Abstract
Molecular profiling has allowed a more precise classification of human cancers. With respect to breast cancer, this approach has been used to identify five subtypes; luminal A, luminal B, HER2-enriched, basal-like and claudin-low. In addition, this approach can be used to determine the type of tumor represented by particular cell lines or transgenic animal models. Therefore, this approach was utilized to classify the mammary tumors that develop in MTB-IGFIR transgenic mice. It was determined that the primary mammary tumors, which develop due to elevated expression of the type I insulin-like growth factor receptor (IGF-IR) in mammary epithelial cells, most closely resemble murine tumors with basal-like or mixed gene expression profiles and with human basal-like breast cancers. Downregulation of IGF-IR transgene in MTB-IGFIR tumor-bearing mice leads to the regression of most of the tumors, followed by tumor reappearance in some of the mice. These tumors that reappear following IGF-IR transgene downregulation do not express the IGF-IR transgene and cluster with murine mammary tumors that express a mesenchymal gene expression profile and with human claudin-low breast cancers. Therefore, IGF-IR overexpression in murine mammary epithelial cells induces mammary tumors with primarily basal-like characteristics, whereas tumors that develop following IGF-IR downregulation express a gene signature that most closely resembles human claudin-low breast tumors.
Keywords: IGF-IR, mammary tumor, DNA microarray, EMT, claudin
Introduction
Molecular profiling has identified five distinct subtypes of human breast tumors; luminal A, luminal B, HER2-enriched, basal-like and claudin-low (Perou et al., 2000; Sorlie et al., 2001, 2003; Fan et al., 2006; Hu et al., 2006). Luminal A tumors are estrogen receptor positive, cytokeratin 8 and 18 positive and have a good prognosis, whereas luminal B tumors express the estrogen receptor and cytokeratins 8 and 18 but have a poorer prognosis than luminal A tumors (Perou et al., 2000; Sorlie et al., 2001, 2003). HER2-enriched tumors, as the name suggests, are characterized by high expression of HER2. Basal-like tumors do not express cytokeratins associated with luminal epithelial cells, such as cytokeratins 8 and 18, but do express cytokeratins 5, 6 and 14, as well as vimentin (Perou et al., 2000; Sorlie et al., 2001, 2003). Claudin-low tumors were identified in 2007 and are characterized by low levels of claudins 3, 4 and 7, as well as other tight junction proteins (Herschkowitz et al., 2007; Prat et al., 2010). Claudin-low tumors express high levels of markers associated with epithelial-to-mesenchymal transition, such as TWIST1, TWIST2, ZEB1, ZEB2, Slug and Snail, while expressing little or no markers of luminal differentiation (Herschkowitz et al., 2007). Claudin-low subtypes have been reported to most closely resemble mammary epithelial stem cells (Prat et al., 2010).
Using the molecular profiles of the various human tumor subtypes, scientist can now determine which type of human mammary tumor their transgenic animal model most closely resembles. This approach has been used for a number of transgenic mammary tumor models, including WAP-Myc, WAP-Tag, MMTV-neu, MMTV-PyVT and MMTV-Wnt transgenic mice (Herschkowitz et al., 2007). One transgenic model that has not been evaluated using this approach is our MTB-IGFIR transgenic mice. These mice overexpress human insulin-like growth factor receptor (IGF-IR) in mammary epithelial cells in a doxycycline-inducible manner (Jones et al., 2007). Transgenic expression of IGF-IR results in the rapid development of mammary tumors. These tumors express variable levels of luminal cytokeratins, such as cytokeratin 8 and 18, and most large tumors contain some cells that express basal cytokeratins, such as cytokeratin 5 and 14 (Jones et al., 2007). Suppression of IGF-IR transgene expression in established mammary tumors in MTB-IGFIR transgenic mice results in the regression of most of the tumors; however, some of the tumors resume growth and these tumors take on a more spindle-like morphology and do not express high levels of the IGF-IR transgene (Jones et al., 2009).
This manuscript describes the molecular profiling of primary mammary tumors (PMTs) that develop in MTB-IGFIR transgenic mice as well as the mammary tumors that become independent of IGF-IR signaling and take on a spindle-shaped morphology (recurrent spindle tumors or RSTs). DNA microarray analysis revealed that PMTs have distinct genetic signatures that segregate from both wild-type (WT) mammary tissue and RSTs. Using the gene signature established by Herschkowitz et al. (2007), it was found that the PMTs clustered most closely with group of human tumors that included 51/58 basal-like tumors, 4/17 claudin-low tumors and 2/31 HER2-enriched tumors. In all, 2/9 RST samples clustered with the same group of tumors as the PMT samples, whereas the remaining 7/9 RSTs clustered most closely with 5/17 claudin-low tumors and 1/15 normal-like sample. Therefore, elevated IGF-IR expression results in relatively homogenous mammary tumors that are genetically similar to human basal-like tumors, whereas downregulation of IGF-IR produces tumors with a gene signature most similar to human claudin-low tumors.
Results
We have developed a transgenic model where IGF-IR transgene expression can be induced in mammary epithelial cells in a doxycycline-inducible manner. Overexpression of IGF-IR in mammary epithelial cells results in the development of mammary tumors and these tumors are designated as PMTs (Jones et al., 2007). Downregulation of IGF-IR transgene (through doxycycline withdrawal) in established mammary tumors resulted in regression of most of the tumors (Jones et al., 2009). A small percentage of these tumors recurred, acquired a spindle-shaped morphology and expressed only low levels of the IGF-IR. These recurrent tumors have been designated as RSTs (Jones et al., 2009).
The activation of intracellular signaling molecules downstream of the IGF-IR in WT, PMT and RST samples has previously been reported (Jones et al., 2007, 2009). We found that the PMT samples had higher levels of IGF-IR, phosphorylated IGF-IR, phosphorylated Akt, phosphorylated Stat3 and total Stat3 compared with normal mammary tissue. Phosphorylated Erk1/2 and phosphorylated p38 MAPK levels were similar in PMT samples and normal mammary tissue (Jones et al., 2007). RST samples had similar levels of phosphorylated Akt, phosphorylated Erk1/2 and phosphorylated Stat3 as the PMT samples (Jones et al., 2009) Analysis of a number of receptors in the WT, PMT and RST samples showed that the PMT samples contain higher levels of IGF-IR and phosphorylated IGF-IR compared with either the WT or RST samples (Figure 1). This finding was consistent with our previous study that showed that the RST samples have much lower levels of IGF-IR than the PMT samples (Jones et al., 2009). The insulin receptor and epidermal growth factor receptor were both expressed at similar levels in all of the samples (Figure 1). The level of phosphorylated epidermal growth factor receptor was also similar in all of the samples (Figure 1).
Figure 1.
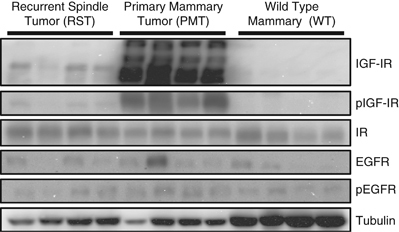
Western blot analysis of the IGF-IR, phosphorylation IGF-IR (pIGF-IR), insulin receptor (IR), epidermal growth factor receptor (EGFR), phosphorylated EGFR (pEGFR) in WT mammary tissue, PMT or RST. β-Tubulin (Tubulin) served as a loading control.
Immunohistochemistry for the luminal cytokeratins, cytokeratin 18 and two basal cytokeratins, cytokeratins 5 and 14, revealed that some of the cells within the PMT samples had intense staining for cytokeratin 18, whereas most of the cells had a lower level of staining (Figure 2). In contrast, most of the cells in the RST samples were negative for cytokeratin 18. The small cluster of cytokeratin 18-positive cells within the RST sample appear to be remnants of a normal mammary duct (Figure 2). Most of the cells within the PMT samples were negative (or stained at very low levels) for cytokeratins 5 and 14; however, small clusters of positive cells could be found in the PMT samples. In contrast, the RST samples displayed increased staining for cytokeratin 5 or 14 compared with the PMT samples (Figure 2).
Figure 2.
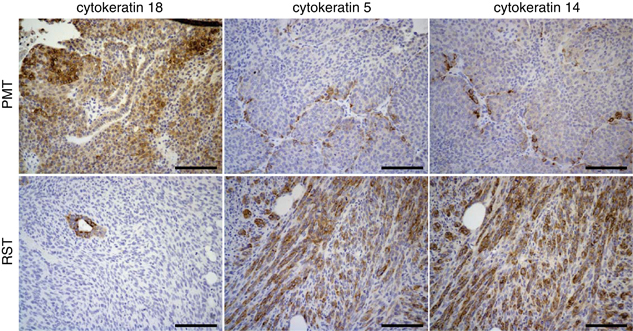
Immunohistochemistry for the epithelial cytokeratin, cytokeratin 18 and two basal cytokeratins, cytokeratins 5 and 14, in PMTs and RSTs. Scale bar, 100 μm.
To gain a better understanding of genetic alterations associated with IGF-IR-induced mammary tumorigenesis and tumor survival following IGF-IR downregulation, DNA microarrays were performed on tissue from WT, PMT and RST samples. Cluster analysis revealed that the WT, PMT and RST samples clustered into discreet groups with one exception, sample RJ427 (Figure 3a). Sample RJ427 was originally classified as an RST but clustered more similarly with the PMT samples than the RST samples. On retrospective histological analysis, this tumor sample had a mixed phenotype containing cells with mesenchymal and epithelial features (Figures 3b-d).
Figure 3.
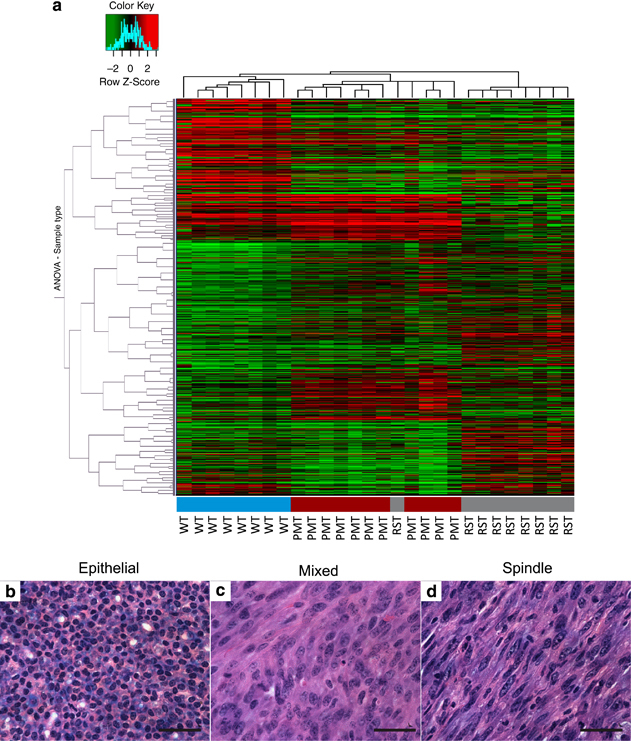
(a) Heat map of the hierarchical clustering of 8 WT mammary glands (blue), 11 PMTs (red) and 9 RSTs (gray). The three tissue types form distinct clusters, except for one of the RST samples that clustered with the PMT samples. The legend shows the z-scores. The values were mean centered, and the colors scaled from −2 to 2 standard deviations. Hematoxylin and eosin-stained sections of a representative (b) PMT sample, (c) RST sample and (d) the one RST sample that contained mixed cell morphology and clustered with the PMT samples. Scale bar, 100 μm.
Comparison of the global gene expression profiles between the PMT and WT samples revealed that 2564 genes were significantly upregulated at least twofold in the PMT samples compared with the WT samples and 2545 genes were significantly downregulated at least twofold in the PMT samples compared with the WT samples. The top 25 upregulated and top 25 downregulated genes are presented in Tables 1 and 2. Quantitative reverse transcriptase PCR (qRT–PCR) was used to confirm the expression of nine genes differentially expressed in the PMT and WT samples (Table 3). As shown in Table 3, all of the genes elevated in the PMT samples compared with the WT samples in the DNA microarray were also elevated as determined by qRT–PCR, except the insulin receptor (Insr). A BLASTN search revealed that the sequence used for the Agilent DNA microarray matched 51 of 59 nucleotides to the human IGF-IR, whereas the forward primer (3-AGATGAGAGGTGCAGTGTGGCT-5) and the reverse primer (3-GGTTCCTTTGGCTCTTGCCACA-5) for murine Insr used for the qRT–PCR do not match human IGF-IR nucleotide sequence. In addition, there was no obvious increase in insulin receptor protein levels in the PMT samples as determined by western blotting (Figure 1). Therefore, the increase in the Insr in the DNA microarray is likely due to the high level of the human IGF-IR transgene being expressed in our MTB-IGFIR mice and not due to the increased expression of murine Insr.
Table 1. Top 25 upregulated genes in PMT versus WT.
| Agilent probe name | Gene name | Gene description | Gene ontology | Fold change | P-value |
|---|---|---|---|---|---|
| A_51_P358765 | Spp1 | Secreted phosphoprotein 1 or osteopontin | Antiapoptosis; biomineral formation; cell adhesion; ossification; osteoblast differentiation | 77.3 | 1.1 × 10−5 |
| A_51_P301435 | B3galt5 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase polypeptide 5 | Protein amino-acid glycosylation | 58.6 | 1.9 × 10−7 |
| A_51_P384894 | Stfa1 | Stefin-A1-like protein | Cysteine-type endopeptidase inhibitor activity | 56.6 | 9.4 × 10−7 |
| A_52_P6490 | Insr | Insulin receptor | Glucose homeostasis | 44.0 | 6.2 × 10−9 |
| A_51_P409694 | Psrc1 | Differential display and activated by p53 | Cell cycle | 35.0 | 4.6 × 10−6 |
| A_52_P646858 | Car12 | Carbonic anhydrase 12 | One-carbon metabolic process | 32.7 | 6.4 × 10−6 |
| A_51_P256827 | S100a8 | S100 calcium-binding protein A8 (calgranulin A) | Chemotaxis; calcium-ion binding | 32.1 | 3.9 × 10−5 |
| A_51_P240501 | 4930583H14 Rik | Ovary-specific acidic protein | Not determined | 30.1 | 5.1 × 10−4 |
| A_51_P184484 | Mmp13 | Matrix metalloproteinase 13 | Metallopeptidase activity; bone mineralization; cartilage development | 25.5 | 3.9 × 10−5 |
| A_52_P387009 | Egln3 | EGL nine homolog 3 | Apoptosis; cell proliferation; response to hypoxia | 22.7 | 1.2 × 10−6 |
| A_51_P268154 | Acsl4 | Acyl-CoA synthetase long-chain family member 4 isoform 2 | Fatty-acid metabolism and transport | 20.5 | 2.3 × 10−9 |
| A_52_P318673 | Saa1 | Serum amyloid A1 | Cholesterol metabolism | 20.2 | 6.5 × 10−5 |
| A_51_P461040 | Crct1 | Cysteine-rich C-terminal 1 | Not determined | 20.0 | 1.8 × 10−4 |
| A_52_P158095 | NAP107172-1 | Not determined | 19.3 | 4.1 × 10−5 | |
| A_52_P232580 | Dyrk3 | Dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 3 | Erythrocyte differentiation; protein kinase activity | 18.8 | 1.7 × 10−6 |
| A_52_P240232 | NAP052665-1 | Not determined | 18.6 | 1.8 × 10−5 | |
| A_51_P499698 | Asprv1 | Aspartic peptidase, retroviral-like 1 | Proteolysis | 18.1 | 5.7 × 10−7 |
| A_51_P302831 | Dbccr1 | Deleted in bladder cancer chromosome region candidate 1 | Cell cycle | 17.2 | 9.8 × 10−5 |
| A_52_P59206 | Cst6 | Cystatin E/M | Epidermis development | 16.3 | 7.5 × 10−6 |
| A_51_P459477 | Col11a1 | Procollagen, type XI, α1 | Cartilage development; cell adhesion; extracellular matrix constituent | 15.9 | 3.3 × 10−7 |
| A_52_P220440 | Slc6a15 | Solute carrier family 6 (neurotransmitter transporter), member 15 | Neurotransmitter transport | 15.6 | 5.1 × 10−4 |
| A_51_P457747 | Krt16 | Keratin 16 | Intermediate filament cytoskeleton organization | 15.0 | 4.2 × 10−5 |
| A_51_P212782 | Il1b | Interleukin 1β | Inflammatory response; angiogenesis; insulin signaling; lipid metabolism; chemotaxis | 14.5 | 1.8 × 10−6 |
| A_51_P147034 | Ica1l | Islet cell autoantigen 1-like | Not determined | 14.1 | 1.5 × 10−5 |
| A_51_P491667 | Derl3 | Der1-like domain family, member 3 | Not determined | 14.1 | 5.6 × 10−6 |
Abbreviations: PMT, primary mammary tumor; WT, wild type.
Table 2. Top 25 Downregulated genes in PMT versus WT.
| Agilent probe name | Gene name | Gene description | Gene ontology | Fold change | P-value |
|---|---|---|---|---|---|
| A_51_P495780 | Plin4 | Perilipin 4 | None listed | 80.0 | 6.0 × 10−5 |
| A_51_P259296 | Lpl | Lipoprotein lipase | Lipid metabolism | 70.4 | 7.6 × 10−6 |
| A_51_P374752 | Rbp4 | Retinol-binding protein 4, cellular | Retinol binding and transport | 57.8 | 5.9 × 10−5 |
| A_51_P420547 | TC1536702 | Not determined | Not determined | 56.2 | 3.8 × 10−8 |
| A_52_P592909 | Dgat2 | Diacylglycerol O-acyltransferase 2 | Glycerol metabolism; lipid metabolism | 49.8 | 8.1 × 10−5 |
| A_51_P412579 | Tmem182 | Transmembrane protein 182 | Not determined | 49.5 | 4.6 × 10−6 |
| A_52_P492989 | Ncan | Neurocan | Neuronal cell-cell adhesion | 48.3 | 2.4 × 10−6 |
| A_52_P258194 | Crtac1 | Cartilage acidic protein 1 | Extracellular matrix | 48.3 | 3.8 × 10−7 |
| A_52_P577533 | AI427515 | Not determined | 46.9 | 4.4 × 10−6 | |
| A_51_P375146 | Cd36 | CD36 antigen | Thrombospondin receptor activity; cell adhesion; fatty acid metabolism | 46.3 | 3.5 × 10−7 |
| A_51_P239236 | Acacb | Acetyl-coenzyme A carboxylase-β | Acetyl-CoA metabolism; fatty acid metabolism | 45.5 | 4.0 × 10−6 |
| A_51_P350453 | Pdk4 | Pyruvate dehydrogenase kinase, isoenzyme 4 | Acetyl-CoA metabolism; carbohydrate metabolism | 45.5 | 3.2 × 10−6 |
| A_52_P572808 | Agpat9 | 1-Acylglycerol-3-phosphate O-acyltransferase 9 | Phospholipid metabolism; triglyceride metabolism | 43.9 | 3.8 × 10−7 |
| A_51_P321126 | Fasn | Fatty acid synthase | Fatty acid metabolism | 41.3 | 3.9 × 10−5 |
| A_52_P306845 | Cav1 | Caveolin1, caveolae protein | Calcium ion homeostasis; cholesterol homeostasis; endocytosis; lipid metabolism; apoptosis; intracellular signaling | 37.0 | 4.6 × 10−5 |
| A_51_P383638 | Amy1 | Amylase 1, salivary | Carbohydrate metabolism | 36.9 | 1.3 × × 10−4 |
| A_51_P253117 | Sorbs1 | Sorbin and SH3 domain containing 1 | Insulin receptor signaling; glucose metabolism; lipid metabolism | 35.7 | 4.4 × 10−4 |
| A_52_P674808 | Chrdl1 | Chordin-like 1 | cell differentiation; ossification | 34.4 | 6.4 × 10−7 |
| A_51_P480020 | 4931408D14Rik | 4931408D15Rik | Not determined | 31.4 | 1.7 × 10−5 |
| A_51_P496432 | Acsl1 | Acyl-CoA synthetase long-chain family member 1 | Fatty acid metabolism; xenobiotic metabolism | 31.2 | 3.7 × 10−4 |
| A_51_P241465 | Gsn | Gelsolin | Actin polymerization–depolymerization, cell adhesion; apoptosis; vesicle- mediated transport | 31.0 | 5.5 × 10−6 |
| A_51_P221062 | Prkar2b | Protein kinase, cyclic AMP-dependent regulatory, type IIβ | Proliferation; fatty acid metabolism; protein amino-acid phosphorylation | 30.4 | 2.8 × 10−6 |
| A_51_P268697 | Slc1a3 | Solute carrier family 1 (glial high-affinity glutamate transporter), member 3 | Amino-acid transport; γ-aminobutyric acid metabolism; glutamate metabolism; response to light and sound; sodium ion export | 29.1 | 1.3 × 10−6 |
| A_51_P520718 | Ptger3 | Prostaglandin E receptor 3 (subtype EP3) | Bicarbonate transport, G-protein-coupled receptor signaling | 28.7 | 6.5 × 10−5 |
| A_51_P114094 | Clstn3 | Calsyntenin 3 | Cell adhesion | 28.2 | 5.9 × 10−7 |
Abbreviations: PMT, primary mammary tumor; WT, wild type.
Table 3. Quantitative reverse transcriptase PCR of genes identified in the DNA microarray, as being elevated in the PMT samples compared with the WT samples.
| Gene | WT | PMT | Fold increase in PMT |
|---|---|---|---|
| Spp1 | 2.00±0.56 | 830±279 | 416 |
| B3galt5 | 0.990.40 | 87.9±43.1 | 88.8 |
| Stfa1 | 2.81±1.53 | 83.2±29.0 | 29.6 |
| Insr | 0.73±0.22 | 1.25±0.20 | 1.72 |
| Car12 | 1.64±0.73 | 948±186 | 577 |
| S100a8 | 0.88±0.11 | 21.3±2.20 | 24.3 |
| MMP13 | 0.83±0.36 | 36.8±15.7 | 44.1 |
| Acsl4 | 0.90±0.10 | 62.3±2.00 | 69.2 |
| Saa1 | 1.20±0.14 | 2880±1975 | 2394 |
Abbreviations: PMT, primary mammary tumor; WT, wild type.
Gene expression is presented relative to hypoxanthine-guanine phosphoribosyltransferase (HPRT) and as mean±s.e.m.
Comparison of the global gene expression profiles between the PMT and RST samples revealed that 1794 genes were significantly upregulated at least twofold in the RST samples compared with the PMT samples and 1515 genes were significantly downregulated at least twofold in the RST samples compared with the PMT samples. The top 25 upregulated and top 25 downregulated genes are presented in Supplementary Tables 2 and 3.
The WT, PMT and RST samples were then clustered with the 13 mouse models of mammary tumorigenesis, initially analyzed by Herschkowitz et al. (2007). This analysis revealed that our WT samples clustered most closely with the normal mammary samples, whereas the PMT samples formed a discrete cluster, which included 11/11 PMT samples and 1/9 RST samples that did not cluster closely with any particular group of mammary tumors. The tumor types closest in gene expression pattern to the PMT samples included 8/11 DMBA-induced tumors, 11/11 TgMMTV-Wnt1 tumors, 5/5 TgWAP-Tag, 6/7 p53+/− IR, 8/8 TgC3(I)-Tag, 7/7 TgWAP-Int3 6/7 tumors, 6/7 TgWAP-T121 tumors, 7/7 BRCA1+/−;p53+/− IR tumors, 3/13 TgWAP-Myc tumors, 7/10 TgMMTV-Cre;BRCA1Co/Co;p53+/− tumors, 4/5 p53−/− transplant tumors and 1/7 TgMMTV-PyMT tumors (Figure 4). The samples that cluster most closely with the RSTs were 3/11 DMBA-induced tumors, 3/10 TgMMTV-Cre;BRCA1Co/Co;p53+/− tumors, 1/7 p53+/− IR tumors, 1/5 p53−/− transplant tumors and 1/10 TgMMTV-neu tumors (Figure 4).
Figure 4.

Unsupervised cluster analysis of murine mammary tumors samples from Herschkowitz et al. (2007) with our WT (blue), PMT (red) and RST (gray) samples. Each sample is identified by a color-coded matrix below the dendrogram. The legend shows the z-scores. The values were mean centered, and the colors scaled from −4 to 4 standard deviations.
When the PMT and RST samples were clustered with human breast cancers, the PMT samples again formed a discrete cluster with 11/11 PMT samples and 1/9 RST samples, and these samples clustered most closely with 51/58 basal-like tumors, 4/17 claudin-low tumors and 2/31 HER2-enriched tumors (Figure 5). As an additional analysis, the samples were clustered with a basal-like tumor gene signature described by Molyneux et al. (2010), and consistent with the clustering with the Herschkowitz data set (Herschkowitz et al., 2007), the PMT samples clustered together and have a gene expression signature similar to basal-like breast tumors (Figure 6).
Figure 5.
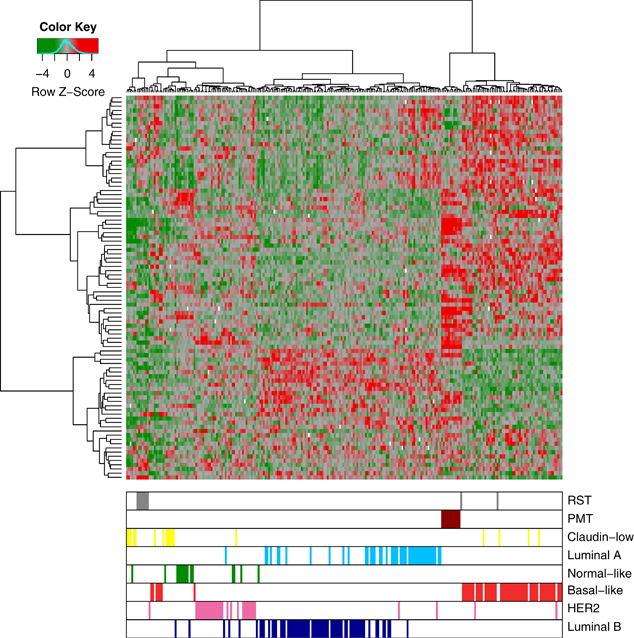
Unsupervised cluster analysis of human breast tumors samples from Herschkowitz et al. (2007) with our WT (blue), PMT (red) and RST (gray) samples. Each sample is identified by a color-coded matrix below the dendrogram. The legend shows the z-scores. The values were mean centered, and the colors scaled from −4 to 4 standard deviations.
Figure 6.
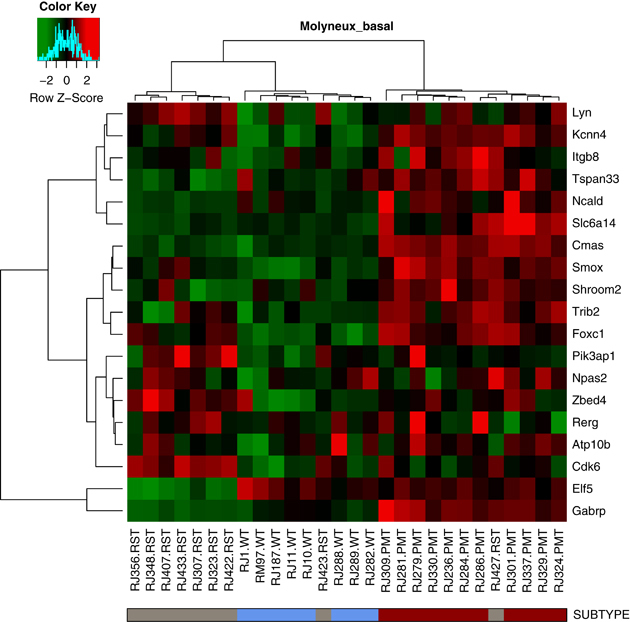
Heat map of WT (blue), PMT (red) and RST (gray) samples using a basal-like gene expression signature from Molyneux et al. (2010). Each sample is identified by a color-coded matrix below the dendrogram. The legend shows the z-scores. The values were mean centered, and the colors scaled from −2 to 2 standard deviations.
Meanwhile, 7/9 RSTs clustered most closely with 5/17 human claudin-low tumors and 1/15 normal samples (Figure 5). To confirm that the RSTs expressed low levels of claudins 3, 4 and 7, qRT–PCR was performed. As shown in Table 4, the RST samples had significantly lower levels of 2/3 claudins compared with the PMT samples. In addition, the gene expression profile of the WT, PMT and RST samples were evaluated using the 30-gene signature used to identify claudin-low tumors by Herschkowitz et al. (2007). When our samples were compared with the Herschkowitz claudin-low gene cluster, it was observed that 7/9 RST samples had low levels of the genes in the claudin-low gene cluster (Figure 7a). Similarly, using the claudin-low predictor described in Prat et al. (2010), 8/9 RST samples had a gene expression pattern consistent with a claudin-low genotype. Genes highly expressed in the claudin-low gene signature are indicated by red bars on the left-hand side of the expression profile (Figure 7b).
Table 4. Claudin expression in PMT and RST samples relative to hypoxanthine-guanine phosphoribosyltransferase (HPRT).
| PMT | RST | |
|---|---|---|
| Claudin 3 | 1.49±0.32 | 0.14±0.13* |
| Claudin 4 | 1.34±0.47 | 0.07±0.07 |
| Claudin 7 | 7.54±1.40 | 0.10±0.09* |
Abbreviations: PMT, primary mammary tumor; RST, recurrent spindle tumor.
*P<0.05.
Figure 7.
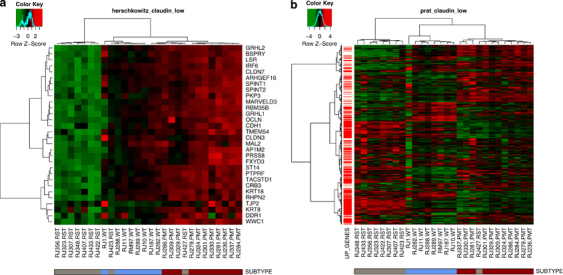
Heat maps of WT (blue), PMT (red) and RST (gray) samples using a claudin-low predictor from (a) Herschkowitz et al. (2007) and (b) Prat et al. (2010). The Herschkowitz predictor set includes 30 genes whose expression is low in claudin-low tumors. The Prat predictor set includes 1667 genes, 1308 of which are elevated in claudin-low tumors and are indicated as red bars on the left-hand side of the heat map. The legend shows the z-scores.
As a final analysis we used gene signature enrichment analysis (GSEA) to compare the gene expression profile of the PMT samples compared with the RST samples. This analysis revealed that 130 gene sets positively correlated with a false discovery rate q-value of <0.05 and 816 negatively correlated with a false discovery rate q-value of <0.05. Table 5 lists the top 20 positively correlated and Table 6 lists the top 20 negatively correlated gene sets based on normalized enrichment scores (all gene sets listed in the table had a false discovery rate q-value of 0.000). A number of the gene sets identified involved comparisons of different breast cancer subtypes or metastatic capacity.
Table 5. Top 20 positive correlations identified by GSEA compared with PMT versus RST.
| Gene Set | Description |
|---|---|
| CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_UP | Genes upregulated in luminal-like breast cancer cell lines compared with the mesenchymal-like ones |
| COLDREN_GEFITINIB_RESISTANCE_DN | Genes downregulated in NSCLC cell lines resistant to gefitinib compared with the sensitive ones |
| CHARAFE_BREAST_CANCER_BASAL_VS_MESENCHYMAL_UP | Genes upregulated in basal-like breast cancer cell lines as compared with the mesenchymal-like ones |
| JAEGER_METASTASIS_DN | Genes downregulated in metastases from malignant melanoma compared with the primary tumors |
| LANDIS_ERBB2_BREAST_TUMORS_324_UP | The 324 genes identified by two analytical methods as changed in the mammary tumors induced by transgenic ERBB2 |
| MCBRYAN_PUBERTAL_BREAST_4_5WK_UP | Genes upregulated during pubertal mammary gland development between weeks 4 and 5 |
| LANDIS_BREAST_CANCER_PROGRESSION_UP | Genes upregulated in preneoplastic mammary tissue and whose expression is maintained in tumors |
| ONDER_CDH1_TARGETS_2_DN | Genes downregulated in HMLE cells (immortalized nontransformed mammary epithelium) after E-cadherin knockdown by RNAi |
| AIGNER_ZEB1_TARGETS | Genes upregulated in MDA-MB-231 cells (breast cancer) after knockdown of ZEB1 by RNAi |
| LIEN_BREAST_CARCINOMA_METAPLASTIC_VS_DUCTAL_DN | Genes downregulated between two breast carcinoma subtypes: metaplastic (MCB) and ductal (DCB) |
| MCBRYAN_PUBERTAL_BREAST_3_4WK_UP | Genes upregulated during pubertal mammary gland development between weeks 3 and 4 |
| REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in tight junction interactions |
| SMID_BREAST_CANCER_ERBB2_UP | Genes upregulated in the ERBB2 subtype of breast cancer samples, characterized by higher expression of ERBB2 |
| WAMUNYOKOLI_OVARIAN_CANCER_GRADES_1_2_UP | Genes upregulated in mucinous ovarian carcinoma tumors of grades 1 and 2 compared with the normal surface epithelium tissue |
| SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_DN | Genes downregulated in IDC relative to DCIS (noninvasive) |
| SENGUPTA_NASOPHARYNGEAL_CARCINOMA_DN | Genes downregulated in nasopharyngeal carcinoma relative to the normal tissue |
| WAMUNYOKOLI_OVARIAN_CANCER_LMP_UP | Genes upregulated in mucinous ovarian carcinoma tumors of LMP compared with normal ovarian surface epithelium tissue |
| ROY_WOUND_BLOOD_VESSEL_DN | Genes downregulated in blood vessel cells from wound site |
| TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_LOBULAR_NORMAL_UP | Genes upregulated in lobular carcinoma versus normal lobular breast cells |
| KEGG_OXIDATIVE_PHOSPHORYLATION | Oxidative phosphorylation |
Abbreviations: DCIS, ductal carcinoma in situ; GSEA, gene signature enrichment analysis; IDC, invasive ductal carcinoma; LMP, low malignant potential; NSCLC; non-small-cell lung carcinoma; PMT, primary mammary tumor; RNAi, RNA interference; RST, recurrent spindle tumor.
False discovery rate (FDR) for all of the above sets was 0.000.
Table 6. Top 20 negative correlations identified by GSEA compared with PMT versus RST.
| SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_UP | Genes upregulated in IDC relative to DCIS (noninvasive) |
| CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_DN | Genes downregulated in luminal-like breast cancer cell lines compared with the mesenchymal-like ones |
| REN_ALVEOLAR_RHABDOMYOSARCOMA_DN | Genes commonly downregulated in human ARMS and its mouse model overexpressing PAX3–FOXO1 fusion |
| LINDGREN_BLADDER_CANCER_CLUSTER_2B | Genes specifically upregulated in cluster IIb of UCC tumors |
| HOSHIDA_LIVER_CANCER_SUBCLASS_s1 | Genes from ‘subtype S1' signature of HCC: aberrant activation of the WNT signaling pathway |
| CLASPER_LYMPHATIC_VESSELS_DURING_METASTASIS_DN | Selected genes downregulated during invasion of lymphatic vessels during metastasis |
| POOLA_INVASIVE_BREAST_CANCER_UP | Genes upregulated in atypical ductal hyperplastic tissues from patients with (ADHC) breast cancer versus those without cancer (ADH) |
| ONDER_CDH1_TARGETS_2_UP | Genes upregulated in HMLE cells (immortalized nontransformed mammary epithelium) after E-cadherin (CDH1) knockdown by RNAi |
| RODWELL_AGING_KIDNEY_UP | Genes whose expression increases with age in normal kidney |
| REACTOME_CELL_CYCLE_MITOTIC | Genes involved in cell cycle, mitotic |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_UP | Genes upregulated in transformed NIH3T3 cells (fibroblasts transformed by activated KRAS) versus normal cells |
| REACTOME_MITOTIC_M_M_G1_PHASES | Genes involved in mitotic M-M/G1 phases |
| NAKAYAMA_SOFT_TISSUE_TUMORS_PCA1_UP | Top 100 probe sets contributing to the positive side of the first principal component; predominantly associated with spindle cell and pleomorphic sarcoma samples |
| VECCHI_GASTRIC_CANCER_ADVANCED_VS_EARLY_UP | Upregulated genes distinguishing between two subtypes of gastric cancer: advanced (AGC) and early (EGC) |
| REACTOME_MITOTIC_PROMETAPHASE | Genes involved in mitotic prometaphase |
| MITSIADES_RESPONSE_TO_APLIDIN_DN | Genes downregulated in the MM1A cells (multiple myeloma) after treatment with aplidin, a marine-derived compound with potential anticancer properties |
| PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_UP | Upregulated genes in AILT compared with normal T lymphocytes |
| WALLACE_PROSTATE_CANCER_RACE_UP | Genes upregulated in prostate cancer samples from African-American patients compared with those from the European-American patients |
| REACTOME_G2_M_TRANSITION | Genes involved in G2/M transition |
| RAMALHO_STEMNESS_DN | Genes depleted in embryonic, neural and hematopoietic stem cells |
Abbreviations: ADH, atypical ductal hyperplasia; ADHC, atypical ductal hyperplastic cancer; AILT, angioimmunoblastic lymphoma; ARMS, alveolar rhabdomyosarcoma; DCIS, ductal carcinoma in situ; GSEA, gene signature enrichment analysis; HCC, hepatocellular carcinoma; IDC, invasive ductal carcinoma; PMT, primary mammary tumor; RNAi, RNA interference; RST, recurrent spindle tumor; UCC, urothelial cell carcinoma.
False discovery rate (FDR) for all of the above sets was 0.000.
Discussion
Expression profiling aids in the characterization and classification of transgenic murine mammary tumor models with respect to other murine mammary tumor models and with human breast tumors. We thus used this approach to further investigate the characteristics of the PMTs and RSTs that developed in the MTB-IGFIR transgenic mice.
When the gene expression profiles of our WT, PMT and RST samples were clustered with the murine mammary tumor models and human breast tumors described in Herschkowitz et al. (2007), it was observed that our WT samples clustered most closely with the WT murine mammary tissue and normal human breast tissue. This finding confirmed the validity of the analysis.
When the PMT samples were clustered with the murine mammary tumor models, it was observed that the PMT samples formed a discrete cluster, indicating that mammary tumors induced by IGF-IR overexpression are relatively homogeneous and possess a gene expression profile that is distinct from other murine mammary tumor models. The murine mammary tumor models that clustered most closely with the PMT samples included TgMMTV-Wnt1, DMBA-induced TgMMTV-Cre;BRCA1Co/Co;p53+/−, TgWAP-T121, p53+/− IR, p53−/− transplant, TgWAP-Myc, BRCA1+/−;p53+/−, TgC3-Tag and TgWAP-Tag tumors. In general, these murine tumor models had characteristics of mesenchymal cells, basal/myoepithelial cells or mixed characteristics (Herschkowitz et al., 2007). The PMT samples did not cluster closely with the models that showed a potential luminal gene expression signature (TgMMTV-neu and TgMMTV-PyMT). This observation was somewhat surprising considering (i) the similarities in the signaling molecules induced downstream of both the IGF-IR and ErbB2 (Huang et al., 2009; Maki, 2010), (ii) the detectable expression of cytokeratin 18 in the PMT samples (however, the level of cytokeratin 18 in the PMTs is typically lower and more variable than in normal mammary epithelial cells, and pockets of cytokeratin-5 and -14-positive cells are found in these tumors) and (iii) the fact that IGF-IR and some of its downstream signaling molecules can be regulated by estrogen (Fagan and Yee, 2008; Lanzino et al., 2008). However, the PMT samples also clustered closely with 51/58 human basal-like tumors. Therefore, in two independent comparisons, the PMT samples clustered with samples expressing a basal-like gene expression profile. In addition, although IGF-IR expression was initially associated with estrogen-receptor-positive (luminal) breast tumors (Peyrat et al., 1988; Koda et al., 2003, 2005) and the signaling via the estrogen receptor can regulate the expression of component involved in IGF-IR signaling (and vice versa; Fagan and Yee, 2008; Lanzino et al., 2008), more recent data has also found that the IGF-IR is expressed in human basal-like breast tumors as well. For example, the IGF-IR has been found to be amplified in human basal breast cancer (Adelaide et al., 2007), and high levels of IGF-IR protein have been detected in human and murine basal mammary tumors (Lerma et al., 2007; Klinakis et al., 2009). Moreover, the IGF-IR promoter can be repressed by genes, such as p53 and BRCA1, which are frequently mutated in basal-like breast cancer, resulting in elevated IGF-IR expression (Werner et al., 1996; Sarfstein et al., 2006).
It is also possible that the PMTs do not cluster with luminal tumors as the PMTs, like most murine mammary tumors, lose expression of estrogen and progesterone receptors in larger tumors (Jones et al., 2007). However, luminal gene signatures usually express high levels of genes such as SPDEF, XBP1, and GATA3, and genes involved in tight junctions, in addition to estrogen-regulated genes (Gruvberger et al., 2001; West et al., 2001; van't Veer et al., 2002; Sotiriou et al., 2003; Herschkowitz et al., 2007). It is the expression of these luminal genes that allows murine models, such as TgMMTV-neu and TgMMTV-PyMT, to cluster with human luminal breast tumors. As the PMT samples did not cluster with TgMMTV-neu or TgMMTV-PyMT tumors, this further supports the finding that PMTs express a mixture of epithelial and basal-like gene or express primarily basal-like genes.
Evaluation of the RST samples revealed that 7/9 RST samples also formed a discrete cluster when these samples were compared with the murine mammary tumor models. This group of seven RST samples clustered most closely with 3/11 DMBA-induced tumors, 3/10 TgMMTV-Cre;BRCA1Co/Co;p53+/− tumors, 1/7 p53+/− IR tumors, 1/5 p53−/− transplant tumors and 1/10 TgMMTV-neu tumors. Most of these mammary tumor models fell into group II as defined by Herschkowitz et al. (2007), which expressed high levels of mesenchymal features. When compared with the human breast cancers, the RST samples clustered most closely with 5/17 claudin-low tumors and 1/15 normal-like samples. In addition, qRT–PCR confirmed the low levels of claudins 3, 4 and 7 in the RST samples, and the RST samples were identified as claudin-low tumors using claudin-low gene identifier sets established by Herschkowitz et al. (2007) and Prat et al. (2010). The claudin-low breast cancer subtype was identified in 2007 based on a unique gene expression signature, which includes low levels of tight junction proteins, such as claudins 3, 4 and 7, as well as E-cadherin and high levels of mesenchymal genes, immune response genes and genes associated with stem cells (Herschkowitz et al., 2007). The prevalence of claudin-low tumors is reported to be 7–14%, and claudin-low tumors have a prognosis similar to luminal B, HER2-enriched and basal-like breast cancers (Prat et al., 2010).
When gene set enrichment analysis was used to evaluate the similarities between our PMT versus RST gene expression profiles with other publically available data sets, 16/40 studies (top 20 positively associated and top 20 negatively associated) involved some aspect of mammary physiology or breast cancer with 2/3 top-ranked positive correlations examining differences between mesenchymal tumors with either luminal or basal-like tumors. Other studies identified included those looking at ZEB1 target genes and genes involved in tight junctions. High levels of ZEB1 have been implicated in the mesenchymal genotype (Yang and Weinberg, 2008), whereas loss of tight junction proteins is consistent with a claudin-low genotype (Herschkowitz et al., 2007; Prat et al., 2010)
In summary, IGF-IR overexpression in MTB-IGFIR transgenic mice produces mammary tumors with a distinct gene expression signature that resembles murine and human mammary tumors with a basal-like genotype, whereas tumors that become independent of IGF-IR signaling cluster with murine mesenchymal tumors and human claudin-low tumors. Therefore, our MTB-IGFIR model can be used to further our understanding of two mammary tumor types, basal like and claudin low, which have a poor prognosis when treated with conventional therapies.
Materials and methods
Tissue collection
The generation of PMTs and RSTs has been previously described (Jones et al., 2007, 2009). Tissue was collected from 8 adult WT female mice (average age 119±10 days), 11 mice with PMTs (average age 121±24 days) and 9 mice with RSTs (average age 265±107 days). Tissue was collected when mice reached the appropriate age or when tumors reached the appropriate size; the collection period spanned several years. All tumors were generated by inducing IGF-IR transgene (through the addition of 2 g of doxycycline/kilogram of food) beginning at 21 days of age. Initial tumor onset for the PMT group was 45±7.5 days after IGF-IR induction, whereas tumor onset in the RST group was 47±8.6 days. Characteristics of the mice used in this study are presented in Supplementary Table 1.
All tumors were collected when the maximum tumor length was approximately 17 mm (the maximum allowed by the Canadian Council of Animal Care). Adult WT mammary glands, PMTs and RSTs were removed from mice and immediately flash frozen by placing the samples on dry ice. RNA was extracted from the tissue using the RNeasy RNA isolation kit (Qiagen, Mississauga, ON, Canada). Two micrograms of RNA from each tissue sample was sent to the Microarray Centre at the University Health Network, Toronto, ON, Canada (http://www.microarray.ca).
Western blotting
Western blotting was performed as described in Jones et al. (2007). All antibodies were obtained from Cell Signalling Technologies (Danvers, MA, USA), except for the IGF-IR antibody, which was obtained from R&D Systems (Minneapolis, MN, USA), and the β-tubulin antibody, which was obtained from Abcam (Cambridge, MA, USA). All antibodies were used at a 1:500 dilution, except for β-tubulin, which was used at a 1:1000 dilution. Appropriate secondary antibodies were obtained from Cell Signalling Technologies and used at a dilution of 1:2000.
Histology
Mammary tumors were collected and processed as previously described (Jones et al., 2007, 2009).
DNA microarray analysis
RNA labeling, hybridization, image acquisition and data analysis were performed by the Microarray Centre using protocols available on their website (http://www.microarray.ca). Briefly, the RNA was labeled using the Agilent Low RNA Input Fluorescent Linear Amplification Kit and hybridized using the In situ Hybridization Kit (Agilent, Santa Clara, CA, USA). Samples were randomized before labeling and the labeled RNA was hybridized on Agilent Whole Mouse Genome 4 × 44k Gene Expression Arrays (product number G4122F, Agilent). All RNA samples were hybridized against a universal mouse reference RNA. Data Analysis was performed using GeneSpring (Agilent), and arrays were first filtered to remove features at or below background in 20 of 28 arrays. Arrays were then filtered further to include only features that had a P-value of <0.05 in 20 of 28 arrays. Analysis of variance was used to determine which genes were statistically significant when comparing the three groups, whereas a Welch's t-test and the Benjamini and Hochberg False Discovery Rate multiple testing correction were used to identify statistically significant genes when only comparing two groups (P<0.05).
Quantitative reverse transcriptase PCR
RNA utilized for qRT–PCR was from the same samples utilized for the DNA microarray analysis. Real-time PCR was performed in accordance with Jones et al. (2009). All primers were obtained from Cedarlane Labs (Burlington, ON, Canada). HPRT (5′-GTTGGATACAGGCCAGACTTTGTTG-3′; (forward) and 5′-GATTCAACTTGCGCTCATCTTAGGC-3′ (reverse)) was used for normalization.
Comparison to human breast tumors
Our 28-mouse whole-genome Agilent arrays were normalized in R using bioconductor's limma package (Gentleman et al., 2004). Arrays were background corrected using the backgroundCorrect function (data.method=‘normexp' and normexp.method=‘rmra') and normalized using the normalizeWithinArrays (data.bg.method=‘loess') and normalizeBetweenArrays functions (data.w.method=‘quantile'). The raw and normalized data is available in the Gene Expression Omnibus (GEO) (GEO:GSE32152).
Mouse and human data sets generated by Herschkowitz et al. (2007) were downloaded from GEO (GSE3165) and mean centered. Our mouse samples were mean centered and clustered with the Herschkowitz mouse models using their 853 gene list and clustered with the Herschkowitz human samples using their 106 gene list. Clustering was performed using Euclidean distance and Ward's algorithm. To further confirm that the RST mouse model is characteristic of the human claudin-low subtype, our mouse models were clustered using two known human claudin-low signatures (Herschkowitz et al., 2007; Prat et al., 2010). To further confirm that the PMT mouse model is characteristic of the human basal-like subtype, our mouse model was clustered using the Molyneux signature (Molyneux et al., 2010). Clustering was performed using Euclidean distance and Ward's algorithm.
Gene signature enrichment analysis
Pathways analysis was performed using GSEA (Subramanian et al., 2005). A unique gene list was created comparing PMT with RST, ranking the genes based on the t-statistic from limma. The latest version of GSEA was used (gsea2-2.07.jar), comparing our ranked list with the ‘all curated gene sets' database (c2.all.v3.0symbols.gmt).
Acknowledgments
This work was funded by operating grants from the Cancer Research Society and the Canadian Institutes of Health Research awarded to RAM.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- Fagan DH, Yee D. Crosstalk between IGF1R and estrogen receptor signaling in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:423–429. doi: 10.1007/s10911-008-9098-0. [DOI] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, et al. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Greer A, Hurlburt W, Han X, Hafezi R, Wittenberg GM, et al. The mechanisms of differential sensitivity to an insulin-like growth factor-1 receptor inhibitor (BMS-536924) and rationale for combining with EGFR/HER2 inhibitors. Cancer Res. 2009;69:161–170. doi: 10.1158/0008-5472.CAN-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RA, Campbell CI, Gunther EJ, Chodosh LA, Petrik JJ, Khokha R, et al. Transgenic overexpression of IGF-IR disrupts mammary ductal morphogenesis and induces tumor formation. Oncogene. 2007;26:1636–1644. doi: 10.1038/sj.onc.1209955. [DOI] [PubMed] [Google Scholar]
- Jones RA, Campbell CI, Wood GA, Petrik JJ, Moorehead RA. Reversibility and recurrence of IGF-IR-induced mammary tumors. Oncogene. 2009;13:407–413. doi: 10.1038/onc.2009.79. [DOI] [PubMed] [Google Scholar]
- Klinakis A, Szabolcs M, Chen G, Xuan S, Hibshoosh H, Efstratiadis A. Igf1r as a therapeutic target in a mouse model of basal-like breast cancer. Proc Natl Acad Sci USA. 2009;106:2359–2364. doi: 10.1073/pnas.0810221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koda M, Przystupa W, Jarzabek K, Wincewicz A, Kanczuga-Koda L, Tomaszewski J, et al. Expression of insulin-like growth factor-I receptor, estrogen receptor alpha, Bcl-2 and Bax proteins in human breast cancer. Oncol Rep. 2005;14:93–98. [PubMed] [Google Scholar]
- Koda M, Sulkowski S, Garofalo C, Kanczuga-Koda L, Sulkowska M, Surmacz E. Expression of the insulin-like growth factor-I receptor in primary breast cancer and lymph node metastases: correlations with estrogen receptors alpha and beta. Horm Metab Res. 2003;35:794–801. doi: 10.1055/s-2004-814152. [DOI] [PubMed] [Google Scholar]
- Lanzino M, Morelli C, Garofalo C, Panno ML, Mauro L, Ando S, et al. Interaction between estrogen receptor alpha and insulin/IGF signaling in breast cancer. Curr Cancer Drug Targets. 2008;8:597–610. doi: 10.2174/156800908786241104. [DOI] [PubMed] [Google Scholar]
- Lerma E, Peiro G, Ramon T, Fernandez S, Martinez D, Pons C, et al. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2/neu (basal-like breast carcinomas) Mod Pathol. 2007;20:1200–1207. doi: 10.1038/modpathol.3800961. [DOI] [PubMed] [Google Scholar]
- Maki RG. Small is beautiful: insulin-like growth factors and their role in growth, development, and cancer. J Clin Oncol. 2010;28:4985–4995. doi: 10.1200/JCO.2009.27.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de RM, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Peyrat JP, Bonneterre J, Beuscart R, Djiane J, Demaille A. Insulin-like growth factor 1 receptors in human breast cancer and their relation to estradiol and progesterone receptors. Cancer Res. 1988;48:6429–6433. [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfstein R, Maor S, Reizner N, Abramovitch S, Werner H. Transcriptional regulation of the insulin-like growth factor-I receptor gene in breast cancer. Mol Cell Endocrinol. 2006;252:241–246. doi: 10.1016/j.mce.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van′t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Werner H, Karnieli E, Rauscher FJ, LeRoith D. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc Natl Acad Sci USA. 1996;93:8318–8323. doi: 10.1073/pnas.93.16.8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


