp90 functions with a set of co-chaperone proteins that modulate the ATPase activity of Hsp90 and/or mediate interactions with client proteins (Pearl et al. 2008). Co-chaperones bind different conformations of Hsp90 and/or compete for the same binding site (Siligardi et al. 2004; Harst et al. 2005). Many co-chaperones contain tetratricopeptide repeat (TPR) domains and compete for binding to the conserved MEEVD sequence found at the carboxy-terminus of Hsp90 (Owens-Grillo et al. 1995; D’Andrea and Regan 2003). In Saccharomyces cerevisiae, there are at least six co-chaperones that bind Hsp90 through their TPR domains: Sti1, Cpr6, Cpr7, Ppt1, Cns1, and Tah1 (Wandinger et al. 2008). Despite competing for the same binding site, TPR-containing co-chaperones have differing effects on Hsp90 activity. Sti1 is a potent inhibitor of Hsp90’s ATPase activity, while Cpr6 and Tah1 weakly stimulate its ATPase activity (Prodromou et al. 1999; Panaretou et al. 2002; Millson et al. 2008).
Differential binding of co-chaperones is linked to conformational changes in Hsp90 as it binds and hydrolyzes ATP (Pearl et al. 2008). Hop/Sti1 preferentially binds Hsp90 in the absence of nucleotide (Prodromou et al. 1999; Richter et al. 2003), while p23/Sba1 binds the ATP-bound, closed conformation (Ali et al. 2006). ATP binding also results in interaction with FKBP51/FKBP52 and Cyp40/Cpr6, which displaces Hop/Sti1 (Johnson and Toft 1994; Prodromou et al. 1999; Johnson et al. 2007). A ternary complex consisting of an Hsp90 dimer bound to both Sti1 and Cpr6 or FKBP51 (occupying different TPR-binding sites) may be a key intermediate in the Hsp90-folding cycle (Li et al. 2011). The structural basis for the ability of TPR-containing co-chaperones such as FKBP51/52 and Cyp40/Cpr6 to specifically recognize the ATP-bound form of Hsp90 is unknown, as the carboxy-terminal domain does not appear to undergo large conformation changes upon ATP binding (Ali et al. 2006).
Recent studies suggest that the co-chaperone Aha1 plays an active role in mediating conformational changes in Hsp90 that are required for ATP hydrolysis (Hessling et al. 2009). To gain insights into other factors governing conformational changes in Hsp90, we determined whether cells expressing Hsp82Δlinker constructs that reduce conformational flexibility but do not result in inviability exhibit synthetic growth defects when combined with deletion of genes encoding co-chaperones. We observed severe growth defects only upon combination of Hsp82Δlinker and deletion of CPR7. Deletion of CPR7 or truncation of the CLR also resulted in loss of nucleotide-specific interaction of Cpr6 with Hsp90. Together, these results suggest that growth defects caused by the loss of CPR7 are linked to a role in mediating conformational or signaling changes in Hsp90.
Materials and Methods
Media, chemicals, antibodies, and plasmids
Standard yeast genetic methods were employed (Burke et al. 2000). Yeast cells were grown in either YPD (1% Bacto yeast extract, 2% peptone, and 2% dextrose) or defined synthetic complete media. Growth was examined by spotting 10-fold serial dilutions of yeast cultures on appropriate media, followed by incubation for 2 days at 25°, 30°, or 37°. 5-FOA was obtained from Toronto Research Chemicals. α-Flag monoclonal antibody, G418, and 5′-adenylylimidodiphosphate (AMP-PNP) were obtained from Sigma. Polyclonal antibodies against Hsc82/Hsp82 have been described (Flom et al. 2006). Polyclonal antisera was raised against keyhole limpet hemocyanin-conjugated peptides corresponding to amino acids 81–96 of Cpr6, 211–225 of Cpr7, 118–131 of Sba1, and 492–509 of Ppt1. Antibodies were validated by establishing that they recognize a protein of the correct size in yeast lysates and fail to recognize a protein of that size in strains lacking that co-chaperone (not shown).
Yeast strains
All strains are isogeneic to W303. Strains JJ816 (hsc82::LEU2hsp82::LEU2/YEp24-HSP82) and JJ832 (sti1::MET2hsc82::LEU2hsp82::LEU2/YEp24-HSP82) have been described (Flom et al. 2006). Additional yeast strains were constructed by transforming the appropriate gene disruption cassette from the yeast genome collection (Open Biosystems) into a hsc82::LEU2hsp82::LEU2/YEp24-HSP82 diploid (JJ928). Colonies that grew in the presence of G418 were sporulated to obtain haploid versions of the desired strain. We constructed the following strains: hsc82::LEU2hsp82::LEU2/YEp24-HSP82 strains lacking PPT1 (JJ146, MATa), CPR6 (JJ110, MATα), CPR7 (JJ149, MATa), SBA1 (JJ94, MATa), AHA1 (JJ95, MATa), HCH1 (JJ111, MATa), or TAH1 (JJ464, MATα). We also constructed the CPR7 disruption strain JJ567 (MATα trp1-1 ura3-1 leu2-3,112 his3-11,15 ade2-1 met2-Δ1 lys2-Δ2 cpr7::kanr).
HSP82 plasmids and constructs
In S. cerevisiae, Hsp90 is encoded by two genes, HSC82 and HSP82, one of which must be present for viability (Borkovich et al. 1989). Plasmids expressing wild-type (WT) HSP82 under the endogenous promoter (YEp24-HSP82) or GPD promoter (pTGPDHSP82) were gifts from Susan Lindquist (Whitehead Institute). Amino acid mutations were constructed using site-directed mutagenesis (QuikChange, Stratagene, La Jolla, Ca). Sequences of mutagenic oligonucleotides are available on request. A plasmid expressing Hsp82 with an amino-terminal 6× His tag and Xpress epitope tag under the constitutive GPD promoter was constructed as described for Hsc82 (Johnson et al. 2007). The following amino acid alterations were constructed within pRS314GPDHISHSP82 and/or pTGPDHSP82: Δ211–259, Δ211–264, and ΔMEEVD, which deletes the last five amino acids of Hsp82 and Δ211-264ΔMEEVD. All mutant constructs were sequenced completely using automated DNA sequencing.
Co-chaperone plasmids
CPR7 was cloned into pRS313 (Sikorski and Hieter 1989) and pRS316ADE3 (Flom et al. 2005) using engineered SacI and XbaI sites at the 5′ and 3′ ends, respectively. CPR6 was cloned into pRS316ADE3 using engineered SacI and BamHI sites at the 5′ and 3′ ends, respectively. Cpr6 was also cloned into pRS416GPD to introduce an amino-terminal 6× His and Xpress epitope tag. Flag-Cpr6 and Flag-Cpr7 were cloned into the XmaI and XhoI sites of pRS413GPD (Mumberg et al. 1995). To create 413GPDFlag-6PPI/7TPR and 413GPDFlag-7PPI/6TPR, ClaI sites were introduced near the end of the peptidyl-prolyl isomerase (PPIase) domains of Cpr6 and Cpr7, resulting in the insertion of isoleucine and aspartic acid after amino acid 197 of Cpr7 and 176 of Cpr6. 413GPDFlag-6TPR expresses amino acids 171–371 of Cpr6 and 413GPDFlag-7TPR expresses amino acids 193–393 of Cpr7. CNS1 was cloned into YEp24 as a 2.0-kb EcoRI fragment. pRS316ADE3STI1 has been previously described (Flom et al. 2005). PPT1 and TAH1 were amplified from WT yeast and cloned into pRS316ADE3 using engineered SacI/XbaI and SacI/BamHI sites at the 5′ and 3′ ends, respectively.
Isolation of His–Hsp82 and His–Cpr6 complexes
His–Hsp82 complexes were isolated as described for His–Hsc82 complexes (Johnson et al. 2007). Briefly, a plasmid expressing His-tagged WT or mutant Hsp82 was transformed into strain JJ816 (or other strain as indicated), and resultant colonies were grown in the presence of 5-FOA to counterselect for the plasmid expressing WT untagged Hsp82 (YEp24-HSP82). Cells were harvested at an OD600 of 1.2–2.0 and lysed in 20 mM Tris (pH 7.5), 100 mM KCl, and 5 mM MgCl2 plus protease inhibitors. As indicated, lysate was adjusted to contain 5 mM AMP-PNP and incubated 5 min at 30∘. Hsp82 complexes were isolated by incubation with nickel resin (1 hr with rocking at 4°) followed by washes with lysis buffer plus 0.1% Tween-20 and 35 mM imidazole. Nickel resin was boiled in SDS-PAGE sample buffer, and protein complexes were separated by gel electrophoresis (10% acrylamide unless noted otherwise) followed by Coomassie Blue staining or immunoblot analysis. Chemiluminescence immunoblots were performed according to the manufacturer’s suggestions (Pierce, Rockford, IL). His–Cpr6 complexes were isolated in a similar manner.
Results
Hsp90 contains two features that are not present in HtpG, the Hsp90 of Escherichia coli: (1) the extended CLR between the amino-terminal and middle domains and (2) the carboxy-terminal pentapeptide MEEVD (Louvion et al. 1996). The MEEVD sequence is the only established binding site for TPR-containing co-chaperones such as Hop/Sti1, Ppt1, Cpr7, and Cpr6. Two recent studies highlight the importance of the CLR in mediating nucleotide-induced changes in Hsp90 structure and co-chaperone interactions. Although partial deletion of the CLR of Hsp82 (Δ211–259) did not significantly affect Hsp90 function, Hsp82 containing a longer deletion (Δ211–266) was unable to support viability. Purified Hsp82 containing deletions in the CLR was able to bind ATP but was unable to undergo amino-terminal dimerization and bind p23/Sba1 in the presence of ATP, indicating that it was conformationally restricted (Hainzl et al. 2009; Tsutsumi et al. 2009).
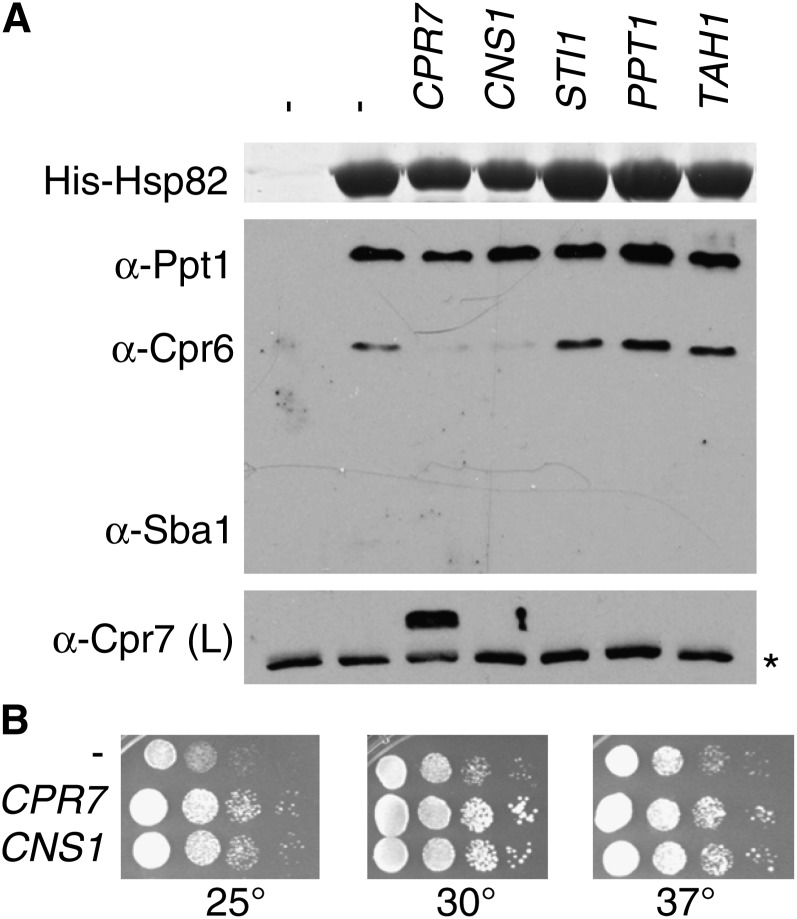
We took a genetic approach to determine whether any co-chaperones become essential when Hsp82 is conformationally restricted due to deletion of the CLR. Starting with strain JJ816 (hsc82hsp82/YEp24-HSP82), we introduced disruptions in genes encoding nonessential co-chaperones. We transformed in a plasmid expressing WT or mutant HSP82 and grew the cells in the presence of 5-FOA so that the introduced form of HSP82 was the only Hsp90 present in the cell. In the starting strain (JJ816), hsp82Δ211-259 exhibited WT growth and hsp82Δ211-264 displayed a growth defect at 37°. Both Hsp82 mutants were expressed at WT levels (Supporting Information, Figure S1). As shown in Figure 1A, there was a strong specific synthetic growth defect upon combination of deletion of CPR7 with hsp82Δ211-259. The combination of hsp82Δ211-264 and cpr7 was inviable. To better illustrate the enhanced growth defect, cells that survived on 5-FOA were tested for growth. As shown in Figure 1B, loss of CPR7 results in decreased growth at 25° and slow growth at 30° and 37°, which is evident by the formation of smaller colonies at those temperatures (Duina et al. 1996). It is unclear why the growth defect of cpr7 cells is more pronounced at lower temperatures. Cells expressing hsp82Δ211-259 grew like cells expressing WT HSP82 (Louvion et al. 1996). Upon combination of hsp82Δ211-259 and cpr7, cells exhibited enhanced growth defects at all three temperatures. We conducted similar tests to examine the growth of hsp82Δ211-259 or hsp82Δ211-264 in the absence of other co-chaperones (Figure 2). No synthetic growth defects were observed upon combination of either hsp82Δ211-259 or hsp82Δ211-264 with deletion of other co-chaperones. This result indicates a specific requirement for CPR7 in cells expressing Hsp82Δlinker constructs.
Figure 1 .
Hsp82Δlinker constructs exhibit specific synthetic growth defects upon loss of CPR7. (A) hsc82hsp82/YEp24-HSP82 strains containing deletions in the indicated co-chaperones were transformed with plasmids expressing His-tagged versions of Hsp82 WT, Hsp82Δ211-259, or Hsp82Δ211-264. Resultant colonies were grown in the presence of 5-FOA for 2 days at 30°. (B) Colonies that appeared in JJ816 (hsc82hsp82) and JJ149 (cpr7hsc82hsp82) strains plated on 5-FOA were grown overnight, serially diluted 10-fold, dropped onto selective media, and grown for 2 days at the indicated temperatures.
Figure 2 .
No synthetic growth defects of Hsp82Δlinker constructs were observed upon loss of other co-chaperones. The indicated hsc82hsp82 co-chaperone deletion strains were transformed with plasmids expressing His-tagged versions of Hsp82Δ211-264 or Hsp82Δ211-259. Growth of colonies that grew in the presence of 5-FOA was assayed as described for Figure 1B.
TPR domain of Cpr7 specifies its in vivo functions
Cpr6 and Cpr7 share 38% amino acid sequence identity, but overexpression of CPR6 does not rescue growth phenotypes or defects in Hsp90 client activity in cells lacking CPR7 (Dolinski et al. 1998; Marsh et al. 1998), indicating that they have different functions in vivo. Cpr6 and Cpr7 contain two main domains: an amino-terminal PPIase domain and a carboxy-terminal TPR domain. The PPIase domain of Cpr7 is dispensable for growth (Duina et al. 1998b). We made chimeras in which the PPIase domains were swapped, creating Cpr6 PPIase domain/Cpr7 TPR domain (6PPI/7TPR) and Cpr7 PPIase domain/Cpr6 TPR domain (7PPI/6TPR). Since Cpr7 is expressed at roughly fivefold lower levels than Cpr6 (Ghaemmaghami et al. 2003), Flag-tagged constructs were expressed under the constitutive GPD promoter to ensure similar levels of expression (Figure 3A and Figure S2). The PPIase domain of Cpr7 is ∼20 amino acids larger than that of Cpr6, accounting for the reduced mobility of those constructs in SDS-PAGE. The growth of cells expressing Flag-Cpr7 expressed under the GPD promoter was indistinguishable from cells expressing CPR7 under its own promoter (not shown). Consistent with prior studies (Tesic et al. 2003), only Flag-Cpr7 or Flag-6PPI/7TPR was able to fully rescue the growth defect of a cpr7 strain (Figure 3B). Similarly, only Flag-Cpr7 or Flag-6PPI/7TPR was able to support viability in the cpr7hsc82hsp82 strain expressing hsp82Δ211-264 (Figure 3C).
Figure 3 .
Expression and function of Cpr6/Cpr7 chimeras. (A) Chimeric versions of Cpr6 and Cpr7 expressed under the GPD promoter and containing an amino-terminal Flag tag were transformed into a WT (JJ762) strain. Cell lysates were separated by SDS-PAGE (7.5% acrylamide) and probed with the α-Flag antibody. The asterisk (*) represents a band that cross-reacts with the Flag antibody and thus serves as an internal loading control. (B) Indicated plasmids were transformed into the cpr7 deletion strain (JJ567) and growth was assayed as described. (C) Indicated plasmids were transformed into the cpr7hsc82hsp82 strain (JJ149) along with a plasmid expressing His-Hsp82Δ211-264. Resultant colonies were grown in the presence of 5-FOA for 2 days at 30°. (D) Plasmids expressing Flag-Cpr6, Flag-Cpr7, or isolated TPR domains of Cpr6 and Cpr7 were assayed for growth as described above.
The TPR domains of Cpr6 and Cpr7 share ∼33% amino acid sequence identity. To determine whether the TPR domain of Cpr7 was sufficient for viability in the cpr7hsc82hsp82 strain expressing hsp82Δ211-264, we constructed plasmids expressing the isolated Flag-tagged TPR domain of Cpr7 (amino acids 193–393) or Cpr6 (amino acids 171–371) under the GPD promoter. These constructs were expressed at levels similar to full-length proteins (Figure S3). Flag-7TPR, but not Flag-6TPR, was able to rescue growth (Figure 3D), indicting that the TPR domain of Cpr7 is necessary and sufficient for viability in the strain expressing hsp82Δ211-264.
Hsp82Δ211-264 exhibits altered interaction with Cpr6 and Cpr7
Cpr6 bound His-Hsc82 in cell lysates only in the presence of nonhydrolyzable ATP, AMP-PNP (Johnson et al. 2007). To examine Cpr7–Hsp82 interaction, cells expressing His-Hsp82 were transformed with the plasmid expressing Flag-Cpr7. His–Hsp82 complexes were isolated by incubating cell extracts with nickel resin, followed by SDS-PAGE and immunoblot analysis using peptide-specific antibodies that we raised against Sba1, Cpr6, Cpr7, or Ppt1. As expected, binding of Cpr6 and Sba1 to His-Hsp82 was observed only in the presence of AMP-PNP (Figure 4A). In contrast, Ppt1 bound Hsp82 in the presence or absence of AMP-PNP. Flag-Cpr7 bound His-Hsp82 only in the presence of AMP-PNP, and the presence of overexpressed Flag-Cpr7 did not affect Ppt1, Cpr6, or Sba1 interaction.
Figure 4 .
Hsp82Δ211-264 exhibits altered interaction with Cpr6 and Cpr7. (A) hsc82hsp82 cells expressing untagged or His-tagged wild-type Hsp82 were transformed with pRS413GPDFlag-Cpr7 (+) or pRS413GPD (−). His–Hsp82 complexes isolated in the presence or the absence of AMP-PNP were subjected to SDS-PAGE and/or immunoblot analysis. (Top) Coomassie Blue-stained gel. (Middle panels) Immunoblot analysis of copurifying proteins. (Bottom) Expression of Flag-Cpr7 in whole-cell lysate. (B) WT and mutant His–Hsp82 complexes were isolated in the presence or absence of AMP-PNP as described. (C) hsc82hsp82 cells expressing His-Hsp82 WT or His-Hsp82Δ211-264 were transformed with pRS413GPD (−), pRS413GPDFlag-Cpr7, or pRS413GPDFlag-Cpr6. (Top) Coomassie Blue-stained gel. (Middle panels) Immunoblot analyses of Cpr6 and Cpr7. The asterisk denotes the migration of Flag-Cpr6 relative to endogenous Cpr6. (Bottom) Expression of Flag-Cpr7 or Flag-Cpr6 in whole-cell extract. All gels were 10% acrylamide except the bottom panel in C comparing the migration of Flag-Cpr6 and Flag-Cpr7 in whole-cell lysates, which was done using a 7.5% acrylamide gel.
Using the same assay, we examined the ability of His-Hsp82Δ211-264 to interact with co-chaperones (Figure 4B). As previously observed, Sba1 exhibited reduced interaction with His-Hsp82Δ211-264 due to the reduced ability of this mutant form to adopt the closed conformation (Ali et al. 2006; Hainzl et al. 2009). In contrast, Cpr6 bound His-Hsp82Δ211-264 in the presence or absence of AMP-PNP. This suggests that deletion of the CLR also disrupts proper signaling of the nucleotide-bound state to the carboxy-terminus, resulting in loss of nucleotide-specific Hsp82–Cpr6 interaction. The shorter CLR truncation, Hsp82Δ211-259, which did not cause notable growth defects, did not exhibit altered Cpr6 interaction (not shown).
To confirm that Cpr6 binding to His-Hsp82 was dependent on the MEEVD sequence, we compared the interaction of Cpr6 with WT His-Hsp82 and His-Hsp82ΔΜEEVD (Figure 4B). As expected, disruption of the TPR acceptor site disrupted binding of Cpr6 and Ppt1 to Hsp90 without altering Sba1 interaction. Hsp82ΔMEEVD also failed to interact with Flag-Cpr7 (data not shown). We also constructed His-Hsp82Δ211-264ΔΜEEVD (ΔΔ). The growth of this mutant was similar to growth of Hsp82Δ211-264 (Figure S1). Cpr6 did not interact with Hsp82Δ211-264ΔΜEEVD, indicating that altered interaction of Cpr6 with Hsp82Δ211-264 was not due to formation of a second binding site that obviates the requirement for the MEEVD binding site.
We next determined whether Cpr6 and Cpr7 exhibit similar binding to His-Hsp82Δ211-264. Flag-Cpr7 or Flag-Cpr6 were expressed in cells expressing His-tagged Hsp82 or His-Hsp82Δ211-264. Endogenous Cpr6 bound WT Hsp82 only in the presence of AMP-PNP, and both Flag-Cpr6 and Flag-Cpr7 bound Hsp82Δ211-264 in the presence or absence of AMP-PNP (Figure 4C). This is consistent with a prior report that Cpr6 and Cpr7 have similar interactions with purified Hsp82 (Mayr et al. 2000).
Cpr6 and Cpr7 form a ternary complex with Hsp90
Li et al. (2011) recently demonstrated that the two TPR acceptor sites within an Hsp90 dimer may be occupied by Sti1 and Cpr6 either in vivo or in vitro. We determined whether a ternary complex forms between Cpr6, Cpr7, and Hsp82 in vivo. Strain JJ110 (cpr6hsc82hsp82/YEp24-HSP82) expressing untagged Hsp82, Hsp82ΔMEEVD, or Hsp82Δ211-264 was transformed with a plasmid expressing WT or His-tagged Cpr6 in addition to a plasmid expressing Flag-Cpr7. His-Cpr6 complexes were isolated in the presence of AMP-PNP to determine whether Flag-Cpr7 and His-Cpr6 bound to the same dimer of Hsp82 (Figure 5). Flag-Cpr7 was isolated in His–Cpr6 complexes when cells expressed WT Hsp82, but not Hsp82ΔMEEVD. The amount of Flag-Cpr7 present in complex with Cpr6 was not significantly affected by the Hsp82Δ211-264 mutation. This indicates the presence of an Hsp82–Cpr6–Cpr7 ternary complex and provides evidence that Cpr6 and Cpr7 do not stably interact in the absence of Hsp82. Under these conditions, very little Sti1 was present in the His–Cpr6 complexes (not shown).
Figure 5 .

Ternary complex between Hsp90, Cpr6, and Cpr7. cpr6hsc82hsp82 cells expressing untagged WT or mutant Hsp82 were transformed with either untagged or His-tagged Cpr6 and pRS413GPD or 413GPDFlag-Cpr7 as indicated. His-Cpr6 was isolated from cells incubated in the presence of AMP-PNP. (Top) Coomassie Blue-stained gel showing isolation of His-Cpr6 and copurifying Hsp82. (Bottom) The asterisk in the lysate samples represents an unknown protein that cross-reacts with the α-Cpr7 antisera, serving as an internal loading control.
Loss of CPR7 results in altered Hsp82–Cpr6 interaction
The synthetic growth phenotype observed upon combination of hsp82Δ211-264 and deletion of CPR7 suggests that the two mutations cause similar defects or affect the same functions of Hsp82. Since Hsp82Δ211-264 exhibited altered Cpr6 interaction, we examined the effect of CPR7 deletion on Hsp82 interaction with other co-chaperones (Figure 6). His–Hsp82 complexes were isolated from strain JJ816 (hsc82hsp82) or JJ149 (cpr7hsc82hsp82) as described. Similar to the results observed with Hsp82Δ211-264, Cpr6 bound to Hsp82 in the presence or absence of AMP-PNP. Unlike Hsp82Δ211-264, no reduction in Sba1 binding to Hsp82 in the presence of AMP-PNP was observed. Thus, Cpr7 may be involved in mediating conformational or signaling changes that result in nucleotide-specific Cpr6 interaction with the carboxy-terminus of Hsp82, but it does not appear to play a role in amino-terminal dimerization and/or Sba1 interaction.
Figure 6 .

Deletion of CPR7 results in altered interaction of Cpr6 with Hsp82. hsc82hsp82 or cpr7hsc82hsp82 cells transformed with plasmids expressing untagged or His-tagged Hsp82 WT were grown on 5-FOA to lose the YEp24-HSP82 plasmid. (Top) Coomassie Blue-stained gel. (Middle panels) Immunoblot analysis of proteins that copurify with His-Hsp82 in the presence or absence of AMP-PNP. (Bottom panels) Immunoblot of whole-cell extract incubated with the α-Cpr7 antisera. The asterisk represents an unknown protein that cross-reacts with the α-Cpr7 antisera, serving as an internal loading control.
Overexpression of CNS1 restores nucleotide-specific Hsp82–Cpr6 interaction in cpr7 cells
Loss of CPR7 resulted in slow growth and defects in activity of clients such as the glucocorticoid receptor and the heat-shock transcription factor Hsf1 (Duina et al. 1996, 1998a). These defects were rescued by the overexpression of CNS1, which encodes an essential TPR-containing co-chaperone (Marsh et al. 1998). We determined whether overexpression of CNS1 or other TPR-containing co-chaperones restored nucleotide-dependent interaction of Cpr6 and Hsp82 in cells lacking CPR7 (Figure 7A). cpr7hsc82hsp82 cells expressing untagged Hsp82 or His-Hsp82 were transformed with a plasmid expressing CPR7, CNS1, PPT1, STI1, or TAH1. His–Hsp82 complexes were isolated from cells in the absence of exogenous nucleotide. In the absence of CPR7, significant amounts of Cpr6 bound Hsp82, but the presence of CPR7 resulted in loss of Cpr6 binding. In the presence of CNS1, the level of Cpr6 binding to Hsp82 was similarly decreased. In contrast, overexpression of STI1, PPT1, or TAH1 had no effect on Hsp82–Cpr6 interaction (Figure 7A). CNS1 overexpression also rescued the growth defect of cpr7hsc82hsp82 cells expressing hsp82Δ211-259 (Figure 7B). Overexpression of STI1, PPT1, or TAH1 had no effect on the slow-growth defect of cpr7 cells or cpr7hsc82hsp82 cells expressing hsp82Δ211-259 (not shown). Thus, the specific ability of CNS1 to restore nucleotide-dependent Hsp82–Cpr6 interaction suggests that this effect is not due to displacement of Cpr6 by binding of another overexpressed TPR-containing protein. This provides additional evidence that the in vivo functions of Cpr7 and Cns1 are related to changes in Hsp90 that regulate Cpr6–Hsp82 interaction.
Figure 7 .
Overexpression of CNS1 restores nucleotide-specific interaction of Hsp82 with Cpr6. (A) cpr7hsc82hsp82 cells expressing His-tagged Hsp82-WT were transformed with pRS316, pRS316ADE3CPR7, YEp24-CNS1, pRS316ADE3STI1, pRS316ADE3PPT1, or pRS316ADE3TAH1. His-Hsp82 was isolated from cells lacking exogenous nucleotide, and complexes were analyzed as described. (Bottom) An immunoblot of whole-cell extract incubated with the α-Cpr7 antisera. The asterisk represents an unknown protein that cross-reacts with the α-Cpr7 antisera, serving as an internal loading control. (B) cpr7hsc82hsp82 cells expressing His-tagged Hsp82Δ211-259 were transformed with pRS316, pRS316ADE3CPR7, or YEp24-CNS1. Growth in selective media was monitored after 2 days on rich media at the indicated temperatures.
Discussion
Ever since the first Hsp90 co-chaperones were identified, a main goal has been to identify their specific roles in the function of the Hsp90 molecular chaperone machine. The Hsp90-binding sites for most co-chaperones have been identified, and most co-chaperones have been analyzed to determine whether they affect the ATPase activity of Hsp90 (Pearl et al. 2008; Wandinger et al. 2008). Despite these advances, there are still many questions about the diverse roles of the 12 known co-chaperones of S. cerevisiae, including questions about whether co-chaperones have overlapping or unique functions. Recent studies suggest that co-chaperones may modulate conformational changes in Hsp90. Aha1 was shown to interact with both the middle and the amino-terminal domains of Hsp90, promoting conformational changes required for ATP hydrolysis. In this way, Aha1 was proposed to play a key role in regulating the timing of the ATP-dependent Hsp90–client interactions (Koulov et al. 2010; Retzlaff et al. 2010).
We took a genetic approach to determining whether any co-chaperones play a role in mediating conformational changes in Hsp90. We started with Hsp82Δlinker mutants that are capable of ATP binding but defective in ATP-dependent dimerization of the amino-termini, resulting in reduced p23/Sba1 binding (Hainzl et al. 2009; Tsutsumi et al. 2009). We found that these “conformationally restricted” forms of Hsp82 exhibited specific growth defects when combined with loss of the gene encoding Cpr7. Deletion of CPR7 or the CLR resulted in loss of ATP-dependent Hsp82–Cpr6 interaction, suggesting that both of these mutations may affect the proper transmission of information about the state of bound nucleotide to the carboxy-terminus. Overexpression of CNS1, which was previously shown to rescue defects of cpr7 cells (Dolinski et al. 1998; Marsh et al. 1998), restored nucleotide-specific interactions between Hsp82 and Cpr6, providing further evidence that this is an important in vivo function of Cpr7. These results do not appear to be a consequence of direct competition between Cpr6 and other co-chaperones for binding to Hsp82, since overexpression of CPR7 did not alter the interaction of Cpr6 with WT or mutant Hsp82. Additionally, overexpression of STI1, TAH1, or PPT1 was unable to restore nucleotide-dependent Hsp82–Cpr6 interactions in cells lacking CPR7. Although the functions of Cpr7 and Cns1 are apparently related to the function of the CLR, overexpression of neither CPR7 nor CNS1 was able to rescue the growth defects of cells expressing hsp82Δ211-264 (not shown).
Hsp90 and Hsp70 contain carboxy-terminal EEVD residues that bind TPR domains in what is described as a “carboxylate clamp” in which the aspartic acid residues bind basic residues within a groove formed by the helical structure of the TPR domains (Scheufler et al. 2000; Pratt and Toft 2003). From structural and mutational evidence, it is clear that differing TPR co-chaperones such as Hop, Cyp40, and FKBP51/52 have similar interactions with the terminal MEEVD residues of Hsp90 and that basic residues within the TPR domains that are required for efficient binding to the MEEVD sequence are highly conserved (Ward et al. 2002; Wu et al. 2003; Carrigan et al. 2004). In yeast lysates as well as in rabbit reticulocyte lysates, Cpr6, Cpr7, and Cyp40 bind Hsp90 preferentially in the presence of AMP-PNP or ATP (Johnson and Toft 1994; Johnson et al. 2007). The structural basis for nucleotide-specific binding of some, but not all, TPR-containing co-chaperones to Hsp90 is unknown.
Sti1/Hop binds Hsp90 in the nucleotide-free form and prevents amino-terminal dimerization. In addition to the MEEVD sequence, Sti1 also contacts the middle domain (Southworth and Agard 2011; Schmid et al. 2012). Displacement of Hsp90–Sti1 complexes by Cpr6, including formation of an Hsp90–Sti1–Cpr6 ternary complex has been proposed to be a key component of the Hsp90-folding cycle (Prodromou et al. 1999; Li et al. 2011). Our results indicate that an Hsp90–Cpr6–Cpr7 ternary complex also exists. In one model (Figure 8A), formation of an Hsp90–Sti1–Cpr6 complex is followed by formation of an Hsp90–Cpr6–Cpr7 complex that is required for Cpr6 and Cpr7 dissociation. A requirement for Cpr7 in dissociating Hsp82–Cpr6 complexes could explain the persistence of Hsp82–Cpr6 complexes in the absence of nucleotide (Figure 8B).
Figure 8 .
Model of Cpr6 and Cpr7 interaction with Hsp90. (A) In the absence of nucleotide, Sti1 binds the open conformation of Hsp90. Upon ATP binding by Hsp90, Cpr6 binds the carboxy-terminus of Hsp90, resulting in formation of an Hsp90–Sti1–Cpr6 complex. Cpr6 binding displaces Sti1, resulting in the closed conformation characterized by Sba1 interaction. Formation of an Hsp90–Cpr6–Cpr7 ternary complex promotes dissociation of Cpr6, allowing for another cycle of Sti1 binding. (B) In the absence of Cpr7, Cpr6 fails to dissociate from Hsp90 after ATP hydrolysis, resulting in the persistence of Hsp82–Cpr6 complexes. N, amino-terminal domain; M, middle domain; C, carboxy-terminal domain.
The above model does not address the important role of ATP in regulating Hsp90 interaction with Cpr6 and Cpr7. It is possible that binding of ATP to the amino-terminus of Hsp90 results in a conformational change or other structural modification near the carboxy-terminus of Hsp90 that results in formation or unmasking of the Cpr6/7 binding site. In cells expressing Hsp82Δ211-264 or lacking CPR7, regulation of this signal is lost, resulting in Hsp82–Cpr6 interaction in the presence or absence of nucleotide. Alternatively, Cpr7 or the CLR of Hsp90 may be required to promote a conformational change in Cpr6 that allows it to distinguish the ATP-bound conformation. It is possible that Hsp90 interaction with another protein or co-chaperone normally prevents Cpr6 interaction in the absence of nucleotide and that other protein is unable to bind Hsp82Δ211-264. However, it is unclear why loss of Cpr7 would cause the same effect since Cpr7 was able to bind Hsp82Δ211-264.
Although both Sba1 and Cpr6 preferentially bind Hsp90 in the presence of AMP-PNP, the two co-chaperones have distinct requirements for Hsp90 binding since we identified mutations that selectively altered either Sba1 or Cpr6 interaction (Johnson et al. 2007). The current study supports our prior results that Cpr6 and Sba1 bind Hsp90 independently. However, the specific requirement for Cpr6 binding to Hsp90 remains unclear. Although Cpr6 stably bound WT Hsp82 only in the presence of AMP-PNP, Hsp82Δ211-264 was able to bind Cpr6 in the absence of AMP-PNP. Our studies are unable to distinguish whether this reflects binding of Cpr6 to the open nucleotide-free form of Hsp82Δ211-264 or an inability of Cpr6 to dissociate from Hsp82Δ211-264 after ATP hydrolysis. Additional studies are required to determine how nucleotide-induced conformational changes in Hsp90 affect the extreme carboxy-terminus. The carboxy-terminal domain of Hsp90 does not appear to undergo large conformational changes upon nucleotide binding. However, the most complete structure of yeast Hsp82 available lacks both part of the charged linker region and the last 32 amino acids, including the MEEVD residues (Ali et al. 2006). Our results suggest that signaling of nucleotide-induced changes in the amino-terminus of Hsp90 that results in stable Cpr6 binding to the carboxy-terminus is disrupted by either deletion of the CLR or loss of Cpr7. Further studies will be necessary to determine the extent of these changes as well as the mechanism of Cpr7 function. Additional studies will be needed to establish the function of the CLR, although a recent report indicates that the specific sequence, rather than the size, of the linker is critical for function (Tsutsumi et al. 2012).
CPR7 deletion leads to growth defects and reduced activity of Hsp90 clients. However, no growth defects or client activity defects are known to occur in cells lacking CPR6 (Duina et al. 1996, 1998a; Lee et al. 2004). Purified Cpr6 and Cpr7 exhibited differences in PPIase activity and molecular chaperone activity yet bound WT Hsp82 with the same affinity (Mayr et al. 2000). We established that sequences outside the PPIase domain are responsible for Cpr7 function. Cpr7-R64A, which alters a residue predicted to be required for catalytic activity (Duina et al. 1998b), was able to support viability in cpr7hsc82hsp82 cells expressing hsp82Δ211-264 (not shown). The chimeric constructs as well as the isolated TPR domain (amino acids 193–393) that supported Cpr7 functions included the TPR domain as well as a linker region postulated to be important for chaperone activity (Mok et al. 2006). Although the importance of Cpr7’s chaperone activity with respect to Hsp90 functions is unclear, it is of interest that Cpr7 was found to have elevated chaperone activity relative to Cpr6 (Mayr et al. 2000). FKBP51 and FKBP52, which share ∼70% sequence similarity, have opposing effects on the activity of steroid hormone receptors and differing patterns of associations with different steroid hormone receptors (Riggs et al. 2004). Prior studies by the Smith lab suggested that the terminal 20–30 amino acids are critical for differences in binding of FKBP51 and FKBP52 to Hsp90 (Cheung-Flynn et al. 2003). Further studies are required to establish the critical residues of Cpr7 and Cns1 that regulate Hsp90–Cpr6 interaction and viability in cells expressing Hsp82Δlinker constructs.
Supplementary Material
Acknowledgments
We thank Gary Flom and Colby Austin for assistance with plasmid and strain construction. We also thank Rick Gaber, Northwestern University, and Sue Lindquist, Whitehead Institute, for reagents. This research was funded by a grant from the National Science Foundation (MCB-0744522).
Footnotes
Communicating editor: S. Fields
Literature Cited
- Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., et al. , 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440: 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S., 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9: 3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D., Dawson D., Stearns T., 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Carrigan P. E., Nelson G. M., Roberts P. J., Stoffer J., Riggs D. L., et al. , 2004. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J. Biol. Chem. 279: 16185–16193 [DOI] [PubMed] [Google Scholar]
- Cheung-Flynn J., Roberts P. J., Riggs D. L., Smith D. F., 2003. C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to Hsp90. J. Biol. Chem. 278: 17388–17394 [DOI] [PubMed] [Google Scholar]
- D’Andrea L. D., Regan L., 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Dolinski K. J., Cardenas M. E., Heitman J., 1998. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol. Cell. Biol. 18: 7344–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A. A., Chang H. C., Marsh J. A., Lindquist S., Gaber R. F., 1996. A cyclophilin function in Hsp90-dependent signal transduction. Science 274: 1713–1715 [DOI] [PubMed] [Google Scholar]
- Duina A. A., Kalton H. M., Gaber R. F., 1998a. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 273: 18974–18978 [DOI] [PubMed] [Google Scholar]
- Duina A. A., Marsh J. A., Kurtz R. B., Chang H. C., Lindquist S., et al. , 1998b. The peptidyl-prolyl isomerase domain of the CyP-40 cyclophilin homolog Cpr7 is not required to support growth or glucocorticoid receptor activity in Saccharomyces cerevisiae. J. Biol. Chem. 273: 10819–10822 [DOI] [PubMed] [Google Scholar]
- Flom G., Weekes J., Johnson J. L., 2005. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr. Genet. 47: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G., Weekes J., Williams J. J., Johnson J. L., 2006. Effect of mutation of the tetratricopeptide repeat and aspartate-proline 2 domains of Sti1 on Hsp90 signaling and interaction in Saccharomyces cerevisiae. Genetics 172: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., et al. , 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Hainzl O., Lapina M. C., Buchner J., Richter K., 2009. The charged linker region is an important regulator of Hsp90 function. J. Biol. Chem. 284: 22559–22567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harst A., Lin H., Obermann W. M., 2005. Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem. J. 387: 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessling M., Richter K., Buchner J., 2009. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 16: 287–293 [DOI] [PubMed] [Google Scholar]
- Jakob U., Lilie H., Meyer I., Buchner J., 1995. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J. Biol. Chem. 270: 7288–7294 [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Toft D. O., 1994. A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J. Biol. Chem. 269: 24989–24993 [PubMed] [Google Scholar]
- Johnson J. L., Halas A., Flom G., 2007. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol. Cell. Biol. 27: 768–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulov A. V., Lapointe P., Lu B., Razvi A., Coppinger J., et al. , 2010. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol. Biol. Cell 21: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P., Shabbir A., Cardozo C., Caplan A. J., 2004. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell 15: 1785–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Richter K., Buchner J., 2011. Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat. Struct. Mol. Biol. 18: 61–66 [DOI] [PubMed] [Google Scholar]
- Louvion J. F., Warth R., Picard D., 1996. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc. Natl. Acad. Sci. USA 93: 13937–13942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J. A., Kalton H. M., Gaber R. F., 1998. Cns1 is an essential protein associated with the hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in cpr7Delta cells. Mol. Cell. Biol. 18: 7353–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Richter K., Lilie H., Buchner J., 2000. Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 275: 34140–34146 [DOI] [PubMed] [Google Scholar]
- McClellan A. J., Xia Y., Deutschbauer A. M., Davis R. W., Gerstein M., et al. , 2007. Diverse cellular functions of the hsp90 molecular chaperone uncovered using systems approaches. Cell 131: 121–135 [DOI] [PubMed] [Google Scholar]
- Millson S. H., Vaughan C. K., Zhai C., Ali M. M., Panaretou B., et al. , 2008. Chaperone ligand-discrimination by the TPR-domain protein Tah1. Biochem. J. 413: 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok D., Allan R. K., Carrello A., Wangoo K., Walkinshaw M. D., et al. , 2006. The chaperone function of cyclophilin 40 maps to a cleft between the prolyl isomerase and tetratricopeptide repeat domains. FEBS Lett. 580: 2761–2768 [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M., 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Owens-Grillo J. K., Hoffmann K., Hutchison K. A., Yem A. W., Deibel M. R., Jr, et al. , 1995. The cyclosporin A-binding immunophilin CyP-40 and the FK506-binding immunophilin hsp56 bind to a common site on hsp90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J. Biol. Chem. 270: 20479–20484 [DOI] [PubMed] [Google Scholar]
- Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J. K., et al. , 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 10: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C., Workman P., 2008. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 410: 439–453 [DOI] [PubMed] [Google Scholar]
- Pratt W. B., Toft D. O., 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228: 111–133 [DOI] [PubMed] [Google Scholar]
- Prodromou C., Siligardi G., O’Brien R., Woolfson D. N., Regan L., et al. , 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18: 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzlaff M., Hagn F., Mitschke L., Hessling M., Gugel F., et al. , 2010. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol. Cell 37: 344–354 [DOI] [PubMed] [Google Scholar]
- Richter K., Buchner J., 2011. Closing in on the Hsp90 chaperone-client relationship. Structure 19: 445–446 [DOI] [PubMed] [Google Scholar]
- Richter K., Muschler P., Hainzl O., Reinstein J., Buchner J., 2003. Sti1 Is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the ATPase cycle. J. Biol. Chem. 278: 10328–10333 [DOI] [PubMed] [Google Scholar]
- Riggs D., Cox M., Cheung-Flynn J., Prapapanich V., Carrigan P., et al. , 2004. Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit. Rev. Biochem. Mol. Biol. 39: 279–295 [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., et al. , 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101: 199–210 [DOI] [PubMed] [Google Scholar]
- Schmid A. B., Lagleder S., Grawert M. A., Rohl A., Hagn F., et al. , 2012. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31: 1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau A. K., Harris S. F., Southworth D. R., Agard D. A., 2006. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127: 329–340 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siligardi G., Hu B., Panaretou B., Piper P. W., Pearl L. H., et al. , 2004. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 279: 51989–51998 [DOI] [PubMed] [Google Scholar]
- Southworth D. R., Agard D. A., 2011. Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol. Cell 42: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street T. O., Lavery L. A., Agard D. A., 2011. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol. Cell 42: 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesic M., Marsh J. A., Cullinan S. B., Gaber R. F., 2003. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in saccharomyces cerevisiae. J. Biol. Chem. 278: 32692–32701 [DOI] [PubMed] [Google Scholar]
- Trepel J., Mollapour M., Giaccone G., Neckers L., 2010. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10: 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S., Mollapour M., Graf C., Lee C. T., Scroggins B. T., et al. , 2009. Hsp90 charged-linker truncation reverses the functional consequences of weakened hydrophobic contacts in the N domain. Nat. Struct. Mol. Biol. 16: 1141–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S., Mollapour M., Prodromou C., Lee C. T., Panaretou B., et al. , 2012. Charged linker sequence modulates eukaryotic heat shock protein 90 (Hsp90) chaperone activity. Proc. Natl. Acad. Sci. USA 109: 2937–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan C. K., Gohlke U., Sobott F., Good V. M., Ali M. M., et al. , 2006. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol. Cell 23: 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger S. K., Richter K., Buchner J., 2008. The Hsp90 chaperone machinery. J. Biol. Chem. 283: 18473–18477 [DOI] [PubMed] [Google Scholar]
- Ward B. K., Allan R. K., Mok D., Temple S. E., Taylor P., et al. , 2002. A structure-based mutational analysis of cyclophilin 40 identifies key residues in the core tetratricopeptide repeat domain that mediate binding to Hsp90. J. Biol. Chem. 277: 40799–40809 [DOI] [PubMed] [Google Scholar]
- Wu B., Li P., Shu C., Shen B., Rao Z., 2003. Crystallization and preliminary crystallographic studies of the C-terminal domain of human FKBP52. Acta Crystallogr. D Biol. Crystallogr. 59: 2269–2271 [DOI] [PubMed] [Google Scholar]
- Zhao R., Davey M., Hsu Y. C., Kaplanek P., Tong A., et al. , 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120: 715–727 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.