Background: DBC1 is a key regulator of SIRT1 activity, although it is unknown how the SIRT1-DBC1 interaction is regulated.
Results: PKA and AMPK activate SIRT1 by disrupting the interaction between SIRT1 and DBC1.
Conclusion: We provide mechanistic evidence on how the SIRT1-DBC1 complex is regulated.
Significance: The SIRT1-DBC1 complex constitutes a target for the development of drugs to activate SIRT1.
Keywords: AMP-activated Kinase (AMPK), NAD, Protein Kinase A (PKA), SIRT1, Sirtuins, CD38, Age-related Diseases, Deleted in Breast Cancer 1, Metabolism, NamPT
Abstract
The NAD+-dependent deacetylase SIRT1 is a key regulator of several aspects of metabolism and aging. SIRT1 activation is beneficial for several human diseases, including metabolic syndrome, diabetes, obesity, liver steatosis, and Alzheimer disease. We have recently shown that the protein deleted in breast cancer 1 (DBC1) is a key regulator of SIRT1 activity in vivo. Furthermore, SIRT1 and DBC1 form a dynamic complex that is regulated by the energetic state of the organism. Understanding how the interaction between SIRT1 and DBC1 is regulated is therefore essential to design strategies aimed to activate SIRT1. Here, we investigated which pathways can lead to the dissociation of SIRT1 and DBC1 and consequently to SIRT1 activation. We observed that PKA activation leads to a fast and transient activation of SIRT1 that is DBC1-dependent. In fact, an increase in cAMP/PKA activity resulted in the dissociation of SIRT1 and DBC1 in an AMP-activated protein kinase (AMPK)-dependent manner. Pharmacological AMPK activation led to SIRT1 activation by a DBC1-dependent mechanism. Indeed, we found that AMPK activators promote SIRT1-DBC1 dissociation in cells, resulting in an increase in SIRT1 activity. In addition, we observed that the SIRT1 activation promoted by PKA and AMPK occurs without changes in the intracellular levels of NAD+. We propose that PKA and AMPK can acutely activate SIRT1 by inducing dissociation of SIRT1 from its endogenous inhibitor DBC1. Our experiments provide new insight on the in vivo mechanism of SIRT1 regulation and a new avenue for the development of pharmacological SIRT1 activators targeted at the dissociation of the SIRT1-DBC1 complex.
Introduction
SIRT1 is an NAD+-dependent deacetylase that regulates gene expression and protein function by deacetylating lysine residues in proteins. It has been shown to regulate many aspects of cell and tissue metabolism, including liver gluconeogenesis (1, 2), insulin secretion (3–5), insulin sensitivity (5), fatty acid oxidation (6), and adipogenesis (7). Although the literature regarding the physiological processes regulated by SIRT1 is vast, our knowledge about how this key enzyme is regulated in the cellular context is scarce. In this regard, several possible regulatory mechanisms have been described.
One of the proposed mechanisms of SIRT1 regulation involves alterations in the intracellular concentration of NAD+. Because SIRT1 enzymatic activity is dependent on NAD+ (8), changes in the concentration of this nucleotide can lead to changes in SIRT1 activity. Indeed, modification of the two main enzymes responsible for the control of intracellular NAD+ levels, namely NamPT (9) and CD38 (10–12), can lead to changes in SIRT1 activity (11, 13, 14). However, the specificity of this mechanism seems low as there are several other NAD+-consuming enzymes in the cell. Moreover, it remains unknown whether global changes in NAD+ are reflected by similar changes in the nuclei where most SIRT1 is localized.
Several authors have shown that SIRT1 can be regulated at the transcriptional level (7, 15, 16). Although this mechanism could certainly explain long term changes in SIRT1 activity, it does not account for transient changes in its activity. Several post-transcriptional modifications can also affect SIRT1 activity. In this regard, it has been described that SUMOylation (17) and phosphorylation by several kinases (18–23) can increase SIRT1 activity. The kinases cyclin-dependent kinase 1 (22), casein kinase, (23, 24), and the c-Jun N-terminal kinase (JNK) (21) have been shown to directly phosphorylate SIRT1. On the other hand, it has been reported that the cAMP-dependent protein kinase (PKA) activates SIRT1 indirectly (19), the effects being mediated by an unidentified kinase. In addition, it has been proposed that AMP-dependent protein kinase (AMPK)6 modulates NAD+ intracellular levels and consequently SIRT1 activity (25). Interestingly, some of the effects of PKA appear to be mediated by AMPK (26, 27). Moreover, PKA activation can lead to a fast activation of AMPK in several tissues and cell models (26, 28–30).
In addition, SIRT1 is regulated by protein-protein interactions. Recently, we and others demonstrated that in vivo SIRT1 is largely associated with its endogenous inhibitor deleted in breast cancer 1 (DBC1) (31–33). DBC1 is a nuclear protein that, in addition to SIRT1, binds to several nuclear receptors and enzymes, including the estrogen receptors α (34) and β (35), the androgen receptor (36), the transcription factor BRCA1 (37), and the deacetylase HDAC3 (38).
SIRT1 and DBC1 form a dynamic complex in cells and in vivo (31). Moreover, the binding between SIRT1 and DBC1 is regulated by the energetic state of the organism (31). So far it is unknown which are the molecular pathways that modulate the interaction between SIRT1 and DBC1 and consequently SIRT1 activity in vivo.
Here, we show that the activation of the cAMP/PKA pathway leads to SIRT1 activation through an AMPK-dependent mechanism. Furthermore, this activation is DBC1-dependent and involves dissociation of the SIRT1-DBC1 complex. We propose that AMPK activation, either pharmacological or induced by PKA, results in the dissociation of SIRT1 from DBC1 and activation of SIRT1. Our results provide insight into the mechanisms that regulate the interaction between SIRT1 and DBC1 and may lead to newer pharmacological approaches to activate SIRT1.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Unless otherwise specified, all reagents and chemicals were from Sigma-Aldrich. Anti-human SIRT1, anti-phosphorylated SIRT1 (Ser-47), anti-phosphorylated AMPK (Thr-172), anti-AMPK antibodies, and AICAR were from Cell Signaling Technology. Anti-DBC1 antibodies were from Bethyl Laboratories. Antibodies for p53, acetylated p53, and tubulin were from Abcam. Anti-actin antibody was from Sigma. PKA activator 6-MB-cAMP and exchange protein activated by cAMP (EPAC) activator 8-(4-chlorophenylthio)-2′-O-methyl-cAMP were from Biolog. A769662 was from Santa Cruz Biotechnology Inc. Resveratrol and recombinant SIRT1 were from Enzo Life Sciences. 1,1,2-Trichloro-1,2,2-trifluroethane was from Fisher.
Cell Culture and Transfections
HepG2, HEK 293T, and DBC1 and AMPK wild type (WT) and knock-out (KO) MEFs were cultured in Dulbecco's modified Eagle's medium (5 g/liter glucose), and A549 cells were maintained in RPMI 1640 medium, all of them supplemented with 10% FBS and penicillin/streptomycin (Invitrogen). For all the experiments, the cultures were serum-starved for 1 h before the treatments. AMPK α1/α2 WT and KO MEFs were a kind gift from Dr. Keith R. Laderoute. SIRT1 KO MEFS were kindly provided by Dr. David Sinclair.
Transient overexpression of SIRT1 and DBC1 was performed under the conditions and using the vectors described previously (31). The dominant-negative mutant of human AMPK α1 catalytic subunit (D157A) was kindly provided by Dr. David Carling. The constitutively active form of AMPK α1 was kindly provided by Dr. Benoit Viollet. Transient overexpression of these constructs was performed using Lipofectamine 2000 (Invitrogen) for 48 h following the manufacturer's instructions.
siRNA
All siRNAs were from Dharmacon (Lafayette, CO). The siRNA duplex against DBC1 was 21 bp as follows: DBC1 siRNA sense strand, 5′-AAACGGAGCCUACUGAACAUU. SMARTpool siRNAs were used to knock down SIRT1. Non-targeting siRNA number 3 was used as a control (D001210-03-20). Transfections were performed with 150 nm siRNA using Darmaphect 1 (Dharmacon) according to the manufacturer's instructions. Cells were harvested 72 h after the transfection.
Immunoprecipitation and Western Blot
Cultured cells were lysed in NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40) supplemented with 5 mm NaF, 50 mm 2-glycerophosphate, 1 mm Na3VO4, and a protease inhibitor mixture (Roche Applied Science). Homogenates were incubated at 4 °C for 30 min under constant agitation and then centrifuged at 11,200 × g for 10 min at 4 °C. For immunoprecipitation, 1–1.5 mg of protein were incubated with 20 μl of Protein A/G (Santa Cruz Biotechnology Inc.) and 1 μg of antibody for 1 h at 4 °C under constant rotation. Nonspecific IgG (Santa Cruz Biotechnology Inc.) was used as a control. Finally, immunoprecipitates were washed two times with cold NETN before addition of 2× Laemmli buffer. Cell lysates and immunoprecipitates were analyzed by Western blot with the indicated antibodies. Western blots were developed using secondary antibodies or protein A-HRP and SuperSignal West Pico chemiluminescent substrate (Pierce).
SIRT1 Activity Measurement
SIRT1 activity was measured with a fluorometric assay (Enzo Life Sciences catalogue number BML-AK555-0001). Cells were extracted with NETN buffer as described above, and then the protein concentration in the lysates was quantified and equalized with deacetylase buffer (50 mm Tris-HCl, pH 8, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, and 1 mg/ml BSA). Samples were incubated for 10 min at 30 °C to allow for NAD+ degradation and incubated for 10 additional min with 2 μm DTT. Finally, 30–50 mg of protein of each sample were transferred to 6 wells of a 96-well plate, and a solution of deacetylase buffer containing 100 μm substrate and 5 μm trichostatin A was added to the wells. Half the wells included 100 μm NAD+. The reaction proceeded for 2 h at room temperature, and then the developer, prepared according to the manufacturer's recommendations, was added for 1 h. Finally, the fluorescence was read with excitation of 360 nm and emission at 460 nm. SIRT1 activity was calculated as NAD+-dependent fluorescence. All the activity measurements were determined in the initial linear portion of the reaction.
NAD+ Extraction and Quantification
Cells were placed on ice, washed with ice-cold PBS twice, harvested, and spun down. The cell pellet was extracted on ice with ice-cold 10% TCA and sonicated three times, and then the TCA was extracted with 2 volumes of an organic phase consisting of 1,1,2-trichloro-1,2,2-trifluroethane and trioctylamine in a 3:1 ratio. Both phases were vigorously vortexed for 15 s and then allowed to separate for 3 min. The pH of the top aqueous layer containing NAD+ was adjusted with 1 m Tris, pH 8. The NAD+ concentration was measured by means of an enzymatic cycling assay. Briefly, the aqueous layer was incubated with a 20 mm Na2HPO4, pH 8 buffer containing 0.76% ethanol, 4 mm flavine mononucleotide (FMN), 27 units/ml alcohol dehydrogenase, 0.4 unit/ml diaphorase, and 8 μm rezarsurin. A standard curve for NAD+ was included. The fluorescence in the samples was followed with excitation at 544 nm and emission at 590 nm. Measurements were made in triplicates.
Autoradiography
293T cells were transfected with FLAG-SIRT1 or with FLAG-SIRT1 and constitutively active (CA) AMPK. After 48 of transfection, the medium was replaced with phosphate-free DMEM supplemented with H332PO4 (0.5 mCi/ml). Cells were incubated in this medium for 2 h. After that, the medium was replaced with regular DMEM. Cells were stimulated with the AMPK activator resveratrol for 2 h. After that, SIRT1 was immunoprecipitated, and 32P incorporation was evidenced by electrophoresis and autoradiography.
Site-directed Mutagenesis
Mutagenesis was performed on FLAG-SIRT1 using a QuikChange II site-directed mutagenesis kit from Agilent Technologies following the manufacturer's instructions.
Statistics
Values are presented as mean ± S.E. of three to five experiments unless otherwise indicated. The significance of differences between means was assessed by ANOVA or two-tailed Student's t test as indicated. A p value less than 0.05 was considered significant.
RESULTS
Measurement of SIRT1 Activity in Cells
Cellular SIRT1 activity and activation were determined with a SIRT1 fluorometric kit (Enzo Life Sciences) according to the manufacturer's instructions. This assay has been extensively characterized in our laboratory (14, 31). The substrate used is a small peptide derived from p53 that includes an acetylated lysine, corresponding to Lys-382 of human p53; this residue is the target of SIRT1 enzymatic activity. Independent investigators have shown that this assay is a reliable tool to measure cellular SIRT1 activity and regulation (19, 31, 39–41) However, a controversy regarding the validity of this assay arose when it was shown that in vitro some compounds produce fluorescence artifacts that could be misinterpreted as changes in SIRT1 activity (42). In fact, the Fluor-de-Lys SIRT1 assay was used to propose that the polyphenol resveratrol is a direct SIRT1 activator (43, 44). It was later shown that the direct activation of SIRT1 by resveratrol was an artifact as a consequence of the interaction between resveratrol and the 7-amino-4-methylcoumarin fluorescent probe linked to the acetylated peptide (42). Nevertheless, these artifacts occur only when SIRT1 activity is measured in vitro and in the presence of small molecules (42, 45, 46) and not when used to measure SIRT1 activity in cell extracts. This has been clearly established by different independent investigators, including us (14, 31, 39–41). Here, we further characterize the assay to demonstrate that indeed it constitutes a highly reliable way to measure SIRT1 activation in cells.
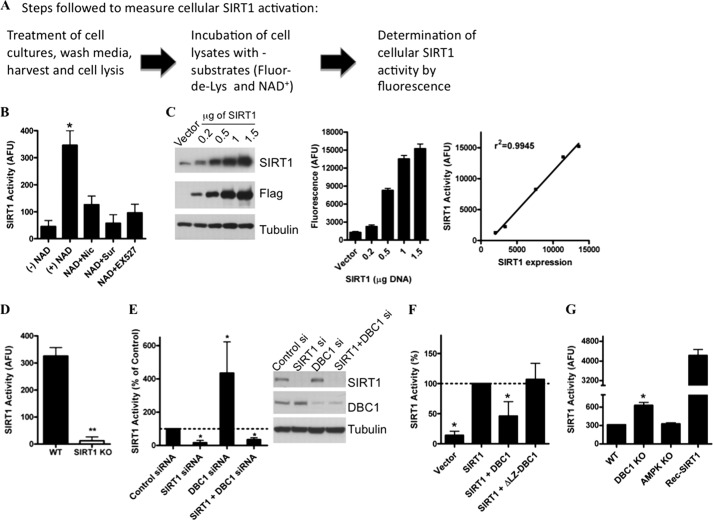
A brief scheme of the basic steps used to measure cellular SIRT1 activity is provided in Fig. 1A. Determination of cellular SIRT1 activity by this method depends on the addition of exogenous NAD+, and this activity is inhibited when the cellular extracts are also incubated with nicotinamide, suramin, or EX527, three very well know inhibitors of SIRT1 (Fig. 1B). The fact that cellular determination of SIRT1 activity by this method requires the addition of NAD+ constitutes at the same time a limitation and an advantage because it provides a way to measure changes in SIRT1 activity regardless of changes in intracellular NAD+ levels. This is especially relevant when one wants to determine whether post-translational modifications like phosphorylation or protein-protein interactions may alter SIRT1 activity.
FIGURE 1.
Characterization of assay used to measure cellular SIRT1 activity. A, basic scheme showing the steps followed to measure cellular SIRT1 activity. Detailed information is provided under “Experimental Procedures.” B, measurement of endogenous cellular SIRT1 activity in 293T cells. Activity was measured in cellular extracts in the absence of exogenous NAD+ (−NAD+), with the addition of 100 μm NAD+ (+NAD), or with NAD+ plus 2 mm nicotinamide (NAD+Nic), with NAD+ plus 100 μm suramin (NAD+Sur), or NAD+ plus 10 μm EX527 (NAD+EX527). C, 293T cells were transfected with different amounts of a FLAG-SIRT1-coding plasmid. Cell lysates were immunoblotted with anti-SIRT1, anti-FLAG, and anti-tubulin antibodies. The graph on the left is the cellular SIRT1 activity measured 24 h after the transfection. The graph on the right shows the relationship between SIRT1 expression levels and cellular SIRT1 activity. D, SIRT1 activity was measured in MEFs obtained from WT and SIRT1 KO mice. E, SIRT1 and DBC1 were knocked down in HepG2 cells with specific siRNAs, and SIRT1 activity was assessed. Activity is shown as -fold change with respect to the control. *, p < 0.05 (ANOVA test, n = 3). Cell lysates were immunoblotted with anti-SIRT1, anti-DBC1, and anti-tubulin antibodies. F, cellular SIRT1 activity was measured in 293T cells transfected with FLAG-SIRT1, FLAG-SIRT1 + Myc-DBC1, or FLAG-SIRT1 + ΔLZ Myc-DBC1, a mutant DBC1 that does not have the leucine zipper domain and does not bind to SIRT1. *, p < 0.05 (ANOVA test, n = 3). G, cellular SIRT1 activity was measured in MEFs from WT, DBC1 KO, and AMPK (α1α2) KO mice. The activity of 1 unit of purified recombinant human SIRT1 was measured in parallel for the same time. *, p < 0.05 (ANOVA test, n = 3). Error bars represent S.D. AFU, arbitrary fluorescence units.
To prove that the cellular activity measured by the Fluor-de-Lys assay quantitatively correlates with the level of SIRT1 expression in the cells, we performed a dose-response curve. For that, we transfected different amounts of a SIRT1-coding plasmid in 293T cells and then measured SIRT1 activity. As can be seen in Fig. 1C, the amount of SIRT1 in the cell lysates increased with increasing concentrations of plasmid. We measured SIRT1 activity in these samples, and we found that the increase in protein levels correlates with the activity measured by the assay. In fact, when we plotted SIRT1 activity versus the expression level of SIRT1, we found that these parameters correlate perfectly with an r2 of 0.9945 (Fig. 1C, right panel). To further prove that the activity measured is linearly dependent on SIRT1 concentration, we assessed different amounts of protein lysates from SIRT1-positive cells. As can be seen in supplemental Fig. S1, SIRT1 activity correlates perfectly with the amount of protein assessed (r2 of 0.994). These experiments show that the activity measured by this method is a quantitative reflection of the SIRT1 content in cells.
Furthermore, to assess the specificity of the assay, we measured SIRT1 activity in WT and SIRT1 KO MEFs. SIRT1 activity was undetectable in SIRT1 KO MEFs (Fig. 1D), clearly showing that the enzymatic activity measured by the Fluor-de-Lys assay is not present in cellular extracts that lack SIRT1. The same result was obtained when SIRT1 was knocked down by siRNA transfection in HepG2 cells (Fig. 1E).
In addition, we evaluated whether the assay was able to detect changes in SIRT1 activity regardless of changes in the expression level of SIRT1. To do so, we evaluated the effect of the SIRT1 negative regulator DBC1 on SIRT1 activity. As we had shown previously (31), knockdown of DBC1 led to an increase in cellular SIRT1 activity, an event that was not observed if SIRT1 was absent (Fig. 1E). Moreover, when SIRT1 was cotransfected with DBC1, we observed inhibition of the SIRT1 cellular activity. This inhibition was lost when SIRT1 was cotransfected with ΔLZ-DBC1, a deletion mutant of DBC1 that was shown previously by us (31, 33) not to bind to SIRT1 (Fig. 1F). Also, cellular SIRT1 activity was higher in DBC1 KO MEFs than in the WT control cells (Fig. 1G). This result is specific for DBC1 KO MEFS because there was no difference in SIRT1 activity between WT cells and AMPK (α1α2) KO cells (Fig. 1G). More importantly, we also found that the cellular SIRT1 activity measured in cell lysates is within the linear range of detection of the assay, and further changes could be determined as shown by the measurement in parallel of recombinant purified SIRT1 activity (Fig. 1G). Taken together, the results shown here plus what we and others have previously and independently shown (31, 39–41) provide very strong evidence that the Fluor-de-Lys SIRT1 assay is an extremely valuable assay to measure cellular SIRT1 activation.
cAMP/PKA Pathway Activates SIRT1
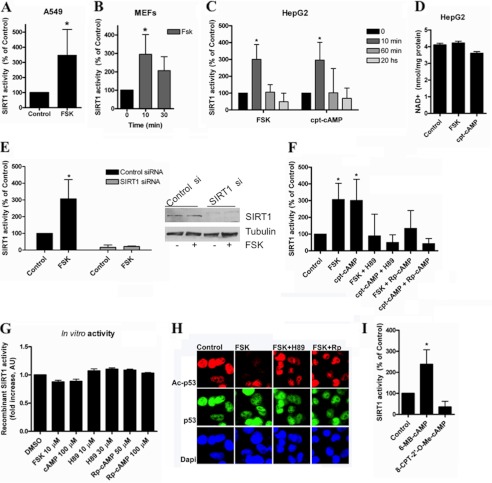
We (31) and other investigators (2, 47) have shown previously that SIRT1 is activated in the liver and in cells upon starvation. In fact, we have shown that starvation increases SIRT1 activity by displacing it from DBC1 (31). However, the molecular pathways that lead to the dissociation of the SIRT1-DBC1 complex are not known. One of the main kinases activated by starvation is PKA (48). In fact, while this manuscript was in preparation, Gerhart-Hines et al. (19) showed that PKA stimulation leads to SIRT1 activation by a not completely elucidated mechanism. Similar to what was found by Gerhart-Hines et al. (19), we observed that treatment of cells with forskolin or cpt-cAMP, a permeant analog of cAMP, produces a rapid and transient activation of SIRT1 in several cell lines (Fig. 2, A–C). Because it has been proposed that changes in NAD+ can lead to changes in SIRT1 activity (49), we measured intracellular NAD+ levels after forskolin and cpt-cAMP treatments. We did not detect changes in the intracellular levels of NAD+ under these experimental conditions (Fig. 2D), suggesting that SIRT1 activation is achieved by a different mechanism.
FIGURE 2.
cAMP/PKA increase SIRT1 activity by mechanism that is independent of changes in NAD levels. SIRT1 activity was measured in A549 cells (A), mouse embryonic fibroblasts (B), and HepG2 cells (C) treated with 10 μm forskolin (FSK) or 100 μm cpt-cAMP for the indicated times. *, p < 0.01 (ANOVA, n = 3–9). SIRT1 activity was normalized to time 0. D, -fold change in NAD+ concentration in HepG2 cells after forskolin (10 μm) or cpt-cAMP (100 μm) incubation for 10 min (n = 3). E, SIRT1 was knocked down in HepG2 cells with specific siRNA, and SIRT1 activity was assessed after stimulation with 10 μm forskolin for 10 min. Activity is shown as -fold change with respect to the control. *, p < 0.05 (ANOVA test, n = 3). Cell lysates were immunoblotted with anti-SIRT1 and anti-tubulin antibodies. F, cells were pretreated for 45 min with the PKA inhibitor H89 (30 μm) or (Rp)-cAMP (100 μm) and then stimulated with 10 μm forskolin or 100 μm cpt-cAMP for 10 min. SIRT1 activity was normalized to the control. * shows significant difference with respect to the control (p < 0.01, ANOVA, n = 3). G, human recombinant purified SIRT1 activity in the presence of different compounds was measured in vitro. 0.2 unit of SIRT1 was incubated with the compounds at the indicated concentrations. H, HepG2 cells were pretreated for 45 min with the PKA inhibitor H89 (30 μm) or (Rp)-cAMP (Rp; 100 μm) and then stimulated with 10 μm forskolin for 10 min. Immunofluorescence for Ac-p53 and total p53 was analyzed using specific antibodies. I, cells were incubated with the PKA activator 6-MB-cAMP (100 μm) and the EPAC activator 8-CPT-2′-O-Me-cAMP; (100 μm) for 10 min before harvesting. SIRT1 activity was measured and normalized to the control. *, p < 0.01 (t test, n = 3). Error bars represent S.D. AU, arbitrary units.
To confirm the specificity of the activity measurements, we transfected HepG2 cells with control and SIRT1 siRNAs. We did not detect any SIRT1 activity in the SIRT1 siRNA-treated cultures either in the control cells or after treatment with forskolin (Fig. 2E).
cAMP can regulate cellular functions by a direct effect on target proteins (50), or it can activate a signal transducer like PKA or the EPACs (50). To further explore the mechanism by which cAMP activates SIRT1, we studied which of these different mechanisms account for the cAMP-mediated SIRT1 activation. The cAMP-induced SIRT1 activation was prevented by the PKA inhibitors H89 and (Rp)-cAMP (Fig. 2F), suggesting that in cells this effect is mediated by PKA. As a control, we tested whether these compounds directly interfered with the SIRT1 assay. We found that none of the PKA activators or inhibitors had a direct effect on SIRT1 activity in vitro (Fig. 2G). To provide additional evidence that SIRT1 is activated by the cAMP/PKA pathway, we performed an immunofluorescence assay for endogenous p53 acetylation. We used an antibody against acetylated Lys-382 on p53, a site that is deacetylated by SIRT1 (51). Although forskolin treatment induced p53 deacetylation, H89 and (Rp)-cAMP prevented the deacetylation induced by forskolin (Fig. 2H), confirming the results obtained using the Fluor-de-Lys assay.
To further assess the role of PKA and to evaluate a possible contribution of EPAC, we treated cell cultures with 6-MB-cAMP, a direct and specific activator of PKA, and with 8-CPT-2′-O-Me-cAMP, a specific activator of EPAC. We observed that only the PKA stimulator produced activation of SIRT1 (Fig. 2I), confirming that the cAMP/PKA pathway can transiently activate SIRT1. Taken together, these results indicate that the cAMP/PKA pathway can activate endogenous cellular SIRT1.
AMPK Mediates Effect of cAMP/PKA on SIRT1 Activation
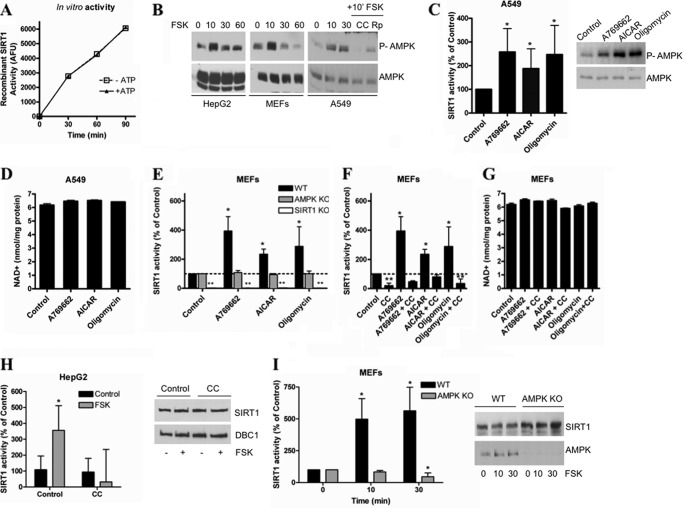
We measured recombinant SIRT1 activity in vitro after a kinase assay that included the catalytic subunit of PKA with or without ATP. We found that PKA does not affect SIRT1 activity directly (Fig. 3A), suggesting that another kinase mediates the effect of PKA on SIRT1 activation. Interestingly, we observed that when cells were treated with forskolin the AMPK was also transiently activated in a pattern that followed the same temporal curve observed for SIRT1 activation (Fig. 3B). This fast AMPK activation by forskolin was observed in several cell lines, including MEFs, HepG2, and A549. Preincubation of A549 cell cultures with the AMPK inhibitor compound C prevented the activation of AMPK, confirming the specificity of the antibody (Fig. 3B). In addition, preincubation of the cells with the PKA inhibitor (Rp)-cAMP also prevented the forskolin-induced increase in AMPK phosphorylation, suggesting that the AMPK activation observed is dependent on PKA (Fig. 3B). In light of these observations, we tested whether AMPK was mediating the PKA-induced SIRT1 activation.
FIGURE 3.
SIRT1 activation by PKA is AMPK-dependent. A, activity of human recombinant purified SIRT1 (0.2 unit) was measured using a fluorometric assay after performing a kinase assay with the catalytic subunit of PKA in the presence or absence of 200 μm ATP. B, AMPK activation was measured by immunoblot using anti-Thr(P)-172 antibody in different cell lines after treatment with 10 μm forskolin (FSK) for different times. Compound C (CC; 10 μm) and (Rp)-cAMP (Rp; 100 μm) were added 2 h prior to the addition of forskolin. C, SIRT1 activity in A549 cells was measured after a 2-h incubation with A769662 (100 μm), AICAR (2 mm), and oligomycin (5 μm). Activity was expressed as the percentage of activity with respect to the control. *, p < 0.05 (ANOVA test, n = 3). AMPK activation by the different compounds was confirmed by Western blot (right) with anti-Thr(P)-172 antibody. D, determination of intracellular NAD+ levels in A549 cells treated as described in C. E, SIRT1 activity was measured in MEFs from WT, AMPK KO, and SIRT1 KO mice. Cells were incubated with A769662 (100 μm), AICAR (2 mm), or oligomycin (5 μm) for 2 h before measuring SIRT1 activity. SIRT1 activity was normalized to the respective control for each cell type. SIRT1 KO cells showed no detectable activity. * and **, p < 0.05 (ANOVA test, n = 3). F, SIRT1 activity was determined in MEFs from WT mice. The AMPK inhibitor compound C (10 μm) was added to the cells 2 h before starting the treatments. Cells were incubated with A769662 (100 μm), A769662 + compound C, AICAR (2 mm), AICAR + compound C, oligomycin (5 μm), and oligomycin + compound C for 2 h before measuring SIRT1 activity. SIRT1 activity was normalized to control. * and **, p < 0.05 (ANOVA test, n = 3). G, intracellular NAD+ levels in WT MEFs treated as described in F. H, AMPK was inhibited in HepG2 cells by a pretreatment with compound C (10 μm) for 2 h, and SIRT1 activity was assessed after stimulation with 10 μm forskolin for 10 min. Activity is shown as -fold change with respect to the control. *, p < 0.05 (ANOVA test, n = 3). I, SIRT1 activity was measured in AMPK WT and KO (α1α2) MEFS treated with 10 μm forskolin for the indicated times. *, p < 0.05 (ANOVA test, n = 3). Error bars represent S.D. AFU, arbitrary fluorescence units; P-AMPK, phosphorylated AMPK.
AMPK is a kinase that has a key role in metabolism and has been shown to regulate SIRT1 activity (25). In fact, it was recently shown that incubation of cells with AMPK activators for long periods (typically 8–12 h) leads to SIRT1 activation through an increase in the intracellular levels of NAD+ (25). Our results showed a fast SIRT1 activation that occurred independently of changes in NAD+ levels, suggesting that SIRT1 is activated through a different mechanism. Therefore, we tested whether short treatments (1–2 h) with AMPK activators can induce an increase in SIRT1 activity without changes in the intracellular levels of NAD+.
To test our hypothesis, we used several AMPK activators: A769662 (52), AICAR (53), and oligomycin (54). We found that a short incubation of A549 cells with all these AMPK activators promoted SIRT1 activation (Fig. 3C) without any changes in NAD+ levels (Fig. 3D). We measured the in vitro recombinant SIRT1 activity in the presence of the AMPK activators and confirmed that the AMPK activators have no direct effect on SIRT1 activity in vitro (supplemental Fig. S2). Resveratrol was excluded from the in vitro SIRT1 activity assay due to the artifact it has been reported to produce in the fluorescence assay. To confirm that the effect of the AMPK activators was indeed AMPK-dependent, we incubated AMPK WT and KO MEFs with AMPK activators and then measured SIRT1 activity. None of the AMPK activators induced SIRT1 activity on the AMPK KO MEFs in contrast to the WT cells (Fig. 3E). As a control for the assay, SIRT1 activity was also measured in SIRT1 KO MEFs (Fig. 3E), and we detected no activity in these cells. Finally, to further confirm our results, we measured the effect of the AMPK activators on SIRT1 activity in WT MEFs pretreated with the AMPK inhibitor compound C. We found that in these conditions the AMPK activators did not induce SIRT1 activation (Fig. 3F). All the observed changes in SIRT1 activity occurred independently of changes in NAD+ levels (Fig. 3G). To demonstrate that the assay we used to measure NAD+ is able to detect even small changes in NAD+, we performed a time course with the NamPT inhibitor FK866. As shown in supplemental Fig. S3, the methodology used in this study was sensitive to small variations in NAD+ concentration. Furthermore, we have previously detected changes in NAD+ and NAD+ metabolites in several cells and tissues using the same method (10, 11, 14, 31, 55–57). Taken together, these results demonstrate that AMPK can activate SIRT1 without detectable changes in cellular NAD+ levels.
Finally, we directly tested the role of AMPK in the cAMP/PKA-mediated SIRT1 activation. For this, we used HepG2 cells pretreated with the AMPK inhibitor compound C. In agreement with our hypothesis, when AMPK activation was blocked by compound C, forskolin no longer induced SIRT1 activation (Fig. 3H). Moreover, the response to forskolin was abrogated in AMPK (α1α2) KO MEFs but not in WT MEFs (Fig. 3I). From these experiments, we concluded that the activation of SIRT1 induced by cAMP/PKA is dependent on AMPK activation.
DBC1 Is Required for SIRT1 Activation Induced by PKA and AMPK
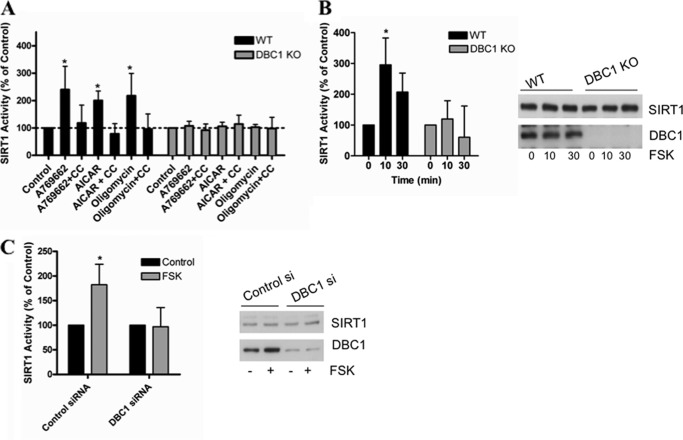
We have observed previously that, in vivo, fasting promotes an increase in SIRT1 activity by disrupting the interaction between SIRT1 and DBC1 without changes in NAD+ levels or in the expression levels of SIRT1 (31). We then explored whether DBC1 was required for the AMPK- and PKA-induced SIRT1 activation. For this, we stimulated AMPK with A769662, AICAR, and oligomycin in DBC1 WT and KO MEFs. As shown in Fig. 4A, none of the AMPK activators were able to induce an increase in SIRT1 activity in cells that lacked DBC1 in contrast with the clear induction of SIRT1 activity in WT cells. We further confirmed that SIRT1 activation was AMPK-mediated in these cells by preincubating them with compound C. Again, the activation promoted by the AMPK activators was prevented by pretreatment with compound C (Fig. 4A).
FIGURE 4.
SIRT1 activation by cAMP/PKA/AMPK pathway depends on DBC1. A, SIRT1 activity was measured in WT and DBC1 KO MEFs. Cells were incubated with A769662 (100 μm) or A769662 + compound C (CC), AICAR (2 mm) or AICAR + compound C, and oligomycin (5 μm) or oligomycin + compound C for 2 h before measuring SIRT1 activity. Compound C was used at 10 μm and was preincubated for 2 h. Activity in the WT cells was normalized to the WT control, and activity in the KO cells was normalized to the KO control. SIRT1 activity in the control was always higher in DBC1 KO than in WT MEFs (see Fig. 1). *, p < 0.05 (ANOVA test, n = 3). B, DBC1 was knocked down in HepG2 cells with siRNA, and SIRT1 activity was assessed after stimulation with 10 μm forskolin (FSK) for 10 min. Activity is shown as -fold change with respect to the control. *, p < 0.05 (ANOVA test, n = 3). C, SIRT1 activity in DBC1 WT and KO MEFS treated with 10 μm forskolin for the indicated times. *, p < 0.05 (ANOVA test, n = 3). Error bars represent S.D.
Next, we investigated whether the SIRT1 response to cAMP/PKA activation was also dependent on DBC1. For this, we stimulated the cAMP/PKA pathway in cells that lacked DBC1 and in cells treated with siRNA specific for DBC1. Our results show that forskolin did not activate SIRT1 in the absence of DBC1 either in DBC1 KO MEFS (Fig. 4B) or in HepG2 cells where DBC1 was knocked down (Fig. 4C). Altogether, our results indicate that DBC1 is required for the AMPK- and PKA-induced SIRT1 activation.
AMPK and PKA Activation Promote Dissociation of SIRT1 from DBC1
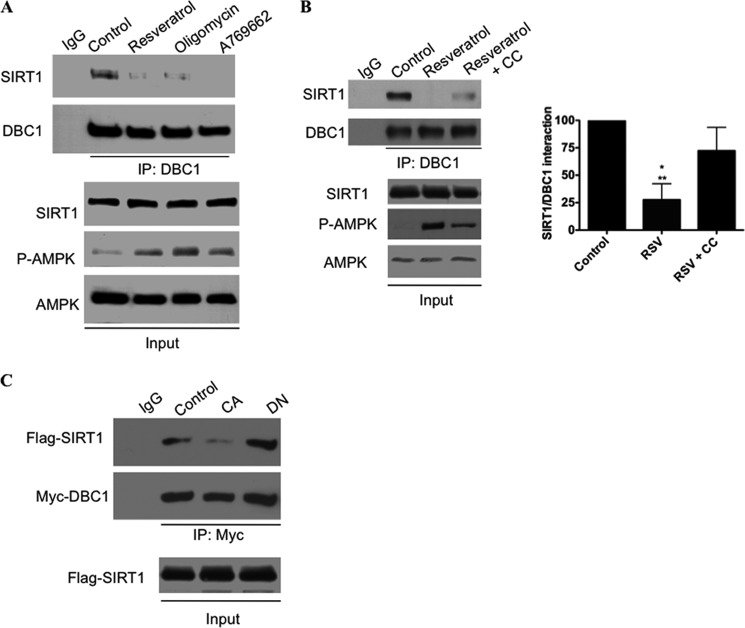
Because DBC1 is required for the AMPK effect on SIRT1, we examined the effect of the AMPK activation on the interaction between endogenous SIRT1 and DBC1. We performed co-immunoprecipitation in A549 cells treated with the AMPK activators A769662, oligomycin, and resveratrol. Resveratrol was used at high concentrations that have been shown previously to activate AMPK (54) due to impairment in mitochondrial function (58) (see also supplemental Fig. S4). As shown in Fig. 5A, all of these compounds promoted a robust dissociation of the SIRT1-DBC1 complex, an event that was partially blocked by preincubation with the AMPK inhibitor compound C (Fig. 5B).
FIGURE 5.
AMPK activation induces dissociation of SIRT1 from DBC1. A and B, SIRT1-DBC1 interaction was evaluated by co-immunoprecipitation in A549 cells. A, cells were treated with resveratrol (100 μm), oligomycin (5 μm), or A769662 (100 μm) for 2 h before performing immunoprecipitation (IP) for DBC1. Proteins were immunoblotted with anti-SIRT1, anti-DBC1, anti-phosphorylated AMPK (P-AMPK) (Thr-172), and anti-AMPK antibodies. B, cells were treated with resveratrol (RSV; 100 μm) or resveratrol + compound C (CC; 10 μm) for 2 h before performing immunoprecipitation. Immunoprecipitates were immunoblotted with anti-SIRT1 and anti-DBC1 antibodies. The graph shows the average of three independent experiments. * Denotes difference to control, and ** denotes difference to RSV+CC. Error bars represent S.D. C, SIRT1-DBC1 interaction was evaluated by co-immunoprecipitation in 293T cells after transfection of FLAG-SIRT1, Myc-DBC1, and CA AMPα or dominant-negative AMPKα (DN). Immunoprecipitates were immunoblotted with anti-FLAG and anti-Myc antibodies.
We also evaluated the effect of a CA form of AMPK or a dominant-negative form of AMPK on the interaction between transfected SIRT1 and DBC1 (Fig. 5C). Our results show that cellular AMPK activity inversely correlates with the amount of interaction between SIRT1 and DBC1. In other words, the transfection of the CA form of AMPK decreased the interaction between SIRT1 and DBC1, whereas the dominant-negative form increased the interaction between these two proteins. This set of experiments demonstrates that AMPK modulates the interaction between SIRT1 and DBC1.
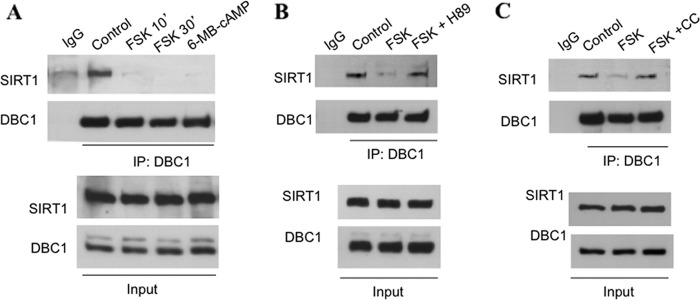
Next, we tested whether the cAMP/PKA pathway also modulates the SIRT1-DBC1 complex. We activated PKA with forskolin and 6-MB-cAMP and evaluated the interaction between SIRT1 and DBC1. Treatment of A549 cells with these compounds resulted in a clear loss of the interaction between SIRT1 and DBC1 (Fig. 6A), an event that was blocked by preincubation with the PKA inhibitor H89 (Fig. 6B). To further prove that PKA activates SIRT1 in an AMPK-dependent manner, we tested the effect of compound C on the dissociation promoted by forskolin. As shown in Fig. 6C, preincubation of cells with compound C blocked the dissociation induced by forskolin, indicating that indeed AMPK is part of the signaling pathway that connects PKA and SIRT1.
FIGURE 6.
cAMP-PKA activation promotes dissociation of SIRT1 from DBC1 by AMPK-dependent mechanism. A–C, co-immunoprecipitation (IP) of SIRT1 with DBC1 in A549 cells. DBC1 was immunoprecipitated from cell lysates, and immunoprecipitates were immunoblotted with anti-SIRT1 and anti-DBC1 antibodies. In A, cells were stimulated with forskolin (FSK) + isobutylmethylxanthine (IBMX) (10 and 70 μm, respectively) for the indicated times or the PKA activator 6-MB-cAMP (100 μm) for 10 min (′). B, cells were pretreated with the PKA inhibitor H89 (30 μm for 45 min) and then stimulated with forskolin and isobutylmethylxanthine (10 and 70 μm, respectively) for 10 min. C, cells were pretreated with compound C (CC; 10 μm) for 2 h and then stimulated as in B.
SIRT1 Is Phosphorylated in Response to AMPK Activation
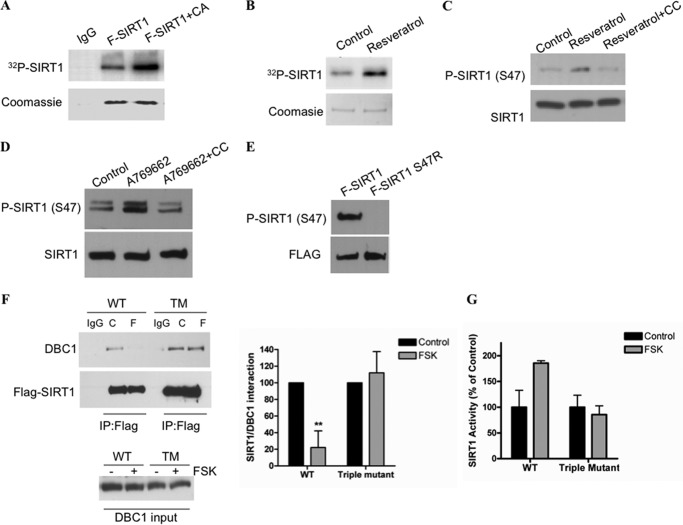
Phosphorylation is a key modification that regulates SIRT1 function as it has been clearly shown in numerous reports (18, 20–24). However, there is no evidence to support that either PKA or AMPK phosphorylates SIRT1 directly in vivo. Gerhart-Hines et al. (19) showed that Ser-434 phosphorylation on mouse SIRT1 is necessary for PKA regulation, although the authors speculate that the site is unlikely to be a direct target of PKA. On the other hand, two independent investigators failed to observe SIRT1 phosphorylation by AMPK in vitro or in cells (25, 59). Therefore, it is unlikely that either PKA or AMPK is directly phosphorylating SIRT1. However, PKA and AMPK activation could result in SIRT1 phosphorylation by an intermediate kinase. To determine whether AMPK activation results in SIRT1 phosphorylation, we performed autoradiography in cells loaded with 32P. As can be seen in Fig. 7A, transfection of a constitutively active form of AMPK resulted in incorporation of 32P into SIRT1. Moreover, treatment of cells with resveratrol also resulted in incorporation of 32P into SIRT1 (Fig. 7B). These data indicate that SIRT1 phosphorylation could be involved in the SIRT1 activation by PKA and AMPK.
FIGURE 7.
Activation of AMPK and PKA leads to SIRT1 phosphorylation. A, 293T cells were transfected with FLAG-SIRT1 or FLAG-SIRT1 plus a CA form of AMPK. The cell cultures were loaded with H332PO4, FLAG-SIRT1 was immunoprecipitated, and the radioactivity incorporated was visualized by autoradiography. B, 293T cells were transfected with FLAG-SIRT1, loaded with H332PO4, and later treated with 10 μm resveratrol for 2 h. SIRT1 phosphorylation was visualized by autoradiography. C, A549 cells were incubated with resveratrol (10 μm) or resveratrol and compound C (CC; 10 μm) for 2 h. Compound C was added 45 min before resveratrol. Samples were immunoblotted with anti-phospho (P)-SIRT1 (Ser-47). D, HepG2 cells were incubated with A769662 (100 μm) or A769662 and compound C (10 μm) for 2 h. E, Western blot with anti-phospho-SIRT1 (Ser-47) in 293T cells transfected with WT SIRT1 (FLAG (F)-SIRT1) or FLAG-S47RSIRT1. F, WT and the triple mutant (TM; S47R,S605R,S615R) SIRT1 were transfected in 293T cells. After 24 h, cells were incubated with forskolin (FSK; 10 μm) for 20 min. FLAG-SIRT1 was immunoprecipitated (IP), and the interaction was evaluated by immunoblot with anti-DBC1 and anti-FLAG antibodies. The graph shows the average of four experiments (**, p < 0.001; ANOVA test). G, SIRT1 activity was measured in the same experimental conditions described in B. Error bars represent S.D.
To date, several phosphorylation sites have been reported on SIRT1. We directly tested the phosphorylation of serine 47 because this site has been suggested to mediate activation of SIRT1 (13). For this, we used a specific antibody against phosphorylated serine 47. We found that treatment with resveratrol or A769662 resulted in an increase in phosphorylation in serine 47 that was blocked by preincubation with compound C (Fig. 7, C and D). The specificity of the phosphoantibody was assessed by immunoblotting of the wild type SIRT1 and the mutant form of SIRT1 in which serine 47 was replaced by arginine (S47R). As can be seen in Fig. 7E, the antibody did not recognize the mutant that lacks serine 47. In light of these findings, we tested the relevance of this site in the regulation of SIRT1 by PKA and AMPK. In particular, we tested the ability of the S47R mutant to dissociate from DBC1 in response to AMPK activation. However, we found that AMPK activators were still able to dissociate the complex between DBC1 and the S47R SIRT1 mutant (data not shown), suggesting that more than one site may be needed to regulate the interaction by AMPK.
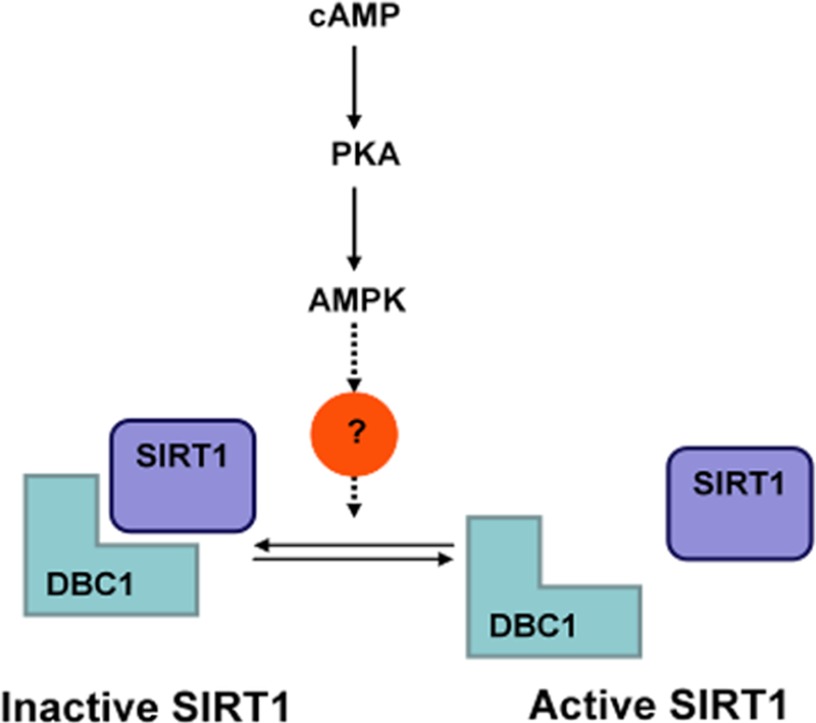
In an attempt to map additional sites on SIRT1 that might be a target of regulation by PKA/AMPK and to evaluate the importance of the dissociation of SIRT1 and DBC1 in the activation of SIRT1 mediated by PKA/AMPK, we performed point mutations of other reported phosphosites (for a complete list of mutations, see supplemental Fig. S5). First, we tested the ability of the single phosphomutants to dissociate from DBC1 upon activation of PKA and AMPK. We found that AMPK activators were still able to dissociate DBC1 from all the SIRT1 single phosphomutants. We therefore speculated that that a combination of phosphosites could be responsible for the regulation of the binding between SIRT1 and DBC1. Interestingly, the triple mutant S47R,S605R,S615R did not dissociates from DBC1 in response to PKA activation (Fig. 7F). Furthermore, the activity of the triple mutant was insensitive to PKA activation (Fig. 7G). The results shown in Fig. 7, F and G, clearly show that the dissociation from DBC1 is required for the SIRT1 activation promoted by the cAMP/PKA pathway. Moreover, our data identified the sites 47, 605, and 615 as residues involved in the regulation of the interaction between SIRT1 and DBC1. Although we do not have evidence that the activation of SIRT1 involves phosphorylation of serines 605 and 615, we provide evidence that suggest that these sites modulate the binding between SIRT1 and DBC1. A scheme representing our main findings is shown in Fig. 8.
FIGURE 8.
Proposed mechanism of SIRT1 regulation by PKA and AMPK.
DISCUSSION
The beneficial effects of SIRT1 activation have been studied extensively (60). Mounting evidence from independent groups shows that SIRT1 activation leads to protection against metabolic syndrome, cardiovascular diseases, and cancer (60) among other diseases. However, a much more complex issue is how to achieve SIRT1 activation in vivo. We have recently shown that, in vivo, most SIRT1 is bound to its regulator DBC1 and that this interaction is dynamic and can be displaced (31). In fact, we also showed that when DBC1 is absent and therefore SIRT1 is more active mice are protected against metabolic syndrome (31). It therefore becomes of key importance to understand how the SIRT1-DBC1 complex is regulated because it may lead to the development of new strategies to activate SIRT1 pharmacologically.
In this work, we attempted to characterize in detail the mechanism involved in the regulation of the interaction between SIRT1 and DBC1 and therefore SIRT1 activity. We found that SIRT1 activity is positively regulated by the protein kinases PKA and AMPK. We showed that PKA activation leads to a fast and transient SIRT1 activation. This activation was AMPK-dependent, involved the dissociation of SIRT1 from DBC1, and occurred independently of changes in NAD+ levels.
While this article was in preparation, Gerhart-Hines et al. (19) also showed SIRT1 activation by PKA. These authors propose that SIRT1 activation by PKA involves phosphorylation of SIRT1 and changes in the affinity of SIRT1 for NAD+. Although our findings do not exclude this possibility, we showed that SIRT1 must dissociate from DBC1 to be activated by PKA. Also, a recent paper shows that the polyphenol resveratrol induces cellular SIRT1 activation via activation of the cAMP/EPAC pathway and AMPK (61). Therefore, our group and two independent groups have shown that an increase in cAMP leads to SIRT1 activation. Our study provides further information about the specific mechanism of SIRT1 activation, which is mediated by modulation of the SIRT1-DBC1 interaction.
Recent publications, including this one, have reported changes in SIRT1 activity that are independent of alterations in the concentration of NAD+ (19, 31). Since our first observation of an increase in SIRT1 activity in vivo without detectable changes in NAD+ concentration (31), evidence has accumulated to prove that it is possible to induce SIRT1 activation without detectable changes in intracellular NAD+ levels (19). During physiological processes, it may be necessary to activate SIRT1 and not other NAD+-consuming enzymes. However, an increase in the cytoplasmic and nuclear levels of NAD+ could result in higher activity of several sirtuins and the poly(ADP-ribosyl) polymerases. Therefore, one can foresee that there must be an alternative, specific mode of SIRT1 regulation.
The results presented here provide mechanistic insight into the PKA-induced SIRT1 activation. Our results point to AMPK as a kinase that is downstream of PKA. In support of this notion, we observed that forskolin induces a transient activation of AMPK that parallels the increase in SIRT1 activity, that the AMPK inhibitor compound C abolishes the effect of forskolin in SIRT1 activity, and that in AMPK KO MEFs SIRT1 is insensitive to cAMP elevations. In line with our observations, others have also reported that PKA can transiently activate AMPK (26–30, 62). This activation seems to occur through the kinase LKB1, which activates AMPK by phosphorylation of serine 172 (63). PKA phosphorylates LKB1 directly at serine 431 (62, 64), and phosphorylation at this site is needed for some of the LKB1 functions (64, 65). Another possible mechanism that may lead to AMPK activation by the cAMP/PKA pathway is through regulation of cAMP degradation. Once cAMP levels increase in cells, the level of this second messenger quickly returns to basal levels due to its degradation by phosphodiesterases. Phosphodiesterases degrade cAMP into 5′-AMP (66), and therefore its action could result in a fast accumulation of AMP. Therefore, it seems plausible that agents that activate the cAMP/PKA pathway could result in activation of AMPK either by activation of LKB1, an increase in AMP levels, or both.
In this work, we present evidence that SIRT1 is phosphorylated upon AMPK activation. However, it remains unknown which kinase is responsible for SIRT1 phosphorylation upon PKA and AMPK activation. Our data identified serines 47, 605, and 615 as key residues involved in the regulation of the interaction between SIRT1 and DBC1. We provide evidence of phosphorylation of serine 47, but it remains to be elucidated whether serines 605 and 615 are also phosphorylated when PKA and AMPK are activated. As mentioned before, two independent groups failed to detect SIRT1 phosphorylation by AMPK (25, 59), which suggests that the regulation of this process is extremely complex. However, sequence analysis of human SIRT1 suggests that SIRT1 could be a target for AMPK phosphorylation. The consensus sequence for AMPK phosphorylation as first proposed by Carling and Hardie (67) consists of an arginine in the −2, −3, or −4 position and a hydrophobic residue at −1 relative to the serine or threonine target of phosphorylation. Later on, a study by Dale et al. (68) described the importance of a leucine at positions −5 and +4 (although other hydrophobic amino acids are accepted too) besides the basic amino acid at −3 or −4. More recently, another study identified that AMPK favors a serine with a valine or arginine at −2 (69). However, it is also important to note that not all known substrates for AMPK have the perfect consensus sequence but a variation of it. FOXO3 for example is phosphorylated by AMPK at Ser-413, which is not flanked by a leucine at position −5 or +4, and Ser-588 that lacks a basic residue at −3 or −4 (59). Using the above data as a reference, we searched the human SIRT1 sequence for serines and threonines in potential AMPK consensus sequence. Interestingly, the serine residue in position 605, one of the sites that is involved in SIRT1 activation by PKA, is flanked by a lysine at −4, a leucine at −5, and a valine at −2 in addition to a hydrophobic amino acid at −1, which is very close to the optimal sequence for AMPK phosphorylation. This fact suggests that further effort should be made to determine whether SIRT1 is a direct target for AMPK.
Of interest is that Kang et al. (70) recently found that the amino acids 631–655 in SIRT1 (what they call the essential for SIRT1 activity region) are necessary for SIRT1 activation. Moreover, these authors proposed that this region is important for the regulation of SIRT1 by DBC1. Interestingly, two of the amino acids, 605 and 615, which are involved in the PKA/AMPK-induced activation of SIRT1, are in close proximity to this region. Therefore, amino acids 605 and 615 may play a role in the activation of SIRT1 by PKA and AMPK by regulating the essential for SIRT1 activity region of SIRT1.
Serine 47 in SIRT1 has been shown to be target of JNK1 (21). Furthermore, Gao et al. (18) recently showed that JNK1 leads to SIRT1 phosphorylation and a fast increase in SIRT1 activity upon glucose treatment that correlates very well with the time course that we observed for PKA-dependent SIRT1 activation. It could be that AMPK and JNK1 are acting in conjunction to promote SIRT1 dissociation form DBC1.
Finally, we want to highlight how important it is to understand how the interaction between SIRT1 and DBC1 is regulated to study the regulation of SIRT1. The fact that in the absence of DBC1 the cAMP/PKA and AMPK pathways are incapable of activating SIRT1 suggests that a possible phosphorylation of SIRT1 per se is not enough to sustain an increase in its activity. Furthermore, our observations place the SIRT1-DBC1 complex as a key physiological target for SIRT1 regulation.
In summary, our results provide a novel mechanism of SIRT1 activation by a cAMP/PKA/AMPK/DBC1-dependent pathway. It also provides the evidence that the interaction between SIRT1 and DBC1 can be regulated by endogenous cell signaling pathways and opens the possibility that other signals may also promote SIRT1 activation through the dissociation of the SIRT1-DBC1 complex. For instance, it was recently shown that the ataxia telangiectasia mutated kinase (ATM) phosphorylates DBC1 and increases the interaction between SIRT1 and DBC1 (71). Finally, mounting evidence indicates that modulation of SIRT1 activity can be achieved without the nonspecific changes in global cellular NAD+ levels. The understanding of specific mechanisms of SIRT1 activation as described here may provide a clearer picture about the regulation of cellular SIRT1 and its physiological consequences.
This work was supported, in whole or in part, by National Institutes of Health Grant DK-084055 from the NIDDK. This work was also supported by grants from the American Federation of Aging Research and the Mayo Foundation and by the Strickland Career Development Award.

This article contains supplemental Figs. S1–S5.
- AMPK
- AMP-activated protein kinase
- DBC1
- deleted in breast cancer 1
- AICAR
- 5-amino-1-β-d-ribofuranosylimidazole-4-carboxamide
- 6-MB-cAMP
- N6-monobutytyl-cAMP
- EPAC
- exchange protein activated by cAMP
- cpt-cAMP
- 8-(4-chlorophenylthio)-adenosine 3,5′-cycle monophosphate-cAMP
- MEF
- mouse embryonic fibroblast
- CA
- constitutively active
- ANOVA
- analysis of variance.
REFERENCES
- 1. Liu Y., Dentin R., Chen D., Hedrick S., Ravnskjaer K., Schenk S., Milne J., Meyers D. J., Cole P., Yates J., 3rd, Olefsky J., Guarente L., Montminy M. (2008) A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 3. Bordone L., Motta M. C., Picard F., Robinson A., Jhala U. S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., Easlon E. J., Lin S. J., Guarente L. (2006) Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic β cells. PLoS Biol. 4, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moynihan K. A., Grimm A. A., Plueger M. M., Bernal-Mizrachi E., Ford E., Cras-Mneur C., Permutt M. A., Imai S. (2005) Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2, 105–117 [DOI] [PubMed] [Google Scholar]
- 5. Schenk S., McCurdy C. E., Philp A., Chen M. Z., Holliday M. J., Bandyopadhyay G. K., Osborn O., Baar K., Olefsky J. M. (2011) Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J. Clin. Investig. 121, 4281–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 9. Revollo J. R., Grimm A. A., Imai S. (2007) The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Curr. Opin. Gastroenterol. 23, 164–170 [DOI] [PubMed] [Google Scholar]
- 10. Aksoy P., White T. A., Thompson M., Chini E. N. (2006) Regulation of intracellular levels of NAD: a novel role for CD38. Biochem. Biophys. Res. Commun. 345, 1386–1392 [DOI] [PubMed] [Google Scholar]
- 11. Barbosa M. T., Soares S. M., Novak C. M., Sinclair D., Levine J. A., Aksoy P., Chini E. N. (2007) The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J. 21, 3629–3639 [DOI] [PubMed] [Google Scholar]
- 12. Chini E. N. (2009) CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr. Pharm. Des. 15, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Revollo J. R., Grimm A. A., Imai S. (2004) The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279, 50754–50763 [DOI] [PubMed] [Google Scholar]
- 14. Aksoy P., Escande C., White T. A., Thompson M., Soares S., Benech J. C., Chini E. N. (2006) Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem. Biophys. Res. Commun. 349, 353–359 [DOI] [PubMed] [Google Scholar]
- 15. Chen D., Bruno J., Easlon E., Lin S. J., Cheng H. L., Alt F. W., Guarente L. (2008) Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 22, 1753–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haigis M. C., Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Y., Fu W., Chen J., Olashaw N., Zhang X., Nicosia S. V., Bhalla K., Bai W. (2007) SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 9, 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao Z., Zhang J., Kheterpal I., Kennedy N., Davis R. J., Ye J. (2011) Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J. Biol. Chem. 286, 22227–22234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerhart-Hines Z., Dominy J. E., Jr., Blttler S. M., Jedrychowski M. P., Banks A. S., Lim J. H., Chim H., Gygi S. P., Puigserver P. (2011) The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD+. Mol. Cell 44, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo X., Williams J. G., Schug T. T., Li X. (2010) DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 285, 13223–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009) JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One 4, e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sasaki T., Maier B., Koclega K. D., Chruszcz M., Gluba W., Stukenberg P. T., Minor W., Scrable H. (2008) Phosphorylation regulates SIRT1 function. PLoS One 3, e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zschoernig B., Mahlknecht U. (2009) Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem. Biophys. Res. Commun. 381, 372–377 [DOI] [PubMed] [Google Scholar]
- 24. Kang H., Jung J. W., Kim M. K., Chung J. H. (2009) CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA damage. PLoS One 4, e6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cant C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yin W., Mu J., Birnbaum M. J. (2003) Role of AMP-activated protein kinase in cyclic AMP-dependent lipolysis in 3T3-L1 adipocytes. J. Biol. Chem. 278, 43074–43080 [DOI] [PubMed] [Google Scholar]
- 27. Wu H. M., Yang Y. M., Kim S. G. (2011) Rimonabant, a cannabinoid receptor type 1 inverse agonist, inhibits hepatocyte lipogenesis by activating liver kinase B1 and AMP-activated protein kinase axis downstream of Gαi/o inhibition. Mol. Pharmacol. 80, 859–869 [DOI] [PubMed] [Google Scholar]
- 28. Kimball S. R., Siegfried B. A., Jefferson L. S. (2004) Glucagon represses signaling through the mammalian target of rapamycin in rat liver by activating AMP-activated protein kinase. J. Biol. Chem. 279, 54103–54109 [DOI] [PubMed] [Google Scholar]
- 29. Williamson D. L., Kubica N., Kimball S. R., Jefferson L. S. (2006) Exercise-induced alterations in extracellular signal-regulated kinase 1/2 and mammalian target of rapamycin (mTOR) signalling to regulatory mechanisms of mRNA translation in mouse muscle. J. Physiol. 573, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Djouder N., Tuerk R. D., Suter M., Salvioni P., Thali R. F., Scholz R., Vaahtomeri K., Auchli Y., Rechsteiner H., Brunisholz R. A., Viollet B., T. P., Wallimann T., Neumann D., Krek W. (2010) PKA phosphorylates and inactivates AMPKα to promote efficient lipolysis. EMBO J. 29, 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Escande C., Chini C. C., Nin V., Dykhouse K. M., Novak C. M., Levine J., van Deursen J., Gores G. J., Chen J., Lou Z., Chini E. N. (2010) Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Investig. 120, 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao W., Kruse J. P., Tang Y., Jung S. Y., Qin J., Gu W. (2008) Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim J. E., Chen J., Lou Z. (2008) DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 34. Trauernicht A. M., Kim S. J., Kim N. H., Boyer T. G. (2007) Modulation of estrogen receptor α protein level and survival function by DBC-1. Mol. Endocrinol. 21, 1526–1536 [DOI] [PubMed] [Google Scholar]
- 35. Koyama S., Wada-Hiraike O., Nakagawa S., Tanikawa M., Hiraike H., Miyamoto Y., Sone K., Oda K., Fukuhara H., Nakagawa K., Kato S., Yano T., Taketani Y. (2010) Repression of estrogen receptor β function by putative tumor suppressor DBC1. Biochem. Biophys. Res. Commun. 392, 357–362 [DOI] [PubMed] [Google Scholar]
- 36. Fu J., Jiang J., Li J., Wang S., Shi G., Feng Q., White E., Qin J., Wong J. (2009) Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J. Biol. Chem. 284, 6832–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hiraike H., Wada-Hiraike O., Nakagawa S., Koyama S., Miyamoto Y., Sone K., Tanikawa M., Tsuruga T., Nagasaka K., Matsumoto Y., Oda K., Shoji K., Fukuhara H., Saji S., Nakagawa K., Kato S., Yano T., Taketani Y. (2010) Identification of DBC1 as a transcriptional repressor for BRCA1. Br. J. Cancer 102, 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chini C. C., Escande C., Nin V., Chini E. N. (2010) HDAC3 is negatively regulated by the nuclear protein DBC1. J. Biol. Chem. 285, 40830–40837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., Sassone-Corsi P. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van der Veer E., Ho C., O'Neil C., Barbosa N., Scott R., Cregan S. P., Pickering J. G. (2007) Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J. Biol. Chem. 282, 10841–10845 [DOI] [PubMed] [Google Scholar]
- 41. Chen Z., Peng I. C., Cui X., Li Y. S., Chien S., Shyy J. Y. (2010) Shear stress, SIRT1, and vascular homeostasis. Proc. Natl. Acad. Sci. U.S.A. 107, 10268–10273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Borra M. T., Smith B. C., Denu J. M. (2005) Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 280, 17187–17195 [DOI] [PubMed] [Google Scholar]
- 43. Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., Scherer B., Sinclair D. A. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 44. Wood J. G., Rogina B., Lavu S., Howitz K., Helfand S. L., Tatar M., Sinclair D. (2004) Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686–689 [DOI] [PubMed] [Google Scholar]
- 45. Beher D., Wu J., Cumine S., Kim K. W., Lu S. C., Atangan L., Wang M. (2009) Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 74, 619–624 [DOI] [PubMed] [Google Scholar]
- 46. Pacholec M., Bleasdale J. E., Chrunyk B., Cunningham D., Flynn D., Garofalo R. S., Griffith D., Griffor M., Loulakis P., Pabst B., Qiu X., Stockman B., Thanabal V., Varghese A., Ward J., Withka J., Ahn K. (2010) SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodgers J. T., Puigserver P. (2007) Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiang G., Zhang B. B. (2003) Glucagon and regulation of glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 284, E671–E678 [DOI] [PubMed] [Google Scholar]
- 49. Cantó C., Auwerx J. (2012) Targeting sirtuin 1 to improve metabolism: all you need is NAD+? Pharmacol. Rev. 64, 166–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murray A. J. (2008) Pharmacological PKA inhibition: all may not be what it seems. Sci. Signal 1, re4. [DOI] [PubMed] [Google Scholar]
- 51. Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 52. Moreno D., Knecht E., Viollet B., Sanz P. (2008) A769662, a novel activator of AMP-activated protein kinase, inhibits non-proteolytic components of the 26S proteasome by an AMPK-independent mechanism. FEBS Lett. 582, 2650–2654 [DOI] [PubMed] [Google Scholar]
- 53. Corton J. M., Gillespie J. G., Hawley S. A., Hardie D. G. (1995) 5-Aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 229, 558–565 [DOI] [PubMed] [Google Scholar]
- 54. Hawley S. A., Ross F. A., Chevtzoff C., Green K. A., Evans A., Fogarty S., Towler M. C., Brown L. J., Ogunbayo O. A., Evans A. M., Hardie D. G. (2010) Use of cells expressing γ subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 11, 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barata H., Thompson M., Zielinska W., Han Y. S., Mantilla C. B., Prakash Y. S., Feitoza S., Sieck G., Chini E. N. (2004) The role of cyclic-ADP-ribose-signaling pathway in oxytocin-induced Ca2+ transients in human myometrium cells. Endocrinology 145, 881–889 [DOI] [PubMed] [Google Scholar]
- 56. Chini E. N., Nagamune K., Wetzel D. M., Sibley L. D. (2005) Evidence that the cADPR signalling pathway controls calcium-mediated microneme secretion in Toxoplasma gondii. Biochem. J. 389, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nagamune K., Hicks L. M., Fux B., Brossier F., Chini E. N., Sibley L. D. (2008) Abscisic acid controls calcium-dependent egress and development in Toxoplasma gondii. Nature 451, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gledhill J. R., Montgomery M. G., Leslie A. G., Walker J. E. (2007) Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. U.S.A. 104, 13632–13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Greer E. L., Oskoui P. R., Banko M. R., Maniar J. M., Gygi M. P., Gygi S. P., Brunet A. (2007) The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 282, 30107–30119 [DOI] [PubMed] [Google Scholar]
- 60. Guarente L. (2011) Franklin H. Epstein Lecture: sirtuins, aging, and medicine. N. Engl. J. Med. 364, 2235–2244 [DOI] [PubMed] [Google Scholar]
- 61. Park S. J., Ahmad F., Philp A., Baar K., Williams T., Luo H., Ke H., Rehmann H., Taussig R., Brown A. L., Kim M. K., Beaven M. A., Burgin A. B., Manganiello V., Chung J. H. (2012) Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Collins S. P., Reoma J. L., Gamm D. M., Uhler M. D. (2000) LKB1, a novel serine/threonine protein kinase and potential tumour suppressor, is phosphorylated by cAMP-dependent protein kinase (PKA) and prenylated in vivo. Biochem. J. 345, 673–680 [PMC free article] [PubMed] [Google Scholar]
- 63. Hardie D. G. (2005) New roles for the LKB1→AMPK pathway. Curr. Opin. Cell Biol. 17, 167–173 [DOI] [PubMed] [Google Scholar]
- 64. Sapkota G. P., Kieloch A., Lizcano J. M., Lain S., Arthur J. S., Williams M. R., Morrice N., Deak M., Alessi D. R. (2001) Phosphorylation of the protein kinase mutated in Peutz-Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90RSK and cAMP-dependent protein kinase, but not its farnesylation at Cys433, is essential for LKB1 to suppress cell growth. J. Biol. Chem. 276, 19469–19482 [DOI] [PubMed] [Google Scholar]
- 65. Shelly M., Cancedda L., Heilshorn S., Sumbre G., Poo M. M. (2007) LKB1/STRAD promotes axon initiation during neuronal polarization. Cell 129, 565–577 [DOI] [PubMed] [Google Scholar]
- 66. Lugnier C. (2006) Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol. Ther. 109, 366–398 [DOI] [PubMed] [Google Scholar]
- 67. Carling D., Hardie D. G. (1989) The substrate and sequence specificity of the AMP-activated protein kinase. Phosphorylation of glycogen synthase and phosphorylase kinase. Biochim. Biophys. Acta 1012, 81–86 [DOI] [PubMed] [Google Scholar]
- 68. Dale S., Wilson W. A., Edelman A. M., Hardie D. G. (1995) Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 361, 191–195 [DOI] [PubMed] [Google Scholar]
- 69. Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kang H., Suh J. Y., Jung Y. S., Jung J. W., Kim M. K., Chung J. H. (2011) Peptide switch is essential for Sirt1 deacetylase activity. Mol. Cell 44, 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yuan J., Luo K., Liu T., Lou Z. (2012) Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 26, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]