Abstract
A critical component for sustaining adequate food production is the protection of local agriculture from invasive pest insects. Essential to this goal is the ability to accurately distinguish foreign from closely related domestic species, a process that has traditionally required identification using diagnostic morphological “keys” that can be both subtle and labor-intensive. This is the case for the Lepidopteran group of insects represented by Spodoptera, a genus of Noctuidae “armyworm” moths that includes several important agricultural pests. Two of the most destructive species, Spodoptera littoralis (Boisduval) (Lepidoptera: Noctuidae) and S. litura (F.) are not yet established in North America. To facilitate the monitoring for these pests, the feasibility of using DNA barcoding methodology for distinguishing between domestic and foreign Spodoptera species was tested. A DNA barcoding database was derived for a subset of Spodoptera species native to Florida, with an emphasis on those attracted to pheromone blends developed for S. litura or S. littoralis. These were then compared to the barcode sequences of S. litura collected from Taiwan and S. littoralis from Portugal. Consistent discrimination of the different species was obtained with phenetic relationships produced that were generally in agreement with phylogenetic studies using morphological characteristics. The data presented here indicate that DNA barcoding has the potential to be an efficient and accurate supplement to morphological methods for the identification of invasive Spodoptera pests in North America.
Keywords : COI, cytochrome-c oxidase subunit I, invasive pests, mitochondrial DNA
Introduction
The genus Spodoptera (Lepidoptera: Noctuidae) includes some of the most important pests of agricultural crops in the world. These are commonly known as “armyworms” because severe infestations can appear as large masses “marching” in search of food; as many as 30 species have been described with members present on six continents. At least nine Spodoptera species are native to Florida (Heppner 1998), including those of greatest economic importance to North American agriculture, i.e., the southern armyworm S. eridania (Stoll), the beet armyworm S. exigua (Hübner), and the fall armyworm S. frugiperda (J. E. Smith).
Because Spodoptera species are of tropical origin, their overwintering ranges are typically limited to areas with mild winters, though some can diapause and thereby survive more extreme conditions. However, many of the native species are capable of extensive migrations and thus can cause seasonal damage well outside their overwintering range. The North American distribution of fall armyworm for example is limited to southern Florida and Texas during the winter months, but infestations extend as far north as Canada during the summer and fall (Luginbill 1928). Therefore, the establishment of invasive and migratory Spodoptera species into Florida is of special concern, as these can serve as source populations for migratory infestations into the rest of the continental United States.
Posing the greatest invasive threat on the USDA-APHIS-PPQ quarantine list are the Egyptian cotton leafworm, S. littoralis (Boisduval), and the tobacco cutworm, S. litura (F.) (Ellis 2004). Both species are highly polyphagous and produce economically significant damage to a range of crops, most notably cotton, soybean, maize, rice, and ornamentals, and each has a wide geographical distribution. Spodoptera littoralis is found in southern Europe, Africa, and the Middle East. The S. litura range includes the Middle East, most of Asia, Australia, and extends into the south Pacific as far west as Hawaii (Ellis 2004; Pogue 2002). From 1985 to 2010 there have been 172 and 663 interceptions of S. littoralis and S. litura in USA ports, respectively, with another 2809 interceptions identified as of the Spodoptera genus but not further classified (USDA-APHIS-PPQ 2010). A 2004 report indicated that most S. littoralis were intercepted in permit cargo flowers from Israel, while the majority of S. litura was found in permit cargo orchids (Ellis 2004). Florida is among the largest producers and distributors of floriculture in the USA, and thus is particularly susceptible to invasion by these Old World Spodoptera species (USDA-National Agricultural Statistics Service 2008). It is predicted that both species have the potential to become established in the southwestern and southeastern USA, reaching as far north as Maryland, and with annual migratory potential extending into Canada (Ellis 2004).
The availability of synthetic pheromone blends that attract S. litura and S. littoralis make possible the use of pheromone trapping as an efficient means of monitoring for the early establishment of these species in vulnerable areas (Neumark et al. 1977; Nemoto et al. 1980). The potential of this method for Florida was tested in areas near orchid nurseries that receive many imported plants (Meagher et al. 2008). Out of almost 200 specimens captured in traps baited with an S. litura pheromone blend, one S. litura specimen was found, with the remainder identified by morphology as belonging to native species. These results indicate that while pheromone trapping can be effective for monitoring large areas for invasive Spodoptera, their limited specificity still requires screening a large number of specimens.
Unfortunately, S. litura and S. littoralis have similar morphology to many domestic Spodoptera species, often making their distinction by classical physical criteria difficult. The most diagnostic morphological keys require microscopic characterization of adult male genital structures, a tedious procedure when screening large numbers, and one that requires substantial sample preparation and undamaged specimens (Pogue 2002). Unambiguous keys are frequently not available for females or immature stages, and substantial overlap in host range and attraction to pheromone blends limit the use of behavioral criteria (reviewed in Pogue 2002; Meagher et al. 2008). Therefore, finding an alternative method to supplement morphometric analyses is of practical interest for the Spodoptera complex.
DNA barcoding has been proposed as a molecular method for assigning individual specimens to known species (Hebert et al. 2003). The barcode involves DNA sequence analysis of a portion (typically between 600–900 bp) of the mitochondrial gene cytochrome c oxidase subunit I (COI). The central assumption is that barcode variation between even closely related species will be substantially greater than that observed within species. This is known as the “barcoding gap”, with a 10-fold difference between mean interspecific and intraspecific variations being frequently mentioned as a minimum threshold for the unambiguous assignment of unknown individuals to a species (Hebert et al. 2004). While this remains a favored criterion of many barcode proponents, even the absence of such gaps may still allow accurate assignment of barcode sequences to species in at least some taxa (Lou and Golding 2010; Virgilio et al. 2010).
To date, barcoding has had mixed success for species assignment in Lepidoptera. There are several examples of inaccurate species assignments using DNA barcoding that identify potential limitations in the methodology (Elias et al. 2007; Dasmahapatra et al. 2010). These were associated with problems arising from incomplete barcode coverage of existing diversity, the apparent absence of consistent barcode gaps in certain taxa, and the potentially confounding effects of incomplete lineage sorting that can be difficult to assess (reviewed in Rubinoff et al. 2006; Silva-Brandao et al. 2009). However, the technique was successfully applied in a study where 150 lepidopteran specimens were correctly assigned using a barcode database of 200 closely allied species (Hebert et al. 2003). In another large survey, a barcode comparison of about 100,000 specimens representing approximately 3500 species of moths, butterflies, flies, and wasps produced no misidentifications when a full barcode was available (Janzen et al. 2009). Perhaps most relevant to this paper, barcode comparisons were able to distinguish between four closely related Helicoverpa species, a complex in the same family (Noctuidae) as Spodoptera (Behere et al. 2007). These observations indicate that while the successful application of barcoding for species assignment may be taxa-dependent, with poorly studied or recently diverging groups being particularly problematic, the method has potential for facilitating the identification of invasive pest arthropods (Armstrong and Ball 2005; Darling and Blum 2007; Floyd et al. 2010).
The accuracy of species assignment by barcode comparisons is in theory dependent upon there being sufficient sampling of the target population and closely related nontarget species to both assess the existence of a barcode gap and to confidently estimate phylogenetic relationships (see for example Wiemers and Fiedler 2007; Darling and Blum 2007). The establishment of representative barcode databases for the exotic population and relevant native species is a potentially major undertaking that could require hundreds of sequences depending upon the genetic variability and similarity of the populations being compared. Therefore, an empirical demonstration of the feasibility of using DNA barcoding for a given taxa and region is prudent.
The objective of this study was to assess the applicability of DNA barcoding to monitor invasive Spodoptera species in Florida. Reference barcode databases were developed for a subset of Spodoptera native to Florida known to be attracted to S. litura pheromone traps (Meagher et al. 2008) commonly found on host plants associated with S. litura and S. littoralis (reviewed in Pogue 2002), and/or are important agricultural pests in North America. These databases were compared to barcode sequences from S. littoralis and S. litura specimens collected from Portugal and Taiwan, respectively, as identified by morphological criteria. The results were assessed for the likelihood of barcode gaps sufficient to discriminate the native from the foreign populations and thereby justify the expansion of the barcode databases for these and other related species. The potential role of DNA barcoding in the monitoring for invasive Spodoptera in Florida is discussed.
Materials and Methods
Specimen collections and sites.
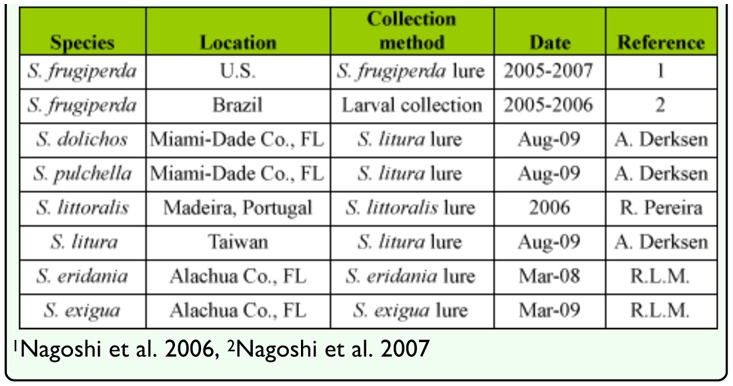
The identification, collection, and processing of adult and larval specimens of S. frugiperda were described in previous studies (Nagoshi et al. 2006; Nagoshi et al. 2007). Other Spodoptera specimens were adult males obtained from pheromone-based traps (Table 1) with captures from each location representing pooled collections from multiple local sites and times. Standard plastic Universal moth traps (Unitraps) were baited with the appropriate commercially available pheromone blends (Suterra LLC, www.suterra.com) and contained insecticide strips (Hercon Environmental, www.herconenviron.com). After collection, specimens were typically stored at -20 °C. The species identity was determined by examination of male genital structures (illustrated in Pogue 2002). Abdomens were dissected and soaked in 70% isopropyl alcohol for indefinite storage. To prepare for examination, the abdomens were cleared in 10% KOH and incubated at 70 °C in a water bath for 45 min until clearing by visual inspection. They were then rinsed twice with isopropyl alcohol and the genitalia cleaned under a dissecting microscope. Morphological examination was done under alcohol or as permanently mounted specimens.
Table 1.
Source locality and host information.
DNA preparation and amplification of the COI region.
Mitochondrial DNA was isolated from adults or larvae as previously described (Nagoshi et al. 2006). The DNA preparation was diluted to a final volume of 40 µl with distilled water. Genomic DNA preparations of fall armyworm samples from previous studies were stored at -20 °C (Table 1). PCR amplification was performed in a 30 µl reaction mix containing 3 µl 10X manufacturer's reaction buffer, 1 µl 10mM dNTP, 0.5 µl 20 µM primer mix, 1 µl DNA template (between 0.05–0.5 µg), 0.5 unit Taq DNA polymerase (New England Biolabs, www.neb.com). The thermocycling program was 94 °C (1 min), followed by 33 cycles of 92 °C (30 sec), 52 °C (45 sec), 72 °C (45 sec), and a final segment of 72 °C for 3 min. Amplification products were analyzed and isolated by agarose gel electrophoresis where 6 µl of 6X gel loading buffer was added to each amplification reaction, and the entire sample run on a 1.5% agarose horizontal gel containing GelRed (Biotium, www.biotium.com) in 0.5X Tris-borate buffer (TBE, 45 mM Tris base, 45 mM boric acid, 1 mM EDTA pH 8.0). A single band corresponding to the expected size of the amplified fragment was obtained from each reaction. To purify the amplified fragment away from excess primers, the fragment was visualized on a long-wave UV light box and cut out from the gel. Fragment isolation was performed using Zymo-Spin I columns (Zymo Research Corporation, www.zymoresearch.com) according to manufacturer's instructions. Primers were synthesized by Integrated DNA Technologies (www.idtdna.com). Spodoptera sequences were amplified using primers derived from earlier characterization of the COI region (Nagoshi et al. 2006; Nagoshi et al. 2007), COI-45F, 5′-TTCGAGCTGAATTAGGRACYC -3′ (Y = C or T; R = A or G) and COI-914R (5′-GCWGATGTYAAATAWGCTCGWG -3′ (W = A or T) that are predicted to produce an 814 bp fragment from coordinate +101 to +914 (Figure 1). Overlapping sequence information was obtained using the same two primers for the internal 771 bp region that does not include the primer sequences.
Figure 1.
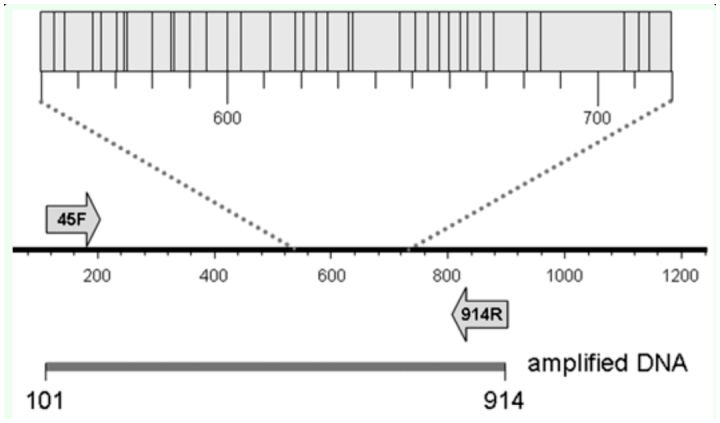
Diagram of the COI region used for barcoding. Arrows identify primers used for PCR amplification and DNA sequencing. Top box describes an approximately 200 bp region with the locations of species diagnostic nucleotide polymorphisms indicated by vertical lines (Table 4). High quality figures are available online.
DNA sequence analysis.
The isolated fragments were analyzed by DNA sequencing performed by the University of Florida Interdisciplinary Center for Biotechnology Research using primers described for the PCR reactions. The quality of the sequence data was confirmed by examination of the chromatographs. In five cases involving S. litura samples, the chromatographs showed dual peaks of similar height at 10–20 nucleotide sites, suggesting that these samples are “heterozygotes” containing two COI sequences with high similarity that differed at the ambiguous sites. In contrast, 18 other S. litura samples gave the expected single sequence for the amplified COI region. The unambiguous sequences were assumed to be representative of the S. litura mitochondrial gene and used to derive the presumptive sequence of the “contaminant”. From the sequence data, two restriction enzyme site polymorphisms were identified, a Dra I site present only in the S. litura COI gene, and an Msp I site present only in the presumptive contaminant. To separate the two templates, the amplified PCR product from the heterozygotes was digested with one or the other restriction enzymes, and in each case the uncut fragment was gel-isolated and analyzed by DNA sequencing. The additional sequence carried in four of the five heterozygous specimens was identical to the S. dolichos-1 haplotype, while the fifth displayed the S. pulchella-1 haplotype. The identity of the additional sequences with that of other specimens analyzed contemporaneously suggests cross-contamination, and these were not included in this study.
DNA comparisons and alignments were performed using the DS Gene program (Accelrys, www.accelrys.com) and the CLUSTAL algorithm. Descriptive DNA sequence statistics and calculations of nucleotide variation based on the Jukes-Cantor (JC) model were performed using DNAsp version 5.1 (Librado and Rozas 2009). Sequence divergences among individuals were calculated using the Kimura 2-Parameter distance model (Kimura 1980) and graphically displayed in a neighbor-joining (NJ) tree (Saitou and Nei 1987). Confidence was assessed by bootstrapping at 2000 replications with the Bombyx mori COI sequence (GenBank #EU141360) and Helicoverpa armigera COI sequence (GenBank #HQ132369) as outliers. All haplotypes obtained in this study have been deposited in GenBank: S. frugiperda (accession nos. HM136586–HM136602), other Spodoptera spp. (accession nos. HM756074–HM756093). Voucher specimens were deposited at CMAVE (Gainesville, FL).
Results and Discussion
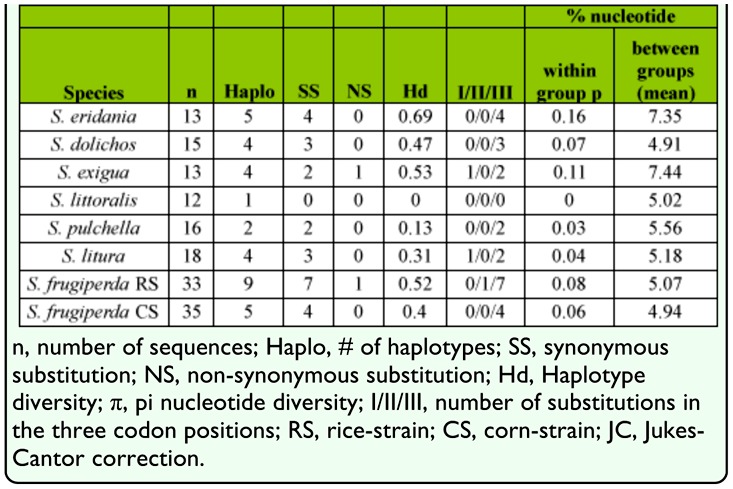
DNA sequences from a portion of the COI region were analyzed from five Spodoptera spp. native to Florida. These included the three major Spodoptera pests in the USA (S. frugiperda (rice-strain and corn-strain), S. eridania, and S. exigua), and two species attracted to S. litura pheromone traps, S. dolichos and S. pulchella (Meagher et al. 2008). These were compared to barcode sequences identified from two potentially invasive species, S. littoralis and S. litura (Table 1). Alignment of the COI sequences found no deletions or insertions and no stop codons, consistent with the amplified DNA arising from functional COI genes. Multiple haplotypes were found for all species except S. littoralis, with the highest haplotype diversity observed in S. eridania (Table 2). The majority of nucleotide substitutions were synonymous (25/27), with all but three mapping to the third codon position.
Table 2.
Descriptive statistics of polymorphisms found in a 771 bp portion of the COI gene from different Spodoptera species.
The existence of a substantial barcode gap within this dataset was evident as the average nucleotide divergence within species was 0.08%, compared to an overall average divergence between species of 5.35% (Table 3). For each pairwise comparison, the mean variation between groups was substantially greater than the intra-group variation, surpassing the 10-fold threshold recommended for the barcode gap. Even the smallest divergence found between populations of 2.13%, observed between the two S. frugiperda strains, was 13-fold greater than the largest intraspecies divergence of 0.16% observed for S. eridania. This indicates that if the genetic variations exhibited by the small sampling groups are representative of the general populations, then COI sequence comparisons should be sufficiently sensitive to distinguish between Spodoptera species. This was confirmed by neighbor-joining phenetic analysis that differentiated at > 75% bootstrap values from the expected species, with S. exigua being most divergent (Figure 2). Closest similarities were found between S. littoralis and S. litura, S. dolichos and S. pulchella, and the two S. frugiperda host strains, but even these pairs segregated at > 95% bootstrap values.
Table 3.
Diagnostic sequence polymorphisms in 160-bp segment of the COI gene.
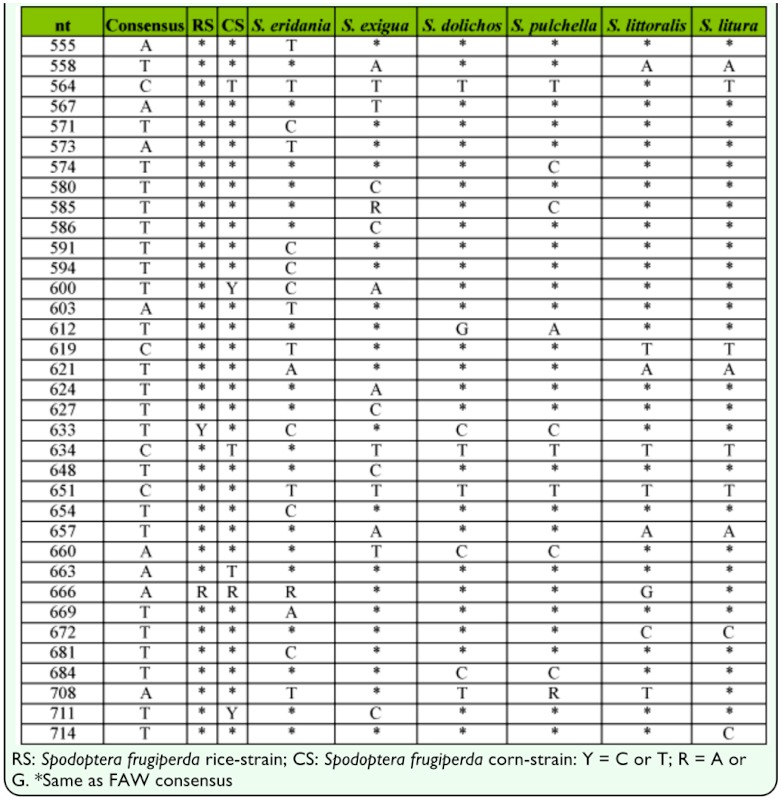
Figure 2.
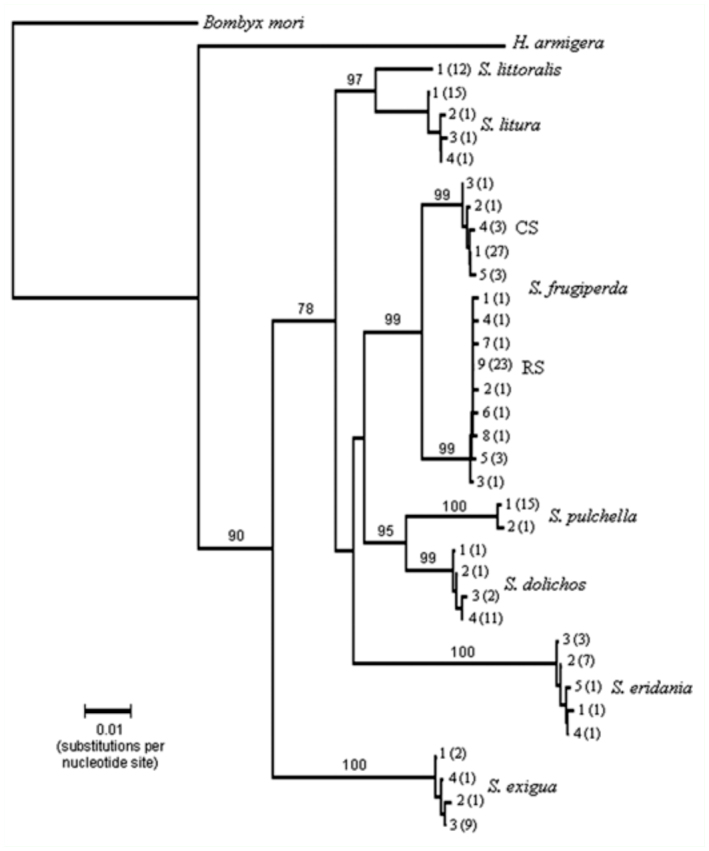
Neighbor-joining tree based on Kimura-2-Parameter distances for COI DNA sequences for different Spodoptera species. DNA sequences representative of the different haplotypes were used. Numbers next to species names identify haplotype category. Number of each haplotype found in parentheses. Numbers at branch points indicate 2000X bootstrap value. COI sequence from Helicoverpa armigera (accession # HQ132369) and Bombyx mori (accession #EU141360) were used as outlier sequences. High quality figures are available online.
These phenetic relationships were generally consistent with the phylogeny derived from morphological characteristics (Pogue 2002). Those cladistic studies identified Spodoptera as a monophyletic group with S. exigua as the most plesiomorphic species and S. littoralis and S. litura as closely related sister species. Discrimination of S. littoralis and S. litura is limited to comparisons of adult genital morphology (Mochida 1973; Ellis 2004). Therefore, the development of diagnostic barcodes will be of use for immature stages. Furthermore, it may be possible to discriminate between many Spodoptera spp. by sequence analysis of relatively short portions of the COI gene. An approximately 200 bp segment contains 36 polymorphisms that in combination readily differentiate between the Spodoptera groups tested in this study (Table 4, Figure 1). This means that even poorer quality specimens that allow only short PCR amplification products can potentially be used to at least delimit, if not completely specify, species identity.
These results support the feasibility of using DNA barcodes to rapidly assess the threat posed by an unknown specimen, either as a complement to morphological analysis or as the primary diagnostic indicator in cases where the requisite morphological keys are unavailable or compromised. This would entail comparing the unknown barcode sequence to barcode databases using pairwise sequence divergence calculations (e.g., the Kimura 2-parameter model) as visualized using a neighbor-joining tree—a methodology used effectively in a case study monitoring for invasive tussock moth species (Armstrong and Ball 2005; Ball and Armstrong 2006). If the unknown displays stronger barcode similarity to a quarantine species than native populations, it would be reason to recommend more extensive monitoring of the relevant areas. Currently, the major limitation of this approach for Spodoptera is the relatively small number of barcode sequences analyzed, both in terms of the range of species characterized and the number of sequences describing each group. The potential of insufficient sampling has challenged the validity of observed barcode gaps in other taxa (Wiemers and Fiedler 2007). Therefore, until more representative databases can be developed, species assignment using this approach should be considered tentative, pending confirmation by other methods. Nevertheless, the results described here suggest that the Spodoptera species complex can be readily differentiated by barcode comparisons and that even the preliminary barcode database from this study can indicate whether a particular specimen merits concern.
DNA barcoding makes possible the use of specimens at developmental stages where morphological keys for species identification are not available or of poor quality. In addition, continued advances in molecular genetic technology will improve the efficiency and economics of barcode analysis, making the screening of even a large number of samples increasingly practical. These benefits combined with the observed applicability of barcoding for species assignment in Spodoptera justify efforts to expand the barcoding database to become broader and more representative of the relevant domestic and exotic species. This could include non-Spodoptera species, such as members of the Mythimna and Helicoverpa species complexes, whose juvenile stages feed on many of the same hosts as S. litura and S. littoralis, and can be difficult to distinguish from Spodoptera by morphological criteria. As the barcoding database expands, so will the accuracy and utility of this approach for assigning species identity to unknown specimens, making it a valuable complement to the morphological methods currently used for the monitoring of invasive Spodoptera and other Lepidopteran pests in the United States.
Acknowledgements
We thank Jane Sharp for excellent technical support, and Dr. Steven Valles (USDA) and Dr. Paul Shirk (USDA) for helpful comments on the manuscript. The use of trade, firm, or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable.
Glossary
Abberviations:
- COI,
cytochrome-c oxidase subunit
References
- Armstrong KF, Ball SL. DNA barcodes for biosecurity: invasive species identification. Philosophical Transactions of the Royal Society B—Biological Sciences. 2005;360:1813–1823. doi: 10.1098/rstb.2005.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SL, Armstrong KF. DNA barcodes for insect pest identification: A test case with tussock moths (Lepidoptera : Lymantriidae). Canadian Journal of Forest Research. 2006;36:337–350. [Google Scholar]
- Behere GT, Russell D, Batterham P, Tay WT. Two species into one: Bottleneck history of Helicoverpa zea from Helicoverpa armigera revealed by DNA barcoding. Journal of Insect Science. 2007;7:29. Available online, http://insectscience.org/7.29. [Google Scholar]
- Darling J, Blum M. DNA-based methods for monitoring invasive species: a review and prospectus. Biological Invasions. 2007;9:751–765. [Google Scholar]
- Dasmahapatra KK, Elias M, Hill RI, Hoffman JI, Mallet J. Mitochondrial DNA barcoding detects some species that are real, and some that are not. Molecular Ecology Resources. 2010;10:264–273. doi: 10.1111/j.1755-0998.2009.02763.x. [DOI] [PubMed] [Google Scholar]
- Elias M, Hill RI, Willmott KR, Dasmahapatra KK, Brower AVZ, Mallet J, Jiggins CD. Limited performance of DNA barcoding in a diverse community of tropical butterflies. Proceedings of the Royal Society of London B—Biological Sciences. 2007;274:2881–2889. doi: 10.1098/rspb.2007.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. E. New Pest Response Guidelines: Noctuidae: Spodoptera. . USDA APHIS PPQ PDMP. 2004. Available online, http://www.aphis.usda.gov/ppq/manuals/
- Floyd R, Lima J, deWaard J, Humble L, Hanner R. Common goals: policy implications of DNA barcoding as a protocol for identification of arthropod pests. Biological Invasions. 2010;12:2947–2954. [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B—Biological Sciences. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biology. 2004;210:e312. doi: 10.1371/journal.pbio.0020312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner JB. Spodoptera armyworms in Florida (Lepidoptera: Noctuidae). Entomology Circular. 1998;390:1–5. [Google Scholar]
- Janzen DH, Hallwachs W, Blandin P, Burns JM, Cadiou JM, Chacon I, Dapkey T, Deans AR, Epstein ME, Espinoza B, Franclemont JG, Haber WA, Hajibabaei M, Hall JPW, Hebert PDN, Gauld ID, Harvey DJ, Hausmann A, Kitching IJ, Lafontaine D, Landry JF, Lemaire C, Miller JY, Miller JS, Miller L, Miller SE, Montero J, Munroe E, Green SR, Ratnasingham S, Rawlins JE, Robbins RK, Rodriguez JJ, Rougerie R, Sharkey MJ, Smith MA, Solis MA, Sullivan JB, Thiaucourt P, Wahl DB, Weller SJ, Whitfield JB, Willmott KR, Wood DM, Woodley NE, Wilson JJ. Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Molecular Ecology Resources. 2009;9:1–26. doi: 10.1111/j.1755-0998.2009.02628.x. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lou M, Golding GB. Assigning sequences to species in the absence of large interspecific differences. Molecular Phylogenetics and Evolution. 2010;56:187–194. doi: 10.1016/j.ympev.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Luginbill P. The fall armyworm. United States Department of Agriculture Technical Bulletin. 1928;34:1–91. [Google Scholar]
- Meagher RL, Brambila J, Hung E. Monitoring for Exotic Spodoptera Species (Lepidoptera: Noctuidae) in Florida. Florida Entomologist. 2008;91:517–522. [Google Scholar]
- Mochida O. Two important insect pests, Spodoptera litura (F.) and S. littoralis (Boisd.) (Lepidoptera:Noctuidae), on various crops-Morphological discrimination of the adult, pupal, and larval stages. Applied Entomology and Zoology. 1973;8:205–214. [Google Scholar]
- Nagoshi RN, Silvie P, Meagher RL., Jr Comparison of haplotype frequencies differentiate fall armyworm (Lepidoptera: Noctuidae) corn-strain populations from Florida and Brazil. Journal of Economic Entomology. 2007;100:954–961. doi: 10.1603/0022-0493(2007)100[954:cohfdf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, Meagher RL, Adamczyk JJ, Braman SK, Brandenburg RL, Nuessly G. New restriction fragment length polymorphisms in the cytochrome oxidase I gene facilitate host strain identification of fall armyworm (Lepidoptera : Noctuidae) populations in the southeastern United States. Journal of Economic Entomology. 2006;99:671–677. doi: 10.1603/0022-0493-99.3.671. [DOI] [PubMed] [Google Scholar]
- Nemoto H, Takahashi K, Kubota A. Reduction of the population-density of Spodoptera litura (F) (Lepidoptera, Noctuidae) using a synthetic sex-pheromone. 1. Experiment in taro field. Japanese Journal of Applied Entomology and Zoology. 1980;24:211–216. [Google Scholar]
- Neumark S, Waters RM, Jacobson M, Teich I. Relative attractiveness of several synthetic formulations of Spodoptera littoralis (Lepidoptera-Noctuidae) pheromone to male moths. Journal of Environmental Science and Health Part A—Environmental Science and Engineering and Toxic and Hazardous Substance Control. 1977;12:259–276. [Google Scholar]
- Pogue MG. A world revision of the genus Spodoptera Guenée (Lepidoptera: Noctuidae). Memoirs of the American Entomological Society. 2002;43:1–202. [Google Scholar]
- Rubinoff D, Cameron S, Will K. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. Journal of Heredity. 2006;97:581–594. doi: 10.1093/jhered/esl036. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor-Joining method - a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Silva-Brandao KL, Lyra ML, Freitas AVL. Barcoding Lepidoptera: Current situation and perspectives on the usefulness of a contentious technique. Neotropical Entomology. 2009;38:441–451. doi: 10.1590/s1519-566x2009000400001. [DOI] [PubMed] [Google Scholar]
- USDA-APHIS-PPQ. Agriculture Quarantine Activity System Pest ID database. 2010.
- USDA-National Agricultural Statistics Service. Floriculture: Top 5 States by Value of Sales, US. 2008. Available online, http://www.nass.usda.gov/Charts_and_Maps/Floriculture_Crops/USDA-NASS.
- Virgilio M, Backeljau T, Nevado B, De Meyer M. Comparative performances of DNA barcoding across insect orders. Bmc Bioinformatics. 2010;11:206. doi: 10.1186/1471-2105-11-206. Available online, http://biomedcentral.com/1471-2105/11/206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M, Fiedler K. Does the DNA barcoding gap exist? — a case study in blue butterflies (Lepidoptera: Lycaenidae). Frontiers in Zoology. 2007;4:8. doi: 10.1186/1742-9994-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


