Abstract
RNA-directed antisense and interference therapeutics are a promising treatment option for cancer. The demonstration of depletion of target proteins within human tumors in vivo using validated methodology will be a key to the application of this technology. Here, we present a flow cytometric-based approach to quantitatively determine protein levels in solid tumor material derived by fiber optic brushing (FOB) of non-small cell lung cancer (NSCLC) patients. Focusing upon the survivin protein, and its depletion by an antisense oligonucleotide (ASO) (LY2181308), we show that we can robustly identify a subpopulation of survivin positive tumor cells in FOB samples, and, moreover, detect survivin depletion in tumor samples from a patient treated with LY2181308. Survivin depletion appears to be a result of treatment with this ASO, because a tumor treated with conventional cytotoxic chemotherapy did not exhibit a decreased percentage of survivin positive cells. Our approach is likely to be broadly applicable to, and useful for, the quantification of protein levels in tumor samples obtained as part of clinical trials and studies, facilitating the proof-of-principle testing of novel targeted therapies.
Keywords: antisense therapy, lung cancer, survivin
Introduction
Therapeutic modalities that take advantage of effects of antisense RNA molecules or the RNA interference pathway hold promise in the treatment of a wide range of conditions, including cancer.1,2 However, numerous pitfalls and potential risks have been identified relating to the therapeutic administration of such agents.2 Primary among these is specificity, where off-target effects may induce adverse effects and immunogenicity could limit efficacy. As part of efforts to determine specificity, it is vital that a RNA therapeutic can be shown to reach its target, in the case of cancer, tumor cells, and act to deplete the factor it is directed against in vivo. For studies in patients with tumors at certain sites, the opportunities to obtain serial samples of such material are very limited, since unlike in animal studies, multiple biopsies and other invasive investigations are usually not possible. Consequently there is an urgent need to develop new approaches to assess target depletion by antisense RNA and related therapeutics.
Although methods exist, most notably immunohistochemistry, for determining protein depletion in a small population of solid tumor cells obtained by biopsy, or similar sampling techniques,3,4 these impose some significant limitations, principally that only a limited number of cells can be analyzed and the data obtained is, at best, semiquantitative.5,6,7 An alternative is quantitative real-time PCR based technology,8 but some methods do not allow ready differentiation between tumor and nontumor cells in a biopsy or sample, or if PCR is performed on a dissected portion of tumor the signal from only a limited number of cells is considered.3,9 In addition, peripheral blood mononuclear cells are very commonly used as a surrogate in such analysis,10,11,12,13,14,15,16,17 but this is not informative regarding the relevant tumor-specific molecular target. Perhaps most critically, correlations between mRNA and protein levels are often not strong.18,19,20 It is therefore clearly desirable to develop more quantitative methods to assess protein expression and its depletion in patient-derived material using methods where tumor cells can be readily identified. Flow cytometric (fluorescence-activated cell sorting (FACS))-based methods are ideal for this purpose, since tens of thousands of cells from each sampling can be analyzed and each expression signal directly quantified. Moreover, the multi-laser capacity of current FACS machines allows several tumor markers to be followed simultaneously (multiplexed).21
Survivin is a small (16.5 KDa) protein, originally identified as an inhibitor of apoptosis factor,22,23,24 existing as a homodimeric complex that is encoded by an essential gene.25,26 There is an increase in survivin expression at mitosis,27 and survivin depletion has been associated with mitotic abnormalities.28,29,30 It is now clear that survivin is, in fact, both a bona fide inhibitor of apoptosis and also an important mitotic progression factor.31,32 Importantly for the present study, survivin is not detectable in most adult human tissues, but is highly expressed, in a cell cycle phase-independent manner, in many human tumors.33 There is evidence that this disregulated survivin protein acts to decrease tumor susceptibility to apoptosis,34 and also to increase angiogenesis and resistance to several anticancer drugs.35,36 For these reasons, there is strong current interest in survivin inhibition and depletion as an anticancer strategy, since survivin inactivation in human cells could increase spontaneous tumor-specific apoptosis and also potentiate the action of other antitumor agents.32 In this regard, a second generation 18-mer 2′-O-methoxyethyl-modified antisense oligonucleotide (ASO) (LY2181308) against survivin has been developed, which targets the translation initiation codon of the survivin transcript, triggering RNase-H–dependent duplex cleavage and subsequent transcript degradation.28 A first-in-human study has been initiated, and we report here a FACS-based strategy to directly quantify survivin protein and its depletion in non-small cell lung cancer (NSCLC) tumor samples obtained from a patient treated with LY2181308.
Results
Proof-of-principle: survivin depletion in NSCLC using cell lines
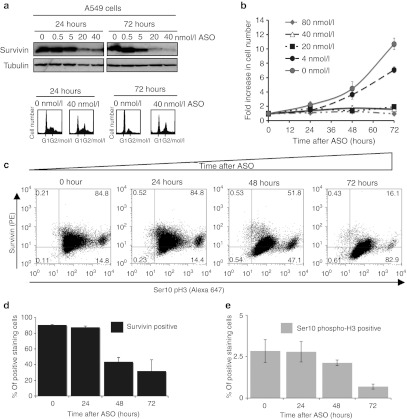
A well-characterized NSCLC cell line was initially evaluated for survivin protein levels, and for survivin depletion by the highly specific anti-survivin ASO LY2181308 (hereafter referred to as ASO).37 Figure 1a shows an anti-survivin immunoblot of whole-cell protein extracted from A549 NSCLC cells at 24 and 72 hours following treatment with ASO, at increasing (and clinically relevant) doses from 0 to 40 nmol/l. It is clear that efficient depletion of survivin is achieved at 20 nmol/l ASO, and increasing the dose to 40 nmol/l does not significantly enhance depletion. Some depletion occurs by 24 hours, but is greater at 72 hours. Cells from this experiment were also fixed and stained with propidium iodide and subjected to FACS analysis (Figure 1a, lower panel). This revealed that treatment with 40 nmol/l ASO produced an enrichment of cells in the G2/M phase of the cell cycle, consistent with the known role for survivin in mitotic progression as a member of the chromosomal passenger complex.38 Moreover, survivin depletion by increasing doses of the ASO resulted in a dose-dependent growth inhibition, again consistent with its role in mitotic progression (Figure 1b).
Figure 1.
Effective survivin depletion can be achieved in NSCLC cell lines at pharmacologically relevant doses of ASO. (a) Upper panel, immunoblot of A549 cells treated with ASO for 24 or 72 hours. Lower panel, cell cycle distribution of treated cells, showing an increase in G2/M population, determined by propidium iodide staining and FACS analysis. (b) Treatment of A549 cells with increasing doses of ASO results in growth inhibition, data is the average of at least four repeats. (c) FACS analysis of 40 nmol/l ASO treated cells with PE-conjugated anti-survivin antibodies, and Alexa Fluor-647 conjugated serine-10 phospho-histone H3 antibodies. The percentage of cells falling in each quadrant is shown. (d,e) The data shown in c is quantified in these graphs, where the results shown are the average of four repeats and error bars show the standard error. ASO, antisense oligonucleotide; FACS, fluorescence-activated cell sorting; NSCLC, non-small cell lung cancer; PE, phycoerythrin.
We next used a flow cytometry/FACS-based assay to analyze and quantify survivin levels in ASO-treated NSCLC cell lines. Briefly, cells were fixed with formaldehyde and immunostained for survivin and the serine-10 phosphorylated form of histone H3 (ser10pH3, a marker of mitotic progression). Consistent with results obtained by immunoblotting, FACS analysis revealed that following ASO treatment, the proportion of cells expressing survivin fell dramatically over the course of 72 hours, from ~90% to under 20% (Figure 1c and quantified in Figure 1d). We conclude that we can detect survivin expression in NSCLC cells by both immunoblot and by FACS analysis. We are also able to demonstrate efficient depletion of survivin by the ASO in NSCLC cell lines by both methods. We observed that ser10pH3 levels are reduced in NSCLC cell lines following survivin depletion, although only a small proportion of cells from these cultures express high levels ser10pH3 (Figure 1e). This reduction is consistent with recent reports that survivin associates with aurora B to promote H3 phosphorylation at mitosis.39 It was therefore judged to be potentially valuable to include analysis of both survivin levels and ser10pH3 in the analysis of tumor samples, since clear changes in ser10pH3 levels would be a potentially informative additional marker of altered mitotic progression following survivin depletion.
Detection of survivin protein expression in human NSCLC tumor samples and normal lung tissue
To determine whether the FACS methodology used above could be adapted for the analysis of patient NSCLC tumor samples obtained by fiber optic-brushing techniques (FOB), we further developed this technique. The chief modifications and additions required to optimize the technique for use with patient FOB samples were: (i) development of methods to disaggregate the tumor material without introducing adventitious cellular damage; and (ii) optimization of the fixing conditions for co-staining with anti-survivin, anti-ser10pH3, and DNA (Hoescht 33258) for such material. Efficient tumor disaggregation was achieved using a “Medimachine” (Becton-Dickinson, Oxford, UK) that gently grinds tissue into a single cell suspension, while introducing little adventitious damage,40 as confirmed by microscopy and FACS analysis assays for increased sub-G1 DNA content material and the induction of apoptosis (data not shown).
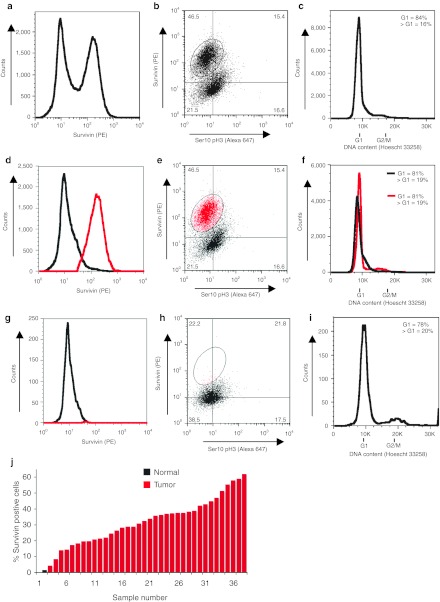
Pre-optimized fixing conditions were then applied to disaggregated FOB NSCLC tumor samples obtained from patients. In Figure 2a, the FACS profile demonstrates a distinct population of cells expressing high levels of survivin, which are highlighted in red in Figure 2d. When survivin expression is plotted against ser10pH3, however, the cells cannot be further stratified into distinct populations, reflecting the overall low expression of ser10pH3 (Figure 2b,e). Figure 2c shows the DNA content-profile of this sample, indicating that the majority of cells are in G0/G1 phase of the cell cycle, and in Figure 2f survivin expression is overlaid on this DNA content plot. This reveals that the distinct high (consistent with tumor) and low (consistent with normal) survivin-expressing populations also have slightly shifted G1 DNA content; with those cells expressing high survivin having a slightly greater DNA content than those with low survivin expression (see Supplementary Figure S1 for a description of how a normal G1 DNA content was ascribed to the samples). Since an increased DNA content (ploidy) is typical of tumor cells,41 as is survivin expression,33 this data suggests that the two populations of cells detected in this patient are: cells with low survivin and low G1 DNA content are likely to be nontumor, and those with high survivin and greater G1 DNA content are likely to be tumor cells.
Figure 2.
Survivin expression and cell cycle profile in NSCLC and normal tissue FOB determined by FACS. (a) Tumor FOB sample, stained for survivin and ser10pH3, where survivin expression alone is plotted. (b) Tumor FOB sample, stained for survivin and ser10pH3, both markers plotted against one another. The percentage of cells falling in each quadrant is shown. (c) Cell cycle profile, obtained by Hoescht 33258 staining. (d) Analysis of survivin positive cells (red) versus negative (black) showing distinct population of positive cells collected in the brushing material. (e) Analysis as in b, where the survivin positive population is highlighted in red. (f) Evidence that G1 peak represents two population, showing survivin positive (red) and negative cells (gray), as defined in b and e. Survivin negative cells have a slightly lower G1 DNA content than the survivin positive population. (g) Cell population obtained from FOB of normal bronchial epithelium. Analysis of survivin staining in normal cells demonstrates the presence of a single population of cells with low survivin expression. (h) These cells cannot be further stratified by plotting ser10pH3 expression against survivin expression. The percentage of cells falling in each quadrant is shown. (i) Cell cycle profile of FOB sample from normal bronchial epithelium. (j) Distribution of samples showing the percentage of survivin positive cells (defined as in e and h) for each sample. Thirty-six NSCLC samples were analyzed (red bars) and two normal bronchial epithelia samples were also obtained (black bars). FACS, fluorescence-activated cell sorting; FOB, fiber optic brushing; NSCLC, non-small cell lung cancer; PE, phycoerythrin; ser10pH3, serine-10 phosphorylated form of histone H3.
To further explore this point, we also obtained a FOB from the unaffected (contralateral) lung of a patient undergoing diagnostic FOB. In contrast to the tumor FOB sample, only a single, low survivin-expressing population of cells was observed (Figure 2g). No distinct subpopulation of cells could be further identified by ser10pH3 staining (Figure 2h), and DNA staining revealed that the majority of these cells were in the G0/G1 phase of the cell cycle (Figure 2i).
In total, we have analyzed 36 NSCLC samples obtained by FOB, and the majority of them have a clearly detectable survivin positive population (with a median 30% survivin positive cells, red bars in Figure 2j). Of the two samples obtained from normal lung epithelia, FACS analysis did not reveal survivin expression markedly above the threshold of detection (1.0 and 1.6%, respectively, black bar in Figure 2j). In summary, cells obtained from normal bronchial epithelia by FOB can be readily distinguished from NSCLC samples due to the absence of survivin expression as determined by FACS analysis.
Note that, as part of our validation and clinical trial analysis, an unstained version of every sample also underwent FACS analysis (negative control). In addition, a control NSCLC cell line (NCI-H647) was stained with all antibodies and isotype negative controls to establish the threshold of detection on the FACS analyzer (see Supplementary Figure S2 for an example).
Analysis of survivin expression in FOB samples obtained from ASO-treated patients
Next, we analyzed a tumor sample from a patient enrolled in the first-in-human clinical trial of the anti-survivin ASO as a single agent in NSCLC. We sought to determine, using our FACS methodology, whether changes in tumor survivin expression could be detected following ASO administration.
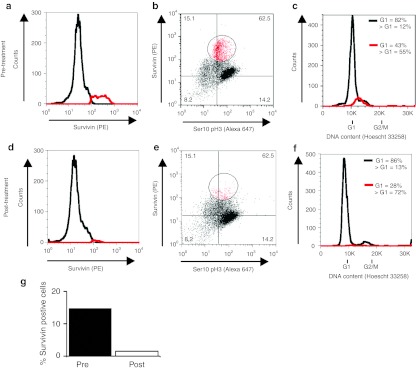
FACS analysis for survivin expression profiles were obtained from FOB samples (taken from the same anatomical location) before (Figure 3a,b), and 48 hours after ASO administration (Figure 3d,e), with survivin alone plotted in Figure 3a,d, and survivin plotted against ser10pH3 in Figure 3b,e. The data is quantified in Figure 3g. An 85% decrease in survivin expression is apparent following ASO treatment, where 14.5% of cells expressed high levels of survivin before treatment, but only 2.2% following treatment. Note that the cell cycle profile of the sample does not radically alter following ASO treatment (Figure 3c,f). Taken together our data suggests that treatment with the ASO has depleted survivin in these tumor cells, or selectively eliminated high survivin-expressing cells within the region of sampling. Regardless, this analysis indicates that we are able to detect changes in survivin expression profile in viable primary tumor material following its targeted depletion.
Figure 3.
Examination of survivin expression pre- and post-ASO treatment in a trial patient. (a) Survivin positive (red) and negative (black) populations defined by FACS, before ASO administration. (b) Survivin expression plotted against ser10pH3 expression, before (pre-) ASO administration. The percentage of cells falling in each quadrant is shown. (c) Cell cycle (DNA content) distribution shown by black line, survivin positive cells (as defined in b) overlaid in red. (d-f) As a-c, but for sample obtained 48 hours following (post-) ASO administration. (g) Quantification of fraction of high survivin-expressing cells pre- and 48 hours post-treatment. ASO, antisense oligonucleotide; FACS, fluorescence-activated cell sorting; PE, phycoerythrin; ser10pH3, serine-10 phosphorylated form of histone H3.
Conventional cytotoxic chemotherapeutics do not deplete survivin
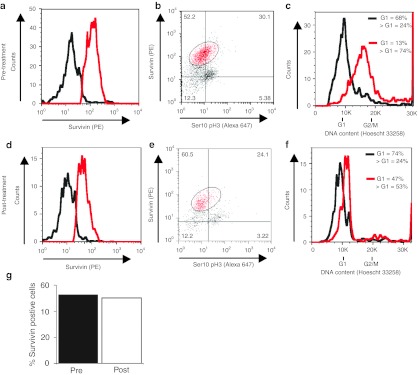
To further explore the specificity of survivin depletion following ASO administration, we also examined survivin expression in a patient treated with conventional cytotoxic chemotherapy (cisplatin-gemcitabine). Tumor FOB samples were taken before (Figure 4a,b), and 48 hours following cisplatin-gemcitabine administration (Figure 4d,e). However, and in contrast to the effects of survivin ASO treatment, survivin levels were only modestly decreased in the NSCLC FOB sample following cisplatin chemotherapy (Figure 4g), from 50.8% expressing high levels of survivin before treatment to 50.2% following treatment. There was some alteration in cell cycle profile in this sample following chemotherapy administration (Figure 4c,f), with a reduction in cells in G2/M phase, consistent with the possibility that the treatment had impacted on the tumor cells and altered their cycling profile.
Figure 4.
Treatment with cisplatin-gemcitabine did not strongly deplete survivin expression in a NSCLC tumor. (a) Survivin positive (red) and negative (black) populations defined by FACS, before cisplatin-gemcitabine chemotherapy administration. (b) Survivin expression plotted against ser10pH3 expression, before (pre-) cisplatin-gemcitabine chemotherapy administration. The percentage of cells falling in each quadrant is shown. (c) Cell cycle (DNA content) distribution, with survivin positive cells (as defined in b) overlaid in red. (d-f) As a-c, but for sample obtained 48 hours following (post-) cisplatin-gemcitabine administration. (g) Quantification of fraction of high survivin-expressing cells pre- and 48 hours post-treatment. FACS, fluorescence-activated cell sorting; NSCLC, non-small cell lung cancer; PE, phycoerythrin; ser10pH3, serine-10 phosphorylated form of histone H3.
Discussion
We have developed a FACS-based approach for evaluating therapeutic protein depletion quantitatively in NSCLC tumor material obtained by FOB. Initially, we established the validity of FACS-based investigations in NSCLC cell lines, and then proceeded to analyze tumor material obtained by FOB from NSCLC patients. The parameters were selected to avoid introducing adventitious damage to the tumor material, because it was clear from pilot experiments that over-treatment in the disaggregation step, or over-fixation with formaldehyde, could have deleterious effects. The resulting methodology has some clear advantages over existing approaches (see Introduction). First, serial FOB sampling is relatively acceptable to patients.42 Second, a sufficient quantity of tumor material can be obtained for the quantitative analysis of many thousands of cells by flow cytometry methods. Third, it is clear that very little adventitious cellular damage is sustained during the FOB procedure and subsequent tumor disaggregation, since we have now analyzed 50 NSCLC FOB samples the vast majority of which were of sufficiently high quality for FACS analysis and/or cell sorting (36 which is 72% of total FOB samples collected, see Figure 2j). Although the conditions we selected appeared to be optimal for NSCLC, we cannot be certain that these conditions would be appropriate for other tumor types.
Having confirmed that FOB coupled to disaggregation is a robust method for obtaining samples that can be analyzed by FACS and cell sorting, we progressed to analysis of survivin expression in NSCLC tumor samples. Notably, survivin is expressed in almost all solid tumors, including NSCLC, but not normal adult tissue, making it an excellent tumor marker with broad applicability.32,33 Based upon this finding, we predict that survivin expression will be an excellent marker for other types of FACS-based clinical studies in NSCLC. For example, the expression of other key biomarkers under investigation in NSCLC can be evaluated in the survivin positive and negative cell populations obtained by FOB, transbronchial needle aspiration, endobronchial ultrasound sampling, and in circulating tumor cells.43,44,45,46 This will permit direct quantification, at the protein level, of markers (for example XPF-ERCC1, EGFR, and K-ras in NSCLC) in patient tumors, and a direct means of comparing their expression in nontumor material obtained from the same anatomical location. Indeed, ongoing studies from our laboratory indicate that the methodology will be an effective way of determining XPF protein expression in NSCLC samples. We conclude that the basic methodology presented here, and identification of survivin as a robust marker of NSCLC tumor cells, will be a powerful tool for performing tumor-specific, quantifiable studies of multiple biomarkers in this disease.44
We then applied this FACS methodology to the analysis of FOB samples from a patient enrolled in a clinical trial of an anti-survivin ASO. The pharmacokinetic/pharmacodynamic model predicts that 750 mg dose of LY2181308 achieves an intratumoral concentration greater than EC50 (half maximal effective concentration) (20 µg/g) that is associated with a >50% survivin protein inhibition.47 Following ASO treatment the number of cells expressing the highest levels of survivin drastically reduced. The methodology developed here is currently being applied to samples from selected NSCLC patients enrolled in the phase 1 clinical trial of the LY2181308 ASO, and the summary data from this cohort of patients has been published elsewhere as part of the phase 1 trial report.47
In summary, we have developed a robust approach to quantitatively assess levels of survivin protein in patient tumor samples derived by FOB. This method can be broadly applied to differentiate tumor from nontumor cells in serial samples from NSCLC patients. Moreover, the method provides a very powerful way of quantifying protein expression within the tumor and nontumor populations of a single sample obtained in clinical trials involving targeted agents such as the ASO presented here.
Materials and Methods
Treatment of cell lines with LY2181308. Cells were transfected with the stated doses of ASO LY2181308 (sequence: 5′-TGTGCTATTCTGTGAATT-3′) dissolved in Lipofectamine transfection reagent and OPTIMEM reduced-serum medium (Invitrogen, Paisley, UK). This second generation ASO contains contains 2′-O-methoxyethyl (2′-MOE) residues (underlined) within a phosphorothioate backbone and the two cytosines (positions 5 and 10) are 5-methyl substituted. Media was removed from cultured cells, which were washed twice in serum-free medium, and then incubated in the ASO transfection mixture for 4 hours. After 4 hours, the transfection mixture was washed off, and cells were placed back into tissue culture media for the appropriate period.
Patient selection and treatment. For the analysis of pre-treated NSCLC tumor as controls, patients undergoing diagnostic FOB as part of their routine care were invited to participate in the Oxford Radcliffe Thoracic Biobank study (Oxford Research Ethics Committee (OxREC) approved study, reference: 09/H0606/5). Endobronchial brushings were sampled and processed immediately according to the methods described below. Patients with NSCLC participating in the first-in-human study of single agent LY2181308 (OxREC reference: 04/MRE02/52) gave informed signed consent to endobronchial tumor sampling by FOB before LY21813008 treatment and at 48 hours after administration of 750 mg intravenously over 3 hours daily for 3 days. For evaluation of the effects of cytotoxic chemotherapy on apoptosis, cell cycle progression, and survivin protein expression in chemotherapy naive patients, patients gave signed informed consent for pre-treatment and post-treatment endobronchial tumor sampling 48 hours following the first cycle of cisplatin (80 mg/M2 intravenous D1) and gemcitabine (1,250 mg/M2 D1) (OxREC reference: C00.082).
LY2181308 solution was supplied by Eli Lilly in glass vials with elastomeric closures containing 4.2 ml of active study drug at a concentration of 25 mg/ml. The drug product formulation contained active LY2181308 and phosphate-buffered saline (PBS) at pH 7.2. The supply and preparation of gemcitabine and cisplatin followed standard operating procedures of the Oxford Radcliffe Hospitals Trust.
FOB. Fiber optic endobronchial tumor sampling is a safe, short, standard procedure used in the diagnosis of pulmonary diseases. Patients who had provided informed, signed consent were lightly sedated, typically with intravenous midazolam, and had topical anesthetic applied to the airways. Endobronchial tumors were sampled by brushing with a 1 mm brush under endoscopic vision.
Tumor disaggregation. The cut FOB brush was placed into 10 ml of cold PBS, and kept at 4 °C for duration of processing. The brush in PBS was vortexed for 5 seconds at maximum speed. The brush was transferred to a fresh 10 ml aliquot of PBS, and the vortexing procedure repeated. The released material was pooled and pelleted by centrifugation 1,000g for 5 minutes at 4 °C, and the pellet resuspended in 1.5 ml cold PBS. The sample was transferred into a 50 mm “Medicon” disaggregation cell, and the unit placed in a “Medicon Medimachine” and subject to a 10 seconds pulse. The suspended sample was removed and the “Medicon” cell rinsed with PBS twice.
Controls for staining made by staining NCI-H647 NSCLC cells that were processed as described.48
Fixation of disaggregated tumor. The tumor cell suspension was concentrated by centrifugation at 1,000g for 5 minutes at 4 °C. The supernatent was removed, and the pellet was resuspended in 1% ice-cold formaldyhyde fixation buffer with gentle agitation. The sample was vortexed for 10 seconds and incubated at 4 °C for 24 hours.
Antibody and DNA staining of FOB sample. The fixative was removed by centrifigation at 1,000g for 5 minutes at 4 °C, followed by washing the pellet with cold PBS. The pellet was resuspended into 50 µl SAP buffer (0.1% (wt/vol) saponin, in Hank's balanced salt solution) containing 5 mg/ml Hoescht 33258. The sample was stained with 2 µl of Alexa fluor 647-conjugated anti-Histone H3 pSer10 antibody (Cell Signaling 9716; Cell Signaling Technology, Hitchin, UK) and 3 µl Phycoerythrin-conjugated anti-Survivin antibody (R&D Systems IC6472P; R&D Systems, Abingdon, UK). Comparable quantities of antibodies were used to stain control cell line NIHC647. IgG isotype controls (the IgG isotype controls for these were phycoerythrin-conjugated mouse IgG (R&D Systems IC002P) and Alexa Fluor-conjugated rabbit IgG (Cell Signaling 3452), respectively) were included. Samples were mixed with gentle agitation and stored in dark for 60 minutes. Samples were pelleted at 1,000g for 5 minutes at 4 °C, and washed with 500 µl PBS containing 5 mg/ml Hoescht 33258, and then stored at 4 °C in dark.
FACS analysis and cell sorting. FACS analysis was performed on a Cyan ADP Flow cytometer, and data analyzed using Summit version 4.3 (Becton Coulter, High Wycombe, UK) and Flowjo version 8 software (TreeStar, Ashland, OR).
SUPPLEMENTARY MATERIAL Figure S1. Shows how cell cycle parameters established. Figure S2. Shows example controls for sample staining.
Acknowledgments
A.L.O. is funded by the Experimental Cancer Centers (ECMC) initiative, work in PMcH's lab is funded by Cancer Research UK. We thank Kevin Clark and Craig Waugh for assistance with FACS analysis. We are very grateful to all members of PMcH's research group, especially Lonnie Swift, for helpful discussions throughout this work, to the patients who contributed to the study, and to Adrian L Harris for helpful comments on the manuscript. D.C.T. has received honoraria and an unrestricted grant from Eli Lilly. The authors declared no conflict of interest.
Supplementary Material
Shows how cell cycle parameters established.
Shows example controls for sample staining.
References
- Rayburn ER., and, Zhang R. Antisense, RNAi, and gene silencing strategies for therapy: mission possible or impossible. Drug Discov Today. 2008;13:513–521. doi: 10.1016/j.drudis.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., and, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi KN, Eisenhauer E, Fazli L, Jones EC, Goldenberg SL, Powers J.et al. (2005A phase I pharmacokinetic and pharmacodynamic study of OGX-011, a 2'-methoxyethyl antisense oligonucleotide to clusterin, in patients with localized prostate cancer J Natl Cancer Inst 971287–1296. [DOI] [PubMed] [Google Scholar]
- Lai SY, Koppikar P, Thomas SM, Childs EE, Egloff AM, Seethala RR.et al. (2009Intratumoral epidermal growth factor receptor antisense DNA therapy in head and neck cancer: first human application and potential antitumor mechanisms J Clin Oncol 271235–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong AS., and, Leong TY. Newer developments in immunohistology. J Clin Pathol. 2006;59:1117–1126. doi: 10.1136/jcp.2005.031179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CR., and, Levenson RM. Quantification of immunohistochemistry–issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–424. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- Walker RA. Quantification of immunohistochemistry–issues concerning methods, utility and semiquantitative assessment I. Histopathology. 2006;49:406–410. doi: 10.1111/j.1365-2559.2006.02514.x. [DOI] [PubMed] [Google Scholar]
- Moulder SL, Symmans WF, Booser DJ, Madden TL, Lipsanen C, Yuan L.et al. (2008Phase I/II study of G3139 (Bcl-2 antisense oligonucleotide) in combination with doxorubicin and docetaxel in breast cancer Clin Cancer Res 147909–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Kolesar J, McNeel DG, Leith C, Schell K, Eickhoff J.et al. (2008A phase I pharmacokinetic and pharmacodynamic correlative study of the antisense Bcl-2 oligonucleotide g3139, in combination with carboplatin and paclitaxel, in patients with advanced solid tumors Clin Cancer Res 142732–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer R, Vidal L, Griffin M, Lesley M, de Bono J, Coulthard S.et al. (2009Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors Clin Cancer Res 153177–3183. [DOI] [PubMed] [Google Scholar]
- Shibata SI, Doroshow JH, Frankel P, Synold TW, Yen Y, Gandara DR.et al. (2009Phase I trial of GTI-2040, oxaliplatin, and capecitabine in the treatment of advanced metastatic solid tumors: a California Cancer Consortium Study Cancer Chemother Pharmacol 641149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi KN, Siu LL, Hirte H, Hotte SJ, Knox J, Kollmansberger C.et al. (2008A phase I study of OGX-011, a 2'-methoxyethyl phosphorothioate antisense to clusterin, in combination with docetaxel in patients with advanced cancer Clin Cancer Res 14833–839. [DOI] [PubMed] [Google Scholar]
- Dritschilo A, Huang CH, Rudin CM, Marshall J, Collins B, Dul JL.et al. (2006Phase I study of liposome-encapsulated c-raf antisense oligodeoxyribonucleotide infusion in combination with radiation therapy in patients with advanced malignancies Clin Cancer Res 121251–1259. [DOI] [PubMed] [Google Scholar]
- O'Dwyer PJ, Stevenson JP, Gallagher M, Cassella A, Vasilevskaya I, Monia BP.et al. (1999c-raf-1 depletion and tumor responses in patients treated with the c-raf-1 antisense oligodeoxynucleotide ISIS 5132 (CGP 69846A) Clin Cancer Res 53977–3982. [PubMed] [Google Scholar]
- Rudin CM, Marshall JL, Huang CH, Kindler HL, Zhang C, Kumar D.et al. (2004Delivery of a liposomal c-raf-1 antisense oligonucleotide by weekly bolus dosing in patients with advanced solid tumors: a phase I study Clin Cancer Res 107244–7251. [DOI] [PubMed] [Google Scholar]
- Stewart DJ, Donehower RC, Eisenhauer EA, Wainman N, Shah AK, Bonfils C.et al. (2003A phase I pharmacokinetic and pharmacodynamic study of the DNA methyltransferase 1 inhibitor MG98 administered twice weekly Ann Oncol 14766–774. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Holmlund J, Fleming GF, Mani S, Stadler WM, Schumm P.et al. (2001Phase I Trial of ISIS 5132, an antisense oligonucleotide inhibitor of c-raf-1, administered by 24-hour weekly infusion to patients with advanced cancer Clin Cancer Res 71214–1220. [PubMed] [Google Scholar]
- Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M.et al. (2003Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays Proc Natl Acad Sci USA 10014229–14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL.et al. (2002Discordant protein and mRNA expression in lung adenocarcinomas Mol Cell Proteomics 1304–313. [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K., and, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Mitchison TJ, Bender A, Young DW., and, Tallarico JA. Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat Rev Drug Discov. 2009;8:567–578. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C., and, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T.et al. (1998IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs Cancer Res 585315–5320. [PubMed] [Google Scholar]
- Deveraux QL., and, Reed JC. IAP family proteins–suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T., and, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7:602–608. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL.et al. (2000Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype Curr Biol 101319–1328. [DOI] [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC.et al. (1998Control of apoptosis and mitotic spindle checkpoint by survivin Nature 396580–584. [DOI] [PubMed] [Google Scholar]
- Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S.et al. (1999Pleiotropic cell-division defects and apoptosis induced by interference with survivin function Nat Cell Biol 1461–466. [DOI] [PubMed] [Google Scholar]
- Lens SM, Wolthuis RM, Klompmaker R, Kauw J, Agami R, Brummelkamp T.et al. (2003Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension EMBO J 222934–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A, Carmena M, Sambade C, Earnshaw WC., and, Wheatley SP. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci. 2003;116 Pt 14:2987–2998. doi: 10.1242/jcs.00612. [DOI] [PubMed] [Google Scholar]
- Altieri DC. New wirings in the survivin networks. Oncogene. 2008;27:6276–6284. doi: 10.1038/onc.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- Fukuda S., and, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5:1087–1098. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- Tran J, Master Z, Yu JL, Rak J, Dumont DJ., and, Kerbel RS. A role for survivin in chemoresistance of endothelial cells mediated by VEGF. Proc Natl Acad Sci USA. 2002;99:4349–4354. doi: 10.1073/pnas.072586399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgillo F, Woo JK, Kim ES, Hong WK., and, Lee HY. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res. 2006;66:10100–10111. doi: 10.1158/0008-5472.CAN-06-1684. [DOI] [PubMed] [Google Scholar]
- Carrasco RA, Stamm NB, Marcusson E, Sandusky G, Iversen P., and, Patel BK. Antisense inhibition of survivin expression as a cancer therapeutic. Mol Cancer Ther. 2011;10:221–232. doi: 10.1158/1535-7163.MCT-10-0756. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M., and, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Chen J, Jin S, Tahir SK, Zhang H, Liu X, Sarthy AV.et al. (2003Survivin enhances Aurora-B kinase activity and localizes Aurora-B in human cells J Biol Chem 278486–490. [DOI] [PubMed] [Google Scholar]
- Ottesen GL, Christensen IJ, Larsen JK, Hansen B., and, Andersen JA. Tissue disaggregation for flow cytometric DNA analysis: comparison of fine-needle aspiration and an automated mechanical procedure. Cytometry. 1996;26:65–68. doi: 10.1002/(SICI)1097-0320(19960315)26:1<65::AID-CYTO10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Merkel DE, Dressler LG., and, McGuire WL. Flow cytometry, cellular DNA content, and prognosis in human malignancy. J Clin Oncol. 1987;5:1690–1703. doi: 10.1200/JCO.1987.5.10.1690. [DOI] [PubMed] [Google Scholar]
- McLean AN, Semple PA, Franklin DH, Petrie G, Millar EA., and, Douglas JG. The Scottish multi-centre prospective study of bronchoscopy for bronchial carcinoma and suggested audit standards. Respir Med. 1998;92:1110–1115. doi: 10.1016/s0954-6111(98)90403-6. [DOI] [PubMed] [Google Scholar]
- Aviel-Ronen S, Blackhall FH, Shepherd FA., and, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8:30–38. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- Coate LE, John T, Tsao MS., and, Shepherd FA. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M., and, Cappuzzo F. Predictive value of EGFR and HER2 overexpression in advanced non-small-cell lung cancer. Oncogene. 2009;28 suppl. 1:S32–S37. doi: 10.1038/onc.2009.199. [DOI] [PubMed] [Google Scholar]
- Vilmar A., and, Sørensen JB. Excision repair cross-complementation group 1 (ERCC1) in platinum-based treatment of non-small cell lung cancer with special emphasis on carboplatin: a review of current literature. Lung Cancer. 2009;64:131–139. doi: 10.1016/j.lungcan.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Talbot DC, Ranson M, Davies J, Lahn M, Callies S, André V.et al. (2010Tumor survivin is downregulated by the antisense oligonucleotide LY2181308: a proof-of-concept, first-in-human dose study Clin Cancer Res 166150–6158. [DOI] [PubMed] [Google Scholar]
- Vindeløv LL., and, Christensen IJ. A review of techniques and results obtained in one laboratory by an integrated system of methods designed for routine clinical flow cytometric DNA analysis. Cytometry. 1990;11:753–770. doi: 10.1002/cyto.990110702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shows how cell cycle parameters established.
Shows example controls for sample staining.