Background: PlaC is a GDSL enzyme and the major GCAT secreted by Legionella pneumophila.
Results: The zinc metalloproteinase ProA processes and activates PlaC. Deletion of regions within a disulfide loop increased GCAT activity.
Conclusion: Inhibitory disulfide loop reduction/deletion by ProA activates PlaC GCAT.
Significance: Here we recognized the postexport GCAT activation mechanism essential for modification of typical eukaryotic sterols.
Keywords: Metalloprotease, Phospholipase A, Protein Processing, Protein Secretion, Proteolytic Enzymes, Legionella, GDSL Enzyme, ProA, Glycerophospholipid:cholesterol Acyltransferase, Proteolytic Activation
Abstract
Enzymes secreted by Legionella pneumophila, such as phospholipases A (PLAs) and glycerophospholipid:cholesterol acyltransferases (GCATs), may target host cell lipids and therefore contribute to the establishment of Legionnaires disease. L. pneumophila possesses three proteins, PlaA, PlaC, and PlaD, belonging to the GDSL family of lipases/acyltransferases. We have shown previously that PlaC is the major GCAT secreted by L. pneumophila and that the zinc metalloproteinase ProA is essential for GCAT activity. Here we characterized the mode of PlaC GCAT activation and determined that ProA directly processes PlaC. We further found that not only cholesterol but also ergosterol present in protozoa was palmitoylated by PlaC. Such ester formations were not induced by either PlaA or PlaD. PlaD was shown here to possess lysophospholipase A activity, and interestingly, all three GDSL enzymes transferred short chain fatty acids to sterols. The three single putative catalytic amino acids (Ser-37, Asp-398, and His-401) proved essential for all PlaC-associated PLA, lysophospholipase A, and GCAT activities. A further four cysteine residues are important for the PLA/GCAT activities as well as their oxidized state, and we therefore conclude that PlaC likely forms at least one disulfide loop. Analysis of cleavage site and loop deletion mutants suggested that for GCAT activation deletion of several amino acids within the loop is necessary rather than cleavage at a single site. Our data therefore suggest a novel enzyme inhibition/activation mechanism where a disulfide loop inhibits PlaC GCAT activity until the protein is exported to the external space where it is ProA-activated.
Introduction
Legionella pneumophila resides in aquatic habitats and replicates in diverse species of protozoa. When inhaled by susceptible humans, the pathogen causes a severe form of pneumonia, termed Legionnaires disease (1–3). Enzymes secreted by Legionella, such as phospholipases A (PLAs),2 may contribute to the establishment of the disease and subsequent lung tissue destruction. So far, three different PLA families, namely the patatin-like, PlaB-like, and GDSL enzymes together with at least 15 distinct proteins, have been identified in L. pneumophila (4, 5).
Patatins were originally identified as a family of plant proteins with acyl hydrolase activity fulfilling important functions in signal transduction processes (6). Bacteria also possess patatin-like proteins. Ten to 11 patatin-like protein genes are typically found in the genome of L. pneumophila (patA to patK). Both bacterial patatin-like proteins and eukaryotic patatins are characterized by a catalytic dyad consisting of a nucleophile serine embedded into the typical lipase consensus motif GXSXG and an aspartate that is often found in a DGG motif (5, 7).
PlaB shows no significant protein homology to any established lipase/phospholipase family and is the most prominent cell-associated PLA/lysophospholipase A (LPLA) of L. pneumophila. PlaB homologues are yet uncharacterized proteins especially found in water bacteria. Instead of the characteristic GXSXG motif, PlaB constitutes the designated THSTG motif surrounding the catalytic serine. Aspartate and histidine, representing the other two members of the catalytic triad, are embedded into the uncommon motifs GSDGVV and SHS, respectively (8).
Three L. pneumophila proteins homologous to the “prototype” of GDSL enzymes, Aeromonas salmonicida/Aeromonas hydrophila SatA, are coded in L. pneumophila genomes and were designated PlaA, PlaC, and PlaD (4, 5, 9–15). GDSL enzymes possess a conserved GDSL signature motif harboring the nucleophile Ser, which together with an Asp and a His (DXXH motif) form the catalytic triad (16, 17). The GDSL proteins share five conserved blocks of amino acid homology, belong to the SGNH hydrolases, and differ in several properties (folding structure, proximity of Ser nucleophile to the N terminus, and close localization of catalytic Asp and His) from the well characterized α/β-hydrolases (17, 18). GDSL family members are found in many bacteria as well as in higher plants and show among others lipase, LPLA, PLA, and/or glycerophospholipid:cholesterol acyltransferase (GCAT) activities (16, 17, 19).
PlaA is the major secreted LPLA of L. pneumophila and contains a predicted signal peptide (20, 21). Because of the absence of PlaA in the culture supernatant of a L. pneumophila type II secretion mutant, it is most likely exported via the Lsp type II pathway (20, 22, 23). During L. pneumophila host cell infection, PlaA promotes membrane destabilization of the Legionella-containing vacuole and activation of the host cell death pathway when the type IVB (Dot/Icm)-secreted effector SdhA is lacking (24).
Lipolytic activity of the PlaD enzyme has not yet been conclusively shown (4, 5). It contains a C-terminal appendage of about 170 amino acids that is missing in PlaA and PlaC. A signal peptide is not predicted for PlaD, implying either a cell-associated localization or its export by a pathway different from the Sec system (4, 5).
PlaC contains a predicted signal peptide and shows 27% sequence identity to PlaA, 24% to PlaD, and 22% to A. salmonicida SatA. A L. pneumophila plaC knock-out mutant exhibits an approximately 20% reduction in secreted PLA and a 10% reduction in secreted LPLA activities (25). Furthermore, L. pneumophila possesses secreted GCAT activity (21). plaC but not plaA knock-out mutants completely lost their ability to transfer long chain fatty acids from dipalmitoylphospholipids to cholesterol. Therefore, PlaC is the major secreted GCAT of L. pneumophila with PLA/LPLA activities (25). Furthermore, the presence of GCAT activity within L. pneumophila culture supernatant depends on the intact Lsp type II secretion system and the type II-secreted zinc metalloproteinase ProA (22, 23, 26). This suggested proteolytic activation of PlaC GCAT activity.
Here we aimed to clarify whether ProA directly (and by which mechanism) activates PlaC GCAT activity. Our data suggest a novel enzyme inhibition/activation mechanism where a disulfide loop inhibits PlaC GCAT and full PLA activity until the protein is exported to the external space and ProA-activated.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
L. pneumophila sg1 strain 130b (ATCC BAA-74) was used to purify the PlaC GCAT-activating factor via anion exchange (AEC) and gel filtration chromatography as described previously (25). L. pneumophila sg1 strain Corby (27) was used for all other experiments, including knock-out mutant construction as well as for recombinant gene expression in Escherichia coli strains DH5α and BL21. The strains used are listed in Table 1 and were grown on buffered charcoal yeast extract agar and in buffered yeast extract broth (both for Legionella) or on Luria-Bertani (LB) agar and in LB broth (both for E. coli) as described previously (25, 28, 29).
TABLE 1.
Overview of strains used in this study
L.pn., L. pneumophila; term., terminal.
| Organism | Mutation(s)/plasmid | Tags | Selection marker(s) | Ref. |
|---|---|---|---|---|
| L.pn. Corby | Wild type | 27 | ||
| L.pn. Corby | plaA knock-out | KmR | This study | |
| L.pn. Corby | plaC knock-out | KmR | This study | |
| L.pn. Corby | plaD knock-out | KmR | This study | |
| L.pn. Corby | proA knock-out | KmR | This study | |
| L.pn. 130b | Wild type | Strain ATCC BAA-74 | ||
| L.pn. 130b | plaC knock-out | KmR | 25 | |
| L.pn. 130b | proA knock-out | KmR | Strain AA200 (30) | |
| E. coli BL21 | pet28a (+) | C-term. His6 tag | KmR | Novagen |
| E. coli BL21 | pMMB2002 | CmR | 23 | |
| E. coli BL21 | pER13 = pBADTOPO plaACorby | C-term. His6 tag | AmpR | This study |
| E. coli BL21 | pMY6 = pMMB2002 plaC130b | CmR | This study | |
| E. coli BL21 | pCL14, pER14 = pet28a (+) plaCCorby | C-term. His6 tag | KmR | This study |
| E. coli BL21 | pER31 = pGEX-6-P1 plaCCorby(−SPa) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL83 = pGEX-6-P1 plaCCorby(−SP)-Strep | N-term. GST tag/C-term. Strep tag | AmpR | This study |
| E. coli BL21 | pCL70 = pGEX-6-P1 plaCCorbyS37N(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL71 = pGEX-6-P1 plaCCorby D398N(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL72 = pGEX-6-P1 plaCCorby H401N(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL73 = pGEX-6-P1 plaCCorby C343N(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL74 = pGEX-6-P1 plaCCorby C388N(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL75 = pGEX-6-P1 plaCCorby C415N(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL76 = pGEX-6-P1 plaCCorby C427N(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL77 = pGEX-6-P1 plaCCorby Δ364–75(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL78 = pGEX-6-P1 plaCCorby Δ361–79(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL79 = pGEX-6-P1 plaCCorby Δ358–79(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL80 = pGEX-6-P1 plaCCorby Δ343–88(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pCL85 = pGEX-6-P1 plaCCorby Δ347–84(−SP) | N-term. GST tag | AmpR | This study |
| E. coli BL21 | pER7 = pET160 plaDCorby | N-term. His6 tag | AmpR | This study |
| E. coli BL21 | pCL15 = pet28a (+) proACorby | C-term. His6 tag | KmR | This study |
a (−SP), constructs cloned without sequence coding for the putative signal peptide (amino acids 1–24).
DNA Techniques and Sequence Analysis
E. coli DH5α or BL21 was used for the propagation of recombinant plasmid DNA with backbones of the following vectors: pMMB2002 (23), pet28a(+) (Novagen), pBADTOPO (Invitrogen), pBCKS (Stratagene), pGEX-6P-1 (GE Healthcare), pGEM-T Easy (Promega GmbH), and pET160 (Invitrogen). Genomic and plasmid DNA were prepared, amplified, and sequenced according to standard protocols. Primers were obtained either from Tib Molbiol or from Eurofins MWG Operon. Restriction enzymes were purchased from New England Biolabs. Foreign DNA was introduced into E. coli or L. pneumophila strains by electroporation with an Invitrogen cell porator according to the manufacturer's specifications as described earlier (25). Nucleotide and translated protein sequences were analyzed using the DNASTAR package, the pedant website, the NCBI website, ExPASy, multiple sequence alignment by Florence Corpet, ClustalW multiple alignment, and BPROM.
Cloning of L. pneumophila plaA, plaC, plaD, and proA into Vectors for Recombinant Expression in E. coli
L. pneumophila Corby plaA (lpc1811) was cloned into the pBADTOPO vector resulting in pER13 (C-terminal 6×His6 tag). L. pneumophila Corby plaC (lpc3121) was cloned into the pet28a(+) and pGEX-6-P1 vectors, resulting in plasmids pCL14 (C-terminal His6 tag) and pER31 (N-terminal GST tag), respectively. To introduce mutations into the plaC gene, the pER31 vector was mutated by means of the QuikChange site-directed mutagenesis kit (Stratagene) with the primers listed in Table 2, resulting in pCL70–pCL76. To create PlaC disulfide loop deletion mutants or a GST-PlaC construct, pER31 was amplified with primers listed in Table 2 and ligated, resulting in pCL77–pCL80 and pCL85 or pCL83, respectively. L. pneumophila 130b plaC was cloned into the pGEM-T Easy vector, yielding pMY4. After digestion of pMY4 by SpeI and SacII, the insert was cloned into pBCKS, yielding in pMY5. This vector was restricted by SacI and KpnI, and the insert was ligated into pMMB2002, yielding pMY6. L. pneumophila Corby plaD (lpc0558) or proA (lpc2877) was cloned into pET160, yielding pER7 (N-terminal His6 tag), or pet28a(+), resulting in pCL15 (C-terminal His6 tag), respectively (for primers, see Table 2).
TABLE 2.
Overview of primers used in this study
term., terminal.
| Plasmid | Gene | Tag | Primer name | Primer sequence |
|---|---|---|---|---|
| pER13 | plaA(−SPa) | C-term. His tag | BADPlaAf | 5′-GGTTTCATGAAACTATTAGCAT-3′ |
| BADPlaAr | 5′-AAACTCTTCATTCTCGGCGAA-3′ | |||
| pER31 | plaC(−SP) | N-term. GST tag | PlaC_ohneSP_Eco | 5′-TGAATTCATGACGTCTATTCCAGTGAAA-3′ |
| PlaC_XhoI_rev | 5′-AGCTCGAGGTATTAGGCGGACTCTGGCAT-3′ | |||
| pCL83 | plaC(−SP)-Strep | N-term. GST tag | pGEX_Strep_fw | 5′-CCGCAGTTCGAAAAATAATACCTCGAGCGGCCG-3′ |
| PlaC_Strep_rv | 5′-GTGGCTCCAGCTAGCGGCGGACTCTGGCAT3–3′ | |||
| pCL14, pER14 | plaC | C-term. His tag | PlaC_EcoRI | 5′-TGAATTCTTATGATCCAAAACAACAGGG-3′ |
| PlaC_XhoI | 5′-AGCTCGAGGTATTGGGCGGACTCTGGCAT-3′ | |||
| pCL70 | plaCS37N(−SP) | N-term. GST tag | PlaC_Serin_f | 5′-AGTGGTGTTTGGAGACAACTTGTCTGATAACGGTAATA-3′ |
| PlaC_Serin_rev | 5′-TATTACCGTTATCAGACAAGTTGTCTCCAAACACCACT-3′ | |||
| pCL71 | plaCD398N(−SP) | N-term. GST tag | PlaC_D398N_fw | 5′-TATTTATTCTGGAATGAAATTCATCCA-3′ |
| PlaC_D398N_rv | 5′-TGGATGAATTTCATTCCAGAATAAATA-3′ | |||
| pCL72 | plaCH401N(−SP) | N-term. GST tag | PlaC_H401N_fw | 5′-TGGGATGAAATTAATCCAACCACGAGA-3′ |
| PlaC_H401N_rv | 5′-TCTCGTGGTTGGATTAATTTCATCCCA-3′ | |||
| pCL73 | plaCC343N(−SP) | N-term. GST tag | PlaC_C343N_fw | 5′-GAAGGATGCCAATATCGATGTGA-3′ |
| PlaC_C343N_rv | 5′-TCACATCGATATTGGCATCCTTC-3′ | |||
| pCL74 | plaCC388N(−SP) | N-term. GST tag | PlaC_C388N_fw | 5′-CTATCACATGAATAGTGAACCAG-3′ |
| PlaC_C388N_rv | 5′-CTGGTTCACTATTCATGTGATAG-3′ | |||
| pCL75 | plaCC415N(−SP) | N-term. GST tag | PlaC_C415N_fw | 5′-TTATGAGGTTAATTTGGCGTTGG-3′ |
| PlaC_C415N_rv | 5′-CCAACGCCAAATTAACCTCATAA-3′ | |||
| pCL76 | plaCC427N(−SP) | N-term. GST tag | PlaC_C427N_fw | 5′-CGAGGTAACTAACAAAATGCCAG-3′ |
| PlaC_C427N_rv | 5′-CTGGCATTTTGTTAGTTACCTCG-3′ | |||
| pCL77 | plaC ΔA364–75(−SP) | N-term. GST tag | PlaC_del375fw | 5′-TTGAGCTCGTAGTTACGAGCAAATGGAG-3′ |
| PlaC_del364rv | 5′-AAGAGCTCAAGCAATTTGCTCCAGGT-3′ | |||
| pCL78 | plaCΔ361–79(−SP) | N-term. GST tag | PlaC_del379fw | 5′-CCAGGTGAAAATAACTATCACATG-3′ |
| PlaC_del361rv | 5′-ACGAGCAAATGGAGAGTGAAT-3′ | |||
| pCL79 | plaCΔ358–79(−SP) | N-term. GST tag | PlaC_del379fw | 5′-CCAGGTGAAAATAACTATCACATG-3′ |
| PlaC_del358rv | 5′-AGAGTGAATGAATGCATTGAAC-3′ | |||
| pCL80 | plaCΔ343–88(−SP) | N-term. GST tag | PlaC_del388fw | 5′-AGTGAACCAGAAGACTATTTATTC-3′ |
| PlaC_del343rv | 5′-GGCATCCTTCACATTATAAAAAC-3′ | |||
| pCL85 | plaCΔ347–84(−SP) | N-term. GST tag | PlaC_del384fw | 5′-TATCACATGTGTAGTGAACCAGAA-3′ |
| PlaC_del347rv | 5′-CACATCGATACAGGCATCCTT-3′ | |||
| pMY4 | PplaC plaC | gdsl2_a1_f | 5′-TGCTTAAAAACCGCTCTGGA-3′ | |
| gdsl2_b1_r | 5′-TTAACGGCATATTGGGTGAA-3′ | |||
| pER7 | plaD | N-term. His tag | PlaD_fTopo | 5′-CACCGCAAATATCATGGCCCAAAAA-3′ |
| PlaD_d1_r | 5′-AGGGACGCATTAAAACTTACCAG-3′ | |||
| pSB2 | plaD | gdsl3_a1_f | 5′-TTTTGCAAGTGGATTAGGTGA-3′ | |
| gdsl3_b1_r | 5′-ACGTCCGATTTGTATGAGTG-3′ | |||
| pCL15 | proA | C-term. His tag | ProA_BamHI_fw | 5′-ATGGATCCATGCACCCAAATTATTAT-3′ |
| ProA_XhoI_rv | 5′-ATCTCGAGATCGACATAACAAGATTG-3′ | |||
| pAF14 | proA:KnR | ProA3a_fw | 5′-ATAACAAACCTGGGCACTCC-3′ | |
| ProA4a_rv | 5′-GACACGATCGCAGGCTATTA-3′ |
a (−SP), without signal peptide.
L. pneumophila GDSL Enzyme Knock-out Mutant Construction
Plasmids pAF3 (21), pBH5 (25), pBH4 (this study), and pAF14 (this study) containing the plaA, plaC, plaD, and proA genes, respectively, disrupted or partially replaced by a KmR cassette were used to introduce a KmR gene insertion mutation into the chromosome of strain Corby via allelic exchange. To assemble pBH4, L. pneumophila 130b plaD was cloned into the pGEM-T Easy vector, yielding pSB2 (for primers, see Table 2). After pSB2 restriction by means of HindIII and NdeI, the insert was cloned into the pBCKS vector, yielding pBH3. pBH3 was digested with AfeI, which cleaves plaD at position 806 bp, to insert the KnR gene cassette, yielding pBH4. To generate pAF14, proA:KnR was cloned from an L. pneumophila 130b proA knock-out mutant (AA200; kindly provided by N. P. Cianciotto) into pGEM-T Easy (for primers, see Table 2) (30). Knock-out mutants were generated by natural transformation and homologous recombination (21, 31). PCR and/or Southern blot analysis was used to examine KmR Legionella for the respective mutation.
Preparations of Culture Supernatants and Cell Lysates
For assessment of hydrolytic activities, bacteria were harvested at the end of exponential growth (if not stated otherwise at an OD660 of 2.0–2.1). Culture supernatants were obtained by centrifugation for 5 min at 5000 × g and in some instances were subsequently concentrated by ultrafiltration with an exclusion size of 10 kDa. Cell lysates were produced as described previously (21, 32). Culture supernatants and cell lysates were either tested immediately for enzymatic activities or were stored overnight at 4 °C. To prove recombinant overexpression of proteins in E. coli, 5 μl of the cell lysate was used for SDS-PAGE with Coomassie Blue staining. For Western blot analysis, Legionella supernatants were complemented with 10 mm 1,10-phenanthroline (metalloproteinase inhibitor) before concentration.
Culture Supernatant Fractionation for Partial Purification of the PlaC-activating Factor
Five liters of culture supernatant of L. pneumophila 130b plaC mutant was 15-fold-concentrated by ultrafiltration with an exclusion size of 30 kDa, and the resulting retentate was directly applied to a 6-ml Resource-Q column (Amersham Biosciences) pre-equilibrated in buffer containing 20 mm Tris-HCl (pH 7.5) for AEC. AEC was carried out at 25 °C, and a gradient of 0–1 m sodium chloride in equilibration buffer was used for elution of proteins followed by gel filtration using a prepacked HiLoad 26/60 Superdex 200 column (Amersham Biosciences). Finally, 10-ml fractions were collected and used for SDS-PAGE with silver staining as well as for evaluation of protease activity by means of paracaseinate precipitation on casein-buffered starch yeast extract agar plates or azocasein hydrolysis and were stored at 4 °C (20, 33–36).
Purification of Recombinant ProA
Recombinant periplasmic ProA was isolated by osmotic shock (37) from E. coli BL21(pCL15). The presence of protease activity was determined as described above (34, 36). To obtain purified ProA, the periplasmic fraction was diluted 1:10 in 20 mm Tris-HCl (pH 9) equilibration buffer and applied to a 1-ml Resource-Q column (Amersham Biosciences) for AEC. AEC was carried out at 25 °C, and a gradient of 0–1 m sodium chloride in equilibration buffer was used. Fractions of interest were combined and concentrated, and buffer exchange with 20 mm Tris-HCl (pH 8) was performed by means of Amicon-15 filter devices (Millipore) and centrifugation. The presence of a single ProA-His protein band of 38 kDa was verified with SDS-PAGE and Coomassie staining.
Overexpression of Recombinant PlaA-His, GST-PlaC Variants, PlaC-His, and His-PlaD and Purification of GST-PlaC-Strep and PlaC-His
For expression of recombinant PlaA-His (from pER13), GST-PlaC variants (from pER31, pCL70–80/83/85), PlaC-His (from pCL14, pER14), and His-PlaD (from pER7), E. coli BL21 cultures were grown at 37 °C to an A600 of 0.8. The GST-PlaC variant-expressing cultures were induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and transferred to 18 °C for 16–17 h. PlaA-His, PlaC-His, and His-PlaD-expressing cultures were induced with 0.2% arabinose for PlaA-His or 1 mm isopropyl 1-thio-β-d-galactopyranoside for all others and collected after 3–4 h of incubation at 37 °C. Cell lysates for lipolytic activities were produced as described previously (21, 32). For the purification of GST-PlaC-Strep (from pCL83), the cell pellet was collected and resuspended in 100 mm Tris-HCl (pH 8.0) containing 100 mm NaCl and 1 mm EDTA. Homogenization was performed using an Emulsi Flex C3 (Avestin) applying 25,000–30,000 p.s.i. The soluble fraction was isolated by centrifugation at 13,000 × g at 4 °C for 60 min. GST-PlaC-Strep was purified using Strep-Tactin® Superflow® high capacity resin (IBA) according to the manufacturer's instructions. To obtain PlaC-Strep, GST-PlaC-Strep bound to the resin was cleaved by PreScission protease (GE Healthcare) according to the manufacturer's instructions. Cells for the purification of PlaC-His (pER14) were resuspended in 20 mm NaH2PO4, 500 mm NaCl, 20 mm imidazole (pH 7.4) and incubated with lysozyme (10 mg/ml) for 30 min on ice followed by ultrasonication and DNase treatment steps. After centrifugation at 13,000 × g at 4 °C for 30 min, PlaC-His was purified using a HisTrap HP column (Amersham Biosciences) according to the manufacturer's instructions and concentrated by Amicon-15 filter devices (Millipore).
Assays for Lipolytic Activities
Lipolytic (PLA, LPLA, lipase, and GCAT) activities were detected as described previously (21, 25, 38) by using the lipid substrates 1-monopalmitoyllysophosphatidylcholine, 1-monopalmitoyllysophosphatidylglycerol, 1-monopalmitoylglycerol, monopalmitoyllysophosphatidylethanolamine, dipalmitoylphosphatidylserine, dipalmitoylphosphatidylethanolamine, 1,2-dipalmitoylphosphatidylglycerol (DPPG; used in most cases as an acyl donor for GCAT detection), 1,2-dipalmitoylphosphatidylcholine (DPPC), cholesterol, and ergosterol (used in amounts comparable with cholesterol). To activate GCAT in cell lysates with expressed recombinant PlaC, 100-μl reaction mixtures (lipid suspension and bacterial product) were incubated with 2–6 milliunits of different ProA preparations, such as culture supernatant from a L. pneumophila plaC mutant or ProA from the periplasmic fraction of BL21(pCL15). All lipids, including standards for thin layer chromatography (TLC), were obtained from Sigma or Avanti Polar Lipids. Detection of reaction products, such as free fatty acids and sterol esters, was performed by lipid extraction and subsequent TLC or by quantitative detection of free fatty acids as described earlier (21, 25).
Western Blot Analysis
Samples were separated on a 12.5% SDS-polyacrylamide gel (39). Proteins were transferred to a nitrocellulose membrane. Primary polyclonal rabbit antibody against PlaC (generated by BioGenes against recombinantly expressed and purified PlaC-Strep) was added at a dilution of 1:1000. α-Rabbit linked to G-horseradish peroxidase (Sigma) was used as a secondary antibody at a dilution of 1:10,000. Proteins were detected by chemiluminescence (ECL kit, Amersham Biosciences).
Proteolysis of PlaC and Peptide Analysis by Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry (Nano-LC/ESI-MS/MS)
Recombinant purified PlaC-His derived from BL21(pER14) was applied to SDS-PAGE, and the ∼51-kDa protein band was cut out, destained (10% acetic acid, 30% ethanol, 60% water (v/v/v)), rinsed with 100 μl of 50 mm ammonium hydrogen carbonate buffer, and subsequently dehydrated with 100 μl of 100% acetonitrile until the fragment became colorless. Next, the fragment was quickly rinsed with ultrapure water, dried in a SpeedVac, and incubated overnight with 0.6 μg of ProA (purified from L. pneumophila supernatant via AEC) in 40 mm Tris-HCl (pH 7.5), 1 μm ZnCl2. Trypsin proteolysis of ProA-digested PlaC-His and determination of generated PlaC peptides via nano-LC/ESI-MS were carried out according to a published protocol (40). The ProA-specific cleavage sites in PlaC were determined by exact peptide mass analysis and fragmentation of the ionized peptides applying collision-induced fragmentation. Peptide identifications were performed using the Mascot search engine (reference genome sequences, L. pneumophila Corby and 130b). The precursor and fragment mass tolerances were set to 0.002 and 0.5 Da, respectively.
Protease-profiling Microarray
For identification of ProA cleavage sites, a peptide microarray (JPT Peptide Technologies) was used. For that purpose, phosphotyrosine-labeled peptide octamers with five overlapping amino acids from the C-terminal part of L. pneumophila 130b PlaC (amino acids 300–433) were synthesized. In short, the peptide microarray (containing three identical subarrays) was incubated with 350 μl of purified L. pneumophila 130b ProA (1 unit/ml) in 1× TBS with 10 μm ZnCl2 for 15 h at 37 °C. As a negative control, 10 mg/ml BSA was applied to an identical microarray. Next, the microarray slides were washed five times with double distilled water followed by treatment with fluorescence-labeled anti-phosphotyrosine antibody. Finally, fluorescence signal intensities of each spot were determined, and the mean value for signal intensities of the spots with identical peptides for the image of control experiment (A) and from images subsequent to ProA incubation (B) was calculated. The calculated ratio (A/B) displayed cleavage efficiency. The highest values (>1) indicated that the spots displaying the peptides were cleaved most effectively by ProA. The assay procedure was carried out according to the manufacturer's instruction.
RESULTS
Chromatographic Separation of the GCAT-activating Factor
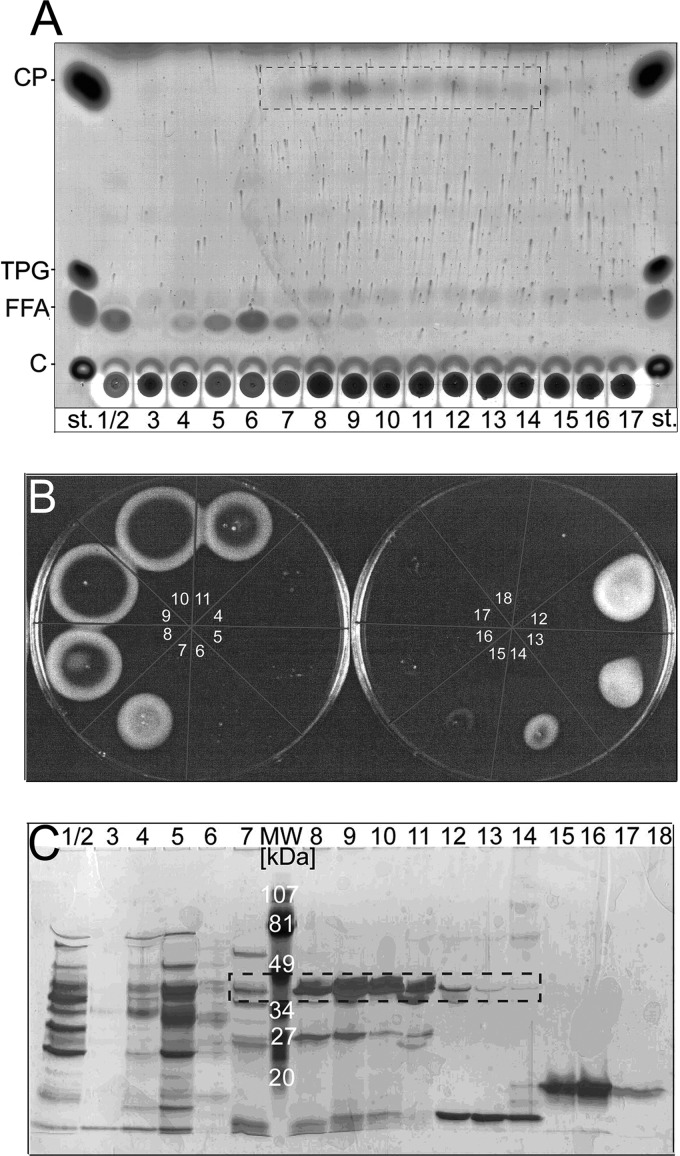
Because L. pneumophila proA knock-out mutant does not transfer palmitic acid to cholesterol and therefore does not exhibit GCAT activity in culture supernatants, it is likely that zinc metalloproteinase ProA directly or indirectly promotes GCAT activation (25). To identify the protein responsible for PlaC GCAT activation, we performed AEC fractionation of L. pneumophila plaC mutant culture supernatant, which contains the activating factor but not PlaC. We subsequently incubated the fractions with cell lysates of an E. coli clone expressing PlaC but not showing GCAT activity without respective activation (25). Fractions 7–14/15 and especially fractions 8 and 9 indeed induced GCAT activity as shown by the formation of cholesterol palmitate (CP) when the mixtures were incubated with DPPG and cholesterol, thereby highlighting the successful fractionation of the GCAT-activating factor (Fig. 1A). Fractions 1–6 and 16–17 did not show CP generation, but instead especially fractions 1/2 and 4–7 showed a marked release of fatty acids, indicating the presence of secreted L. pneumophila PLAs (Fig. 1A) (4, 5, 25). Next, the fractions were tested for protease activity on casein agar, and protease-positive fractions 7–15 were exactly overlapping the fractions inducing cholesterol acylation (Fig. 1B). Reducing SDS-PAGE showed the consistent presence of a prominent ∼38-kDa protein in the fractions of interest especially in fractions 8–11 (Fig. 1C). The 38-kDa protein was N-terminally sequenced, and the sequence “EKVQA” was determined that corresponds to mature ProA starting from amino acid 208 after cleavage of the signal peptide (amino acids 1–24) and the N-terminal propeptide (amino acids 25–207) (41).
FIGURE 1.
Chromatographic separation of an L. pneumophila ∼38-kDa protease as the GCAT-activating factor of PlaC. A, AEC fractions (1–17) from culture supernatant of L. pneumophila 130b plaC mutant were assayed for GCAT activation of plaC-expressing DH5α (pMMB2002 plaC = pMY6). DH5α(pMY6) cell lysates were incubated with the respective AEC fraction, DPPG/cholesterol, and 2 mm ZnCl2 for 20 h at 37 °C. Subsequently, lipids were extracted and subjected to TLC. Accumulation of cholesterol palmitate indicates GCAT activity in fractions 7–14/15 (dashed frame). Results are representative for at least two additional experiments. st., standard; TPG, tripalmitoylglycerol; FFA, free fatty acids; C, cholesterol. B, AEC fractions 4–18 were applied to an agar plate containing casein and incubated for 8–18 h at 37 °C. Precipitation of paracaseinate indicates the presence of proteinase activity in the fractions showing GCAT activation. C, silver-stained reducing SDS-PAGE of AEC fractions showed enrichment of an ∼38-kDa protein in the GCAT-activating fractions. Results are representative of at least one additional experiment.
ProA Directly Activates PlaC GCAT Activity
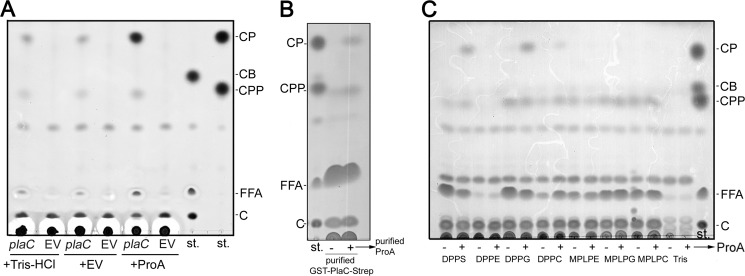
As judged from our biochemical analysis, it was likely that ProA directly activated PlaC GCAT activity. To confirm that observation, L. pneumophila proA and plaC were cloned into vector pet28a(+), resulting in C-terminal His6-tagged gene fusions. Subsequently, the periplasmic fraction of E. coli BL21 expressing proA was incubated together with cell lysates of the plaC-expressing E. coli strain. The plaC-expressing E. coli clones or the empty vector controls alone showed only very minor or no GCAT activity. Only when plaC- and proA-expressing bacterial products were combined was a marked increase in CP generation observed (Fig. 2A). Also, recombinant purified GST-PlaC-Strep led to cholesterol acylation when incubated with purified ProA-His (Fig. 2B). In addition to the CP spot, another compound was formed due to PlaC activity (Fig. 2, A and B). This spot, running to a lower height than CP during TLC analysis, was also mentioned in earlier reports describing L. pneumophila GCAT activity expressed both naturally in L. pneumophila and recombinantly in E. coli. There it was designated “unknown substance” whose development depends on the presence both of cholesterol and PlaC; therefore, it might represent another cholesterol ester (21, 25, 33). We tested several defined cholesterol esters with different acyl chain lengths, such as cholesterol propionate (CPP) and butyrate, for their running behavior and found that the additional compound represents CPP (Fig. 2, A and B). To summarize, we found that PlaC GCAT is directly activated by ProA and that it also transfers fatty acids with a shorter chain length.
FIGURE 2.
L. pneumophila zinc metalloproteinase ProA directly activates PlaC GCAT activity transferring both palmitic and propionic acid to cholesterol and analysis of acyl donor substrate specificities. A, E. coli BL21 cell lysates from clones expressing plaC (BL21(pet28a plaC = pCL14)) or harboring the empty vector (EV) (BL21(pet28a)) were incubated with DPPG/cholesterol and supplemented with Tris-HCl as the buffer control, the periplasmic fraction of BL21(pet28a) as the empty vector control, or the protease-active periplasmic fraction of E. coli expressing proA (BL21(pet28a proA = pCL15)). B, recombinantly expressed and purified GST-PlaC-Strep was incubated with DPPG/cholesterol and supplemented with Tris-HCl or recombinantly expressed and purified ProA-His. C, for determination of donor substrate specificities of GCAT activity, L. pneumophila Corby proA mutant 10-fold-concentrated late logarithmic phase culture supernatant was incubated with different diacyl- and lysophospholipids and cholesterol and supplemented either with Tris-HCl or protease-active periplasmic fractions of BL21(pet28a proA = pCL15). After 3 (B) or 16 h (A and C) of incubation, lipids were extracted and analyzed for the formation of cholesterol esters by means of TLC. All results shown are representative of at least two additional experiments. st., standard; CB, cholesterol butyrate; C, cholesterol; MPLPC, 1-monopalmitoyllysophosphatidylcholine; MPLPG, 1-monopalmitoyllysophosphatidylglycerol; MPLPE, monopalmitoyllysophosphatidylethanolamine; DPPS, dipalmitoylphosphatidylserine; DPPE, dipalmitoylphosphatidylethanolamine.
Acyl Donor Substrate Specificities of L. pneumophila GCAT
To assess the nature of the phospholipid acyl donor for L. pneumophila GCAT activity and its dependence upon activation by ProA, we monitored CP generation after incubation of L. pneumophila Corby proA mutant culture supernatant (containing secreted not GCAT-activated PlaC) with cholesterol and different diacyl- and lysophospholipids, such as dipalmitoylphosphatidylserine, dipalmitoylphosphatidylethanolamine, DPPG, DPPC, monopalmitoyllysophosphatidylethanolamine, 1-monopalmitoyllysophosphatidylglycerol, and 1-monopalmitoyllysophosphatidylcholine. Here we show that only incubations with diacylphospholipids dipalmitoylphosphatidylserine and DPPG as well as to a lesser extent with DPPC and concomitant addition of recombinant ProA developed CP (Fig. 2C). This means that L. pneumophila GCAT activity preferentially transfers palmitic acid from dipalmitoylphosphatidylserine, DPPG, and DPPC to cholesterol, and it depends on enzyme activation by ProA. In contrast, development of CPP was observed for a variety of diacyl- and lysophospholipids and did not seem to be strictly dependent on ProA. This therefore suggests that enzymes in addition to PlaC may be responsible for transferring fatty acids with shorter chain lengths (Fig. 2C). We also performed experiments with 16:0 18:1 diacylphosphatidylglycerol and 16:0 18:1 diacylphosphatidylcholine as well as Acanthamoeba and U937 cell lysates (the latter two without external sterol addition) and observed that ProA-activated PlaC uses a variety of acyl lipid donors for sterol acylation but not unesterified fatty acids, such as propionic and palmitic acids (data not shown).
PlaC Is the Major L. pneumophila GCAT Transferring Palmitic Acid to Both Cholesterol and Ergosterol, and Together with PlaA and PlaD It Transfers Short Chain Fatty Acid to Sterols
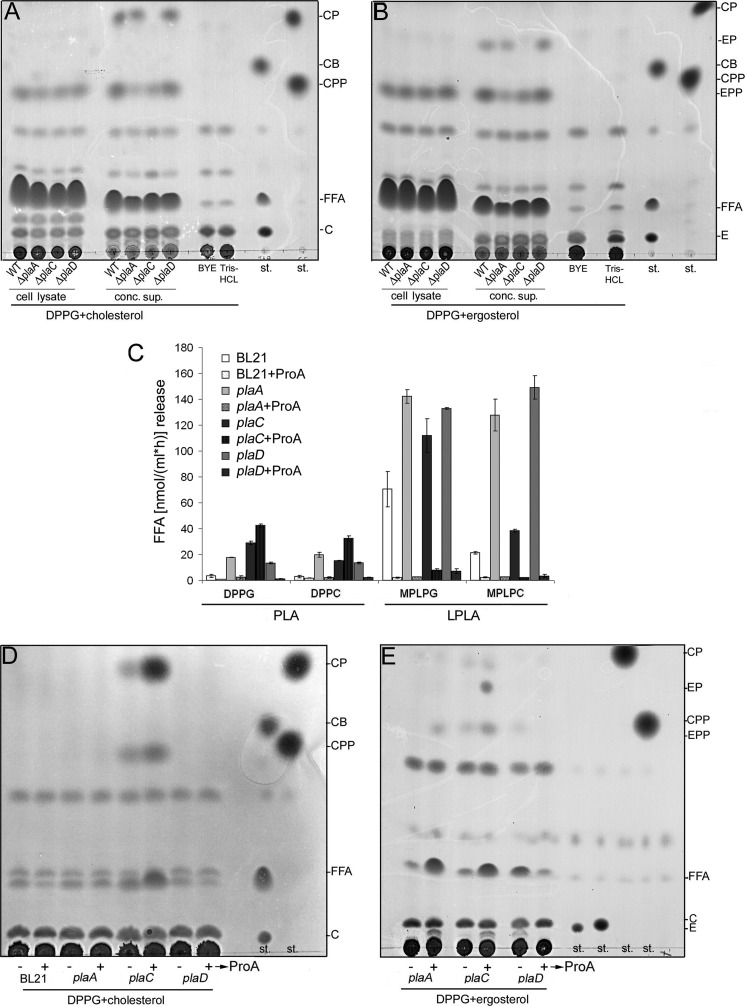
It has been shown earlier that secreted GCAT activity of a L. pneumophila plaC knock-out mutant was negligible with respect to CP generation and that plaA does not contribute to that activity (25). Here we examined the three L. pneumophila single GDSL enzyme knock-out mutants for their GCAT activity with respect to CP and CPP development. To that end, we incubated cell lysates and concentrated culture supernatants of wild type L. pneumophila and single GDSL enzyme knock-out mutants with DPPG and cholesterol. Fig. 3A confirms that PlaC is the major secreted GCAT inducing CP production, whereas PlaA and PlaD do not contribute to this. However, because both plaA and plaC mutant culture supernatants showed reduced generation of CPP, we conclude that PlaA and PlaC transfer short fatty acids (such as propionic acid) to cholesterol (Fig. 3A). Interestingly, generation of CPP was also induced by cell lysates of the wild type and all mutants. Therefore, L. pneumophila may contain another cell-associated enzyme, different from the three GDSL enzymes introduced here, that promotes cholesterol acylation.
FIGURE 3.
PlaC is the major L. pneumophila GCAT transferring palmitic acid to both cholesterol and ergosterol, and together with PlaA and PlaD it transfers propionic acid to sterols. Formation of different cholesterol (A) or ergosterol esters (B) by late logarithmic phase cell lysates and 8-fold-concentrated culture supernatants from L. pneumophila Corby wild type versus single GDSL protein knock-out mutants was studied after 16-h incubation at 37 °C with DPPG/sterol, lipid extraction, and TLC. Cell lysates of E. coli clones expressing plaA, plaC, and plaD (BL21(pBADTOPO plaA = pER13), BL21(pet28a plaC = pCL14), and BL21(pET160 plaD = pER27)) were incubated with DPPG, DPPC, 1-monopalmitoyllysophosphatidylglycerol (MPLPG), and 1-monopalmitoyllysophosphatidylcholine (MPLPC) with Tris-HCl or the periplasmic fraction of BL21(pet28a proA = pCL15) to determine PLA/LPLA activities via quantification of FFA (C). The results represent the means and standard deviations of duplicate cultures and three reactions each and are representative of three independent experiments. The E. coli preparations were further incubated with DPPG/cholesterol (D) or DPPG/ergosterol (E) with Tris-HCl or the periplasmic fraction of BL21(pet28a proA = pCL15) for 16 h, and then lipids were extracted and analyzed for sterol ester formation by means of TLC. All results shown are representative of at least one additional experiment. st., standard; CB, cholesterol butyrate; C, cholesterol; E, ergosterol; BYE, buffered yeast extract; EP, ergosterol palmitate; EPP, ergosterol propionate.
L. pneumophila is predominantly an environmental bacterium that successfully replicates in protozoa, such as amoebae. Because amoebae do not contain cholesterol but instead a variety of sterol species, including ergosterol (4, 25, 42–45), we were interested to know whether the Legionella GDSL enzymes acylate this potential acceptor lipid. We therefore incubated cell lysates and concentrated culture supernatants of L. pneumophila wild type and single GDSL enzyme knock-out mutants with DPPG and ergosterol. A result very similar to the above described cholesterol acylation was observed here; i.e. for the wild type supernatants, a compound spot that likely represents ergosterol palmitate and a spot likely representing ergosterol propionate were detected. Only the plaC mutant culture supernatant exhibited severely reduced production of ergosterol palmitate, and the plaA and plaC mutants exhibited reduced development of ergosterol propionate (Fig. 3B). The cell lysates of all tested strains showed the presence of a so far unknown cell-associated activity responsible for esterification of sterols (both ergosterol and cholesterol) (Fig. 3B).
To confirm the mutant studies and to get further insight into the L. pneumophila GDSL enzyme activities, plaA, plaC, and plaD were expressed in E. coli. All three genes were found to be expressed as shown by the presence of PLA/LPLA activities with and without addition of recombinant ProA (Fig. 3C). Recombinantly expressed PlaA and PlaD showed some PLA activity but major LPLA activity as already known for PlaA (21). For both enzymes, PLA and LPLA activities were reduced by ProA addition. Mainly, LPLA activity and some PLA activity were also found to be associated with the clone expressing plaC. As shown before, PlaC-dependent PLA activity but not LPLA activity increased by concomitant incubation with ProA (Fig. 3C) (25). Next, clones expressing the three genes were tested for GCAT activity. PlaC possessed activity for transfer of palmitic acid and propionic acid to both cholesterol and ergosterol that was activated by ProA (Fig. 3, D and E). The generation of sterol palmitate was not observed for plaA- and plaD-expressing clones; however, both induced generation of ergosterol propionate. Interestingly, in the case of PlaA, this ester was only formed with ProA, and in the case of PlaD, a weak spot of ergosterol propionate was observed only without ProA incubation (Fig. 3E). From those experiments, we conclude that not only PlaC but also PlaA and PlaD may transfer fatty acids to host cell sterols, and therefore all three enzymes possess acyltransferase activity. In this context, it seems that PlaA and PlaD may have a preference for the transfer of short chain fatty acids, such as propionic acid, whereas PlaC in addition transfers long chain fatty acids, such as palmitic acid.
PlaC Processing Depends on ProA
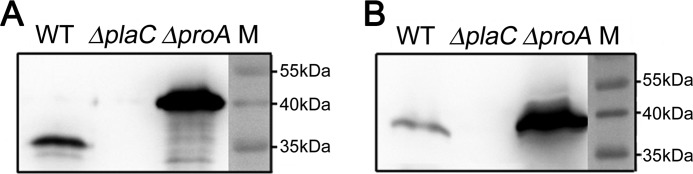
We have shown that ProA directly activates PlaC GCAT activity. Now it was of interest to determine whether PlaC is indeed processed by ProA. Therefore we analyzed the protein size of PlaC present in the culture supernatant of L. pneumophila wild type versus a proA mutant. Whereas in the wild type a protein of about 36 kDa was detected in the culture supernatant after reducing SDS-PAGE and Western blotting, in a proA mutant, an approximately 40–45-kDa protein was found (Fig. 4A). The unprocessed protein present in the proA mutant likely represents PlaC without a signal peptide (calculated mass, 47 kDa). To further evaluate the nature of processing, the PlaC protein sizes were compared under non-reducing SDS-PAGE. Here no considerable difference in the protein size was observed (Fig. 4B). It was further found that after overexpression of PlaC-Strep in L. pneumophila wild type and affinity extraction of PlaC-Strep from the culture supernatant protein bands of ∼38–39 and ∼8–9 kDa, corresponding to processing at about amino acids 360–370, were detected after reducing SDS-PAGE, whereas only the full-length 47-kDa protein was found in a proA mutant (data not shown). This suggests processing within a disulfide loop where a disulfide bond still keeps the separated protein chains connected. PlaC contains four cysteine residues in its C-terminal half, and those might form disulfide loops involved in GCAT activation. But before clarifying the importance of cysteines, we aimed to characterize essential catalytic amino acids.
FIGURE 4.
Secreted PlaC is processed by ProA in L. pneumophila. 10-fold-concentrated culture supernatants of L. pneumophila Corby wild type, plaC mutant, and proA mutant were applied to reducing (360 mm β-mercaptoethanol) (A) and non-reducing (B) SDS-PAGE, Western blot analysis, and detection with an α-PlaC antibody. Results are representative of at least one additional experiment. WT, wild type; M, molecular weight marker.
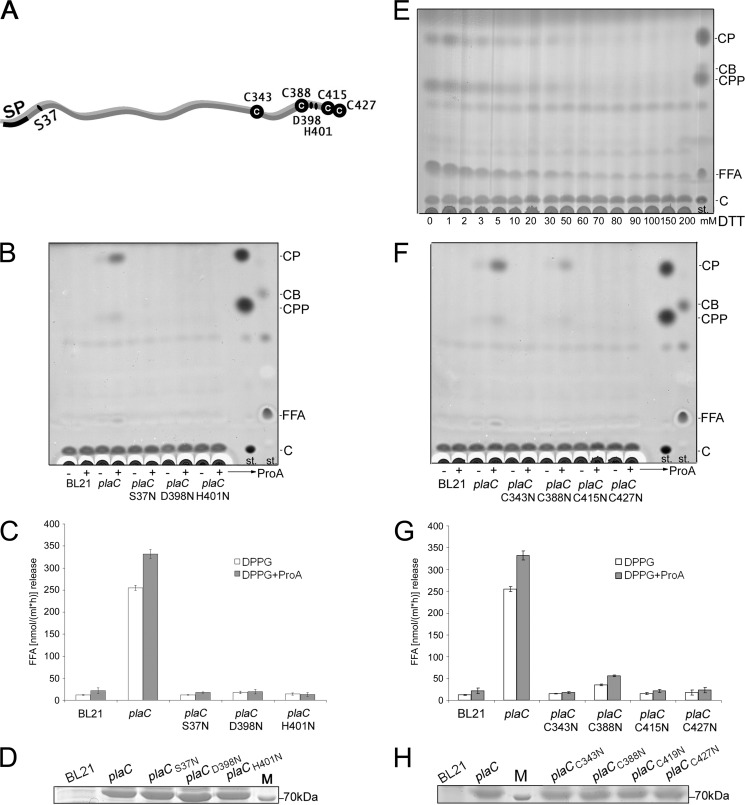
The Individual Members of the Putative Catalytic Triad Ser-37/Asp-398/His-401 Are Essential for the PlaC-associated GCAT and PLA Activities
Amino acid homology to A. hydrophila SatA and the presence of defined protein regions characteristic of GDSL enzymes and SGNH hydrolases within PlaC suggested the importance of Ser-37, Asp-398, and His-401 for enzymatic activity (Figs. 5 and 6A) (15, 19, 46, 47). To determine the significance of these three amino acids for PlaC PLA and GCAT activities, point mutations (S37N, D398N, and H401N) were introduced in GST-PlaC (Table 1). The constructs were expressed in E. coli and tested for the above mentioned activities. Fig. 6B shows that only the wild type but not the mutated construct possesses GCAT activity. PLA activity of the mutants was severely reduced compared with the wild type using DPPG as substrate (Fig. 6C), although the proteins were expressed at similar levels (Fig. 6D). Our data verify that Ser-37, Asp-398, and His-401 likely constitute the catalytic triad and that PlaC possesses identical catalytic residues for both GCAT and PLA activities.
FIGURE 5.
Conserved domains and catalytic triad (bold) of L. pneumophila Corby GDSL enzymes PlaA, PlaC, and PlaD compared with SatA of A. salmonicida. The digits highlight the number of amino acids before and after the conserved regions.
FIGURE 6.
The individual members of the putative catalytic triad Ser-37/Asp-398/His-401 and cysteines Cys-343, Cys-388, Cys-415, and Cys-427 are essential for the PlaC-associated GCAT and PLA activities. The localization of the three catalytic amino acids, the four cysteine residues, and the predicted signal peptide (SP) of L. pneumophila PlaC is depicted in A. Cell lysates of E. coli clones expressing plaC (BL21(pGEX plaC = pER32)); the catalytic triad mutants (BL21(pGEX plaCS37N = pCL70), BL21(pGEX plaCD398N = pCL71), and BL21(pGEX plaCH401N = pCL72)); or C343N, C388N, C415N, and C427N mutants (BL21(pGEX plaCC343N = pCL73), BL21(pGEX plaCC388N = pCL74), BL21(pGEX plaCC415N = pCL75), and BL21(pGEX plaCC427N = pCL76)) were incubated with DPPG/cholesterol with Tris-HCl or the periplasmic fraction of BL21(pet28a proA = pCL15) for 3 h, and then lipids were extracted and analyzed for sterol ester formation by means of TLC (B and F). The E. coli preparations were further incubated solely with DPPG supplemented with Tris-HCl or the periplasmic fraction of BL21(pet28a proA = pCL15) to determine PLA/LPLA activities via quantification of FFA (C and G). The results represent the means and standard deviations of duplicate cultures and three reactions each and are representative of three independent experiments. Comparable expression of the different plaC constructs was shown by detection of the overexpressed 74-kDa GST-PlaC protein in reducing SDS-PAGE and Coomassie Blue staining (D and H). Cell lysates of BL21(pGEX plaC = pER32) were incubated with DPPG/cholesterol supplemented with the periplasmic fraction of BL21(pet28a proA = pCL15) and with different concentrations of the reducing agent DTT for 3 h. Subsequently, the lipids were extracted and analyzed for sterol ester formation by means of TLC (E). All results shown are representative of at least one additional experiment. st., standard; CB, cholesterol butyrate; C, cholesterol, M, molecular weight marker.
Cysteines Cys-343, Cys-388, Cys-415, and Cys-427 Are Essential for PlaC-associated PLA and GCAT Activities
It has been shown that the GCAT activity of A. salmonicida SatA requires activation by a bacterial serine proteinase (48). Additionally, it was published that trypsin is able to activate A. hydrophila SatA GCAT expressed in A. salmonicida by means of processing the protein between two cysteines (Cys-225 and Cys-281), forming a disulfide bridge (49). L. pneumophila PlaC contains four cysteines in the C-terminal region that may form one or two disulfide bridges and thereby may assure integrity of the catalytic triad after possible proteolytic cleavage (Fig. 6A). Indeed, the program DiANNA predicted disulfide bridge formation (a) between the PlaC residues Cys-343 and Cys-388 and (b) the residues Cys-415 and Cys-427 (50). Insulin, chymotrypsinogen, prothrombin, and diverse bacterial toxins like the diphtheria toxin are good examples of enzymes that undergo processing and concomitant activation in between their disulfide bridge-forming residues (51–55) Therefore, to get insight into the importance of the four cysteine residues (Cys,343, Cys,388, Cys,415, and Cys,427), we incubated the wild type plaC-expressing clone with increasing amounts of the reducing agent dithiothreitol (DTT) and observed that GCAT activity for transfer of both palmitic and propionic acid was inhibited by 50 mm DTT (Fig. 6E). Additionally, the four cysteines were individually mutated in GST-PlaC and expressed in E. coli. Fig. 6, F, G, and H, illustrate that after mutation of Cys-343, Cys-388, Cys-415, and Cys-427, although expressed at a similar level, both GCAT and PLA activities were severely reduced or even completely lost as compared with that of the wild type construct. Collectively, these data imply that disulfide bridge formation between Cys-343 and Cys-388 as well as Cys-415 and Cys-427 may be essential for GCAT activity.
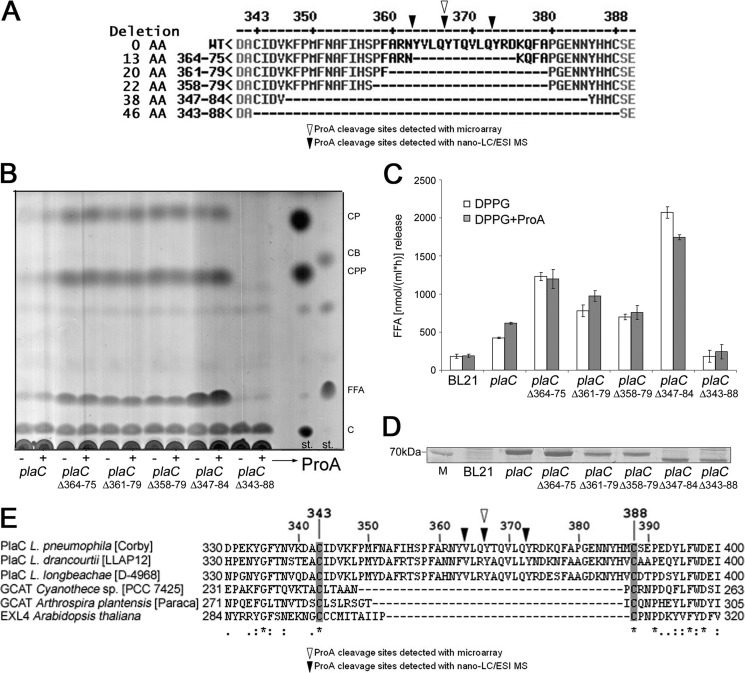
Determination of ProA-specific Cleavage Sites in PlaC: Disulfide Loop Reduction/Deletion Independently of ProA Activates PlaC-dependent PLA and GCAT Activities
Three different experimental strategies were followed to identify the potential ProA cleavage site within PlaC. Strategy 1 used an array of phosphotyrosine-labeled overlapping peptides of 8 amino acids, representing the last 134 amino acids (amino acids 300–433) of PlaC. Cleavage of a specific peptide was detected by comparison of spot intensities from identical peptides on chips incubated with ProA or BSA, respectively. One unambiguous cleavage site was found between Gln-366 and Tyr-367 located within the potential loop region between Cys-343 and Cys-388 (Fig. 7A). An additional, not clearly defined, potential cleavage site was found between the amino acids Phe-315 and Asn-319. Processing at this site, i.e. in front of the potentially disulfide bridge-forming cysteines, however, would separate the three members of the catalytic triad. Because we have shown that the lack of even one of the triad renders the enzyme inactive, we attribute the cleavage rather to the nature of peptides used in the assay and therefore did not include it for further analysis. No cleavage was observed in the region between Cys-415 and Cys-427. Strategy 2 purified denatured PlaC, which was digested with ProA and trypsin, and the resulting peptides were mapped by means of LC/ESI-MS. Here three major cleavage sites within the potential Cys-343 and Cys-388 loop, namely between Tyr-363 and Val-364, Gln-366 and Tyr-367, and Gln-372 and Tyr-373, were detected (Fig. 7A). The second proposed cleavage site (Gln-366 and Tyr-367) was also found by strategy 1. Additional cleavage was observed between Asp-314 and Phe-315, Phe-315 and Leu-316, Leu-316 and Phe-317, and Phe-317 and Val-318. This cleavage region matches a region also identified in strategy 1, but the site was not further analyzed due to the expected inactivation of the enzyme. Because several potential cleavage sites were observed for the region between Cys-343 and Cys-388 but no potential cleavage sites were observed between Cys-415 and Cys-427, we focused on a more detailed analysis of the first region. In strategy 3, five single (V364G, Q366A, Y367F, Y367W, and Y367A), five triple (P358A/F359A, R361A/N362A/Y363A, V364A/L365A/Q366A, Y367A/T368A/Q369A, and Y373A/R374A/D375A), and five double triple (P358A/F359A + Y367A/T368A/Q369A, P358A/F359A+ V364A/L365A/Q366A, R361A/N362A/Y363A + Y367A/T368A/Q369A, V364A/L365A/Q366A + Y373A/R374A/D375A, Y367A/T368A/Q369A + Y373A/R374A/D375A) amino acid changes especially in the potential cleavage sites were applied to the region between Cys-343 and Cys-388; however, none of the PlaC mutants showed defective GCAT activation (data not shown). We then introduced deletions of different lengths within the loop structure, i.e. from amino acids 364 to 375, 361 to 379, 358 to 379, 347 to 384, and 343 to 388 (Fig. 7A). Except the latter deletion, all others showed a very interesting enzymatic phenotype: instead of the expected loss of ProA-induced activation of GCAT, these constructs possessed extraordinarily high GCAT and PLA activities even without activation by ProA (Fig. 7, B and C). GCAT activation of those deletion mutants was completely independent of ProA. This was observed despite the detection of comparable or even lower amounts of the respective proteins (Fig. 7D). This implies that reduction/deletion of the potential loop region, for example via proteolytic cleavage at several sites, rather than cleavage at a single specific site induces GCAT activity.
FIGURE 7.
Disulfide loop deletion independently of ProA activates PlaC-dependent PLA and GCAT activities. An overview of the PlaC region between amino acids 341 and 390 highlighting the potential ProA cleavage sites as identified by means of the peptide cleavage microarray and nano-LC/ESI-MS as well as PlaC loop-deletion mutants is shown (A). Cell lysates of E. coli clones expressing plaC (BL21(pGEX plaC = pER32)) and/or mutants (BL21(pGEX plaCΔ364–75 = pCL77), BL21(pGEX plaCΔ361–79 = pCL78), BL21(pGEX plaCΔ358–79 = pCL79), BL21(pGEX plaCΔ347–84 = pCL85, and BL21(pGEX plaCΔ343–88 = pCL80)) were incubated with DPPG/cholesterol supplemented with Tris-HCl or the periplasmic fraction of BL21(pet28a proA = pCL15) for 3 h, and then lipids were extracted and analyzed for sterol ester formation by means of TLC (B). The E. coli preparations were further incubated solely with DPPG supplemented with Tris-HCl or the periplasmic fraction of BL21(pet28a proA = pCL15) to determine PLA/LPLA activities via quantification of FFA (C). The results represent the means and standard deviations of duplicate cultures and three reactions each and are representative of three independent experiments. Comparable expression of the different plaC constructs was shown by detection of the overexpressed 74-kDa GST-PlaC proteins in reducing SDS-PAGE and Coomassie Blue staining (D). The disulfide loop region (PlaC amino acids 330–400) ClustalW2 multiple alignment of different PlaC-like GDSL proteins of Legionella ssp., different cyanobacterial species, and A. thaliana extracellular lipase EXL4 highlighting the potential ProA cleavage sites is shown (E). st., standard; CB, cholesterol butyrate; C, cholesterol; M, molecular weight marker; AA, amino acids; * indicates positions which have a single, fully conserved residue, : indicates conservation between groups of strongly similar properties, and . indicates conservation between groups of weakly similar properties.
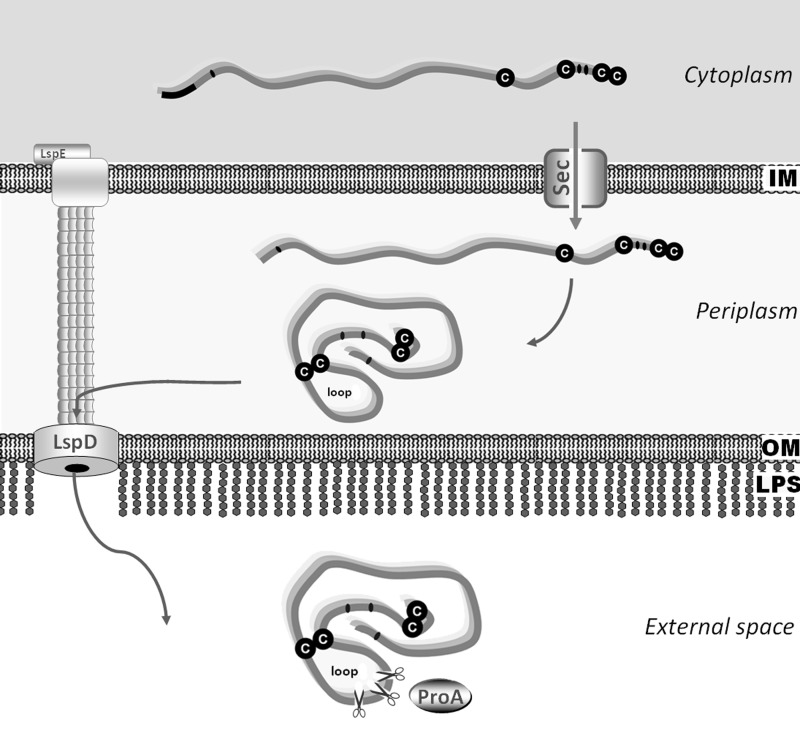
When an alignment of the L. pneumophila PlaC Cys-343/Cys-388 loop region with homologous proteins was performed, Legionella ssp. PlaC-like GDSL proteins were found to contain similar regions, whereas several cyanobacterial GDSL proteins and Arabidopsis thaliana extracellular lipase EXL4 lack the domain (Fig. 7E). This suggests that Legionella bacteria might possess specific means for postexport activation of GCAT and PLA activities by the described ProA-dependent mechanism (Fig. 8).
FIGURE 8.
Proposed model for ProA-dependent activation of L. pneumophila PlaC GCAT activity. Within the cytoplasm, PlaC and ProA are present in their inactive preproform. After transport of PlaC and ProA through the inner bacterial membrane via the Sec system and concomitant signal peptide cleavage, ProA is present in its inactive proform. In the periplasm, a disulfide bridge(s) may be formed within PlaC, which is then still found in its GCAT-inactive proform. After subsequent export of ProA and PlaC via the Lsp secretion system, ProA acquires its mature active form via autoactivation by means of N-terminal propeptide cleavage. PlaC is then processed and thereby activated by mature ProA and subsequently shows GCAT and full PLA activity. IM, inner bacterial membrane; OM, outer bacterial membrane; LPS, lipopolysaccharide.
DISCUSSION
We postulate a novel enzyme activation mechanism by which cells may control a per se toxic enzymatic activity until it is released from the cell (Fig. 8). Such a mechanism might be of extraordinary importance to prevent damage of Legionella via its PlaC-dependent PLA activity that targets phospholipids typically contained in prokaryotes, such as phosphatidylglycerol or as shown for some intracellular bacteria phosphatidylcholine (56–58). Although the here described ProA-dependent increase in PlaC-derived PLA after expression in E. coli was not as prominent as seen for GCAT, we showed earlier that secreted PLA is reduced by two-thirds in a L. pneumophila proA knock-out mutant compared with the wild type (20).
We have now mechanistically elucidated GCAT activation of a bacterial GDSL protein by a bacterial metalloproteinase. So far, only bacterial serine proteinase-dependent activation in A. salmonicida SatA and eukaryotic RhoA-dependent activation for Salmonella enterica SseJ have been described (48, 49, 59–61).
The L. pneumophila zinc metalloproteinase ProA fulfils a variety of functions that may trigger bacterial pathogenicity. It degrades host extracellular matrix and immunity proteins, supports Hartmanella vermiformis amoebae and guinea pig infection, and is indeed found in the L. pneumophila phagosome (62–64). Here we showed that the enzyme also regulates lipolytic activities of the pathogen that might contribute to the observed loss of hemolytic activities in a L. pneumophila proA knock-out mutant (65, 66).
As shown in the case of PlaC GCAT activation, disulfide loop reduction/deletion rather than cleavage at a single site as a precondition for enzyme activation to our knowledge has not yet been described. There are other examples of eukaryotic and bacterial enzymes where proteolytic processing within a protein loop structure and often subsequent release of individual protein domains result in increased activity. For example, RipA, an endopeptidase of Mycobacterium tuberculosis, is expressed as a zymogen and activated via proteolytic processing of an exposed loop connecting the prodomain with the catalytic domain, resulting in the release of the catalytic domain (67). In other cases, such as Pseudomonas exotoxin A, enzyme activation is triggered by proteolytic cleavage at Arg-279 between two disulfide bridge-forming cysteines, and after a following reduction of the disulfide bridge, the activity domain is released (55, 68). Several bacterial toxins, including diphtheria toxin, Shiga toxin, cholera toxin, and E. coli heat-labile toxin, require activation by bacterial or host proteinases (55, 69–71). For example, the C-terminal region of Shiga toxin subunit A contains two disulfide bridge-linked cysteines forming a loop that contains the trypsin and furin proteinase recognition sequence Arg-Val-Ala-Arg (72). Cleavage at a single site results in enzymatically active disulfide bridge-linked A1 and A2 fragments (70).
Conformational changes associated with loop cleavage seem important for the regulation of enzyme activity and substrate binding. In some cases, such as RipA of M. tuberculosis, the loop binds tightly to the catalytic cleft, for example by means of several hydrogen bridges (67). It is possible that reduction/deletion of the possible Cys-343/Cys-388 loop structure in PlaC may eliminate a sterically inhibiting structure and then results in an “open gate” conformation for more efficient fatty acid substrate donor and acceptor binding. Because a reduced level of PLA and full LPLA activity was observed before ProA activation (Fig. 3C), we speculate that the more voluminous diacylphospholipid substrates might get better access to the substrate binding or catalytic domains after loop reduction/removal and that access to the acceptor molecule might only be possible after proteolytic processing. Interestingly, similar PlaC Cys-343/Cys-388 loop protein sequences were only found in PlaC-like Legionella enzymes, suggesting that the protein region targeted by ProA for GCAT activation is perhaps genus-specific. However, as our own experiments suggest, the mechanism of activation by loop reduction/deletion may be more universal. A further possible target of a similar activation is L. pneumophila PlaA acyltransferase whose activation here was shown to be ProA-dependent (Fig. 3E). Importantly, the localization of potentially loop-forming cysteine residues in many other GDSL enzymes, including PlaA, is conserved as they are generally located between homology block IV and V.3 Even more intriguing, we found several GDSL enzymes of different cyanobacteria annotated as phosphatidylcholine-sterol O-acyltransferases and plant enzymes, such as the extracellular A. thaliana GDSL esterase/lipase EXL4, that only showed a very small protein region of about 8–10 amino acids between the potentially disulfide bridge-forming cysteines (Fig. 7E), suggesting that those enzymes may be active even without proteinase activation.
Here we found that the three potential catalytic amino acids of PlaC, namely Ser-37, Asp-396, and His-401, were all required for both PLA and GCAT activities. The three catalytic amino acids are conserved in the three L. pneumophila GDSL enzymes and are strictly conserved in other GDSL enzymes, suggesting that all enzymes of this family additionally possess acyltransferase activity. Indeed, here we showed that, in addition to PlaC, PlaA and PlaD also contribute to sterol acylation (Fig. 3). The latter two were not able to transfer palmitic acid but rather short chain fatty acids. Although the origin of the short chain fatty acid, especially propionic acid, substrates is not clear, they might be a minor constituent of the DPPG substrate used.
PlaC, PlaA, and to some extent PlaD acylate ergosterol, a typical membrane lipid of protozoa (including amoebae), fungi, and microalgae (Fig. 3, B and E) (43–45). Free living amoebae inhabiting aquatic systems are the predominant host for Legionella intracellular growth in natural environments (73). Ergosterol substitutes for mammalian cholesterol as a major sterol in many protozoan/amoebal cells, including Acanthamoeba (42, 44). Sterols are essential components of the membranes modulating membrane fluidity, signal transduction, and activity of membrane-bound enzymes (45). Therefore, acylation of sterols present not only in protozoans but also in the accidental human host provides Legionella with an important tool for host cell manipulation. However, specific processes that are targeted by the L. pneumophila GCATs remain to be established in the future.
Acknowledgments
We thank Stefan Stevanovic and Anne Müller for protein identification via mass spectrometry and Mayte Bewersdorff for construction of pMY6. We further acknowledge Karan Kaval for critical reading of the manuscript.
This work was supported by German Research Foundation (Deutsche Forschungsgemeinschaft) Grant DFG FL 359/6-1.
C. Lang and A. Flieger, unpublished observation.
- PLA
- phospholipase A
- LPLA
- lysophospholipase A
- GCAT
- glycerophospholipid:cholesterol acyltransferase
- DPPG
- 1,2-dipalmitoylphosphatidylglycerol
- DPPC
- 1,2-dipalmitoylphosphatidylcholine
- AEC
- anion exchange chromatography
- ESI
- electrospray ionization
- CP
- cholesterol palmitate
- CPP
- cholesterol propionate.
REFERENCES
- 1. Fields B. S. (1996) The molecular ecology of legionellae. Trends Microbiol. 4, 286–290 [DOI] [PubMed] [Google Scholar]
- 2. Hilbi H., Jarraud S., Hartland E., Buchrieser C. (2010) Update on Legionnaires' disease: pathogenesis, epidemiology, detection and control. Mol. Microbiol. 76, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winn W. C., Jr., Myerowitz R. L. (1981) The pathology of the Legionella pneumonias. A review of 74 cases and the literature. Hum. Pathol. 12, 401–422 [DOI] [PubMed] [Google Scholar]
- 4. Banerji S., Aurass P., Flieger A. (2008) The manifold phospholipases A of Legionella pneumophila—identification, export, regulation, and their link to bacterial virulence. Int. J. Med. Microbiol. 298, 169–181 [DOI] [PubMed] [Google Scholar]
- 5. Lang C., Flieger A. (2011) Characterisation of Legionella pneumophila phospholipases and their impact on host cells. Eur. J. Cell Biol. 90, 903–912 [DOI] [PubMed] [Google Scholar]
- 6. Holk A., Rietz S., Zahn M., Quader H., Scherer G. F. (2002) Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 130, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banerji S., Flieger A. (2004) Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150, 522–525 [DOI] [PubMed] [Google Scholar]
- 8. Bender J., Rydzewski K., Broich M., Schunder E., Heuner K., Flieger A. (2009) Phospholipase PlaB of Legionella pneumophila represents a novel lipase family: protein residues essential for lipolytic activity, substrate specificity, and hemolysis. J. Biol. Chem. 284, 27185–27194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buckley J. T., Halasa L. N., Lund K. D., MacIntyre S. (1981) Purification and some properties of the hemolytic toxin aerolysin. Can. J. Biochem. 59, 430–435 [DOI] [PubMed] [Google Scholar]
- 10. Cazalet C., Rusniok C., Brüggemann H., Zidane N., Magnier A., Ma L., Tichit M., Jarraud S., Bouchier C., Vandenesch F., Kunst F., Etienne J., Glaser P., Buchrieser C. (2004) Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 11. Chien M., Morozova I., Shi S., Sheng H., Chen J., Gomez S. M., Asamani G., Hill K., Nuara J., Feder M., Rineer J., Greenberg J. J., Steshenko V., Park S. H., Zhao B., Teplitskaya E., Edwards J. R., Pampou S., Georghiou A., Chou I. C., Iannuccilli W., Ulz M. E., Kim D. H., Geringer-Sameth A., Goldsberry C., Morozov P., Fischer S. G., Segal G., Qu X., Rzhetsky A., Zhang P., Cayanis E., De Jong P. J., Ju J., Kalachikov S., Shuman H. A., Russo J. J. (2004) The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305, 1966–1968 [DOI] [PubMed] [Google Scholar]
- 12. D'Auria G., Jiménez-Hernández N., Peris-Bondia F., Moya A., Latorre A. (2010) Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glöckner G., Albert-Weissenberger C., Weinmann E., Jacobi S., Schunder E., Steinert M., Hacker J., Heuner K. (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298, 411–428 [DOI] [PubMed] [Google Scholar]
- 14. Hilton S., Buckley J. T. (1991) Action of a microbial lipase/acyltransferase on phospholipid monolayers. Biochemistry 30, 6070–6074 [DOI] [PubMed] [Google Scholar]
- 15. Hilton S., Buckley J. T. (1991) Studies on the reaction mechanism of a microbial lipase/acyltransferase using chemical modification and site-directed mutagenesis. J. Biol. Chem. 266, 997–1000 [PubMed] [Google Scholar]
- 16. Brick D. J., Brumlik M. J., Buckley J. T., Cao J. X., Davies P. C., Misra S., Tranbarger T. J., Upton C. (1995) A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett. 377, 475–480 [DOI] [PubMed] [Google Scholar]
- 17. Upton C., Buckley J. T. (1995) A new family of lipolytic enzymes? Trends Biochem. Sci. 20, 178–179 [DOI] [PubMed] [Google Scholar]
- 18. Mølgaard A., Kauppinen S., Larsen S. (2000) Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 8, 373–383 [DOI] [PubMed] [Google Scholar]
- 19. Arpigny J. L., Jaeger K. E. (1999) Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343, 177–183 [PMC free article] [PubMed] [Google Scholar]
- 20. Flieger A., Gong S., Faigle M., Stevanovic S., Cianciotto N. P., Neumeister B. (2001) Novel lysophospholipase A secreted by Legionella pneumophila. J. Bacteriol. 183, 2121–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flieger A., Neumeister B., Cianciotto N. P. (2002) Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine. Infect. Immun. 70, 6094–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DebRoy S., Dao J., Söderberg M., Rossier O., Cianciotto N. P. (2006) Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U.S.A. 103, 19146–19151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossier O., Starkenburg S. R., Cianciotto N. P. (2004) Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72, 310–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Creasey E. A., Isberg R. R. (2012) The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc. Natl. Acad. Sci. U.S.A. 109, 3481–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banerji S., Bewersdorff M., Hermes B., Cianciotto N. P., Flieger A. (2005) Characterization of the major secreted zinc metalloprotease-dependent glycerophospholipid:cholesterol acyltransferase, PlaC, of Legionella pneumophila. Infect. Immun. 73, 2899–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hales L. M., Shuman H. A. (1999) Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect. Immun. 67, 3662–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jepras R. I., Fitzgeorge R. B., Baskerville A. (1985) A comparison of virulence of two strains of Legionella pneumophila based on experimental aerosol infection of guinea-pigs. J. Hyg. 95, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bertani G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edelstein P. H. (1981) Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14, 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moffat J. F., Black W. J., Tompkins L. S. (1994) Further molecular characterization of the cloned Legionella pneumophila zinc metalloprotease. Infect. Immun. 62, 751–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stone B. J., Kwaik Y. A. (1999) Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181, 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Flieger A., Rydzewski K., Banerji S., Broich M., Heuner K. (2004) Cloning and characterization of the gene encoding the major cell-associated phospholipase A of Legionella pneumophila, plaB, exhibiting hemolytic activity. Infect. Immun. 72, 2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Broich M., Rydzewski K., McNealy T. L., Marre R., Flieger A. (2006) The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32. J. Bacteriol. 188, 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Conlan J. W., Baskerville A., Ashworth L. A. (1986) Separation of Legionella pneumophila proteases and purification of a protease which produces lesions like those of Legionnaires' disease in guinea pig lung. J. Gen. Microbiol. 132, 1565–1574 [DOI] [PubMed] [Google Scholar]
- 35. Prestidge L., Gage V., Spizizen J. (1971) Protease activities during the course of sporulation on Bacillus subtilis. J. Bacteriol. 107, 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thorpe T. C., Miller R. D. (1981) Extracellular enzymes of Legionella pneumophila. Infect. Immun. 33, 632–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neu H. C., Heppel L. A. (1965) The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J. Biol. Chem. 240, 3685–3692 [PubMed] [Google Scholar]
- 38. Flieger A., Gongab S., Faigle M., Mayer H. A., Kehrer U., Mussotter J., Bartmann P., Neumeister B. (2000) Phospholipase A secreted by Legionella pneumophila destroys alveolar surfactant phospholipids. FEMS Microbiol. Lett. 188, 129–133 [DOI] [PubMed] [Google Scholar]
- 39. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 40. Ahrends R., Pieper S., Neumann B., Scheler C., Linscheid M. W. (2009) Metal-coded affinity tag labeling: a demonstration of analytical robustness and suitability for biological applications. Anal. Chem. 81, 2176–2184 [DOI] [PubMed] [Google Scholar]
- 41. Black W. J., Quinn F. D., Tompkins L. S. (1990) Legionella pneumophila zinc metalloprotease is structurally and functionally homologous to Pseudomonas aeruginosa elastase. J. Bacteriol. 172, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Korn E. D., Wright P. L. (1973) Macromolecular composition of an amoeba plasma membrane. J. Biol. Chem. 248, 439–447 [PubMed] [Google Scholar]
- 43. Raederstorff D., Rohmer M. (1985) Sterol biosynthesis de nova via cycloartenol by the soil amoeba Acanthamoeba polyphaga. Biochem. J. 231, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith F. R., Korn E. D. (1968) 7-Dehydrostigmasterol and ergosterol: the major sterols of an amoeba. J. Lipid Res. 9, 405–408 [PubMed] [Google Scholar]
- 45. Volkman J. K. (2003) Sterols in microorganisms. Appl. Microbiol. Biotechnol. 60, 495–506 [DOI] [PubMed] [Google Scholar]
- 46. Akoh C. C., Lee G. C., Liaw Y. C., Huang T. H., Shaw J. F. (2004) GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552 [DOI] [PubMed] [Google Scholar]
- 47. Brumlik M. J., Buckley J. T. (1996) Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophila. J. Bacteriol. 178, 2060–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eggset G., Bjornsdottir R., McQueen Leifson R., Arnesen J. A. (1994) Extracellular glycerophospholipid:cholesterol acyltransferase from Aeromonas salmonicida: activation by serine protease. J. Fish Dis. 17, 17–29 [Google Scholar]
- 49. Hilton S., McCubbin W. D., Kay C. M., Buckley J. T. (1990) Purification and spectral study of a microbial fatty acyltransferase: activation by limited proteolysis. Biochemistry 29, 9072–9078 [DOI] [PubMed] [Google Scholar]
- 50. Ferrè F., Clote P. (2005) DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 33, W230–W232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Choe S., Bennett M. J., Fujii G., Curmi P. M., Kantardjieff K. A., Collier R. J., Eisenberg D. (1992) The crystal structure of diphtheria toxin. Nature 357, 216–222 [DOI] [PubMed] [Google Scholar]
- 52. Downing M. R., Butkowski R. J., Clark M. M., Mann K. G. (1975) Human prothrombin activation. J. Biol. Chem. 250, 8897–8906 [PubMed] [Google Scholar]
- 53. Duckworth W. C., Stentz F. B., Heinemann M., Kitabchi A. E. (1979) Initial site of insulin cleavage by insulin protease. Proc. Natl. Acad. Sci. U.S.A. 76, 635–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fasman G. D., Foster R. J., Beychok S. (1966) The conformational transition associated with the activation of chymotrypsinogen to chymotrypsin. J. Mol. Biol. 19, 240–253 [DOI] [PubMed] [Google Scholar]
- 55. Gordon V. M., Leppla S. H. (1994) Proteolytic activation of bacterial toxins: role of bacterial and host cell proteases. Infect. Immun. 62, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Geiger O. (2010) in Handbook of Hydrocarbon and Lipid Microbiology, pp. 3195–3202, Springer, New York [Google Scholar]
- 57. Goldfine H. (1982) Lipids of prokaryotes—structure and distribution. Curr. Top. Membr. Transp. 17, 1–43 [Google Scholar]
- 58. Sturt H. F., Summons R. E., Smith K., Elvert M., Hinrichs K. U. (2004) Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18, 617–628 [DOI] [PubMed] [Google Scholar]
- 59. Lossi N. S., Rolhion N., Magee A. I., Boyle C., Holden D. W. (2008) The Salmonella SPI-2 effector SseJ exhibits eukaryotic activator-dependent phospholipase A and glycerophospholipid:cholesterol acyltransferase activity. Microbiology 154, 2680–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nawabi P., Catron D. M., Haldar K. (2008) Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Mol. Microbiol. 68, 173–185 [DOI] [PubMed] [Google Scholar]
- 61. Christen M., Coye L. H., Hontz J. S., LaRock D. L., Pfuetzner R. A., Megha, Miller S. I. (2009) Activation of a bacterial virulence protein by the GTPase RhoA. Sci. Signal. 2, ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moffat J. F., Edelstein P. H., Regula D. P., Jr., Cirillo J. D., Tompkins L. S. (1994) Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol. Microbiol. 12, 693–705 [DOI] [PubMed] [Google Scholar]
- 63. Rossier O., Dao J., Cianciotto N. P. (2008) The type II secretion system of Legionella pneumophila elaborates two aminopeptidases, as well as a metalloprotease that contributes to differential infection among protozoan hosts. Appl. Environ. Microbiol. 74, 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Okamoto T., Akuta T., Tamura F., van Der Vliet A., Akaike T. (2004) Molecular mechanism for activation and regulation of matrix metalloproteinases during bacterial infections and respiratory inflammation. Biol. Chem. 385, 997–1006 [DOI] [PubMed] [Google Scholar]
- 65. Quinn F. D., Tompkins L. S. (1989) Analysis of a cloned sequence of Legionella pneumophila encoding a 38 kD metalloprotease possessing haemolytic and cytotoxic activities. Mol. Microbiol. 3, 797–805 [DOI] [PubMed] [Google Scholar]
- 66. Keen M. G., Hoffman P. S. (1989) Characterization of a Legionella pneumophila extracellular protease exhibiting hemolytic and cytotoxic activities. Infect. Immun. 57, 732–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ruggiero A., Marasco D., Squeglia F., Soldini S., Pedone E., Pedone C., Berisio R. (2010) Structure and functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation. Structure 18, 1184–1190 [DOI] [PubMed] [Google Scholar]
- 68. Ogata M., Fryling C. M., Pastan I., FitzGerald D. J. (1992) Cell-mediated cleavage of Pseudomonas exotoxin between Arg279 and Gly280 generates the enzymatically active fragment which translocates to the cytosol. J. Biol. Chem. 267, 25396–25401 [PubMed] [Google Scholar]
- 69. Giannelli V., Fontana M. R., Giuliani M. M., Guangcai D., Rappuoli R., Pizza M. (1997) Protease susceptibility and toxicity of heat-labile enterotoxins with a mutation in the active site or in the protease-sensitive loop. Infect. Immun. 65, 331–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kurmanova A., Llorente A., Polesskaya A., Garred O., Olsnes S., Kozlov J., Sandvig K. (2007) Structural requirements for furin-induced cleavage and activation of Shiga toxin. Biochem. Biophys. Res. Commun. 357, 144–149 [DOI] [PubMed] [Google Scholar]
- 71. Tsuneoka M., Nakayama K., Hatsuzawa K., Komada M., Kitamura N., Mekada E. (1993) Evidence for involvement of furin in cleavage and activation of diphtheria toxin. J. Biol. Chem. 268, 26461–26465 [PubMed] [Google Scholar]
- 72. Garred O., van Deurs B., Sandvig K. (1995) Furin-induced cleavage and activation of Shiga toxin. J. Biol. Chem. 270, 10817–10821 [DOI] [PubMed] [Google Scholar]
- 73. Winiecka-Krusnell J., Linder E. (1999) Free-living amoebae protecting Legionella in water: the tip of an iceberg? Scan. J. Infect. Dis. 31, 383–385 [DOI] [PubMed] [Google Scholar]