Abstract
Healthy oysters are inhabited by abundant microbial communities that vary with environmental conditions and coexist with immunocompetent cells in the circulatory system. In Crassostrea gigas oysters, the antimicrobial response, which is believed to control pathogens and commensals, relies on potent oxygen-dependent reactions and on antimicrobial peptides/proteins (AMPs) produced at low concentrations by epithelial cells and/or circulating hemocytes. In non-diseased oysters, hemocytes express basal levels of defensins (Cg-Defs) and proline-rich peptides (Cg-Prps). When the bacterial load dramatically increases in oyster tissues, both AMP families are driven to sites of infection by major hemocyte movements, together with bactericidal permeability/increasing proteins (Cg-BPIs) and given forms of big defensins (Cg-BigDef), whose expression in hemocytes is induced by infection. Co-localization of AMPs at sites of infection could be determinant in limiting invasion as synergies take place between peptide families, a phenomenon which is potentiated by the considerable diversity of AMP sequences. Besides, diversity occurs at the level of oyster AMP mechanisms of action, which range from membrane lysis for Cg-BPI to inhibition of metabolic pathways for Cg-Defs. The combination of such different mechanisms of action may account for the synergistic activities observed and compensate for the low peptide concentrations in C. gigas cells and tissues. To overcome the oyster antimicrobial response, oyster pathogens have developed subtle mechanisms of resistance and evasion. Thus, some Vibrio strains pathogenic for oysters are equipped with AMP-sensing systems that trigger resistance. More generally, the known oyster pathogenic vibrios have evolved strategies to evade intracellular killing through phagocytosis and the associated oxidative burst.
Keywords: innate immunity, invertebrate, mollusk, host pathogen interaction, mode of action, selective pressure, molecular diversity, antimicrobial peptide
Introduction
The Pacific oyster Crassostrea gigas is a marine invertebrate belonging to the family Ostreidae (Mollusca, Bivalvia) with a worldwide distribution from Japan to occidental countries in Europe and America (Guo et al., 2008). Oysters are filter-feeders cultured in bays, lagoons, and estuaries (Buestel et al., 2009; Dumbauld et al., 2009) where they are exposed to abundant and changing microbial communities. To date, the oyster-associated microflora, which in some regions of the world includes human pathogens, has been essentially studied for food safety purposes. Although the microbial communities associated to healthy oysters remain poorly studied, they were shown to be abundant and to vary with environmental conditions. This abundance in non-diseased animals could be relevant of a protective function and raises several questions on oyster biology, immunity, and homeostasis: Does an oyster-specific microbial community exist? How does the oyster microflora coexist with immunocompetent cells in the circulatory system? How can the oyster immune system discriminate between pathogens and commensals? In invertebrates, pathogen recognition, and disease control are mediated by an innate immune system, in which reactive oxygen species (ROS) and antimicrobial peptides play a key defense function (Lemaitre and Hoffmann, 2007). In this review, we give a brief summary on oyster environmental and resident microflora and present a state of the art on the antimicrobial defense of C. gigas oysters, its regulation, diversity, and its molecular effectors, with a special focus on the antimicrobial peptides and proteins.
Oysters and the Environmental Microflora
The health status of marine organisms is uniquely related to their immediate environments, which can contain very high concentrations of microorganisms. When conditions become favorable for multiplication, both the saprophytic and pathogenic microorganisms from their environment are capable of infecting marine organisms (Ellis, 2001). However, under normal conditions, marine invertebrates like oysters maintain a healthy status by defending themselves against these potential invaders using a repertoire of innate defenses (Bachère et al., 2004; Schmitt et al., 2012b).
The environmental microflora in lagoons and estuaries
Human activities are concentrated along the coasts worldwide. As a result, marine coastal systems are under increasing anthropic and environmental pressures. The high densities of human populations along the coasts as well as the human exploitation of the sea, lagoons, and estuaries for recreational activities, fishing, and shellfish farming are important factors that modify marine coastal systems. Moreover, due to the shallow water of these systems, they are highly impacted by seasonal variations and long term climate changes. Thus, both drastic and rapid changes are observed in sea water temperature, salinity, oxygenation, sunlight, pH, and content in organic matter. Those variations, which occur both temporarily and spatially, influence the viability/culturability, the abundance, and the ecology of bacteria (Pruzzo et al., 2005).
For instance, changes in the microflora of lagoons resulting from human activities have been observed following major rain events with a massive input in fecal coliforms and streptococci. It was shown that floods massively drained bacteria entrapped in the sediment of coastal rivers down to the lagoons (Chu et al., 2011). Additional sources of bacterial loads during rainfall events were also attributed to overflow from sewage systems and the failure of septic tanks (Al Bakri et al., 2008; Astrom et al., 2009).
Such natural events not only modify the abundance but also the composition of the coastal microflora by favoring the development of natural inhabitants of coastal sea waters better adapted to a given salinity or temperature. Thus, vibrios, which are autochthonous in riverine, estuarine, and coastal waters throughout both temperate and tropical regions of the world, are highly dependent on environmental parameters such as temperature, salinity, concentration of organic matter and the presence of chitinous organism (e.g., zooplankton), which are in turn controlled by larger-scale climate variability (Lipp et al., 2002). For instance, the prevalence of the human pathogen Vibrio cholerae is inversely correlated with salinity (Jiang, 2001), and a statistically significant relationship could be established between the time series for cholera and rainfall anomalies (Constantin de Magny et al., 2008).
Numerous surveys of coastal marine systems have shown that temperature is favoring the spread of vibrios including species pathogenic for marine invertebrates. Thus, as a consequence of ocean warming, Vibrio-associated diseases have increased causing mass mortalities in both wild and farmed species of marine invertebrates such as corals (Vezzulli et al., 2010), abalone (Travers et al., 2009), and oysters (Garnier et al., 2007). It is still difficult to determine how far the increase of the Vibrio load in marine coastal water contributes to animal disease. Indeed, environmental factors such as temperature also modulate the virulence status of bacteria (Vezzulli et al., 2010) and the physiological status of the animal species cultured in these systems, in particular oysters, whose reproductive status is dependent on an elevated sea water temperature.
Oyster commensal microflora and homeostasis
Animals including marine invertebrates carry numerous species of bacteria and it is becoming increasingly clear that most of these microorganisms constitute a host-specific community. However, we understand relatively little about the mechanisms involved in the reciprocal host–bacterial signaling because of the multiple interactions existing between and among the typically multispecies microbial consortia of animals (Ruby, 1999).
As filtering animals, oysters need high volumes of water for their nutrition and respiration. Consequently, they are in continuous contact with abundant microorganisms from their marine environment, which include both pathogens and commensals. Generally, oyster body surfaces harbor a dense and diverse natural microbiota, dominated by Gram-negative bacteria of the genera Pseudomonas and Vibrionaceae (Ortigosa et al., 1994). Like the human gut, oyster bacteria-enriched epithelia offer a naturally competitive environment that can prevent the establishment of potential pathogens (Salzman et al., 2007). Surprisingly, in healthy oysters, many organs inside the body, including the hemolymph, are also populated by a natural microbiota. Among others, it is composed of species from the genus Vibrio, Pseudomonas, Flavobacterium, Cytophaga, Bacillus, and Micrococcus (Lauckner, 1983; Paillard et al., 2004).
The existence of a host-specific microflora in oysters is still questioned. The microflora associated to oysters varies according to the environmental microflora. Thus, depuration, which is used after collection and prior to commercialization of oysters, results in a major decrease of coliforms and other transient residents of oyster tissues. However, bacterial communities in oyster hemolymph and surrounding sea water display different composition (de Lorgeril et al., unpublished data), indicating the existence of communities better adapted to oyster hemolymph (see below). Among these, the Vibrio species, which have a long co-evolution story with marine invertebrates resist rather efficiently to depuration procedures of edible bivalves, having a worldwide incidence on Vibrio-related seafood-borne diseases (Pruzzo et al., 2005). Pruzzo et al. (2005) therefore proposed that this resistance to depuration could indicate that vibrios belong to the persistent microflora of oysters.
Homeostasis is the maintenance of a stable state within the body in response to a changing external environment. Without homeostasis, no life form could adapt to their environment. Therefore, maintaining this equilibrium is essential for any organism to remain healthy and stay alive. As mentioned above, the oyster environment is subjected to drastic and rapid changes. Therefore, oysters must have an efficient system to maintain homeostasis. Oyster homeostasis is maintained through layered defenses. The first line of defense is determined by physical protective barriers that prevent damage of the underlying tissues and infections by pathogenic microorganisms. The main physical barriers are the shell and mucus (Glinski and Jarosz, 1997). However, when pathogens breach these protective barriers, they must confront the oyster immune response. For this, oysters have developed complex mechanisms of recognition and immune regulation to discriminate advantageous microorganisms from pathogenic ones, control infections, and maintain homeostasis.
It is generally assumed that the circulatory system of healthy animals is sterile, whereas the presence of bacteria is considered as infection. The abundant microflora of healthy oysters is therefore puzzling. The coexistence of immunocompetent cells and microorganisms in the circulatory system of oysters appears intriguing and impressive. It has undoubtedly been decisive for the evolution of the oyster immune system and its interaction with pathogens. One can assume that the oyster microflora plays a protective role by stimulating oyster immunity and/or competing with potential pathogens, and is therefore essential for the maintenance of homeostasis. The production of antimicrobial peptides (bacteriocins) is widespread in bacteria associated to marine organisms (Desriac et al., 2010). Together with the host antimicrobial defenses, which themselves are enhanced upon pathogen recognition (see immune responses section), antimicrobials from bacterial origin likely have a protective effect against pathogens. Thus, as in corals, a dynamic relationship between commensal microorganisms and oysters could select for a holobiont (host and associated microorganisms) better suited for resistance to specific pathogens. This probiotic hypothesis has been referred to as the hologenome theory of evolution (Rosenberg et al., 2007).
Infectious agents and diseases in oysters
While oyster-associated microorganisms can be significantly beneficial for the maintenance of oyster homeostasis, some can be highly virulent for given species of oysters resulting in massive mortalities or even the extinction of some oyster populations. Oyster pathogens include protozoans, viruses, and bacteria, which affect all stages of development, from larvae to juveniles and adult oysters (for review see Schmitt et al., 2012b).
Considering bacteria, the difficulties encountered in identifying the pathogenic ones are due to the normal accumulation of a very rich bacterial commensal microbiota composed of different species. Some of these bacteria, mainly opportunistic and pathogenic, eventually colonize and invade the host, depending on the environmental factors influencing host–bacteria interactions (Paillard et al., 2004). As an example, oyster mortalities are frequently associated to the presence of bacterial pathogens of the Vibrio genus, particularly strains of V. splendidus and V. aestuarianus (Le Roux et al., 2002; Garnier et al., 2008). However, it remains unclear whether these bacteria act as primary pathogens or as opportunists (Paillard et al., 2004). Besides, many vibrios potentially pathogenic to humans, inhabitants of coastal waters, are found in bivalves and can be transmitted to humans through their raw consumption. Those human pathogens do not cause disease in oyster and are likely normal residents of the oyster microflora. The ability of Vibrio species to persist within oyster tissues is responsible for their high concentration within this host that can act as an environmental reservoir of human pathogens (Pruzzo et al., 2005).
Oyster Immune Defenses
Oyster immune cells
As marine invertebrates, oysters have an immune system that resembles the innate immune system of vertebrates (for review see Schmitt et al., 2012b). In oysters, both hemocytes (circulating blood cells) and surface epithelia display immune functions, expressing both non-self recognition receptors and immune effectors involved in the control of the resident/pathogenic microflora. Still, to date, most of the studies have been dedicated to the description of hemocyte immune functions.
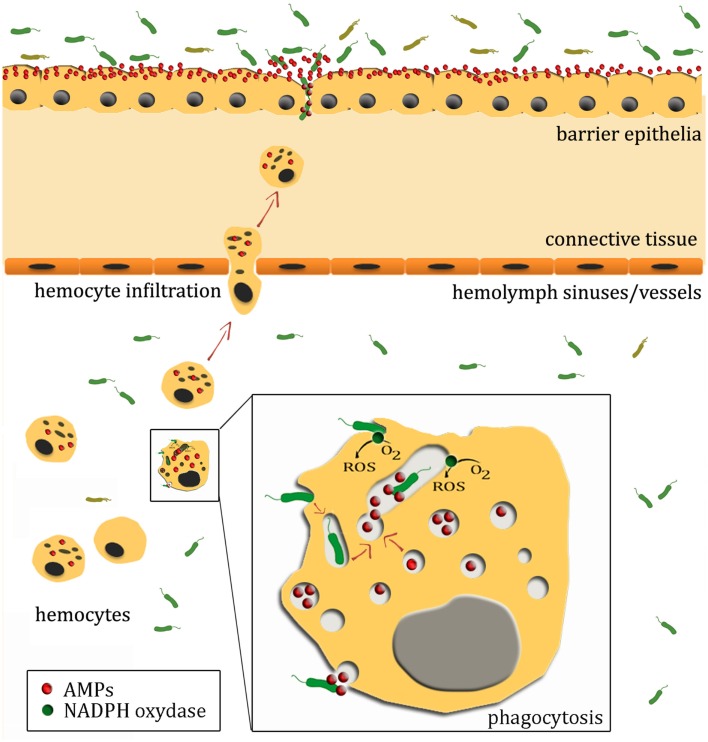
Oysters possess a semi-open circulatory system where the blood, called hemolymph, flows largely through a system of sinuses. The hemolymph bathes the organs directly providing oxygen and nutrients, and is composed of fluid plasma and hemocytes. The circulatory system contributes to the hemocyte ability to migrate toward connective tissues (Figure 1). They migrate by diapedesis from blood sinuses and vessels to the surface body, particularly to surface epithelia such as mantle and gills (Cheng, 1996).
Figure 1.
A not-to-scale schematic illustration of oyster antimicrobial defenses. Oyster epithelial surfaces are in contact with an abundant microflora that can offer a competitive environment that prevents the establishment of potential pathogens. In healthy oysters, surface epithelia constitutively express AMPs, which also contribute to prevent invasion by pathogens. Upon wounding or infection, damage- or pathogen-associated molecular patterns trigger hemocyte infiltration to the site of infection. Infiltrating hemocytes serve as vehicles for AMP families that display synergistic activities with AMPs from epithelial surfaces. Upon recognition by pattern recognition receptors/proteins, microorganisms present in tissues and hemolymph can also be phagocytosed and further eliminated by the production of reactive oxygen species (ROS), hydrolytic enzymes, and AMPs. It is still unknown whether AMP-containing hemocyte granules fuse with the phagosome, undergo exocytosis or both.
The classification of oyster hemocytes has been the subject of extensive studies since early 1970s. Based on different criteria (morphology, ultrastructure, physicochemical features, biological functions), they have been classified into main two cell types (granular and agranular cells), which are subdivided into several subgroups (Cheng, 1981; Hine, 1999; Bachère et al., 2004). Whereas granulocytes form a distinct group composed of both basophilic and eosinophilic granulocytes, agranular hemocytes are heterogeneous in appearance and ultrastructure, characterized by the absence or the presence of few cytoplasmic granules (Bachère et al., 1988). Three types of agranular hemocytes have been identified: blast-like cells, basophilic macrophage-like cells, and hyalinocytes. However, the distinction between types and/or lineages, the origin, maturation, and the life span of oyster hemocytes has never been demonstrated.
At present, there is considerable information about the role of hemocytes in oyster defense. Hemocytes are capable of non-self recognition, chemotaxis, and active phagocytosis (Cheng, 1981; Bachère et al., 2004). They are implicated in cytotoxic reactions by the production of hydrolytic enzymes (Cheng and Rodrick, 1975), ROS (Bachère et al., 1991a; Lambert et al., 2007) and antimicrobial peptides/proteins (Gueguen et al., 2006, 2009; Gonzalez et al., 2007a,b; Rosa et al., 2011). They also produce soluble factors to protect oyster against infections (Rinkevich and Muller, 1996; Pruzzo et al., 2005). Thus, as immunocompetent cells, infiltrating hemocytes are present in all cavities and tissues of oyster body where they can be able to phagocyte and to destroy infectious agents.
Although less studied for their immune functions, surface epithelial cells also express receptors dedicated to pathogen recognition (Itoh and Takahashi, 2008) as well as hydrolytic enzymes (Matsumoto et al., 2006; Itoh and Takahashi, 2007; Itoh et al., 2007, 2010a; Xue et al., 2010) and antimicrobial peptides/proteins involved in the control of infections (Gueguen et al., 2006; Gonzalez et al., 2007b).
Immune responses
Recent advance in our knowledge of the oyster immune response has been made through the development of genomic-based approaches (Gueguen et al., 2003; Tanguy et al., 2004; Fleury et al., 2009; Lang et al., 2009; Roberts et al., 2009; Taris et al., 2009; de Lorgeril et al., 2011). The main defense mechanisms described in oysters have been recently reviewed in (Schmitt et al., 2012b). They include: (i) recognition of microbe/pathogen-associated molecular patterns (M/PAMPs) or damage-associated molecular patterns (DAMPs) by both soluble and cellular pattern recognition receptors/proteins (PRRs and PRPs), (ii) hemocyte signaling and activation, (iii) hemocytes mediated reactions (phagocytosis, encapsulation, and tissue infiltration), and (iv) production of reactive species of oxygen and nitrogen and host defense molecules (Figure 1).
Recognition is mediated by a series of PRPs and PRRs. In oysters, the known PRPs/PRRs include peptidoglycan recognition proteins (PGRPs), which recognize specifically peptidoglycans (PG) from bacteria (Itoh and Takahashi, 2008), carbohydrate-binding proteins such as the β-1,3-glucan binding protein (βGBP), which binds to β-1,3-glucans from fungi cell walls (Tanguy et al., 2004; Itoh and Takahashi, 2009) and lectins (gigalins, C-type lectins, ficolins, and galectins) (Olafsen et al., 1992; Gueguen et al., 2003; Tasumi and Vasta, 2007; Yamaura et al., 2008). Over expression of C-type lectins and a rhamnospondin-like (l-rhamnose-binding lectin) in C. gigas hemocytes has been associated to a successful immune response to an experimental infection with virulent Vibrio strains as compared to an infection with an avirulent Vibrio (de Lorgeril et al., 2011). Additionally to those classical PRPs/PRRs, other proteins were also described as participating in oyster non-self recognition by their LPS-binding properties. These include the major plasma protein Cu/Zn extracellular superoxide dismutase (Cg-EcSOD) and the antimicrobial protein Cg-BPI for bactericidal/permeability-increasing (BPI) protein that will be further discussed below (Gonzalez et al., 2005, 2007b).
Intracellular signaling cascades are initiated upon M/PAMP or DAMP recognition. Highly conserved in the animal kingdom, one major signaling cascade identified in oyster is the NF-κB pathway (for review see Schmitt et al., 2012b). In Drosophila, it plays a major role in the regulation of the antimicrobial response (Lemaitre and Hoffmann, 2007). In oysters, although many elements of the NF-κB pathway have been identified, its direct role in immunity remains unexplored. Signaling events induce several defense responses, such as hemocyte activation, migration, and the expression of immune mediators that elicit defense reactions which coordinate the elimination of pathogens and infected cells. Some families of oyster antimicrobial peptides/proteins including big defensins and BPI protein are among the known effectors of oyster immunity that respond to a microbial challenge or an injury (see Regulation of AMP Expression in Healthy and Diseased Animals). Nonetheless, the signaling cascades controlling their gene expression are still unknown.
The control of pathogens relies on a combination of cellular and molecular defenses. Thus, following injury, hemocytes are recruited massively toward the sites of infection where they contribute to a local defense (Cochennec-Laureau et al., 2003; Bachère et al., 2004). Hemocyte migration leads to a local concentration of immune effectors likely to prevent host invasion (Schmitt et al., 2012a). May invasion occur, circulating hemocytes are capable of readily initiating phagocytosis as a major defense reaction in oyster (Figure 1). The crucial role of phagocytosis in C. gigas immune response has been highlighted by the overrepresentation of transcripts of phagocytosis-related genes in hemocytes of oysters surviving infections by virulent Vibrio strains as compared to oysters receiving the same doses of an avirulent Vibrio (de Lorgeril et al., 2011). Not surprisingly, phagocytosis is consequently either avoided by extracellular pathogens such as V. aestuarianus (Labreuche et al., 2006a,b) or impaired in elimination of intracellular pathogens such as V. splendidus (Duperthuy et al., 2011).
To be phagocytosed, the foreign agent is opsonized by plasma molecules and further engulfed within a phagosome. As soon as they come in contact with hemocyte membranes, phagocytosed microorganisms are exposed to the respiratory burst, a major microbicidal reaction in oysters (see below). The phagosome maturation then leads to acidification and fusion with cytoplasmic vesicles/granules (including lysosomes), which in oyster were shown to contain diverse families of antimicrobial peptides/proteins (Gueguen et al., 2009; Schmitt et al., 2012a). The release into the phagosome of this arsenal of microbicidal compounds leads to the rapid neutralization/degradation of the microorganisms non-adapted to intracellular life (Figure 1). Not surprisingly, lysosome-related genes (enzymes and transport proteins) have been associated to survival in oysters infected with virulent Vibrio strains (de Lorgeril et al., 2011). On the opposite, as far as intravacuolar pathogens are concerned, the success of infection appears to rely on a restricted respiratory burst and/or the development of mechanisms of resistance to the oyster antimicrobial response. This has been observed for two oyster pathogens, V. splendidus and Perkinsus marinus, which resist to intracellular elimination after phagocytosis (Schott et al., 2003; Duperthuy et al., 2010, 2011).
Effectors of the Antimicrobial Response
Production of reactive oxygen species
Immediately following phagocytosis, there is a significant increase in the intracellular oxygen consumption (respiratory burst), resulting in the production of a variety of intermediate reactive oxygen species (ROS) (Bachère et al., 1991b; Torreilles and Romestand, 2001; Lambert et al., 2003, 2007).
The production of ROS (superoxides and peroxides) is one major inducible mechanism of the antimicrobial response in oysters (Bachère et al., 1991b; Lambert et al., 2003). It results from the activation of the transmembrane NADPH oxidase, which generates the highly toxic and reactive superoxide anion . The production of ROS can cause serious injuries to the host tissues. Interestingly, parallel to the well known NADPH oxidase, a sequence homologous to another transmembrane enzyme termed DUOX for (DUal OXidase) involved in the production of ROS in Drosophila melanogaster gut epithelial cells was evidenced in the C. gigas genomic resource database (Fleury et al., 2009). In C. gigas, the expression of a DUOX-like gene was associated, together with other genes related to oxidative stress, to a successful response to virulent vibrios as compared to avirulent vibrios (de Lorgeril et al., 2011). Interestingly, DUOX expression was shown to be involved in the tolerance of resident microflora in the gut of Drosophila and to be over expressed when the gut bacterial load dramatically increases (for review see Royet, 2011). Therefore, as in Drosophila, DUOX, which is expressed in the digestive gland of oysters, could be involved in the epithelial defense against food-borne pathogens and in the maintenance of homeostasis.
To avoid self-damage, the host relies on effective antioxidant defense systems that involve antioxidant enzymes such as a superoxide dismutase (SOD), which converts into hydrogen peroxide H202, and peroxidases (catalase and glutathione peroxidase), which convert H202 into hypochlorous acid in the presence of chloride ions. As a sign of its contribution to an effective immune response in oysters, the C. gigas extracellular SOD was shown to be higher expressed in an oyster line selected for its resistance to in situ mortalities than in a susceptible line (Fleury and Huvet, 2012). It was also higher expressed in oysters surviving an infection with virulent Vibrio strains than in oysters challenged with avirulent vibrios (de Lorgeril et al., 2011). Other antioxidant enzymes are also produced in oysters such as peroxiredoxins, glutathione S-transferases, and others (Huvet et al., 2004; Tanguy et al., 2004; Vertuani et al., 2004; Gonzalez et al., 2005; Jo et al., 2008; Green et al., 2009; Park et al., 2009; Itoh et al., 2010b; Yu et al., 2011). As with Cg-EcSOD, transcripts of glutathione S-transferases (microsomal glutathione S-transferase, glutathione S-transferase theta and omega class glutathione S-transferase) were more abundant in hemocytes of oysters capable to survive an injection with virulent Vibrio strains than in oysters challenged with an avirulent Vibrio strain (de Lorgeril et al., 2011).
Hydrolytic enzymes
After phagocytosis, microorganisms are entrapped in phagosomes that fuse with lysosomes and/or intracellular granules. These granules are characterized by their acidic pH and the presence of a wide variety of hydrolytic enzymes and antimicrobial effectors (Figure 1). Among the hydrolytic enzymes, the lysozymes play an important role in microbial destruction due to their lytic properties on the peptidoglycan of the bacteria cell wall (Hancock and Scott, 2000). Oyster lysozymes form a diverse group and are expressed in many tissues, such as hemocytes (CGL-1 or Cg_lysoz1), digestive diverticula (CGL-2 or Cg_lysoz2), and mantle (CGL-3 or Cg_lysoz3) (Matsumoto et al., 2006; Itoh and Takahashi, 2007; Itoh et al., 2007, 2010a; Xue et al., 2010). In hemocytes, lysozyme transcript abundance was shown to be higher in oysters capable to survive an injection of virulent Vibrio strains than in oyster challenged with an avirulent Vibrio (de Lorgeril et al., 2011). Recombinant lysozymes from C. gigas (CGL-1 and CGL-3) were found to be active only against Gram-positive bacteria (Itoh et al., 2010a).
Antimicrobial peptides and proteins
Antimicrobial peptides and proteins (AMPs) are key host defense effectors found in virtually all kingdoms of life (Yeaman and Yount, 2003). They display multifunctional roles in immunity, including microbicidal activities and selective immunomodulatory effects (Yount et al., 2006; Guaní-Guerra et al., 2010). AMPs are classically described as small cationic (less than 10 kDa), amphipathic, gene-encoded peptides that differ considerably in amino acid sequence and structural conformation (Bulet et al., 2004; Brogden, 2005). Several categories of AMPs have been described based on common structural features or conserved sequence motifs. More recently, other groups of antimicrobial molecules referred to as “non-conventional” have been identified that do not fit into this classical definition. These are (i) anionic peptides, (ii) cationic proteins larger than 10 kDa, and (iii) multifunctional proteins that contain antimicrobial sub-domains that are cleaved under certain conditions and generate fragments that behave as antimicrobial peptides (Brogden, 2005; Yount et al., 2006). At present, several families of AMPs or polypeptides sharing common molecular features with the currently known AMP families have been characterized in oysters. These include defensin-like peptides (defensins and big defensins), proline-rich AMPs, and BPI proteins (Table 1).
Table 1.
Oyster antimicrobial peptides and proteins (AMPs).
| Oyster AMP | Variants | MW (kDa) | pI | Localization (expression) |
|---|---|---|---|---|
| CSαβ-containing defensins | Cg-Defm | 4.6 | 8.7 | Mantle (constitutive) |
| Cg-Defh1 | 4.7 | 8.5 | Hemocytes (constitutive) | |
| Cg-Defh2 | 4.6 | 8.5 | Hemocytes (constitutive) | |
| Big defensins | Cg-BigDef1 | 10.7 | 9.2 | Hemocytes (inducible) |
| Cg-BigDef2 | 9.8* | 8.6 | Hemocytes (inducible) | |
| Cg-BigDef3 | 9.7* | 8.8 | Hemocytes (constitutive) | |
| Proline-rich peptides | Cg-lgPrp | 1.8* | 12.1 | Hemocytes (repressed) |
| Cg-stPrp | 1.5* | 12.0 | Hemocytes (repressed) | |
| Bactericidal/permeability-increasing protein | Cg-BPI | 50.1 | 9.3 | Hemocytes (inducible) |
| Epithelia (constitutive) |
MW: observed or theorical (*) molecular weight; pI: theoretical isoelectric point.
Structure and classification of C. gigas AMPs
Defensins
Defensins were identified in most plants, fungi, and both invertebrate and vertebrate animals (Bulet et al., 2004). They are the best characterized AMPs to date. The term “defensin” was first introduced by Ganz et al. (1985) after the discovery of antimicrobial cysteine-rich peptides in human neutrophils. All vertebrate defensins are small (3–5 kDa) cationic molecules containing six cysteine residues engaged in three intramolecular disulfide bridges. Vertebrate defensin family falls into three subfamilies, namely the open-ring α- and β-defensins as well as the head-to-tail θ-defensins, which differ in terms of cysteine array and secondary structure (for review see Lehrer and Lu, 2012). Thus, the cysteine pairing of β-defensins differs from that of α- and θ-vertebrate defensins.
Unlike vertebrate defensins, which adopt a three-stranded antiparallel β-sheet structure, the defensins from plants, fungi, and invertebrates are composed of an α-helix linked to an antiparallel two-stranded β-sheet by disulfide bridges, making the so-called cysteine-stabilized α-helix/β-sheet motif (CSαβ; Zhu et al., 2005). They contain from six to eight cysteine residues. This motif is widespread in fungi and in invertebrate defensins, like in arthropods and mollusks (Yang et al., 2000; Bulet and Stöcklin, 2005; Mygind et al., 2005), but the presence of eight cysteine residues was only reported in species of bivalve mollusks (Hubert et al., 1996; Gueguen et al., 2006; Gonzalez et al., 2007a). CSαβ-containing defensins have no evident phylogenetic relationships with the vertebrate defensins and big defensins (Rosa et al., 2011).
CSαβ-containing defensins (Cg-Defs)
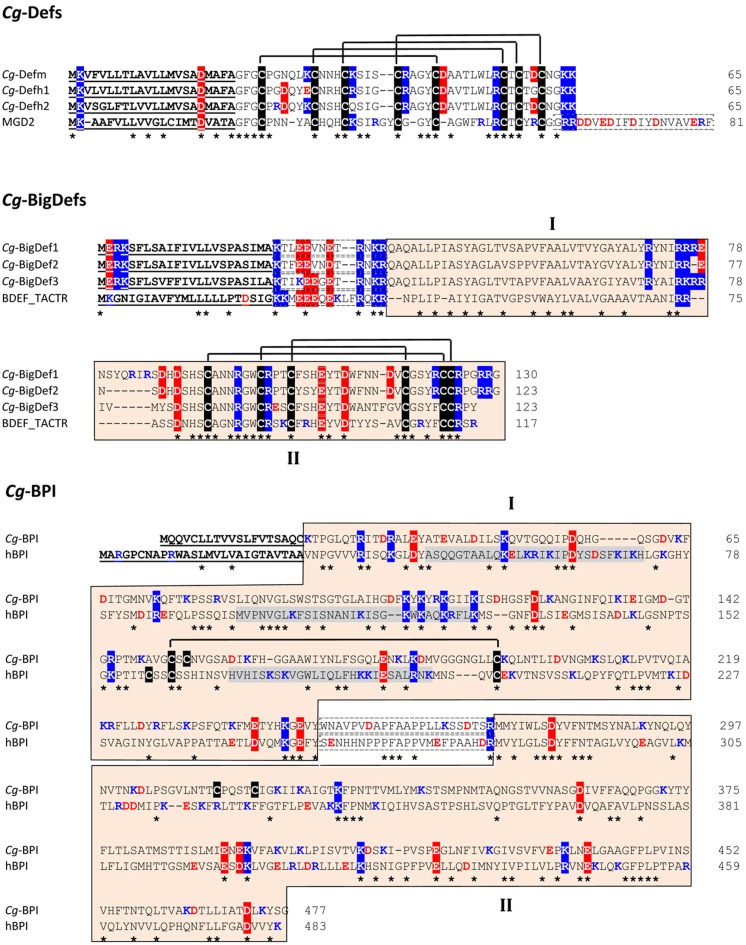
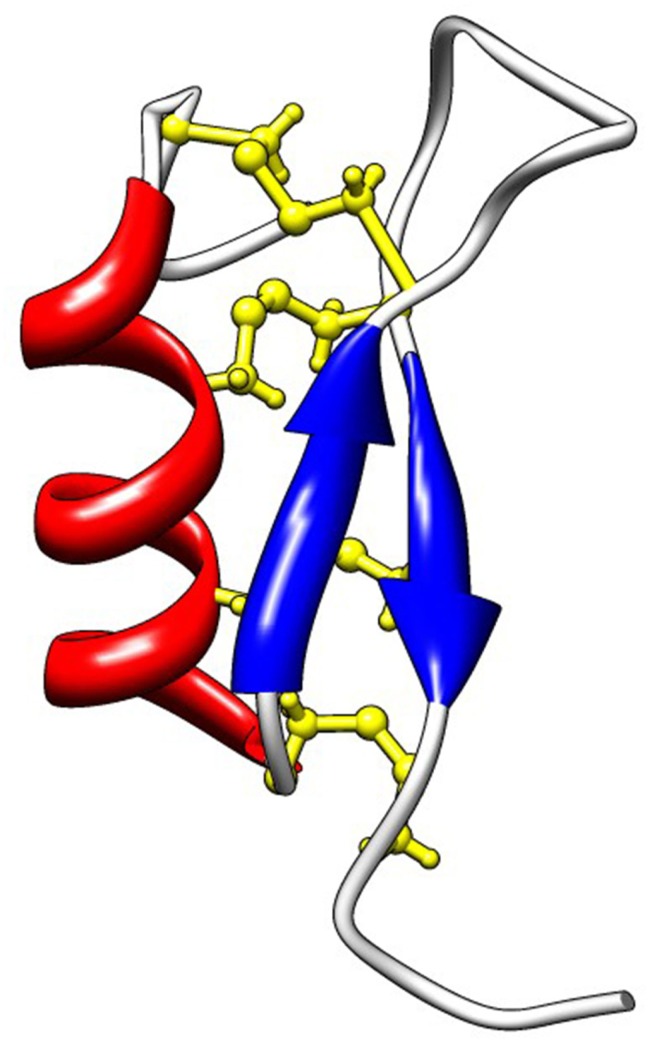
In C. gigas, members of the invertebrate CSαβ-containing defensin family were identified by screening EST libraries of oyster immune cells (hemocytes and surface epithelia). A broad diversity of CSαβ-containing defensins was found in C. gigas (see diversity section below). Still, three representative members, namely Cg-Defm, Cg-Defh1, and Cg-Defh2, were characterized in more details in terms of structure, antimicrobial activity, and mechanism of action. Cg-Defm was identified from the oyster mantle whereas Cg-Defh1 and Cg-Defh2 were identified from the hemocytes (Gueguen et al., 2006; Gonzalez et al., 2007a). All oyster defensin precursors consist in a hydrophobic signal peptide (prepeptide) immediately followed by the 4.6–4.7-kDa cationic mature peptide (pI 8.5–8.7), which contains eight cysteine residues (Figure 2). The tridimensional structure of the recombinant Cg-Defm was solved (PDB: 2B68; Figure 3), showing that the CSαβ motif that characterizes this AMP family is stabilized by four disulfide bridges (cysteine pattern: C1–5C2–6C3–7C4–8; Gueguen et al., 2006; Table 1). The fourth disulfide bridge, only found in oyster and mussel defensins, has been proposed to be implicated in the stabilization of the mature peptide to the high osmolarity environment found in the sea water (Yang et al., 2000).
Figure 2.
Amino acid alignment of oyster antimicrobial precursors with similar proteins/polypeptides from other species. Cg-Def precursors are composed of a 22-residue signal peptide followed by a 43-residue mature peptide. By comparison, the Mytilus galloprovincialis MGD2 (GenBank AAD45118.1) carries an additional anionic proregion at C-terminal position. The amino acid sequences displayed here for Cg-Defm, Cg-Defh1, and Cg-Defh2 correspond to GenBank AJ565499, ACQ73009, and ACQ76280, respectively. The cysteine array is based on the 3D structure of Cg-Defm (PDB: 2B68). Cg-BigDef precursors are composed of a predicted signal peptide (23 residues) followed by 13-residue anionic proregion and the putative mature BigDefs (87–94 residues). Mature BigDefs are multi-domain polypeptides composed of a hydrophobic N-terminal domain (I) and a C-terminal cysteine-rich domain (β-defensin-like domain, II) shown here as orange boxes. A putative cleavage motif (RXKR) for furin-like enzymes separates both domains. Sequences of big defensins from Crassostrea gigas (Cg-BigDef1: GenBank AEE92768, Cg-BigDef2: GenBank AEE92775, Cg-BigDef3: GenBank AEE92778) are aligned here with Tachypleus tridentatus big defensin (BDEF_TACTR, GenBank: P80957). The cysteine array is based on the 3D structure of the horseshoe crab big defensin (PDB: 2RNG). Cg-BPI precursor is composed of a 19-residue signal peptide followed by a 458 amino acid protein. Its amino acid sequence (GenBank AY165040) is aligned here with human BPI (hBPI) sequence (GenBank J04739). The N-terminal (domain I) and C-terminal (domain II) barrel type domains characterized for hBPI as well as the corresponding sequences in Cg-BPI are in orange boxes (domains I and II, respectively). The proline-rich central domain is boxed with a dashed line. The three functional regions of hBPI, which display LPS-binding activity, are highlighted in gray. The conserved disulfide bridge characterized in hBPI 3D structure (PDB: 1BP1) is displayed. Amino acids are numbered on the right. Signal peptides are underlined. Cysteines are highlighted in black and disulfide bridges are displayed as black lines. Conserved residues are shown by an asterisk. Lys/Arg residues are in blue. Asp/Glu residues are in red. Positively and negatively charged residues conserved in more than 50% of the sequences are highlighted in blue and red, respectively. In Cg-Def and Cg-BigDef alignments, anionic proregions are in dashed boxes. In Cg-BPI alignment, the central proline-rich region is in dashed boxes.
Figure 3.
Solution structure of Cg-Defm (PDB: 2B68). Cg-Defs display the CSαβ motif also found in defensins from plants, fungi, and invertebrates, which is composed of an α-helix (red) linked to an antiparallel two-stranded β-sheet (blue) by disulfide bridges (yellow). In Cg-Defs, eight cysteines are involved in four disulfide bridges as follows: C1–5C2–6C3–7C4–8.
Each Cg-Def is encoded by a separate gene with different genomic organization. The mantle defensin genes (Cg-defm) present two structures (i) two exons separated by a unique intron (Gueguen et al., 2006), a similar genomic organization to that of the mussel and scorpion defensin genes (Froy and Gurevitz, 2003), and (ii) three exons separated by two introns. Hemocyte defensin genes (Cg-defhs) only display the latter structure, in which the second intron separates the two last residues of the mature peptide apart from the rest of the sequence (Schmitt et al., 2010a). The number of Cg-def gene copies was shown to be highly variable (14–53 copies) among individual oysters (Schmitt et al., 2010a). At the transcriptional level, Cg-Defs appear to be constitutively expressed in each specific tissue.
Until now, native defensins could not be purified from C. gigas oysters in sufficient amounts to enable complete biochemical characterization and determine their spectrum of activity. Such data were obtained with recombinant oyster defensins (Gueguen et al., 2006; Schmitt et al., 2010b, 2012a). All oyster defensins were shown to be mainly active against Gram-positive bacteria against which they displayed very low minimal inhibitory concentrations (MIC; 0.01–6 μM) as determined by liquid growth inhibition assay. Conversely, they did not display significant antimicrobial activity against Gram-negative bacteria including oyster pathogens (>40 μM).
Big defensins (Cg-BigDefs)
Big defensins (BigDefs) are antimicrobial polypeptides that were initially purified from the hemolymph cells (amoebocytes) of the horseshoe crab Tachypleus tridentatus (Chelicerata; Saito et al., 1995). The solution structure of the horseshoe crab BigDef (PDB: 2RNG) revealed a N-terminal globular domain connected to a C-terminal domain containing β-sheet structures and folded by three disulfide bridges whose arrangement is identical to that of vertebrate β-defensins (β-defensin domain: C1–5C2–4C3–6; Kouno et al., 2008). The horseshoe crab BigDef is produced as a precursor molecule that is further processed into a 8.6-kDa mature polypeptide (Saito et al., 1995). Big defensin encoding sequences were also evidenced in amphioxus (Cephalochordata) and in several mollusk species including C. gigas oysters (for review see Rosa et al., 2011). Oyster big defensins (Cg-BigDefs) share 47–55% amino acid sequence identity with the big defensin from horseshoe crab and amphioxus species, and within mollusks, they are 25–28% identical to gastropod sequences and 39–67% identical to bivalve big defensins (Rosa et al., 2011).
Oyster big defensins form a diverse AMP family composed of three representative members, namely Cg-BigDef1, Cg-BigDef2, and Cg-BigDef3. Big defensin precursors are expressed as prepropeptides, which start with a predicted 23-residue signal peptide (prepeptide), followed by a propeptide region of 13 residues and a cationic 94- or 87-residue mature polypeptide of 10.7 kDa (Cg-BigDef1), 9.8 kDa (Cg-BigDef2), or 9.7 kDa (Cg-BigDef3; Table 1). The mature polypeptides have theorical pI ranging from 8.6 to 9.2. Based on mass spectrometry data, it has been proposed that after elimination of the, the N-terminal glutamine residue of the native Cg-BigDef1 is converted into a pyroglutamic acid (Rosa et al., 2011). Cg-BigDefs are chimeric molecules that display the structural feature of the horseshoe crab BigDef composed by a hydrophobic N-terminal region and a cationic C-terminal region containing the conserved six cysteine residues (Figure 2).
Each of the three Cg-BigDefs is encoded by a separate gene whose expression is restricted to hemocytes, both circulating and infiltrating oyster tissues. The genomic organization of Cg-bigdef1 and Cg-bigdef2 genes is similar, with two exons interrupted by a single intron. In contrast, in Cg-bigdef3, additional intron and exon are observed upstream the first exon common to the other Cg-bigdefs. In all Cg-bigdef genes, the β-defensin domain is exclusively encoded by the last exon (Rosa et al., 2011).
The antimicrobial activities of Cg-BigDefs are still unknown, but in another bivalve mollusk, Argopecten irradians, and in the horseshoe crab T. tridentatus, BigDefs were reported to be active against both Gram-positive and Gram-negative bacteria and fungi (Saito et al., 1995; Zhao et al., 2007). Besides, the native horseshoe crab BigDef was shown to display a significant LPS-binding activity (Saito et al., 1995).
Proline-rich AMPs (Cg-Prps)
Like defensins, proline-rich AMPs (Prps) have been identified in vertebrates including mammals and amphibians as well as invertebrates such as insects and crustaceans (Scocchi et al., 2011). They form a group of diverse peptides that share some structural features, like a high content in proline and arginine residues (typically from 25 to 50%). They display short Pro-Arg-Pro motifs, which have been proposed to be implicated in their antimicrobial activity. Prps are also characterized by their mode of action, which does not involve the lysis of bacterial membranes but rather the penetration into cells, where they then act intracellularly (Scocchi et al., 2011).
In oysters, a cDNA sequence showing homologies to proline-rich AMPs was identified as expressed in hemocytes by the exploration of C. gigas EST libraries (Gueguen et al., 2009). The cDNA sequence encoded a 37-amino acid peptide, which is composed of an N-terminal acidic region (putative propeptide) and a C-terminal cationic proline-rich region, containing two repetitions of a Pro-Arg-Pro motif (Figure 4). This highly cationic peptide (theorical pI 12.1), which displays the typical features of proline-rich AMPs, was named Cg-Prp. A high number of Cg-Prp forms were then identified as expressed by oyster hemocytes. They display two lengths, a shorter peptide being evidenced, which derives from the original Cg-Prp form by the deletion of a Arg-Pro dipeptide (Schmitt et al., 2010a). Thereby, we renamed the original form as long Cg-Prp (Cg-lgPrp; 1.8 kDa) and the new form as short Cg-Prp (Cg-stPrp; 1.5 kDa) (Schmitt et al., 2012a; Table 1; Figure 4).
Figure 4.
Amino acid alignment of two sequences representative of long and short Cg-Prps. Precursors of Cg-Prps are composed of a 24-residue signal peptide (underlined) followed by a 19-residue anionic proregion (dashed box), and a 16–18 residue putative mature peptide. In long forms (referred to as Cg-lgPrps), two repeats of the Pro-Arg-Pro motif (orange boxes) are found, while in short forms (Cg-stPrps), one motif is missing. While Pro-Arg-Pro motifs are common in proline-rich AMPs across species, no further structure similarity has been evidenced until now between oyster Prps and other proline-rich peptides. The amino acid sequences displayed here for Cg-lgPrp and Cg-stPrp correspond to GenBank FJ669361 and FJ669381, respectively. Amino acids are numbered on the right. Conserved residues are shown by an asterisk. Conserved Lys/Arg and Asp/Glu residues are highlighted in blue and red, respectively. Pro residues are shown in green.
Cg-Prps are a multigenic family with genes containing or not an intron. The presence/absence of an intron is observed in the genes encoding both the long and short peptide forms. The number of Cg-prp gene copies was shown to vary among individuals from 4 to 18 copies (Schmitt et al., 2010a).
Native Cg-Prps could not be purified from oyster hemocyte extracts until now. Therefore, synthetic Cg-Prps were used to explore their sprectrum of antimicrobial activity. Both long and short Cg-Prps were poorly active against Gram-positive bacteria and were not active against Gram-negative bacteria. However, Cg-lgPrp displayed strong synergy with Cg-Defs and Cg-BPI against both Gram-positive and Gram-negative bacteria (Gueguen et al., 2009; Schmitt et al., 2012a).
Bactericidal/permeability-increasing protein (Cg-BPI)
Bactericidal/permeability-increasing proteins and lipopolysaccharide-binding proteins (LBPs) are components of the immune system that have been mainly characterized in mammals. Both proteins show 45% sequence identity, sharing basic structural features and residues involved in LPS-binding (Krasity et al., 2011). Human LBP is an acute phase plasma protein constitutively secreted by liver that induces cellular responses (Thomas et al., 2002). In particular, LBP participates in the acute mobilization of circulating neutrophils to sites of tissue injury. In contrast, human BPI is a 55-kDa cationic protein specifically active against Gram-negative bacteria. Stored in neutrophils, human bactericidal/permeability-increasing (hBPI) contributes to the elimination of bacteria and increases the permeability of the bacterial membranes (Levy, 2004). Accumulated extracellularly, it also opsonizes bacteria, enhancing neutrophil phagocytosis (Iovine et al., 1997). The antibacterial BPI also displays LPS-neutralizing properties and suppresses LPS inflammatory activity (Weiss, 2003). By genomic approaches, LBP/BPI-related genes have been found in a number of non-mammalian vertebrate species and several invertebrates like nematodes and mollusks (Krasity et al., 2011).
A homolog of the hBPI protein was identified in C. gigas oysters by a screening of a hemocyte EST library (Gonzalez et al., 2007b). As deduced by molecular modeling, Cg-BPI displays the typical structural features of hBPI 3D structure (PDB: 2RNG) with an N- and C-terminal β-barrel type domains connected by a proline-rich central domain. In addition, the N-terminal domain of Cg-BPI contains the LPS-binding regions characterized in hBPI and the Lys and Arg residues required for LPS-binding. This domain also contains the cysteine bridge of hBPI at conserved position, three extra cysteines being identified (both in the N- and C-terminal domains) whose folding remains unknown. The recombinant Cg-BPI was shown to be a monomeric protein (50.1 kDa) and to display both LPS- and Lipid A-binding activities (Gonzalez et al., 2007b). It was highly active against the short-chain LPS E. coli SBS363, against which it displayed bactericidal activity, but was at least 30 times less active against the long-chain LPS E. coli ML35. Like hBPI, recombinant Cg-BPI permeabilized both the inner and outer membranes of E. coli ML35 (Gonzalez et al., 2007b). Like Cg-BPI, the recently identified Cg-BPI2 was also shown to be exclusively active against Gram-negative bacteria (Zhang et al., 2011).
Diversity of oyster antimicrobial peptides
High levels of sequence diversity were reported to be a characteristic of several genes belonging to the innate immunity system of invertebrates (Schulenburg et al., 2007). Similarly, oyster families of antimicrobials were recently shown to be highly diverse (Schmitt et al., 2010a). Phylogenetic analyses showed that Cg-Defs are represented by three distinct groups, of which Cg-Defh1, Cg-Defh2, and Cg-Defm forms are the most representatives. Cg-Prp is represented by two major forms, Cg-lgPrp and Cg-stPrp, but its phylogeny showed more diverse groups of sequences and suggested an ongoing or recent process of neo- or sub-functionalization (Schmitt et al., 2010a). In contrast to both AMPs, Cg-BPI was mostly represented by the only one sequence originally identified, but the recently discovery of Cg-BPI2 suggests a higher diversity for this antimicrobial (Zhang et al., 2011). Additionally, the in silico analysis of the oyster EST database (http://www.sigenae.org/aquafirst/) revealed that Cg-BigDefs are a diverse family with several isoforms for the three Cg-BigDef1, -2, and -3 forms (Rosa et al., 2011).
Evolutionary analyses showed that Cg-Defs and Cg-Prps have been subjected to several genetic mechanisms of diversification and that directional selection pressures have shaped their sequence variations. Noteworthy, directional selection driving evolution of invertebrate immune system has been addressed for several genes (Cohuet et al., 2008; Lazzaro, 2008; Roger et al., 2008) and it has been related with the general hypothesis of co-evolution or “arms race” between host and pathogens (Van Valen, 1974).
Specifically, the diversification of AMPs by the accumulation of multiple variants around an initial form has been previously associated to the gain of specific antimicrobial functions (Litman et al., 2005; Rosenfeld and Shai, 2006; Yang et al., 2006). Notably, it has been shown that the different forms of each oyster AMP family could display different antimicrobial potency (Schmitt et al., 2012a). The long Cg-Prp forms, which display an extra Pro-Arg-Pro motif in the active region are more potent that the short forms (Schmitt et al., 2012a). When referring to Cg-Defs, although the three forms show a similar spectrum of activity, they display different potency against Gram-positive bacteria. In particular, Cg-Defh2 is the most potent, showing MIC values 2- to 40-fold lower than Cg-Defh1 and Cg-Defm against most of the Gram-positive strains tested (Table 2; Schmitt et al., 2010a, 2012a). Interestingly, the differences in potency are much higher against the marine strain B. stationis (40-fold) than against all other bacteria tested (less than eightfold), which unlikely belong to the oyster environmental microflora and have unlikely co-evolved with the oyster immune system. These evidences tempt us to hypothesize that the diversity of oyster AMPs may have been shaped by the co-evolution between the oyster immune system and species of oyster pathogens, generating new AMP variants with enhanced potency. In support of this hypothesis, the more active defensin forms, Cg-Defm and Cg-Defh2, display a charged residue under positive selection (Lys and Arg, respectively) where Cg-Defh1 displays an uncharged Gly. Because those residues are highly exposed at the surface of oyster defensins, it was inferred they could improve defensin antibacterial activity by promoting a better binding to bacterial membranes. Therefore, as far as oyster defensins are concerned, diversifying selection is affecting charge distribution, which is known as one major determinant of antimicrobial activity in AMPs. Thus, although solid evidences are still missing in oysters, the selection of specific repertoires of AMPs with enhanced activity may have consequences on the oyster capacity to survive infections, as proposed in other species (Tennessen et al., 2009).
Table 2.
Antimicrobial activities of Cg-Defs, Cg-Prps, and Cg-BPI against selected microorganisms.
| MIC (μM) | Cg-Defh2 | Cg-Defm | Cg-Defh1 | Cg-lgPrp | Cg-stPrp | Cg-BPI |
|---|---|---|---|---|---|---|
| GRAM-POSITIVE BACTERIA | ||||||
| Micrococcus lysodeikticus CIP5345 | 0.01 | 0.01 | 0.03 | 80 | >80 | >20 |
| Bacilllus megaterium CIP 6620 | 0.03 | 0.03 | 0.06 | 20 | >20 | nt |
| Staphylococcus aureus CIP 103428 | 0.25 | 2 | 2 | >20 | >20 | nt |
| Staphylococcus aureus SG511 | 0.12 | 0.25 | 0.5 | >20 | >20 | nt |
| Microbacterium maritypicum CIP 105733T | 1 | 1 | 2 | >20 | >20 | nt |
| Brevibacterium stationis CIP 101282 | 0.1 | 0.2 | 4 | >20 | >20 | nt |
| Staphylococcus haemolyticus | 2 | 2 | 6 | >20 | >20 | nt |
| GRAM-NEGATIVE BACTERIA | ||||||
| Escherichia coli SBS 363 | 20 | 20 | 40 | >80 | >80 | 0.7 |
| Vibrio splendidus LGP32 CIP 107715 | >40 | >40 | >40 | >40 | >40 | 10 |
| Vibrio aestuarianus LPi 02/41 | >40 | >40 | >40 | nt | nt | nt |
| Vibrio anguillarum ATCC 19264 | >40 | >40 | >40 | nt | nt | nt |
| Vibrio nigripulchritudo CIP103195 | >40 | >40 | >40 | >40 | >40 | nt |
| FILAMENTOUS FUNGI | ||||||
| Fusarium oxysporum | nt | 9 | nt | >200 | nt | 0.7 |
| Botrytis cinerea | nt | >20 | nt | >200 | nt | nt |
| Penicillium crustosum | nt | >20 | nt | nt | nt | nt |
Mechanisms of antimicrobial action and synergism
The mechanisms of action of antimicrobial peptides have been extensively reviewed. Many families of AMPs, which carry a positive net charge and adopt an amphipathic structure, have been shown to be membrane-disruptive. Their physicochemical properties determine their initial electrostatic interaction with the negatively charged bacterial membranes and further insertion into the bacterial membrane lipid bilayer. Several models have been proposed for membrane-disruptive mechanisms of action, ranging from pore formation to detergent-like activities (for review see Brogden, 2005). Still, not all antimicrobials are membrane-disruptive and alternative mechanisms of action are increasingly described in which intracellular molecules involved in vital processes (e.g., enzymes, precursors …) are the target of AMPs. Among others, such non-lytic mechanisms of action have been reported in proline-rich AMPs (Scocchi et al., 2011) as well as in CSαβ defensins (Wilmes et al., 2011). The mechanism of action and consequently the target specificity of oyster antimicrobials are very diverse. At present, Cg-Defs and Cg-BPI are the best described oyster antimicrobials in terms of mechanisms of action.
Cg-Defs are potent inhibitors of the peptidoglycan biosynthesis in Gram-positive bacteria, against which they display strong antibacterial activity (in the nanomolar range). As shown on strains of Staphylococcus aureus, antibacterial activity is observed in absence of membrane disruption and results from the high affinity binding of Cg-Defs to the cell wall precursor lipid II, which cannot be further incorporated into peptidoglycan biosynthesis and accumulates in the bacterial cytoplasm (Schmitt et al., 2010b). Interestingly, at high concentrations (10 μM and above), Cg-Defs can also present membrane-disruptive properties as observed against the Gram-negative V. splendidus LGP32 (Duperthuy et al., 2010).
Cg-BPI, as a member of the BPI family, was shown to be both bactericidal and membrane-permeabilizing against Gram-negative bacteria. Its mechanism of action was mainly studied on E. coli strains. Bactericidal activity (loss of culturability) against E. coli SBS363 was recorded at 1 μM. Besides, a dose-dependent inner membrane permeabilization was evidenced by the extracellular cleavage in a lactose-permease deficient strain (E. coli ML35) of a β-galactosidase (a cytoplasmic enzyme) substrate (Gonzalez et al., 2007b).
Cg-Prps and Cg-BigDefs remain poorly studied in terms of mechanism of action. It can be speculated that, like many proline-rich AMPs (Kragol et al., 2001; Otvos, 2002; Scocchi et al., 2011), Cg-Prps interact with intracellular targets in bacteria. Concerning big defensins, a mechanism of action was proposed for the horseshoe crab BigDef, which is based on the penetration of the hydrophobic N-terminal domain into the microbial cell membrane (Kouno et al., 2009).
As a result of their different mechanism of action, oyster antimicrobials show a diverse range of antimicrobial activities: Cg-Defs are mainly active against Gram-positive bacteria, Cg-Prps are barely active alone, and Cg-BPI is only active against Gram-negative bacteria. Noteworthy, the combination of oyster antimicrobials produces strong synergistic activities that enlarge their spectra of activity (Schmitt et al., 2012a). Synergistic activities between oyster antimicrobials were evidenced between different families such as Cg-Defs and Cg-Prps (Gueguen et al., 2009) but also Cg-Defhs and Cg-BPI (Schmitt et al., 2012a). Interestingly, synergies were also shown to occur between forms of the same AMP family, different potencies of synergism being observed (as measured through fractional inhibitory concentration [FIC] indexes) according to the combination of forms (Schmitt et al., 2012a; Table 3). It is therefore tempting to speculate that the production of a large number of structurally similar AMPs is evolutionary relevant in that it helps oyster antimicrobials cover a broader spectrum of antimicrobial activities through combinations of peptide isoforms which produce synergistic effects.
Table 3.
Synergistic activities between Cg-Defs, Cg-Prps, and Cg-BPI against selected bacteria expressed as a fractional inhibitory concentration (FIC index*).
| FIC index | Gram-positive bacteria | Gram-negative bacteria |
|
|---|---|---|---|
| M. lysodeikticus CIP5345 | E. coli SBS 363 | V. splendidus CIP 107715 | |
| Cg-Defh1 + Cg-Defh2 | 0.50 | 0.75 | 0.75 |
| Cg-Defh1 + Cg-Defm | 0.75 | 0.75 | 0.75 |
| Cg-Defh2 + Cg-Defm | 0.37 | 0.50 | 0.50 |
| Cg-lgPrp + Cg-stPrp | 2 | 2 | 2 |
| Cg-Defh1 + Cg-lgPrp | 0.35 | 0.50 | 2 |
| Cg-Defh2 + Cg-lgPrp | 0.30 | 0.28 | 2 |
| Cg-Defm + Cg-lgPrp | 0.45 | 0.75 | 2 |
| Cg-BPI + Cg-lgPrp | nd | 0.50 | 2 |
| Cg-BPI + Cg-stPrp | nd | 0.75 | 2 |
| Cg-BPI + Cg-Defh1 | 0.45 | 0.26 | 2 |
| Cg-BPI + Cg-Defh2 | 0.23 | 0.27 | 2 |
| Cg-BPI + Cg-Defm | 0.23 | 0.31 | 2 |
Adapted from Schmitt et al. (2012a).
*FIC index = [A]/MICA + [B]/MICB, where MICA and MICB are the MICs of peptides A and B alone and [A] and [B] are the MICs of peptides A and B in combination. The MICs for the peptides alone are as given in Table 2. When MIC values are superior to the highest concentration we tested, we chose this value as the MIC in the calculation of the FIC index. FIC index were interpreted as follows: ≤0.5, strong synergy; 0.5−1 synergy; ≥1; additive effect; =2, no effect; ≥2, antagonism. nd, not determined.
The mechanisms underlying the synergistic effects among members of a given family of antimicrobials, as evidenced for Cg-Defs and Cg-Prps (Schmitt et al., 2012a), are still not understood. However, with the increasing knowledge on oyster antimicrobial mechanisms of action, a model can be proposed for the synergism between different families of antimicrobials. Thus, the combination of membrane-active antimicrobials such as Cg-BPI with non-lytic peptides such as Cg-Defs (and potentially Cg-Prps), which interact with an intracellular target, can be highly effective in terms of antimicrobial activity. Thus, Cg-BPI, which was shown to permeabilize bacterial membranes (Gonzalez et al., 2007b), likely facilitates the access of Cg-Defs to their intracellular target (lipid II), in particular in Gram-negative bacteria where lipid II is located at both the inner and outer leaflets of the cytoplasmic membranes, protected from Cg-Def interaction by the inner and outer membranes. Although the existence of an intracellular target for Cg-Prps is still speculative, synergy with Cg-BPI could also rely on such a combination with a membrane-disruptive antimicrobial. As a consequence of synergism, the peptide concentrations required to kill bacteria are much lower than when peptides are acting alone.
Regulation of AMP expression in healthy and diseased animals
Oyster antimicrobials exhibit a wide variety of expression profiles in hemocyte populations and epithelia, which vary according to the families and the oyster health status. Cg-Defhs, Cg-Prps, and Cg-BigDef3 are expressed in hemocytes of healthy oysters. Besides, Cg-Defm and Cg-BPI are constitutively expressed in surface epithelia. While Cg-Defm is specifically expressed in the mantle, Cg-BPI is expressed in a broad series of tissues including mantle, gills, digestive tract, digestive gland diverticula, and gonad follicles. Comparatively, transcripts of Cg-BPI and Cg-BigDef1 and 2 are barely detected in hemocytes from healthy oysters (Gueguen et al., 2006, 2009; Gonzalez et al., 2007a,b; Rosa et al., 2011; Schmitt et al., 2012a).
After a microbial infection, each one of these antimicrobials follows a different pattern of gene expression. The expression of Cg-BPI as well as Cg-BigDef1 and 2 is strongly induced by a bacterial challenge in hemocytes, both circulating and infiltrating massively the oyster connective tissues of the gills, gonads, or digestive gland (Gonzalez et al., 2007b; Rosa et al., 2011). On the opposite, Cg-Prp gene expression appears down-regulated in hemocytes upon a Vibrio infection (Schmitt et al., 2012a). Based on immunocytochemistry data, Cg-BPI, Cg-Defhs, and Cg-Prps would be stored in cytoplasmic granules of oyster hemocytes (Gueguen et al., 2009; Schmitt et al., 2012a). It is still unknown whether Cg-BigDefs are also stored in granules, as in horseshoe crab amoebocytes, or secreted into oyster hemolymph. In surface epithelia, genes encoding Cg-BPI as well as Cg-Defm remain constitutively expressed (Gueguen et al., 2006; Gonzalez et al., 2007b). Whether the antimicrobials are stored or secreted remains to be established.
Interestingly, with the massive infiltration of given hemocyte populations at sites of infection, some AMPs constitutively expressed in oyster hemocytes show a decrease in transcript abundance in circulating hemocyte populations after a Vibrio challenge. Thus, it has been shown that upon injection of V. splendidus in the adductor muscle of oysters, Cg-Defh-expressing hemocytes migrate toward the damaged tissues as evidenced by the concomitant and significant increase of Cg-Defh transcript abundance at the Vibrio injection site (Schmitt et al., 2012a). As a consequence, hemocytes seem to serve as a vehicle for given peptides like Cg-Defhs, driving them to tissues exposed to microorganisms where other peptides like Cg-Defm or Cg-BPI are constitutively expressed.
How AMPs may participate in oyster defense
The constitutive expression of Cg-Defm and Cg-BPI in barrier epithelia of healthy oysters probably contributes to the control of their endobiont microbiota, which at a basal level could be beneficial for outcompeting potential pathogens from the environment. Indeed, in mammals, the constitutive expression of AMPs in tissues exposed to microbes regulates the numbers of colonizing microbes by creating an antimicrobial environment that prevents heavy colonization and helps to shape the composition of the colonizing microflora (Salzman et al., 2007; Duerkop et al., 2009; Bevins and Salzman, 2011).
Upon infection, additional families of AMPs expressed by hemocytes, namely Cg-BPI, Cg-BigDefs, and Cg-Defhs, are rapidly and massively colocalized in oyster tissues. While the constitutively expressed Cg-Defhs are transported through the migratory behavior of hemocytes, Cg-BPI, Cg-BigDef1 and 2 are massively transcribed in hemocytes infiltrating tissues (Rosa et al., 2011; Schmitt et al., 2012a). This contributes to colocalize the antimicrobials which in turn can express synergism. As a consequence, by migration of antimicrobial-expressing hemocyte populations and continuous tissue expression, oyster antimicrobials are likely merged in tissues upon exposure to pathogens. For instance, the strong synergy observed between hemocyte defensins (Cg-Defh1 and 2) and mantle defensin (Cg-Defm) against the oyster pathogen Vibrio splendidus LGP32 (Table 3) can only take place upon co-localization, which is dependent on hemocyte infiltration of the infected tissue.
Co-localization of oyster antimicrobials not only occurs in barrier epithelia but also in some populations of circulating hemocyte as demonstrated for Cg-Defhs and Cg-Prps on the one hand, and Cg-Defhs and Cg-BPI on the other hand (Gueguen et al., 2009; Schmitt et al., 2012a). The subsequent synergism which potentially takes place could be of prime importance in the intracellular elimination of phagocytosed microorganisms. However, data are still missing on the behavior of hemocyte granules after infection. Do they fuse with the phagosome as described for human neutrophil defensins or are they exocytosed in the oyster plasma as shown for human cathelicidin LL-37? In other species of bivalve mollusks such as the mussel, in crustaceans, and in chelicerates, the contact with microorganisms or their cell wall components induces the degranulation of hemocytes and consequently the release of stored host defense molecules (Mitta et al., 2000b; Muñoz et al., 2004; Kawabata, 2011). Comparatively, in insects, the gene expression and secretion of large amounts of AMPs is induced in epithelial cells and in the fat body by the presence of microorganisms (Hoffmann et al., 1999). Because C. gigas antimicrobial response seems to be highly based on phagocytic processes (Cheng, 1996; Hine, 1999; Takahashi and Mori, 2000), oyster AMPs could be involved in the elimination of invading pathogens during phagocytosis, as proposed for mussel mytilins (Mitta et al., 2000a) and human neutrophil peptides (HNP; Ganz et al., 1985). Intracellularly, AMPs could act synergistically through the co-localization of diverse families and forms; they could also display antimicrobial activities in a coordinate manner with additional microbicidal reactions that involve lysosomial enzymes and ROS.
Unlike in many other marine invertebrates including species of bivalve mollusks, C. gigas AMPs are produced at low concentrations and have not been purified to homogeneity until now. Unsuccessful attempts to purify native oyster AMPs by the conventional biochemical methods normally used for the purification of cationic gene-encoded AMPs were documented (Bachère et al., 2004). Besides, after a bacterial challenge, increase of AMP concentration in plasma has never been reported. The lack of release of large amounts of AMPs within hemolymph upon infections greatly suggests the absence of a systemic humoral antimicrobial response in C. gigas oysters.
In summary, recent studies suggest that the production of a large number of AMPs within the same animal is an evolutionary strategy to increase the spectrum of antimicrobial activities by using combinations of peptide isoforms (Mangoni and Shai, 2009). Through the migratory properties of oyster hemocytes expressing diverse families of antimicrobials and the basal expression of antimicrobials in tissues, synergistic activities could take place and contribute to control microbial invasion. We propose that such a synergy, which was evidenced both within and between families of antimicrobials (Schmitt et al., 2012a), is essential during the oyster immune response, and that the high degree of sequence diversity in oyster antimicrobials may compensate through synergic activities their low concentration in oyster tissues.
How oyster pathogens evade the antimicrobial response
The continuous interplay between hosts and pathogens has evolved over millions of years. Thus, the success of a microbial infection or the achievement of host strategies to overcome diseases is a consequence of an active co-evolutionary process (Peschel and Sahl, 2006). In this way, pathogens have placed diverse mechanisms of resistance against antimicrobial responses. Particularly, several strategies are used by bacteria to evade host antimicrobials. They can resist AMPs by their inactivation by peptidases and proteases, or modify the net anionic charge of their cell envelope to reduce the affinity for AMPs; or even, AMPs can be actively extruded from bacterial cells (Yeaman and Yount, 2003).
Both commensals and pathogens are found in the hemolymph of oysters from which they can be isolated in a culturable state. Among them, many bacterial strains belong to the Vibrio genus (Gay et al., 2004). Interestingly, as commented above, the C. gigas plasma (cell-free hemolymph) is poor in antimicrobials. This could make oyster plasma adapted to bacterial life. Whether or not AMP-resistance is required for life in plasma is uncertain. However, in a recent study, one mechanism of AMP-resistance in vibrios, which is mediated by the major outer membrane protein OmpU (Mathur and Waldor, 2004; Mathur et al., 2007), was shown to endow the oyster pathogen V. splendidus LGP32 with (i) an increased resistance to oyster AMPs and (ii) a growth advantage in oyster plasma (Duperthuy et al., 2010).
While AMP-resistance mechanisms are still poorly documented in oyster pathogens, evasion of the cellular hemocyte reactions appears essentials. Indeed, both bacterial and protozoan parasites have developed strategies to avoid intracellular elimination by oyster phagocytes. Mechanisms of immune evasion have been reported in pathogenic strains of P. marinus, V. splendidus, and V. aestuarianus (Schott et al., 2003; Labreuche et al., 2006b, Duperthuy et al., 2011). Both P. marinus and V. splendidus invade oyster hemocytes as part of their pathogenic process. Therefore, after hemocyte invasion through subversion of hemocyte membrane receptors (galectins and β-integrins, respectively; Tasumi and Vasta, 2007; Duperthuy et al., 2011), they have developed strategies to resist to and/or suppress hemocyte defense reactions. Thus, P. marinus fails to elicit ROS production when entering hemocytes and displays intrinsic resistance to ROS (Schott et al., 2003). Similarly, V. splendidus LGP32 is able to manipulate phagosome maturation and to survive in oyster hemocytes by avoiding acidic vacuole formation and ROS production (Duperthuy et al., 2011). Although it is still unknown whether V. splendidus LGP32 invades hemocyte types that produce AMPs, by impairing phagosome maturation, it could prevent contact with host AMPs stored in hemocyte cytoplasmic granules.
Unlike V. splendidus LGP32, which is also associated to the summer mortality syndrome of oysters, V. aestuarianus strain 01/32 is described as an extracellular pathogen (Labreuche et al., 2006a) and evades hemocyte reactions in a totally different manner. The evasion of V. aestuarianus 01/32 from oyster immune response relies on the avoidance of phagocytosis by the secretion of inhibitory extracellular products (ECPs; Labreuche et al., 2006a,b).
Thus, bacterial oyster pathogens have developed different strategies that can eventually reduce the probability to encounter oyster AMPs within the hemocytes. In addition, we have evidence that Vibrio spp. display high in vitro resistance to oyster AMPs (Duperthuy et al., 2010; Schmitt et al., 2010b). From our current knowledge, such a resistance could be conferred by (i) the OmpU-mediated sensing of membrane perturbations and further signaling of the envelope stress response (Mathur and Waldor, 2004; Mathur et al., 2007), (ii) the VexAB-mediated efflux of AMPs (Bina et al., 2008), or (iii) other structural properties such as capsule formation that could also protect vibrios from the oyster antimicrobial response.
Conclusion
Knowledge has been acquired on the antimicrobial response of the oyster over the past years. Still, much remains to be explored regarding its role in the oyster defenses and host–pathogen interactions. From our current knowledge, phagocytosis and the oxidative burst would be major reactions of the defense of C. gigas oysters against infections. Remarkably in this species, conventional AMPs such as those reviewed in the present article are found at very low concentrations in tissues. This is not a common feature in species of bivalve mollusks nor in invertebrates more generally. It is therefore reasonable to ask whether AMPs play a significant role in the oyster defense. In support of such a role are the major selective pressures that have been shown to shape AMP sequences at positions required for an efficient mechanism of action (electrostatic interactions with bacterial membranes, interaction with intracellular molecular targets …). Still, there is no positive correlation evidenced between AMP expression and resistance to disease. We have shown here how the diversity of sequences and/or expression patterns of AMPs together with their co-localization in surface epithelia, which is mediated by hemocyte infiltration, can give rise to synergistic activities that likely contribute to the control of infections. We therefore believe that in this species, which lives in permanent contact with and is inhabited by a great variety of microorganisms, the diversity of AMPs may compensate for their scarcity in the regulation of the resident/pathogenic microflora, and as a consequence in the maintenance of oyster homeostasis. Future studies on the microbial communities that constitute the oyster microflora and their interaction with the oyster immune system will be required to better understand how oysters discriminate between commensal and pathogenic bacteria and to identify which host factors are involved in the regulation of this microflora.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Al Bakri D., Rahman S., Bowling L. (2008). Sources and management of urban stormwater pollution in rural catchments, Australia. J. Hydrol. 356, 299–311 10.1016/j.jhydrol.2008.04.017 [DOI] [Google Scholar]
- Astrom J., Pettersson T. J. R., Stenstrom T. A., Bergstedt O. (2009). Variability analysis of pathogen and indicator loads from urban sewer systems along a river. Water Sci. Technol. 59, 203–212 10.2166/wst.2009.860 [DOI] [PubMed] [Google Scholar]
- Bachère E., Boulo V., Godin P., Goggin L., Hervio D., Le Gall G., Morvan A., Mialhe E. (1991a). In vitro chemiluminescence studies of marine bivalve defence mechanisms and responses against specific pathogens. Dev. Comp. Immunol. 15, S102. 10.1016/0145-305X(91)90330-2 [DOI] [Google Scholar]
- Bachère E., Hervio D., Mialhe E. (1991b). Luminol-dependent chemiluminescence by hemocytes of two marine bivalves, Ostrea edulis and Crassostrea gigas. Dis. Aquat. Org. 11, 173–180 10.3354/dao011173 [DOI] [Google Scholar]
- Bachère E., Chagot D., Grizel H. (1988). Separation of Crassostrea gigas hemocytes by density gradient centrifugation and counterflow centrifugal elutriation. Dev. Comp. Immunol. 12, 549–559 10.1016/0145-305X(88)90071-7 [DOI] [PubMed] [Google Scholar]
- Bachère E., Gueguen Y., Gonzalez M., de Lorgeril J., Garnier J., Romestand B. (2004). Insights into the antimicrobial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunol. Rev. 198, 149–168 10.1111/j.0105-2896.2004.00115.x [DOI] [PubMed] [Google Scholar]
- Bevins C. L., Salzman N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 10.1038/nrmicro2546 [DOI] [PubMed] [Google Scholar]
- Bina X. R., Provenzano D., Nguyen N., Bina J. E. (2008). Vibrio cholerae RND-family efflux systems are required for antimicrobial resistance, optimal virulence factor production and colonization of the infant mouse small intestine. Infect. Immun. 76, 3595–3605 10.1128/IAI.01620-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- Buestel D., Ropert M., Prou J., Goulletquer P. (2009). History, status and future of oyster culture in France. J. Shellfish Res. 28, 813–820 10.2983/035.028.0410 [DOI] [Google Scholar]
- Bulet P., Stöcklin R. (2005). Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 12, 3–11 10.2174/0929866053406011 [DOI] [PubMed] [Google Scholar]
- Bulet P., Stöcklin R., Menin L. (2004). Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198, 169–184 10.1111/j.0105-2896.2004.0124.x [DOI] [PubMed] [Google Scholar]
- Cheng T. C. (1981). “Invertebrates blood cells,” in Bivalves, eds Ratcliffe A. N., Rowley F. A. (London: Academic Press; ), 233–301 [Google Scholar]
- Cheng T. C. (1996). “Hemocytes: forms and functions,” in The Eastern Oyster Crassostrea virginica, eds Kennedy V. S., Newell R. I. E., Eble A. E. (College Park, MD: Sea Grant College, University Of Maryland; ), 299–333 [Google Scholar]
- Cheng T. C., Rodrick G. E. (1975). Lysozyme and other enzymes in the hemolymph of Crassostrea virginica and Mercenaria mercenaria. Comp. Biochem. Physiol. 52, 443. [DOI] [PubMed] [Google Scholar]
- Chu Y., Salles C., Tournoud M. G., Got P., Troussellier M., Rodier C., Caro A. (2011). Faecal bacterial loads during flood events in Northwestern Mediterranean coastal rivers. J. Hydrol. 405, 501–511 10.1016/j.jhydrol.2011.05.047 [DOI] [Google Scholar]
- Cochennec-Laureau N., Auffret M., Renault T., Langlade A. (2003). Changes in circulating and tissue-infiltrating hemocyte parameters of European flat oysters, Ostrea edulis, naturally infected with Bonamia ostreae. J. Invertebr. Pathol. 83, 23–30 10.1016/S0022-2011(03)00015-6 [DOI] [PubMed] [Google Scholar]
- Cohuet A., Krishnakumar S., Simard F., Morlais I., Koutsos A., Fontenille D., Mindrinos M., Kafatos F. C. (2008). SNP discovery and molecular evolution in Anopheles gambiae, with special emphasis on innate immune system. BMC Genomics 9, 227. 10.1186/1471-2164-9-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin de Magny G., Murtugudde R., Sapiano M. R., Nizam A., Brown C. W., Busalacchi A. J., Yunus M., Nair G. B., Gil A. I., Lanata C. F., Calkins J., Manna B., Rajendran K., Bhattacharya M. K., Huq A., Sack R. B., Colwell R. R. (2008). Environmental signatures associated with cholera epidemics. Proc. Natl. Acad. Sci. U.S.A. 105, 17676–17681 10.1073/pnas.0809654105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorgeril J., Zenagui R., Rosa R. D., Piquemal D., Bachère E. (2011). Whole transcriptome profiling of successful immune response to Vibrio infections in the oyster Crassostrea gigas by digital gene expression analysis. PLoS ONE 5, e23142. 10.1371/journal.pone.0023142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desriac F., Defer D., Bourgougnon N., Brillet B., Le Chevalier P., Fleury Y. (2010). Bacteriocin as weapons in the marine animal-associated bacteria warfare: inventory and potential applications as an aquaculture probiotic. Mar. Drugs 8, 1153–1177 10.3390/md8041153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop B. A., Vaishnava S., Hooper L. V. (2009). Immune responses to the microbiota at the intestinal mucosal surface. Immunity 31, 368–376 10.1016/j.immuni.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Dumbauld B. R., Ruesink J. L., Rumrill S. S. (2009). The ecological role of bivalve shellfish aquaculture in the estuarine environment: a review with application to oyster and clam culture in West Coast (USA) estuaries. Aquaculture 290, 196–223 10.1016/j.aquaculture.2009.02.033 [DOI] [Google Scholar]
- Duperthuy M., Binesse J., Le Roux F., Romestand B., Caro A., Got P., Givaudan A., Mazel D., Bachère E., Destoumieux-Garzón D. (2010). The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ. Microbiol. 12, 951–963 10.1111/j.1462-2920.2009.02138.x [DOI] [PubMed] [Google Scholar]
- Duperthuy M., Schmitt P., Garzón E., Caro A., Rosa R. D., Le Roux F., Lautredou-Audouy N., Got P., Romestand B., de Lorgeril J., Kieffer-Jaquinod S., Bachère E., Destoumieux-Garzón D. (2011). Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl. Acad. Sci. U.S.A. 108, 2993–2998 10.1073/pnas.1015326108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis A. E. (2001). Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 25, 827–839 10.1016/S0145-305X(01)00038-6 [DOI] [PubMed] [Google Scholar]
- Fleury E., Huvet A. (2012). Microarray analysis highlights immune response of Pacific oysters as a determinant of resistance to summer mortality. Mar. Biotechnol. 14, 203–217 10.1007/s10126-011-9403-6 [DOI] [PubMed] [Google Scholar]
- Fleury E., Huvet A., Lelong C., de Lorgeril J., Boulo V., Gueguen Y., Bachère E., Tanguy A., Moraga D., Fabioux C., Lindeque P., Shaw J., Reinhardt R., Prunet P., Davey G., Lapegue S., Sauvage C., Corporeau C., Moal J., Gavory F., Wincker P., Moreews F., Klopp C., Mathieu M., Boudry P., Favrel P. (2009). Generation and analysis of a 29,745 unique expressed sequence tags from the Pacific oyster (Crassostrea gigas) assembled into a publicly accessible database: the gigasdatabase. BMC Genomics 10, 341–356 10.1186/1471-2164-10-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O., Gurevitz M. (2003). Arthropod and mollusk defensins-evolution by exon-shuffling. Trends Genet. 19, 684–687 10.1016/j.tig.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Ganz T., Selsted M. E., Szklarek D., Harwig S. S. L., Daher K., Bainton D. F., Lehrer R. I. (1985). Defensins: natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76, 1427–1435 10.1172/JCI112120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier M., Labreuche Y., Garcia C., Robert M., Nicolas J. L. (2007). Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb. Ecol. 53, 187–196 10.1007/s00248-006-9061-9 [DOI] [PubMed] [Google Scholar]
- Garnier M., Labreuche Y., Nicolas J. L. (2008). Molecular and phenotypic characterization of Vibrio aestuarianus subsp. francensis subsp. nov., a pathogen of the oyster Crassostrea gigas. Syst. Appl. Microbiol. 31, 358–365 10.1016/j.syapm.2008.06.003 [DOI] [PubMed] [Google Scholar]
- Gay M., Berthe F. C., Le Roux F. (2004). Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis. Aquat. Org. 59, 49–56 10.3354/dao059049 [DOI] [PubMed] [Google Scholar]
- Glinski Z., Jarosz J. (1997). Molluscan immune defenses. Arch. Immunol. Ther. Exp. (Warsz.) 45, 149–155 [PubMed] [Google Scholar]
- Gonzalez M., Gueguen Y., Desserre G., de Lorgeril J., Romestand B., Bachère E. (2007a). Molecular characterization of two isoforms of defensin from hemocytes of the oyster Crassostrea gigas. Dev. Comp. Immunol. 31, 332–339 10.1016/j.dci.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Gonzalez M., Gueguen Y., Destoumieux-Garzón D., Romestand B., Fievet J., Pugniere M., Roquet F., Escoubas J. M., Vandenbulcke F., Levy O., Saune L., Bulet P., Bachère E. (2007b). Evidence of a bactericidal permeability increasing protein in an invertebrate, the Crassostrea gigas Cg-BPI. Proc. Natl. Acad. Sci. U.S.A. 104, 17759–17764 10.1073/pnas.0702281104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., Romestand B., Fievet J., Huvet A., Lebart M. C., Gueguen Y., Bachère E. (2005). Evidence in oyster of a plasma extracellular superoxide dismutase which binds LPS. Biochem. Biophys. Res. Commun. 338, 1089–1097 10.1016/j.bbrc.2005.10.075 [DOI] [PubMed] [Google Scholar]
- Green T. J., Dixon T. J., Devic E., Adlard R. D., Barnes A. C. (2009). Differential expression of genes encoding anti-oxidant enzymes in Sydney rock oysters, Saccostrea glomerata (Gould) selected for disease resistance. Fish Shellfish Immunol. 26, 799–810 10.1016/j.fsi.2009.03.003 [DOI] [PubMed] [Google Scholar]
- Guaní-Guerra E., Santos-Mendoza T., Lugo-Reyes S. O., Teran L. M. (2010). Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin. Immunol. 135, 1–11 10.1016/j.clim.2010.03.009 [DOI] [PubMed] [Google Scholar]
- Gueguen Y., Cadoret J. P., Flament D., Barreau-Roumiguiere C., Girardot A. L., Garnier J., Hoareau A., Bachère E., Escoubas J. M. (2003). Immune gene discovery by expressed sequence tags generated from hemocytes of the bacteria-challenged oyster, Crassostrea gigas. Gene 303, 139–145 10.1016/S0378-1119(02)01149-6 [DOI] [PubMed] [Google Scholar]
- Gueguen Y., Herpin A., Aumelas A., Garnier J., Fievet J., Escoubas J. M., Bulet P., Gonzalez M., Lelong C., Favrel P., Bachère E. (2006). Characterization of a defensin from the oyster Crassostrea gigas. Recombinant production, folding, solution structure, antimicrobial activities, and gene expression. J. Biol. Chem. 281, 313–323 10.1074/jbc.M510850200 [DOI] [PubMed] [Google Scholar]
- Gueguen Y., Romestand B., Fievet J., Schmitt P., Destoumieux-Garzón D., Vandenbulcke F., Bulet P., Bachère E. (2009). Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol. Immunol. 46, 516–522 10.1016/j.molimm.2008.07.021 [DOI] [PubMed] [Google Scholar]
- Guo X., Wang Y., Wang L., Lee J. H. (2008). “Oysters,” in Genome Mapping and Genomics in Fishes and Aquatic Animals, eds Kocher T., Kole C. (Berlin: Springer Berlin Heidelberg; ), 163–175 [Google Scholar]
- Hancock R. E., Scott M. G. (2000). The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. U.S.A. 97, 8856–8861 10.1073/pnas.97.16.8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine P. M. (1999). The inter-relationships of bivalve haemocytes. Fish Shellfish Immunol. 9, 367–385 10.1006/fsim.1998.0205 [DOI] [Google Scholar]
- Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. (1999). Phylogenetic perspectives in innate immunity. Science 284, 1313–1318 10.1126/science.284.5418.1313 [DOI] [PubMed] [Google Scholar]
- Hubert F., Noel T., Roch P. (1996). A member of the arthropod defensin family from edible Mediterranean mussels (Mytilus galloprovincialis). Eur. J. Biochem. 240, 302–306 10.1111/j.1432-1033.1996.0302h.x [DOI] [PubMed] [Google Scholar]
- Huvet A., Herpin A., Degremont L., Labreuche Y., Samain J. F., Cunningham C. (2004). The identification of genes from the oyster Crassostrea gigas that are differentially expressed in progeny exhibiting opposed susceptibility to summer mortality. Gene 343, 211–220 10.1016/j.gene.2004.09.008 [DOI] [PubMed] [Google Scholar]
- Iovine N. M., Elsbach P., Weiss J. (1997). An opsonic function of the neutrophil bactericidal/permeability-increasing protein depends on both its N- and C-terminal domains. Proc. Natl. Acad. Sci. U.S.A. 94, 10973–10978 10.1073/pnas.94.20.10973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N., Okada Y., Takahashi K. G., Osada M. (2010a). Presence and characterization of multiple mantle lysozymes in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 29, 126–135 10.1016/j.fsi.2010.02.027 [DOI] [PubMed] [Google Scholar]
- Itoh N., Xue Q. G., Schey K. L., Li Y., Cooper R. K., La Peyre J. F. (2010b). Characterization of the major plasma protein of the eastern oyster, Crassostrea virginica, and a proposed role in host defense. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 158 9–22 10.1016/j.cbpb.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Itoh N., Takahashi K. G. (2007). cDNA cloning and in situ hybridization of a novel lysozyme in the Pacific oyster, Crassostrea gigas. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 148, 160–166 10.1016/j.cbpb.2007.07.076 [DOI] [PubMed] [Google Scholar]
- Itoh N., Takahashi K. G. (2008). Distribution of multiple peptidoglycan recognition proteins in the tissues of Pacific oyster, Crassostrea gigas. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 150, 409–417 10.1016/j.cbpb.2008.04.011 [DOI] [PubMed] [Google Scholar]
- Itoh N., Takahashi K. G. (2009). A novel peptidoglycan recognition protein containing a goose-type lysozyme domain from the Pacific oyster, Crassostrea gigas. Mol. Immunol. 46, 1768–1774 10.1016/j.molimm.2009.01.022 [DOI] [PubMed] [Google Scholar]
- Itoh N., Xue Q. G., Li Y., Cooper R. K., La Peyre J. F. (2007). cDNA cloning and tissue expression of plasma lysozyme in the eastern oyster, Crassostrea virginica. Fish Shellfish Immunol. 23, 957–968 10.1016/j.fsi.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Jiang S. C. (2001). Vibrio cholerae in recreational beach waters and tributaries of Southern California. Hydrobiologia 460, 157–164 10.1023/A:1013152407425 [DOI] [Google Scholar]
- Jo P. G., Choi Y. K., Choi C. Y. (2008). Cloning and mRNA expression of antioxidant enzymes in the Pacific oyster, Crassostrea gigas in response to cadmium exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 147, 460–469 10.1016/j.cbpc.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Kawabata S. (2011). Immunocompetent molecules and their response network in horseshoe crabs. Adv. Exp. Med. Biol. 708, 122–136 10.1007/978-1-4419-8059-5_7 [DOI] [PubMed] [Google Scholar]
- Kouno T., Fujitani N., Mizuguchi M., Osaki T., Nishimura S., Kawabata S., Aizawa T., Demura M., Nitta K., Kawano K. (2008). A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria. Biochemistry 47, 10611–10619 10.1021/bi800957n [DOI] [PubMed] [Google Scholar]
- Kouno T., Mizuguchi M., Aizawa T., Shinoda H., Demura M., Kawabata S., Kawano K. (2009). A novel beta-defensin structure: big defensin changes its N-terminal structure to associate with the target membrane. Biochemistry 48, 7629–7635 10.1021/bi900756y [DOI] [PubMed] [Google Scholar]
- Kragol G., Lovas S., Varadi G., Condie B. A., Hoffmann R., Otvos L., Jr. (2001). The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40, 3016–3026 10.1021/bi002656a [DOI] [PubMed] [Google Scholar]
- Krasity B. C., Troll J. V., Weiss J. P., McFall-Ngai M. J. (2011). LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem. Soc. Trans. 39, 1039–1044 10.1042/BST0391039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labreuche Y., Lambert C., Soudant P., Boulo V., Huvet A., Nicolas J. L. (2006a). Cellular and molecular hemocyte responses of the Pacific oyster, Crassostrea gigas, following bacterial infection with Vibrio aestuarianus strain 01/32. Microbes Infect. 8, 2715–2724 10.1016/j.micinf.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Labreuche Y., Soudant P., Gonçalves M., Lambert C., Nicolas J. L. (2006b). Effects of extracellular products from the pathogenic Vibrio aestuarianus strain 01/32 on lethality and cellular immune responses of the oyster Crassostrea gigas. Dev. Comp. Immunol. 30, 367–379 10.1016/j.dci.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Lambert C., Soudant P., Choquet G., Paillard C. (2003). Measurement of Crassostrea gigas hemocyte oxidative metabolism by flow cytometry and the inhibiting capacity of pathogenic vibrios. Fish Shellfish Immunol. 15, 225–240 10.1016/S1050-4648(02)00160-2 [DOI] [PubMed] [Google Scholar]
- Lambert C., Soudant P., Jegaden M., Delaporte M., Labreuche Y., Moal J., Boudry P., Jean F., Huvet A., Samain J. (2007). In-vitro modulation of reactive oxygen and nitrogen intermediate (ROI/RNI) production in Crassostrea gigas hemocytes. Aquaculture 270, 413–421 10.1016/j.aquaculture.2007.03.016 [DOI] [Google Scholar]
- Lang R. P., Bayne C. J., Camara M. D., Cunningham C., Jenny M. J., Langdon C. J. (2009). Transcriptome profiling of selectively bred Pacific oyster Crassostrea gigas families that differ in tolerance of heat shock. Mar. Biotechnol. 11, 650–668 10.1007/s10126-009-9181-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauckner G. (1983). “Diseases of mollusca: bivalvia,” in Diseases of Marine Animals, ed. Kinne O. (Hamburg: Biologishe Anstalt Helgoland; ), 477–963 [Google Scholar]
- Lazzaro B. P. (2008). Natural selection on the Drosophila antimicrobial immune system. Curr. Opin. Microbiol. 11, 284–289 10.1016/j.mib.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux F., Gay M., Lambert C., Waechter M., Poubalanne S., Chollet B., Nicolas J. L., Berthe F. (2002). Comparative analysis of Vibrio splendidus-related strains isolated during Crassostrea gigas mortality events. Aquat. Living Resour. 15, 251–258 10.1016/S0990-7440(02)01176-2 [DOI] [Google Scholar]
- Lehrer R. I., Lu W. (2012). alpha-Defensins in human innate immunity. Immunol. Rev. 245, 84–112 10.1111/j.1600-065X.2011.01082.x [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- Levy O. (2004). Antimicrobial proteins and peptides: anti-infective molecules of mammalian leukocytes. J. Leukoc. Biol. 76, 909–925 10.1189/jlb.0604320 [DOI] [PubMed] [Google Scholar]
- Lipp E. K., Huq A., Colwell R. R. (2002). Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15, 757–770 10.1128/CMR.15.4.757-770.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman G. W., Cannon J. P., Rast J. P. (2005). New insights into alternative mechanisms of immune receptor diversification. Adv. Immunol. 87, 209–236 10.1016/S0065-2776(05)87006-3 [DOI] [PubMed] [Google Scholar]
- Mangoni M. L., Shai Y. (2009). Temporins and their synergism against Gram-negative bacteria and in lipopolysaccharide detoxification. Biochim. Biophys. Acta 1788, 1610–1619 10.1016/j.bbamem.2009.04.021 [DOI] [PubMed] [Google Scholar]
- Mathur J., Davis B. M., Waldor M. K. (2007). Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 63, 848–858 10.1111/j.1365-2958.2006.05544.x [DOI] [PubMed] [Google Scholar]
- Mathur J., Waldor M. K. (2004). The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 72, 3577–3583 10.1128/IAI.72.6.3577-3583.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., Nakamura A. M., Takahashi K. G. (2006). Cloning of cDNAs and hybridization analysis of lysozymes from two oyster species, Crassostrea gigas and Ostrea edulis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 145, 325–330 10.1016/j.cbpb.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Mitta G., Vandenbulcke F., Noel T., Romestand B., Beauvillain J. C., Salzet M., Roch P. (2000a). Differential distribution and defence involvement of antimicrobial peptides in mussel. J. Cell Sci. 113, 2759–2769 [DOI] [PubMed] [Google Scholar]
- Mitta G., Vandenbulcke F., Roch P. (2000b). Original involvement of antimicrobial peptides in mussel innate immunity. FEBS Lett. 486, 185–190 10.1016/S0014-5793(00)02192-X [DOI] [PubMed] [Google Scholar]
- Muñoz M., Vandenbulcke F., Garnier J., Gueguen Y., Bulet P., Saulnier D., Bachère E. (2004). Involvement of penaeidins in defense reactions of the shrimp Litopenaeus stylirostris to a pathogenic vibrio. Cell. Mol. Life Sci. 61, 961–972 10.1007/s00018-003-3441-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mygind P. H., Fischer R. L., Schnorr K. M., Hansen M. T., Sonksen C. P., Ludvigsen S., Raventos D., Buskov S., Christensen B., De Maria L., Taboureau O., Yaver D., Elvig-Jorgensen S. G., Sorensen M. V., Christensen B. E., Kjaerulff S., Frimodt-Moller N., Lehrer R. I., Zasloff M., Kristensen H. H. (2005). Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 437, 975–980 10.1038/nature04051 [DOI] [PubMed] [Google Scholar]
- Olafsen J. A., Fletcher T. C., Grant P. T. (1992). Agglutinin activity in Pacific oyster (Crassostrea gigas) hemolymph following in vivo Vibrio anguillarum challenge. Dev. Comp. Immunol. 16, 123–138 10.1016/0145-305X(92)90013-3 [DOI] [PubMed] [Google Scholar]
- Ortigosa M., Garay E., Pujalte M. J. (1994). Numerical taxonomy of Vibrionaceae isolated from oysters and seawater along an annual cycle. Syst. Appl. Microbiol. 17, 216–225 10.1016/S0723-2020(11)80011-1 [DOI] [Google Scholar]
- Otvos L., Jr. (2002). The short proline-rich antibacterial peptide family. Cell. Mol. Life Sci. 59, 1138–1150 10.1007/s00018-002-8493-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard C., Le Roux F., Borreg J. J. (2004). Bacterial disease in marine bivalves, a review of recent studies: trends and evolution. Aquat. Living Resour. 17, 477–498 10.1051/alr:2004054 [DOI] [Google Scholar]
- Park M. S., Jo P. G., Choi Y. K., An K. W., Choi C. Y. (2009). Characterization and mRNA expression of Mn-SOD and physiological responses to stresses in the Pacific oyster Crassostrea gigas. Mar. Biol. Res. 5, 451–461 10.1080/17451000802626554 [DOI] [Google Scholar]
- Peschel A., Sahl H. G. (2006). The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4, 529–536 10.1038/nrmicro1441 [DOI] [PubMed] [Google Scholar]
- Pruzzo C., Gallo G., Canesi L. (2005). Persistence of vibrios in marine bivalves: the role of interactions with haemolymph components. Environ. Microbiol. 7, 761–772 10.1111/j.1462-2920.2005.00792.x [DOI] [PubMed] [Google Scholar]
- Rinkevich B., Muller W. E. G. (1996). Invertebrate Immunology Progress in Molecular and Subcellular Biology. New York: Springer Verlag, 250 [Google Scholar]
- Roberts S., Goetz G., White S., Goetz F. (2009). Analysis of genes isolated from plated hemocytes of the Pacific oyster, Crassostrea gigas. Mar. Biotechnol. 11, 24–44 10.1007/s10126-008-9117-6 [DOI] [PubMed] [Google Scholar]
- Roger E., Grunau C., Pierce R. J., Hirai H., Gourbal B., Galinier R., Emans R., Cesari I. M., Cosseau C., Mitta G. (2008). Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata). PLoS Negl. Trop. Dis. 2, e330. 10.1371/journal.pntd.0000330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa R. D., Santini A., Fievet J., Bulet P., Destoumieux-Garzón D., Bachère E. (2011). Big defensins, a diverse family of antimicrobial peptides that follows different patterns of expression in hemocytes of the oyster Crassostrea gigas. PLoS ONE 6, e25594. 10.1371/journal.pone.0025594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. 5, 355–362 10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Rosenfeld Y., Shai Y. (2006). Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim. Biophys. Acta 1758, 1513–1522 10.1016/j.bbamem.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Royet J. (2011). Epithelial homeostasis and the underlying molecular mechanisms in the gut of the insect model Drosophila melanogaster. Cell. Mol. Life Sci. 68, 3651–3660 10.1007/s00018-011-0828-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby E. G. (1999). The Euprymna scolopes-Vibrio fischeri symbiosis: a biomedical model for the study of bacterial colonization of animal tissue. J. Mol. Microbiol. Biotechnol. 1, 13–21 [PubMed] [Google Scholar]
- Saito T., Kawabata S., Shigenaga T., Takayenoki Y., Cho J., Nakajima H., Hirata M., Iwanaga S. (1995). A novel big defensin identified in horseshoe crab hemocytes: isolation, amino acid sequence, and antibacterial activity. J. Biochem. 117, 1131–1137 10.1093/jb/117.2.276 [DOI] [PubMed] [Google Scholar]
- Salzman N. H., Underwood M. A., Bevins C. L. (2007). Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin. Immunol. 19, 70–83 10.1016/j.smim.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Schmitt P., de Lorgeril J., Gueguen Y., Destoumieux-Garzón D., Bachère E. (2012a). Expression, tissue localization, and synergy of antimicrobial peptides, and proteins in the immune response of the oyster Crassostrea gigas. Dev. Comp. Immunol. 10.1016/j.dci.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Schmitt P., Duperthuy M., Montagnani C., Bachère E., Destoumieux-Garzón D. (2012b). “Immune responses in the Pacific oyster Crassostrea gigas an overview with focus on summer mortalities,” in Oysters Physiology, Ecological Distribution and Mortality, ed. Qin J. (New York: Nova Science Publishers, Inc.). [Google Scholar]
- Schmitt P., Gueguen Y., Desmarais E., Bachère E., de Lorgeril J. (2010a). Molecular diversity of antimicrobial effectors in the oyster Crassostrea gigas. BMC Evol. Biol. 10, 23. 10.1186/1471-2148-10-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt P., Wilmes M., Pugnière M., Aumelas A., Bachère E., Sahl H. G., Schneider T., Destoumieux-Garzón D. (2010b). Insight into invertebrate defensin mechanism of action: oyster defensins inhibit peptidoglycan biosynthesis by binding to lipid II. J. Biol. Chem. 285, 29208–29216 10.1074/jbc.M110.147017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott E. J., Pecher W. T., Okafor F., Vasta G. R. (2003). The protistan parasite Perkinsus marinus is resistant to selected reactive oxygen species. Exp. Parasitol. 105, 232–240 10.1016/j.exppara.2003.12.012 [DOI] [PubMed] [Google Scholar]
- Schulenburg H., Boehnisch C., Michiels N. K. (2007). How do invertebrates generate a highly specific innate immune response? Mol. Immunol. 44, 3338–3344 10.1016/j.molimm.2007.02.019 [DOI] [PubMed] [Google Scholar]
- Scocchi M., Tossi A., Gennaro R. (2011). Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. 68, 2317–2330 10.1007/s00018-011-0721-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. G., Mori K. (2000). Functional profiles of hemocytes in the bio-defense process of the Pacific oyster, Crassostrea gigas. Tohoku J. Agric. Res. 51, 15–27 [Google Scholar]
- Tanguy A., Guo X., Ford S. E. (2004). Discovery of genes expressed in response to Perkinsus marinus challenge in Eastern (Crassostrea virginica) and Pacific (C. gigas) oysters. Gene 338, 121–131 10.1016/j.gene.2004.05.019 [DOI] [PubMed] [Google Scholar]
- Taris N., Lang R. P., Reno P. W., Camara M. D. (2009). Transcriptome response of the Pacific oyster (Crassostrea gigas) to infection with Vibrio tubiashii using cDNA AFLP differential display. Anim. Genet. 40, 663–677 10.1111/j.1365-2052.2009.01894.x [DOI] [PubMed] [Google Scholar]
- Tasumi S., Vasta G. R. (2007). A galectin of unique domain organization from hemocytes of the Eastern oyster (Crassostrea virginica) is a receptor for the protistan parasite Perkinsus marinus. J. Immunol. 179, 3086–3098 [DOI] [PubMed] [Google Scholar]
- Tennessen J. A., Woodhams D. C., Chaurand P., Reinert L. K., Billheimer D., Shyr Y., Caprioli R. M., Blouin M. S., Rollins-Smith L. A. (2009). Variations in the expressed antimicrobial peptide repertoire of northern leopard frog (Rana pipiens) populations suggest intraspecies differences in resistance to pathogens. Dev. Comp. Immunol. 33, 1247–1257 10.1016/j.dci.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. J., Kapoor M., Sharma S., Bausinger H., Zyilan U., Lipsker D., Hanau D., Surolia A. (2002). Evidence of a trimolecular complex involving LPS, LPS binding protein and soluble CD14 as an effector of LPS response. FEBS Lett. 531, 184–188 10.1016/S0014-5793(02)03499-3 [DOI] [PubMed] [Google Scholar]
- Torreilles J., Romestand B. (2001). In vitro production of peroxynitrite by haemocytes from marine bivalves: C-ELISA determination of 3-nitrotyrosine level in plasma proteins from Mytilus galloprovincialis and Crassostrea gigas. BMC Immunol. 2, 1. 10.1186/1471-2172-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers M. A., Basuyaux O., Le Goïc N., Huchette S., Nicolas J. L., Koken M., Paillard C. (2009). Influence of temperature and spawning effort on Haliotis tuberculata susceptibility to Vibrio harveyi: an example of emerging vibriosis linked to global warming. Glob. Chang. Biol. 15, 1365–1376 10.1111/j.1365-2486.2008.01764.x [DOI] [Google Scholar]
- Van Valen L. (1974). Molecular evolution as predicted by natural selection. J. Mol. Evol. 3, 89–101 10.1007/BF01796554 [DOI] [PubMed] [Google Scholar]
- Vertuani S., Angusti A., Manfredini S. (2004). The antioxidants and pro-antioxidants network: an overview. Curr. Pharm. Des. 10, 1677–1694 10.2174/1381612043384655 [DOI] [PubMed] [Google Scholar]
- Vezzulli L., Previati M., Pruzzo C., Marchese A., Bourne D. G., Cerrano C. (2010). Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ. Microbiol. 12, 2007–2019 10.1111/j.1462-2920.2010.02209.x [DOI] [PubMed] [Google Scholar]
- Weiss J. (2003). Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochem. Soc. Trans. 31, 785–790 10.1042/BST0310785 [DOI] [PubMed] [Google Scholar]
- Wilmes M., Cammue B. P., Sahl H. G., Thevissen K. (2011). Antibiotic activities of host defense peptides: more to it than lipid bilayer perturbation. Nat. Prod. Rep. 28, 1350–1358 10.1039/c1np00022e [DOI] [PubMed] [Google Scholar]
- Xue Q., Hellberg M. E., Schey K. L., Itoh N., Eytan R. I., Cooper R. K., La Peyre J. F. (2010). A new lysozyme from the eastern oyster, Crassostrea virginica, and a possible evolutionary pathway for i-type lysozymes in bivalves from host defense to digestion. BMC Evol. Biol. 10, 213. 10.1186/1471-2148-10-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaura K., Takahashi K. G., Suzuki T. (2008). Identification and tissue expression analysis of C-type lectin and galectin in the Pacific oyster, Crassostrea gigas. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 149, 168–175 10.1016/j.cbpb.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Yang W. Y., Wen S. Y., Huang Y. D., Ye M. Q., Deng X. J., Han D., Xia Q. Y., Cao Y. (2006). Functional divergence of six isoforms of antifungal peptide Drosomycin in Drosophila melanogaster. Gene 379, 26–32 10.1016/j.gene.2006.03.017 [DOI] [PubMed] [Google Scholar]
- Yang Y. S., Mitta G., Chavanieu A., Calas B., Sanchez J. F., Roch P., Aumelas A. (2000). Solution structure and activity of the synthetic four-disulfide bond Mediterranean mussel defensin (MGD-1). Biochemistry 39, 14436–14447 10.1021/bi000061+ [DOI] [PubMed] [Google Scholar]
- Yeaman M. R., Yount N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55 10.1124/pr.55.1.2 [DOI] [PubMed] [Google Scholar]
- Yount N. Y., Bayer A. S., Xiong Y. Q., Yeaman M. R. (2006). Advances in antimicrobial peptide immunobiology. Biopolymers 84, 435–458 10.1002/bip.20543 [DOI] [PubMed] [Google Scholar]
- Yu Z., He X., Fu D., Zhang Y. (2011). Two superoxide dismutase (SOD) with different subcellular localizations involved in innate immunity in Crassostrea hongkongensis. Fish Shellfish Immunol. 31, 533–539 10.1016/j.fsi.2011.06.022 [DOI] [PubMed] [Google Scholar]
- Zhang Y., He X., Li X., Fu D., Chen J., Yu Z. (2011). The second bactericidal permeability increasing protein (BPI) and its revelation of the gene duplication in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 30, 954–963 10.1016/j.fsi.2010.12.023 [DOI] [PubMed] [Google Scholar]
- Zhao J., Song L., Li C., Ni D., Wu L., Zhu L., Wang H., Xu W. (2007). Molecular cloning, expression of a big defensin gene from bay scallop Argopecten irradians and the antimicrobial activity of its recombinant protein. Mol. Immunol. 44, 360–368 10.1016/j.molimm.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Zhu S., Gao B., Tytgat J. (2005). Phylogenetic distribution, functional epitopes and evolution of the CSalphabeta superfamily. Cell. Mol. Life Sci. 62, 2257–2269 10.1007/s00018-005-5200-6 [DOI] [PMC free article] [PubMed] [Google Scholar]