Abstract
Background
Bai Ku Yao is a special subgroup of the Yao minority in China. The present study was undertaken to detect the association of rs5888 single nucleotide polymorphism (SNP) in the scavenger receptor class B type 1 (SCARB1) gene and several environmental factors with serum lipid levels in the Guangxi Bai Ku Yao and Han populations.
Methods
A total of 598 subjects of Bai Ku Yao and 585 subjects of Han Chinese were randomly selected from our stratified randomized cluster samples. Genotypes of the SCARB1 rs5888 SNP were determined by polymerase chain reaction and restriction fragment length polymorphism combined with gel electrophoresis, and then confirmed by direct sequencing.
Results
The levels of total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), apolipoprotein (Apo) AI were lower but ApoB was higher in Bai Ku Yao than in Han (P < 0.05-0.001). The frequencies of C and T alleles were 78.3% and 21.7% in Bai Ku Yao, and 73.7% and 26.3% in Han (P < 0.01); respectively. The frequencies of CC, CT and TT genotypes were 60.0%, 36.6% and 3.4% in Bai Ku Yao, and 54.2%, 39.0% and 6.8% in Han (P < 0.01); respectively. The subjects with TT genotype in both ethnic groups had lower HDL-C and ApoAI levels than the subjects with CC or CT genotype (P < 0.05 for all). Subgroup analyses showed that the subjects with TT genotype in Bai Ku Yao had lower HDL-C and ApoAI levels in males than the subjects with CC or CT genotype (P < 0.05 for all), and the T allele carriers had higher TC, LDL-C and ApoB levels in females than the T allele noncarriers (P < 0.05 for all). The participants with TT genotype in Han also had a lower tendency of HDL-C and ApoAI levels in males than the participants with CC or CT genotype, but the difference did not reach statistically significant (P = 0.063 and P = 0.086; respectively). The association of serum HDL-C and ApoAI levels and genotypes was confirmed by the multiple linear regression analysis in both ethnic groups. Serum lipid parameters were also correlated with several environmental factors.
Conclusions
The differences in serum lipid levels between the two ethnic groups might partially attribute to the differences in the SCARB1 rs5888 SNP and several environmental factors.
Introduction
Coronary heart disease (CHD) remains a major cause of worldwide morbidity and mortality despite therapeutic advances that control many risk factors such as low-density lipoprotein cholesterol (LDL-C) to levels lower than previously possible [1]. Plasma high-density lipoprotein cholesterol (HDL-C) levels are considered as a major determinant of susceptibility to coronary atherosclerosis in the general population [2,3]. A low plasma HDL-C level is the most common lipid abnormality found in families with premature coronary atherosclerosis [2]. Epidemiological studies showed that plasma HDL-C levels are inversely associated with the risk of CHD: an increase of 1 mg/dl of HDL-C levels is associated with a 2% to 3% decrease of the risk for CHD [4,5]. The metabolism of HDL-C is complex, with many factors influencing its circulating plasma levels, both genetic and non-genetic. It has been reported that variation in HDL-C levels is at least 50% genetically determined [6]. A number of variants in candidate genes have been implicated in the regulation of plasma HDL-C levels [7,8].
The scavenger receptor class B type 1 (SCARB1) is the HDL receptor which binds HDL-C with high affinity. It is expressed primarily in liver and nonplacental steroidogenic tissues and mediates selective cholesterol uptake of HDL by a mechanism distinct from the classic LDL-C receptor pathway, and plays an important role in reverse cholesterol translation (RCT) [9]. In mouse models, studies have clearly demonstrated the crucial role of SCARB1 gene in HDL-C metabolism. Hepatic overexpression of SCARB1 markedly reduced HDL-C and apolipoprotein (Apo) AI levels, and increased biliary cholesterol [10]. In contrast, targeted disruption of the SCARB1 gene in mice reduced selective uptake of cholesterol ester from HDL into the liver and significantly increased plasma HDL-C [11,12]. As a multilipoprotein receptor, SCARB1 also regulates the concentrations of LDL-C and very-low-density lipoprotein (VLDL-C) [13,14]. Mice with hepatic overexpression of SCARB1 have lower concentrations of VLDL and LDL [15,16]. Conversely, mice with attenuated expression of SCARB1 display elevated concentration of LDL-C [17]. Despite the obvious functional evidence for an influence of SCARB1 on altered serum lipid profile in animal models, it remains to be determined whether this receptor has an equally important function in humans.
The human SCARB1 gene encodes a 509 amino acid protein with a molecular weight of 82 kDa [13] and located on chromosome 12q24, a region showing significant linkage to plasma HDL-C levels [18]. Previous studies have investigated the relationship between variants in the SCARB1 gene and alterations on serum lipid profile in diverse human populations [19-24]. The single nucleotide polymorphism (SNP) of rs5888, a“C” to “T” substitution at amino acid 350 in exon 8, has been associated with the lipid concentrations of HDL-C [19,21,23,24] and LDL-C [20,21,23]. The C allelic frequency of SCARB1 rs5888 was varied from 40% to 60% in white population [19-23]. However, in Korean subjects the C allelic frequency of SCARB1 rs5888 was 67% in control and 83% in CHD [24]. These studies suggest that the polymorphism of SCARB1 rs5888 might be different among diverse ethnic groups. Thus, the SCARB1 rs5888 polymorphism in other racial groups still needs to be determined [24].
China is a multi-ethnic country. There are 56 ethnic groups. Han is the largest ethnic group and Yao is the eleventh largest minority among the 55 minority groups according to the population size. Bai Ku Yao (White-trouser Yao), an isolated branch of the Yao minority, is named so because all of the men wear white knee-length knickerbockers. The population size is about 30000. Because of isolation from the other ethnic groups, the special customs and cultures including their clothing, intra-ethnic marriages, dietary habits, and corn wine and rum intakes are still completely preserved to the present day. They are currently in a transitional period from the matriarchal society to patriarchal society. In several previous epidemiological studies, we showed that several serum lipid phenotypes were lower in Bai Ku Yao than in Han Chinese from the same region [25,26]. This ethnic difference in serum lipid profiles is still not well known. We hypothesized that some genetic factors may be different between the two ethnic groups. The SNP of rs5888 in SCARB1 gene had not been discussed in the Chinese population. The aim of the present study was to determine the association of SCARB1 rs5888 SNP and several environmental factors with serum lipid levels in the Guangxi Bai Ku Yao and Han populations.
Materials and methods
Study population
This study included 598 subjects of Bai Ku Yao who reside in Lihu and Baxu villages in Nandan County, Guangxi Zhuang Autonomous Region, People’s Republic of China and 585 subjects of Han Chinese reside in the same villages. The subjects of Bai Ku Yao consisted of 286 (47.8%) males and 312 (52.2%) females, ranged in age from 16 to 80 years, with a mean age of 40.22 ± 15.18 years. The subjects of Han consisted of 277 (47.4%) males and 308 (52.6%) females, aged 16–92 years, with a mean age of 41.87 ± 16.35 years. All of the subjects were randomly selected from our previous stratified randomized cluster samples [25,26]. The study subjects were healthy rural agricultural workers and without evidence of any chronic illness, including hepatic, renal, or thyroid. The participants with a history of heart attack or myocardial infarction, stroke, congestive heart failure, diabetes have been excluded. The participants were not on any lipid-lowering treatment. The present study was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University. Informed consent was obtained from all subjects after they received a full explanation of the study.
Epidemiological survey
The survey was carried out using internationally standardized methods [27]. Information on demographics, socioeconomic status, and lifestyle factors were collected with standardized questionnaires. The alcohol information included questions about the number of liangs (about 50 g) of rice wine, corn wine, rum, beer, or liquor consumed during the preceding 12 months. At the physical examination, several parameters such as height, weight, and waist circumference were measured. Sitting blood pressure was measured using a mercury sphygmomanometer on 3 separated intervals after the subjects had a 5-minute rest, and the average of the three measurements was used for the level of blood pressure. Body mass index (BMI) was calculated as weight in kg divided by the square of height in meters (kg/m2).
Biochemical analysis
Venous blood samples were obtained from all subjects after at least 12 hours of fasting. The levels of serum TC, triglyceride (TG), HDL-C, and LDL-C in samples were determined by enzymatic methods with commercially available kits. Serum ApoAI and ApoB levels were detected by the immunoturbidimetric immunoassay using a commercial kit [25,26] .
DNA amplification and genotyping
Genomic DNA was extracted from peripheral blood leukocytes using the phenol-chloroform method [28]. Genotyping of the SCARB1 rs5888 SNP was performed by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP). A pair of primers was designed to introduce a HinI1 restriction site (GACGCC) by changing a base from A to G. Forward 5'-CCTTGTTTCTCTCCCATCCTCACTTCCTCGACGC-3', Reverse primer 5'-CACCACCCCAGCCCACAGCAGC-3' (Sangon, Shanghai, People’s Republic of China). Each 20 μL PCR reaction mixture consisted of 1 μL of genomic DNA, 0.5 μL of each primer (10 pmol/L), 10 μL of 2 × Taq PCR Mastermix (constituent: 20 mM Tris–HCl, pH 8.3, 100 mM KCl, 3 mM MgCl2, 0.1 U Taq Polymerase/μL, 500 μM dNTP each; Tiangen, Beijing, People’s Republic of China), and 8 μL of ddH2O (DNase/RNase-free). The reaction mixture was subjected to denaturation at 95°C for 5 min, followed by 33 cycles at 95°C for 45 s, 71.5°C for 30 s, 72°C for 50 s, then by a final extension at 72°C for 8 min. The quality of PCR products was controlled by electrophoresis on 2% agarose gel and visualized with ethidium-bromide staining ultraviolet illumination. Then 5 μL of amplification products were digested at 37°C overnight with 5 U of HinI1 restriction enzyme (Fermentas Co. Canada). After restriction enzyme digestion of the amplified DNA, the fragments were separated by electrophoresis on 3% agarose gels stained with ethidium bromide, photographed in ultraviolet light. Genotypes were scored by an experienced reader blinded to epidemiological data and serum lipid levels. Six samples (CC, CT and TT genotypes in two; respectively) detected by the PCR-RFLP were also confirmed by direct sequencing.
Diagnostic criteria
The normal values of serum TC, TG, HDL-C, LDL-C, ApoAI, ApoB levels, and the ratio of ApoAI to ApoB in our Clinical Science Experiment Center were 3.10-5.17, 0.56-1.70, 0.91-1.81, 2.70-3.20 mmol/L, 1.00-1.78, 0.63-1.14 g/L, and 1.00-2.50; respectively[25,26]. Hypertension was diagnosed according to the criteria of 1999 World Health Organization-International Society of Hypertension Guidelines for the management of hypertension [29]. Normal weight, overweight and obesity were defined as a BMI < 24, 24–28, and > 28 kg/m2; respectively [25,26].
Statistical analyses
Epidemiological data were recorded on a pre-designed form and managed with Excel software. All statistical analyses were carried out using the statistical software package SPSS 13.0 (SPSS Inc., Chicago, Illinois). Qualitative variables were expressed as raw count and percentage. Mean ± standard deviation was used for the presentation of quantitative variables. Genotypic and allelic frequencies were calculated by direct counting, the Hardy-Weinberg equilibrium (HWE) was tested by chi-square, using the observed genotypic frequencies obtained from the data and expected genotypic frequency obtained using the HWE. A chi-square analysis was also used to evaluate the difference in genotype distribution and sex ratio between the groups. The difference in general characteristics between Bai Ku Yao and Han was tested by the Student’s unpaired t-test. The association of genotypes and serum lipid parameters was tested by analysis of covariance (ANCOVA). Sex, age, BMI, blood pressure, alcohol consumption, cigarette smoking were adjusted for the statistical analysis. All significant associations were corrected for multiple testing by applying a Bonferroni correction. In order to evaluate the association of serum lipid levels with genotypes (CC = 1, CT = 2, TT = 3) and several environment factors, multiple linear regression analysis with stepwise modeling was also performed in the combined population of Bai Ku Yao and Han, Bai Ku Yao, Han, males and females; respectively. A two-tailed P value less than 0.05 was considered statistically significant.
Results
General characteristics and serum lipid levels
The general characteristics and serum lipid levels between the Bai Ku Yao and Han populations are summarized in Table 1. As comparison with the population of Han, Bai Ku Yao has lower levels of height, weight, systolic blood pressure, diastolic blood pressure, serum TC, HDL-C, LDL-C, ApoAI, and higher serum ApoB levels, percentages of subjects who consumed alcohol or smoked cigarettes (P < 0.05-0.001). There were no significant differences in the levels of BMI, pulse pressure, serum TG, the ratio of ApoAI to ApoB, age structure, or the ratio of male to female between the two ethnic groups (P > 0.05 for all).
Table 1.
The general characteristics and serum lipid levels
| Parameter | Bai Ku Yao | Han Chinese | t (χ2) | P |
|---|---|---|---|---|
| Number |
598 |
585 |
– |
– |
| Male/female |
286/312 |
277/308 |
0.027 |
0.870 |
| Age (years) |
40.22 ± 15.18 |
41.87 ± 16.35 |
−1.795 |
0.073 |
| Height (cm) |
152.64 ± 7.37 |
156.33 ± 7.90 |
−8.318 |
0.000 |
| Weight (kg) |
51.69 ± 7.23 |
54.57 ± 9.74 |
−5.742 |
0.000 |
| Body mass index (kg/m2) |
22.15 ± 2.38 |
22.29 ± 3.46 |
−0.814 |
0.416 |
| Systolic blood pressure (mmHg) |
119.26 ± 17.44 |
122.78 ± 18.76 |
−3.352 |
0.001 |
| Diastolic blood pressure (mmHg) |
75.46 ± 9.69 |
77.58 ± 11.52 |
−3.430 |
0.001 |
| Pulse pressure (mmHg) |
43.80 ± 13.23 |
45.20 ± 12.79 |
−1.855 |
0.064 |
| Cigarette smoking [n (%)] | ||||
| Nonsmoker |
414 (69.2) |
437 (74.7) |
|
|
| < 20 cigarettes/day |
84 (14.0) |
57 (9.7) |
|
|
| ≥ 20 cigarettes/day |
100 (16.7) |
91 (15.6) |
6.074 |
0.048 |
| Alcohol consumption [n (%)] | ||||
| Nondrinker |
329 (55.0) |
458 (78.3) |
|
|
| < 25 g/day |
172 (28.8) |
80 (13.7) |
71.959 |
0.000 |
| ≥ 25 g/day |
97 (16.2) |
47 (8.0) |
|
|
| Total cholesterol (mmol/L) |
4.35 ± 0.92 |
4.71 ± 0.99 |
−6.551 |
0.000 |
| Triglyceride (mmol/L) |
1.00 (0.65) |
1.00 (0.79) |
−1.734 |
0.083 |
| HDL-C (mmol/L) |
1.67 ± 0.42 |
1.73 ± 0.46 |
−2.431 |
0.015 |
| LDL-C (mmol/L) |
2.57 ± 0.76 |
2.72 ± 0.79 |
−3.502 |
0.000 |
| Apolipoprotein (Apo) AI (g/L) |
1.31 ± 0.32 |
1.35 ± 0.31 |
−2.110 |
0.035 |
| ApoB (g/L) |
0.85 ± 0.23 |
0.82 ± 0.20 |
2.314 |
0.021 |
| ApoAI/ApoB | 1.67 ± 0.75 | 1.73 ± 0.53 | −1.382 | 0.167 |
HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol. The value of triglyceride was presented as median (interquartile range). The difference between the two ethnic groups was determined by the Wilcoxon-Mann–Whitney test.
Results of electrophoresis and genotyping
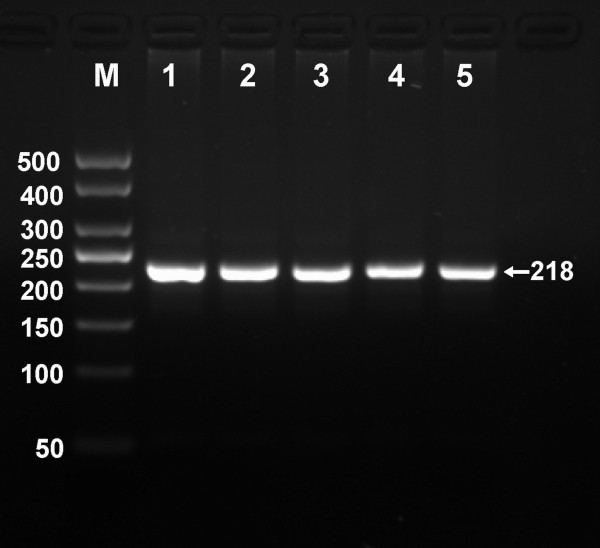
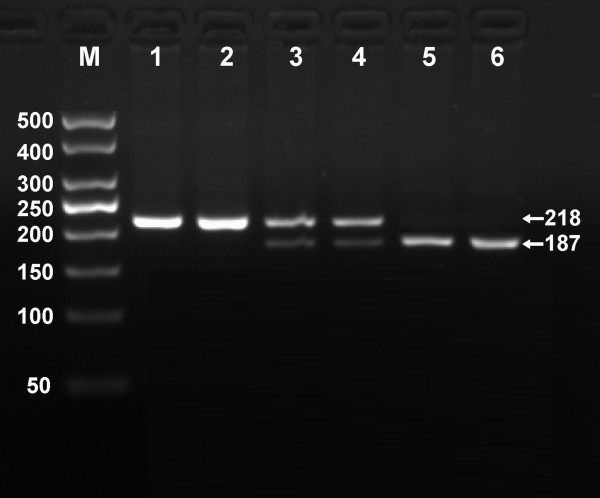
After the genomic DNA of the samples was amplified by PCR and imaged by 2.0% agarose gel electrophoresis, the PCR products of 218 bp nucleotide sequences could be found in all samples (Figure 1). The genotypes identified were named according to the presence or absence of the enzyme restriction sites, when a C to T transversion at amino acid 350 of the SCARB1 gene. The presence of the cutting site indicates the C allele, while its absence indicates the T allele (cannot be cut). Thus, the TT genotype is homozygote for the absence of the site (band at 218 bp), CT genotype is heterozygote for the absence and presence of the site (bands at 218-, 187- and 31-bp), and CC genotype is homozygote for the presence of the site (bands at 187- and 31-bp; Figure 2). The 31 bp fragment was invisible in the gel owing to its fast migration speed.
Figure 1.
Electrophoresis of PCR products of the samples. Lane M, 50 bp marker ladder; lanes 1–5, samples. The 218 bp bands are the PCR products.
Figure 2.
Genotyping of rs5888 SNP in the SCARB1 gene. Lane M, 50 bp marker ladder; lanes 1–2, TT genotype (218 bp); lanes 3 and 4, CT genotype (218-, 187- and 31-bp); and lanes 5 and 6, CC genotype (187- and 31-bp). The 31 bp fragment was invisible in the gel owing to its fast migration speed.
Genotypic and allelic frequencies
The genotypic and allelic frequencies of SCARB1 rs5888 SNP are shown in Table 2. The frequency of C and T alleles was 78.3% and 21.7% in Bai Ku Yao, and 73.7% and 26.3% in Han (P < 0.01); respectively. The frequency of CC, CT and TT genotypes was 60.0%, 36.6% and 3.3% in Bai Ku Yao, and 54.2%, 39.0% and 6.8% in Han (P < 0.01); respectively. There was no significant difference in the genotypic and allelic frequencies between males and females in both ethnic groups.
Table 2.
The genotype and allele frequencies of the SCARB1 rs5888 SNP [n (%)]
| Group | n | Genotype |
Allele |
|||
|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||
| Bai Ku Yao |
598 |
359 (60.0) |
219 (36.6) |
20 (3.4) |
937 (78.3) |
259 (21.7) |
| Han Chinese |
585 |
317 (54.2) |
228 (39.0) |
40 (6.8) |
862 (73.7) |
308 (26.3) |
|
χ2 |
– |
9.316 |
7.076 |
|||
|
P |
– |
0.009 |
0.008 |
|||
| Bai Ku Yao | ||||||
| Male |
286 |
176 (61.5) |
99 (34.6) |
11 (3.8) |
451 (78.8) |
121 (21.2) |
| Female |
312 |
183 (58.7) |
120 (38.5) |
9 (2.9) |
486 (77.9) |
138 (22.1) |
|
χ2 |
– |
1.222 |
0.163 |
|||
|
P |
– |
0.543 |
0.687 |
|||
| Han Chinese | ||||||
| Male |
277 |
145 (52.3) |
114 (41.2) |
18 (6.5) |
404 (72.9) |
150 (27.1) |
| Female |
308 |
172 (55.8) |
114 (37.0) |
22 (7.1) |
458 (74.4) |
158 (25.6) |
|
χ2 |
– |
1.060 |
0.306 |
|||
| P | – | 0.589 | 0.580 | |||
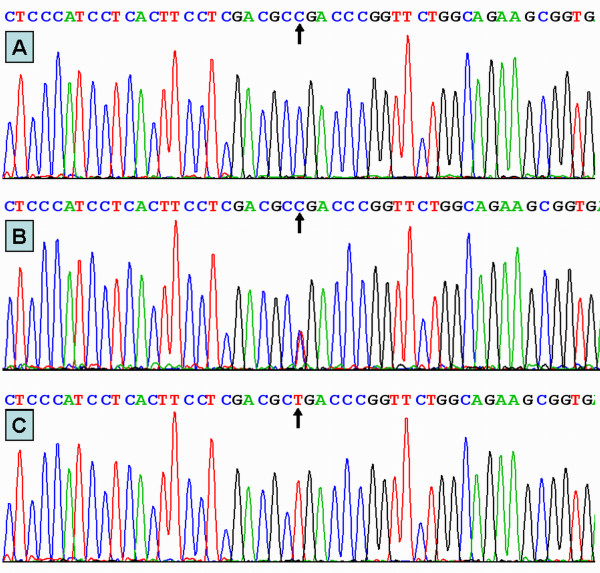
Results of sequencing
The results were shown as CC, CT and TT genotypes by PCR-RFLP, the CC, CT and TT genotypes were also confirmed by sequencing (Figure 3); respectively.
Figure 3.
A part of the nucleotide sequence of rs5888 SNP in the SCARB1 gene. (A) CC genotype; (B) CT genotype; (C) TT genotype
Genotypes and serum lipid levels
As shown in Table 3, the levels of HDL-C and ApoAI in the Bai Ku Yao and Han subjects were different among the three genotypes (P < 0.05 for all), the subjects with TT genotype had lower serum HDL-C and ApoAI levels than the subjects with CT or CC genotype. When analysis of covariance was stratified according to sex in both ethnic groups, we found that the subjects with TT genotype in Bai Ku Yao had lower HDL-C and ApoAI levels in males than the subjects with CC or CT genotype (P < 0.05 for all), and the T allele carriers had higher TC, LDL-C and ApoB levels in females than the T allele noncarriers (P < 0.05 for all). The participants with TT genotype in Han also had a lower tendency of HDL-C and ApoAI levels in males than the participants with CC or CT genotype, but the difference did not reach statistically significant (P = 0.063 and P = 0.086; respectively).
Table 3.
The genotypes of SCARB1 rs5888 SNP and serum lipid levels
| Genotype | n | TC (mmol/L) | TG (mmol/L) | HDL-C (mmol/L) | LDL-C (mmol/L) | ApoAI (g/L) | ApoB (g/L) | ApoAI/ApoB |
|---|---|---|---|---|---|---|---|---|
| Bai Ku Yao | ||||||||
| CC |
359 |
4.30 ± 0.94 |
0.98 (0.69) |
1.66 ± 0.42a |
2.54 ± 0.76 |
1.31 ± 0.32a |
0.84 ± 0.23 |
1.68 ± 0.76 |
| CT |
219 |
4.44 ± 0.89 |
1.02 (0.62) |
1.69 ± 0.41a |
2.64 ± 0.75 |
1.33 ± 0.33a |
0.87 ± 0.22 |
1.67 ± 0.72 |
| TT |
20 |
4.04 ± 0.96 |
1.10 (0.52) |
1.43 ± 0.36 |
2.42 ± 0.88 |
1.14 ± 0.27 |
0.78 ± 0.27 |
1.66 ± 1.00 |
|
F |
– |
2.770 |
0.388 |
4.001 |
1.625 |
3.878 |
1.871 |
0.023 |
|
P |
– |
0.063 |
0.824 |
0.019 |
0.198 |
0.021 |
0.155 |
0.977 |
| Male | ||||||||
| CC |
176 |
4.39 ± 1.09 |
1.18 (0.95) |
1.70 ± 0.49a |
2.54 ± 0.92 |
1.38 ± 0.37a |
0.83 ± 0.23 |
1.82 ± 0.95 |
| CT |
99 |
4.43 ± 1.01 |
1.05 (0.69) |
1.74 ± 0.46b |
2.55 ± 0.86 |
1.39 ± 0.40a |
0.83 ± 0.23 |
1.86 ± 0.92 |
| TT |
11 |
3.97 ± 1.07 |
1.25 (0.61) |
1.30 ± 0.45 |
2.35 ± 0.99 |
1.10 ± 0.35 |
0.76 ± 0.30 |
1.74 ± 1.32 |
|
F |
– |
0.985 |
1.822 |
4.743 |
0.253 |
3.769 |
0.559 |
0.117 |
|
P |
– |
0.375 |
0.402 |
0.009 |
0.776 |
0.024 |
0.572 |
0.889 |
| CC |
176 |
4.39 ± 10.9 |
1.18 (0.95) |
1.70 ± 0.49 |
2.54 ± 0.92 |
1.38 ± 0.37 |
0.83 ± 0.23 |
1.82 ± 0.95 |
| CT + TT |
110 |
4.38 ± 1.02 |
1.06(0.65) |
1.70 ± 0.47 |
2.53 ± 0.87 |
1.36 ± 0.40 |
0.82 ± 0.24 |
1.85 ± 0.96 |
|
F |
– |
0.003 |
−1.246 |
0.024 |
0.014 |
0.222 |
0.106 |
0.072 |
|
P |
– |
0.959 |
0.213 |
0.877 |
0.907 |
0.638 |
0.745 |
0.788 |
| Female | ||||||||
| CC |
183 |
4.22 ± 0.76 |
0.90 (0.51) |
1.63 ± 0.33 |
2.53 ± 0.58 |
1.24 ± 0.25 |
0.84 ± 0.22 |
1.56 ± 0.48 |
| CT |
120 |
4.45 ± 0.78c |
1.00 (0.61) |
1.65 ± 0.35 |
2.72 ± 0.64c |
1.27 ± 0.25 |
0.90 ± 0.19 |
1.49 ± 0.44 |
| TT |
9 |
4.19 ± 0.84 |
1.05 (0.38) |
1.51 ± 0.24 |
2.57 ± 0.74 |
1.16 ± 0.16 |
0.84 ± 0.24 |
1.46 ± 0.31 |
|
F |
– |
3.562 |
2.102 |
0.655 |
3.881 |
1.198 |
2.601 |
1.121 |
|
P |
– |
0.030 |
0.350 |
0.520` |
0.022 |
0.303 |
0.076 |
0.327 |
| CC |
183 |
4.22 ± 0.76 |
0.90 (0.51) |
1.63 ± 0.33 |
2.53 ± 0.58 |
1.24 ± 0.25 |
0.84 ± 0.22 |
1.56 ± 0.48 |
| CT + TT |
129 |
4.43 ± 0.78 |
1.01(0.60) |
1.64 ± 0.34 |
2.71 ± 0.65 |
1.26 ± 0.24 |
0.89 ± 0.20 |
1.48 ± 0.44 |
|
F |
– |
6.103 |
−1.429 |
0.054 |
7.234 |
0.542 |
4.473 |
2.224 |
|
P |
– |
0.014 |
0.153 |
0.817 |
0.008 |
0.462 |
0.035 |
0.137 |
| Han Chinese | ||||||||
| CC |
317 |
4.73 ± 0.96 |
0.97 (0.82) |
1.76 ± 0.49a |
2.75 ± 0.76 |
1.36 ± 0.31a |
0.82 ± 0.19 |
1.74 ± 0.54 |
| CT |
228 |
4.71 ± 1.01 |
1.05 (0.78) |
1.72 ± 0.44a |
2.73 ± 0.85 |
1.35 ± 0.31a |
0.82 ± 0.21 |
1.74 ± 0.54 |
| TT |
40 |
4.54 ± 1.05 |
0.99 (0.97) |
1.55 ± 0.35 |
2.58 ± 0.67 |
1.23 ± 0.21 |
0.81 ± 0.18 |
1.56 ± 0.35 |
|
F |
– |
0.821 |
1.764 |
4.125 |
0.965 |
3.824 |
0.044 |
2.313 |
|
P |
– |
0.441 |
0.414 |
0.017 |
0.381 |
0.022 |
0.957 |
0.100 |
| Male | ||||||||
| CC |
145 |
4.73 ± 0.94 |
1.11 (0.89) |
1.68 ± 0.54 |
2.74 ± 0.72 |
1.34 ± 0.36 |
0.83 ± 0.18 |
1.67 ± 0.50 |
| CT |
114 |
4.70 ± 1.07 |
1.13 (0.90) |
1.64 ± 0.39 |
2.74 ± 0.91 |
1.32 ± 0.32 |
0.84 ± 0.23 |
1.67 ± 0.54 |
| TT |
18 |
4.59 ± 1.22 |
1.23 (1.79) |
1.43 ± 0.26 |
2.47 ± 0.65 |
1.18 ± 0.18 |
0.83 ± 0.20 |
1.49 ± 0.37 |
|
F |
– |
0.233 |
1.500 |
2.793 |
1.070 |
2.474 |
0.036 |
1.160 |
|
P |
– |
0.793 |
0.472 |
0.063 |
0.344 |
0.086 |
0.965 |
0.315 |
| CC |
145 |
4.73 ± 0.94 |
1.11 (0.89) |
1.68 ± 0.54 |
2.74 ± 0.72 |
1.34 ± 0.36 |
0.83 ± 0.18 |
1.67 ± 0.50 |
| CT + TT |
132 |
4.69 ± 1.09 |
1.15(0.94) |
1.61 ± 0.38 |
2.71 ± 0.88 |
1.30 ± 0.30 |
0.84 ± 0.22 |
1.65 ± 0.52 |
|
F |
– |
0.212 |
−1.071 |
1.746 |
0.139 |
1.216 |
0.041 |
0.124 |
|
P |
– |
0.646 |
0.284 |
0.187 |
0.709 |
0.271 |
0.839 |
0.725 |
| Female | ||||||||
| CC |
172 |
4.74 ± 0.98 |
0.91 (0.80) |
1.83 ± 0.43 |
2.76 ± 0.80 |
1.38 ± 0.27 |
0.81 ± 0.19 |
1.80 ± 0.57 |
| CT |
114 |
4.69 ± 0.94 |
0.96 (0.60) |
1.78 ± 0.48 |
2.70 ± 0.78 |
1.37 ± 0.31 |
0.80 ± 0.19 |
1.80 ± 0.53 |
| TT |
22 |
4.54 ± 0.91 |
0.94 (0.69) |
1.68 ± 0.38 |
2.68 ± 0.69 |
1.29 ± 0.23 |
0.80 ± 0.15 |
1.64 ± 0.34 |
|
F |
– |
0.559 |
0.914 |
1.343 |
0.373 |
1.139 |
0.115 |
0.937 |
|
P |
– |
0.572 |
0.633 |
0.263 |
0.689 |
0.322 |
0.891 |
0.393 |
| CC |
172 |
4.74 ± 0.98 |
0.91 (0.80) |
1.83 ± 0.43 |
2.76 ± 0.80 |
1.38 ± 0.27 |
0.81 ± 0.19 |
1.80 ± 0.57 |
| CT + TT |
136 |
4.66 ± 0.93 |
0.96(0.60) |
1.76 ± 0.46 |
2.69 ± 0.77 |
1.36 ± 0.30 |
0.80 ± 0.18 |
1.77 ± 0.50 |
|
F |
|
0.644 |
−0.657 |
1.841 |
0.730 |
0.602 |
0.227 |
0.255 |
| P | 0.423 | 0.511 | 0.176 | 0.394 | 0.439 | 0.634 | 0.614 | |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoAI, apolipoprotein AI; ApoB, apolipoprotein B. The value of TG was presented as median (interquartile range), the difference among the genotypes was determined by the Kruskal-Wallis test or the Wilcoxon-Mann–Whitney test. aP <0.05, bP <0.01 in comparison with the TT genotype of the same ethnic group and cP <0.05 in comparison with the CC genotype of the same ethnic group.
Relative factors for serum lipid parameters
Multiple linear regression analysis showed that serum HDL-C and ApoAI levels were correlated with genotypes in the combined populations of Bai Ku Yao and Han, Bai Ku Yao, and Han; respectively (P < 0.05 for all). When multiple linear regression analysis was performed according to sex in both ethnic groups, we found that the levels of HDL-C and ApoAI in Bai Ku Yao were correlated with genotypes in males (P < 0.05 for each, Table 4) but not in females. The levels of TC and LDL-C in Bai Ku Yao were also associated with genotypes in females but not in males. The levels of TG and HDL-C in Han were correlated with genotypes in males (P < 0.05 for each). Serum lipid parameters were also correlated with sex, age, BMI, alcohol consumption, cigarette smoking, and blood pressure in both ethnic groups (Tables 5 and 4).
Table 4.
Correlative factors for serum lipid parameters between males and females
| Lipid parameter | Relative factor | Unstandardized coefficient | Std. error | Standardized coefficient | t | P |
|---|---|---|---|---|---|---|
| Bai Ku Yao/male | ||||||
| TC |
Body mass index |
0.120 |
0.028 |
0.245 |
4.268 |
0.000 |
| TG |
Body mass index |
0.112 |
0.033 |
0.200 |
3.436 |
0.001 |
| HDL-C |
Alcohol consumption |
0.192 |
0.036 |
0.304 |
5.274 |
0.000 |
| |
Age |
0.006 |
0.002 |
0.189 |
3.302 |
0.001 |
| |
Genotype |
−0.416 |
0.137 |
−0.166 |
−3.036 |
0.003 |
| |
Body mass index |
−0.026 |
0.012 |
−0.116 |
−2.116 |
0.035 |
| LDL-C |
Body mass index |
0.098 |
0.024 |
0.236 |
4.092 |
0.000 |
| ApoAI |
Alcohol consumption |
0.170 |
0.028 |
0.342 |
6.048 |
0.000 |
| |
Age |
0.004 |
0.001 |
0.155 |
2.743 |
0.006 |
| |
Genotype |
−0.287 |
0.107 |
−0.145 |
−2.690 |
0.008 |
| ApoB |
Body mass index |
0.029 |
0.006 |
0.267 |
4.677 |
0.000 |
| ApoAI/ApoB |
Alcohol consumption |
0.376 |
0.071 |
0.301 |
5.274 |
0.000 |
| |
Body mass index |
−0.069 |
0.025 |
−0.157 |
−2.752 |
0.006 |
| Bai Ku Yao/female | ||||||
| TC |
Systolic blood pressure |
0.007 |
0.003 |
0.145 |
2.602 |
0.010 |
| |
Body mass index |
0.036 |
0.017 |
0.120 |
2.138 |
0.033 |
| |
Genotype |
0.165 |
0.078 |
0.118 |
2.118 |
0.035 |
| TG |
Alcohol consumption |
0.311 |
0.079 |
0.219 |
3.961 |
0.000 |
| LDL-C |
Genotype |
0.151 |
0.061 |
0.135 |
2.465 |
0.014 |
| |
Body mass index |
0.034 |
0.014 |
0.141 |
2.519 |
0.012 |
| |
Age |
0.005 |
0.003 |
0.122 |
1.997 |
0.047 |
| ApoAI |
Age |
0.002 |
0.001 |
0.141 |
2.509 |
0.013 |
| ApoB |
Systolic blood pressure |
0.002 |
0.001 |
0.175 |
3.134 |
0.002 |
| |
Body mass index |
0.012 |
0.005 |
0.143 |
2.572 |
0.011 |
| ApoAI/ApoB |
Diastolic blood pressure |
−0.007 |
0.003 |
−0.139 |
−2.466 |
0.014 |
| Han/male | ||||||
| TC |
Diastolic blood pressure |
0.019 |
0.005 |
0.227 |
3.783 |
0.000 |
| |
Age |
0.013 |
0.003 |
0.239 |
4.243 |
0.000 |
| |
Alcohol consumption |
0.249 |
0.071 |
0.186 |
3.494 |
0.001 |
| |
Body mass index |
0.031 |
0.015 |
0.120 |
2.108 |
0.036 |
| TG |
Body mass index |
0.119 |
0.021 |
0.320 |
5.729 |
0.000 |
| |
Cigarette smoking |
0.282 |
0.092 |
0.172 |
3.069 |
0.002 |
| |
Genotype |
0.292 |
0.132 |
0.125 |
2.216 |
0.027 |
| HDL-C |
Alcohol consumption |
0.177 |
0.034 |
0.287 |
5.223 |
0.000 |
| |
Age |
0.006 |
0.001 |
0.237 |
4.082 |
0.000 |
| |
Body mass index |
−0.029 |
0.007 |
−0.243 |
−4.147 |
0.000 |
| |
Diastolic blood pressure |
0.006 |
0.002 |
0.156 |
2.523 |
0.012 |
| |
Genotype |
−0.082 |
0.041 |
−0.108 |
−2.011 |
0.045 |
| LDL-C |
Age |
0.009 |
0.003 |
0.210 |
3.450 |
0.001 |
| |
Diastolic blood pressure |
0.009 |
0.004 |
0.142 |
2.181 |
0.030 |
| |
Body mass index |
0.026 |
0.013 |
0.128 |
2.082 |
0.038 |
| ApoAI |
Alcohol consumption |
0.161 |
0.023 |
0.369 |
6.960 |
0.000 |
| |
Age |
0.005 |
0.001 |
0.281 |
5.301 |
0.000 |
| ApoB |
Body mass index |
0.015 |
0.003 |
0.283 |
5.122 |
0.000 |
| |
Age |
0.003 |
0.001 |
0.245 |
4.418 |
0.000 |
| |
Alcohol consumption |
0.035 |
0.015 |
0.132 |
2.384 |
0.018 |
| ApoAI/ApoB |
Body mass index |
−0.036 |
0.008 |
−0.272 |
−4.721 |
0.000 |
| |
Alcohol consumption |
0.161 |
0.039 |
0.240 |
4.150 |
0.000 |
| Han/female | ||||||
| TC |
Age |
0.020 |
0.004 |
0.292 |
5.413 |
0.000 |
| |
Body mass index |
0.074 |
0.017 |
0.232 |
4.308 |
0.000 |
| TG |
Body mass index |
0.057 |
0.016 |
0.196 |
3.499 |
0.001 |
| HDL-C |
Cigarette smoking |
0.479 |
0.198 |
0.137 |
2.425 |
0.016 |
| LDL-C |
Age |
0.016 |
0.003 |
0.299 |
5.612 |
0.000 |
| |
Body mass index |
0.066 |
0.014 |
0.254 |
4.769 |
0.000 |
| ApoAI |
Age |
0.004 |
0.001 |
0.209 |
3.761 |
0.000 |
| |
Cigarette smoking |
0.316 |
0.124 |
0.142 |
2.559 |
0.011 |
| ApoB |
Body mass index |
0.022 |
0.003 |
0.344 |
6.530 |
0.000 |
| |
Age |
0.003 |
0.001 |
0.223 |
4.228 |
0.000 |
| ApoAI/ApoB |
Body mass index |
−0.042 |
0.010 |
−0.236 |
−4.335 |
0.000 |
| Cigarette smoking | 0.883 | 0.232 | 0.207 | 3.810 | 0.000 | |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoAI, apolipoprotein AI; ApoB, apolipoprotein B.
Table 5.
Correlative factors for serum lipid parameters
| Lipid parameter | Relative factor | Unstandardized coefficient | Std. error | Standardized coefficient | t | P |
|---|---|---|---|---|---|---|
| Both Yao and Han | ||||||
| TC |
Age |
0.011 |
0.002 |
0.181 |
6.402 |
0.000 |
| |
Body mass index |
0.059 |
0.009 |
0.180 |
6.419 |
0.000 |
| |
Ethnic group |
0.343 |
0.054 |
0.177 |
6.368 |
0.000 |
| |
Diastolic blood pressure |
0.010 |
0.003 |
0.115 |
3.850 |
0.000 |
| |
Alcohol consumption |
0.089 |
0.039 |
0.064 |
2.285 |
0.022 |
| TG |
Body mass index |
0.078 |
0.010 |
0.211 |
7.567 |
0.000 |
| |
Sex |
−0.257 |
0.071 |
−0.117 |
−3.631 |
0.000 |
| |
Alcohol consumption |
0.207 |
0.052 |
0.133 |
3.981 |
0.000 |
| |
Ethnic group |
0.167 |
0.063 |
0.076 |
2.657 |
0.008 |
| HDL-C |
Age |
0.005 |
0.001 |
0.174 |
6.163 |
0.000 |
| |
Alcohol consumption |
0.172 |
0.021 |
0.275 |
8.177 |
0.000 |
| |
Sex |
0.155 |
0.029 |
0.176 |
5.435 |
0.000 |
| |
Ethnic group |
0.115 |
0.025 |
0.130 |
4.548 |
0.000 |
| |
Body mass index |
−0.018 |
0.004 |
−0.119 |
−4.252 |
0.000 |
| |
Genotype |
−0.049 |
0.020 |
−0.066 |
−2.389 |
0.017 |
| LDL-C |
Body mass index |
0.051 |
0.008 |
0.193 |
6.678 |
0.000 |
| |
Age |
0.008 |
0.001 |
0.167 |
5.731 |
0.000 |
| |
Ethnic group |
0.103 |
0.045 |
0.066 |
2.313 |
0.021 |
| |
Diastolic blood pressure |
0.007 |
0.002 |
0.089 |
2.894 |
0.004 |
| |
Alcohol consumption |
−0.065 |
0.032 |
−0.059 |
−2.029 |
0.043 |
| ApoAI |
Alcohol consumption |
0.156 |
0.015 |
0.346 |
10.638 |
0.000 |
| |
Age |
0.004 |
0.001 |
0.194 |
7.111 |
0.000 |
| |
Ethnic group |
0.086 |
0.018 |
0.136 |
4.852 |
0.000 |
| |
Genotype |
−0.132 |
0.039 |
−0.092 |
−3.420 |
0.001 |
| |
Sex |
0.065 |
0.020 |
0.103 |
3.285 |
0.001 |
| ApoB |
Body mass index |
0.018 |
0.002 |
0.249 |
8.686 |
0.000 |
| |
Age |
0.002 |
0.000 |
0.148 |
5.150 |
0.000 |
| |
Ethnic group |
−0.038 |
0.012 |
−0.089 |
−3.237 |
0.001 |
| |
Diastolic blood pressure |
0.002 |
0.001 |
0.080 |
2.669 |
0.008 |
| ApoAI/ApoB |
Alcohol consumption |
0.267 |
0.031 |
0.286 |
8.523 |
0.000 |
| |
Body mass index |
−0.042 |
0.006 |
−0.190 |
−6.777 |
0.000 |
| |
Ethnic group |
0.142 |
0.038 |
0.108 |
3.755 |
0.000 |
| |
Sex |
0.105 |
0.043 |
0.080 |
2.458 |
0.014 |
| Bai Ku Yao | ||||||
| TC |
Body mass index |
0.067 |
0.016 |
0.172 |
4.214 |
0.000 |
| |
Age |
0.007 |
0.002 |
0.109 |
2.655 |
0.008 |
| |
Diastolic blood pressure |
0.008 |
0.004 |
0.083 |
1.972 |
0.049 |
| TG |
Alcohol consumption |
0.258 |
0.067 |
0.198 |
3.872 |
0.000 |
| |
Body mass index |
0.054 |
0.016 |
0.132 |
3.302 |
0.001 |
| |
Sex |
−0.330 |
0.106 |
−0.169 |
−3.116 |
0.002 |
| |
Cigarette smoking |
−0.153 |
0.070 |
−0.120 |
−2.185 |
0.029 |
| HDL-C |
Alcohol consumption |
0.126 |
0.022 |
0.228 |
5.777 |
0.000 |
| |
Age |
0.004 |
0.001 |
0.142 |
3.586 |
0.000 |
| |
Genotype |
−0.245 |
0.090 |
−0.106 |
−2.714 |
0.007 |
| LDL-C |
Body mass index |
0.062 |
0.013 |
0.194 |
4.855 |
0.000 |
| |
Age |
0.005 |
0.002 |
0.098 |
2.437 |
0.015 |
| ApoAI |
Alcohol consumption |
0.143 |
0.016 |
0.333 |
8.708 |
0.000 |
| |
Age |
0.003 |
0.001 |
0.140 |
3.655 |
0.000 |
| |
Genotype |
−0.177 |
0.068 |
−0.099 |
−2.611 |
0.009 |
| ApoB |
Body mass index |
0.019 |
0.004 |
0.205 |
5.064 |
0.000 |
| |
Age |
0.002 |
0.001 |
0.104 |
2.564 |
0.011 |
| |
Alcohol consumption |
−0.032 |
0.012 |
−0.107 |
−2.611 |
0.009 |
| |
Diastolic blood pressure |
0.002 |
0.001 |
0.097 |
2.306 |
0.021 |
| ApoAI/ApoB |
Alcohol consumption |
0.311 |
0.039 |
0.309 |
7.940 |
0.000 |
| |
Body mass index |
−0.044 |
0.012 |
−0.137 |
−3.522 |
0.000 |
| Han Chinese | ||||||
| TC |
Age |
0.016 |
0.002 |
0.260 |
6.443 |
0.000 |
| |
Body mass index |
0.050 |
0.011 |
0.174 |
4.374 |
0.000 |
| |
Diastolic blood pressure |
0.012 |
0.004 |
0.142 |
3.341 |
0.001 |
| |
Alcohol consumption |
0.164 |
0.061 |
0.101 |
2.697 |
0.007 |
| TG |
Body mass index |
0.094 |
0.013 |
0.270 |
6.949 |
0.000 |
| |
Cigarette smoking |
0.357 |
0.063 |
0.221 |
5.697 |
0.000 |
| HDL-C |
Age |
0.006 |
0.001 |
0.219 |
5.533 |
0.000 |
| |
Sex |
0.244 |
0.040 |
0.264 |
6.074 |
0.000 |
| |
Alcohol consumption |
0.190 |
0.033 |
0.250 |
5.702 |
0.000 |
| |
Body mass index |
−0.022 |
0.005 |
−0.163 |
−4.119 |
0.000 |
| |
Genotype |
−0.075 |
0.028 |
−0.101 |
−2.635 |
0.009 |
| LDL-C |
Age |
0.012 |
0.002 |
0.245 |
5.873 |
0.000 |
| |
Body mass index |
0.043 |
0.009 |
0.186 |
4.533 |
0.000 |
| |
Diastolic blood pressure |
0.006 |
0.003 |
0.090 |
2.062 |
0.040 |
| ApoAI |
Age |
0.005 |
0.001 |
0.251 |
6.600 |
0.000 |
| |
Alcohol consumption |
0.165 |
0.022 |
0.326 |
7.621 |
0.000 |
| |
Sex |
0.134 |
0.026 |
0.217 |
5.096 |
0.000 |
| |
Genotype |
−0.037 |
0.019 |
−0.074 |
−1.979 |
0.048 |
| ApoB |
Body mass index |
0.018 |
0.002 |
0.323 |
8.524 |
0.000 |
| |
Age |
0.003 |
0.000 |
0.228 |
5.967 |
0.000 |
| |
Cigarette smoking |
0.028 |
0.010 |
0.105 |
2.807 |
0.005 |
| ApoAI/ApoB |
Body mass index |
−0.039 |
0.006 |
−0.256 |
−6.450 |
0.000 |
| |
Sex |
0.232 |
0.047 |
0.218 |
4.931 |
0.000 |
| Alcohol consumption | 0.184 | 0.039 | 0.211 | 4.724 | 0.000 | |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoAI, apolipoprotein AI; ApoB, apolipoprotein B.
Discussion
The current study shows that the levels of serum TC, HDL-C, LDL-C, ApoAI were lower, whereas the levels of ApoB was higher in Bai Ku Yao than in Han. There was no significant difference in the levels of TG and the ratio of ApoAI to ApoB between the two ethnic groups. It is well known that dyslipidemia is a complex trait caused by both environmental and genetic factors. Bai Ku Yao is an isolated subgroup of the Yao minority in China. Strict intra-ethnic marriages have been performed in this population from time immemorial. Therefore, we are confident that the hereditary characteristics and genotypes of some lipid metabolism-related genes in this population may be different from those in Han Chinese.
In the present study, we showed that there was significant difference in the allelic and genotypic frequencies of SCARB1 rs5888 SNP between the two ethnic groups. The frequencies of CC genotypes were higher in Bai Ku Yao than in Han. The frequency of C allele was also higher in Bai Ku Yao than in Han (78.3% vs 73.7%, P < 0.01). To the best of our knowledge, this is the first study to report the association of SCARB1 rs5888 SNP and serum lipid levels in the Bai Ku Yao population. The genotypic and allelic frequency of SCARB1 rs5888 SNP in Han population observed in our research was also consistent with the frequency of Han Chinese from Beijing published on NCBI database (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=5888). Several similar researches have carried out in European and North American populations. In a initial study in Spain population, Acton et al.[20] determined the polymorphism of Exon 8 (C/T) (rs5888) and showed that the frequency of C allele was 56.22% in a general white Spanish population. In another study, McCarthy et al.[22] reported the frequency of C allele was 44.7% in Israeli, 60.2% in Finn and 52.5% in Swede; respectively. In an American population of Central European ancestry, the allele frequency of C in the SCARB1 gene was 59.0% [19]. In a population of Caucasian in America, the frequency of C allele of rs5888 polymorphism in the SCARB1 gene was 51.39% in men and women [23]. In Korean subjects, however, the frequency of the common C allele of SCARB1 HaeIII (rs5888) was 67% in control and 83% in CHD [24] which was similar to our study in Chinese population. Thus, our findings, coupled with reports in the literature, provide evidence for the prevalence of the C allele variation of rs5888 in the SCARB1 gene may have an ethnic specificity.
Several ethnic distinct populations have been previously reported that the T allele of SCARB1 rs5888 is correlated with an increase of serum HDL-C concentration and a decrease of LDL-C concentration. In a population of healthy Spanish, Acton et al.[20] reported that the T allele carriers have significant lower concentration of LDL-C than the T allele noncarriers in females but not in males. They did not find the association of exons 8 (rs5888) variant in SCARB1 with HDL-C levels. In a population of Caucasian in North America, Osgood et al.[23] showed that the T allele was associated with an increase in HDL particle size in men and women, serum HDL-C levels in men, and a decrease in LDL-C levels in women. In a cohort of the Amish Family Diabetes, Roberts et al.[19] documented that the rs5888 variant was significantly associated with higher HDL-C level in women younger than 50 years but not in women aged 50 years or older. In a case–control study including 137 subject with CHD and 124 age-matched controls, the concentrations of plasma HDL-C and ApoAI varied significantly among HaeIII (rs5888) genotypes in the CHD patients but not in the controls, the common genotype (CC) was associated with a lower concentrations of plasma HDL-C and ApoAI [24]. Morabia et al.[21] also found that the T allele has higher levels of HDL-C and HDL-C/LDL-C ratio in men. Two other studies carried out by McCarthy et al.[20,30] did not find the single rs5888 variant was correlated with lipid profiles, when combined with other polymorphisms in the SCARB1 gene, the combined genotypes were associated with TG, TG/HDL-C ratio and HDL-C in the women. However, some researches found that the rs5888 T allele was associated with increased levels of TC, LDL-C and TG. In a research on subjects with heterozygous familial hypercholesterolemia, Tai et al.[31] found that the exon 8 (rs5888) polymorphism was associated with increased levels of TC, VLDL-C, LDL-C and TG. Morabia et al.[21] also showed that the genotype of TT had higher levels of TC and LDL-C than the CC genotype in females.
It is well documented that an overexpression of SCARB1 in the animal model could induce a decreased in the levels of HDL-C, LDL-C, and ApoB [10,15] and mice with attenuated expression of SCARB1 display elevated concentrations of LDL-C as well as HDL-C [11,12]. In human, a novel 11-base pair deletion mutation in the promoter region of the SCARB1 gene explained some of the interindividual variation in plasma HDL-C levels in Chinese Taiwanese [32]. A study of women with hyperalphalipoproteinemia also showed that SCARB1 protein was inversely correlated with HDL-C levels and HDL size [33], which suggesting that the SCARB1 in human may partly play the similar role as in the animals. Furthermore, a recent research reported that the rs5888 variant in the SCARB1 affected SCARB1 RNA secondary structure, protein translation, and was significantly associated with reduced SCARB1 protein expression and function in vitro[34]. In the present research, however, we showed that the subjects with TT genotype in Bai Ku Yao had lower HDL-C and ApoAI levels in males than the subjects with CC or CT genotype, and the T allele carriers had higher TC, LDL-C and ApoB levels in females than the T allele noncarriers. The participants with TT genotype in Han also had a lower tendency of HDL-C and ApoAI levels in males than the participants with CC or CT genotype, but the difference did not reach statistically significant. The association of serum HDL-C and ApoAI levels and genotypes was confirmed by the multiple linear regression analysis in both ethnic groups. It is difficult to explain these contradictory findings. The possible reasons may be as follows. Firstly, in our study the frequency of C allele was 78.3% in Bai Ku Yao and 73.7% in Han. In white populations, however, the C allelic frequency of SCARB1 rs5888 was varied from 40% to 60%, which suggests that our study populations had different genetic background from white populations. Moreover, the C-T change at exon 8 did not affect the amino acid sequence of the protein, the SNP might be in linkage disequilibrium with a functional mutation in the SCARB1 gene, or alternatively with another nearby functional variant at the chromosomal region of 12q24 where several other candidate genes involved in lipid metabolism have been localized (i.e. ACACB, PLA2, CLTA, MVK-MMAB, ACADS, and TCF1) [20,23]. Thus, our findings contradict with previous papers may attribute to different functional mutation of the SCARB1 rs5888 or to be in linkage disequibibrium with other SNPs. Secondly, it might because of difference in study designs, sample size, as well as gene-enviromental interactions. Lastly, the size of our study population is a bit small, and the number of subjects with TT genotype in both ethnic groups is also small, which might not have had the power to detect the association of TT genotype and serum lipid levels.
In addition to the influence of genetic factors, environmental factors also play an important role in determining serum lipid levels [25,26] . In the present study, we also found that serum lipid parameters were correlated with several environmental factors. Although Bai Ku Yao and Han reside in the same region, the diet and lifestyle were significant difference between the two ethnic groups. For the Bai Ku Yao population, corn was the staple food and rice, soy, buckwheat, sweet potato, and pumpkin products were the subsidiary foods. Approximately 90% of the beverages were corn wine and rum. The alcohol content is about 15% (v/v). They are also accustomed to drink Hempseed soup. In contrast, rice was the staple food and corn, broomcorn, potato, and taro products were the subsidiary foods in the Han population. About 90% of the beverage was rice wine. The content of alcohol is about 30% (v/v). These differences in diet and lifestyle could partly account for the differences in serum lipid profiles between the two ethnic groups. Corn contains abundant dietary fiber and plant protein [35]. Consumption of dietary fiber, specifically the soluble type, such as pectins and guar gum can decrease serum TC levels [36,37]. Plant protein might promote the transportation and excretion of free cholesterol. Dietary soy protein has well-documented beneficial effects on serum lipid concentrations [38,39]. Buckwheat protein products have a potent hypocholesterolemic activity [40,41]. Ludvik et al.[42] found that ingestion of 4 g/day caiapo (the extract of white-skinned sweet potato) for 6 weeks reduced TC and LDL-C in type 2 diabetic patients previously treated by diet alone. Studies have demonstrated that pumpkin is a useful therapy for hypercholesterolemia through reducing oxidative stress and cholesterol levels [43]. There are more than 29 fat soluble constituents in Hempseed, among which saturated and unsaturated fatty acid methyl esters account for 12.36% and 86.96%; respectively [44]. Previous experimental and clinical studies have demonstrated that Hempseed or Hempseed oil can decrease TC, TG and LDL-C levels, reduce atherogenic index, and increase HDL-C concentration [25,26,45].
Conclusion
The present study shows that the frequency of T allele of SCARB1 rs5888 SNP is lower in Bai Ku Yao than in Han Chinese. The subjects with TT genotype in Bai Ku Yao had lower HDL-C and ApoAI levels in males than the subjects with CC or CT genotype, and the T allele carriers had higher TC, LDL-C and ApoB levels in females than the T allele noncarriers. The participants with TT genotype in Han also had a lower tendency of HDL-C and ApoAI levels in males than the participants with CC or CT genotype, but the difference did not reach statistically significant. The levels of HDL-C and ApoAI were correlated with genotypes in the both ethnic groups. These results suggest that the differences in serum lipid levels between the two enthnic groups might partially attribute to the differences in the SCARB1 rs5888 SNP and several environmental factors.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DFW participated in the design, undertook genotyping, and drafted the manuscript. RXY conceived the study, participated in the design, carried out the epidemiological survey, collected the samples, and helped to draft the manuscript. XJH, LHHA, XLC, LM, QL and TTY collaborated to the genotyping. JZW and SLP carried out the epidemiological survey and collected the samples. All authors read and approved the final manuscript.
Contributor Information
Dong-Feng Wu, Email: wulove26@tom.com.
Rui-Xing Yin, Email: yinruixing@yahoo.com.cn.
Xi-Jiang Hu, Email: huxijianght@126.com.
Lynn Htet Htet Aung, Email: lifeinnivana@gmail.com.
Xiao-Li Cao, Email: maten1996@gmail.com.
Lin Miao, Email: caotianshuangmu@163.com.
Qing Li, Email: liqing000116dsl@163.com.
Ting-Ting Yan, Email: yan083199@163.com.
Jin-Zhen Wu, Email: yinrx2003@yahoo.com.cn.
Shang-Ling Pan, Email: bitterlypan@yahoo.com.cn.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No: 30660061)
References
- Natarajan P, Ray KK, Cannon CP. High-density lipoprotein and coronary heart disease: current and future therapies. J Am Coll Cardiol. 2010;55:1283–99. doi: 10.1016/j.jacc.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Genest JJ, Martin-Munley SS, McNamara JR, Ordovas JM, Jenner J, Myers RH, Silberman SR, Wilson PW, Salem DN, Schaefer EJ. Familial lipoprotein disorders in patients with premature coronary artery disease. Circulation. 1992;85:2025–33. doi: 10.1161/01.CIR.85.6.2025. [DOI] [PubMed] [Google Scholar]
- Ballantyne CM, Olsson AG, Cook TJ, Mercuri MF, Pedersen TR, Kjekshus J. Influence of low high-density lipoprotein cholesterol and elevated triglyceride on coronary heart disease events and response to simvastatin therapy in 4 S. Circulation. 2001;104:3046–51. doi: 10.1161/hc5001.100624. [DOI] [PubMed] [Google Scholar]
- Linsel-Nitschke P, Tall AR. HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat Rev Drug Discov. 2005;4:193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- Wilson PW. High-density lipoprotein, low-density lipoprotein and coronary artery disease. Am J Cardiol. 1990;66:7A–10A. doi: 10.1016/0002-9149(90)90562-F. [DOI] [PubMed] [Google Scholar]
- Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–6. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- Chen SN, Cilingiroglu M, Todd J, Lombardi R, Willerson JT, Gotto AM, Ballantyne CM, Marian AJ. Candidate genetic analysis of plasma high-density lipoprotein-cholesterol and severity of coronary atherosclerosis. BMC Med Genet. 2009;10:111. doi: 10.1186/1471-2350-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes E, Coassin S, Kollerits B, Heid IM, Kronenberg F. Genetic-epidemiological evidence on genes associated with HDL cholesterol levels: a systematic in-depth review. Exp Gerontol. 2008;44:136–60. doi: 10.1016/j.exger.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–20. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- Kozarsky KF, Donahee MH, Rigotti A, Iqbal SN, Edelman ER, Krieger M. Overexpression of the HDL receptor SR-BI alters plasma HDL and bile cholesterol levels. Nature. 1997;387:414–7. doi: 10.1038/387414a0. [DOI] [PubMed] [Google Scholar]
- Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci USA. 1997;94:12610–5. doi: 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varban ML, Rinninger F, Wang N, Fairchild-Huntress V, Dunmore JH, Fang Q, Gosselin ML, Dixon KL, Deeds JD, Acton SL, Tall AR, Huszar D. Targeted mutation reveals a central role for SR-BI in hepatic selective uptake of high density lipoprotein cholesterol. Proc Natl Acad Sci USA. 1998;95:4619–24. doi: 10.1073/pnas.95.8.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo D, Gomez-Coronado D, Lasuncion MA, Vega MA. CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler Thromb Vasc Biol. 1997;17:2341–9. doi: 10.1161/01.ATV.17.11.2341. [DOI] [PubMed] [Google Scholar]
- Swarnakar S, Temel RE, Connelly MA, Azhar S, Williams DL. Scavenger receptor class B, type I, mediates selective uptake of low density lipoprotein cholesteryl ester. J Biol Chem. 1999;274:29733–9. doi: 10.1074/jbc.274.42.29733. [DOI] [PubMed] [Google Scholar]
- Wang N, Arai T, Ji Y, Rinninger F, Tall AR. Liver-specific overexpression of scavenger receptor BI decreases levels of very low density lipoprotein ApoB, low density lipoprotein ApoB, and high density lipoprotein in transgenic mice. J Biol Chem. 1998;273:32920–6. doi: 10.1074/jbc.273.49.32920. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Royer L, Gong E, Zhang J, Cooper PN, Francone O, Rubin EM. Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J Biol Chem. 1999;274:7165–71. doi: 10.1074/jbc.274.11.7165. [DOI] [PubMed] [Google Scholar]
- Huszar D, Varban ML, Rinninger F, Feeley R, Arai T, Fairchild-Huntress V, Donovan MJ, Tall AR. Increased LDL cholesterol and atherosclerosis in LDL receptor-deficient mice with attenuated expression of scavenger receptor B1. Arterioscler Thromb Vasc Biol. 2000;20:1068–73. doi: 10.1161/01.ATV.20.4.1068. [DOI] [PubMed] [Google Scholar]
- Feitosa ME, Rice T, Borecki IB, Rankinen T, Leon AS, Skinner JS, Despres JP, Blangero J, Bouchard C, Rao DC. Pleiotropic QTL on chromosome 12q23-q24 influences triglyceride and high-density lipoprotein cholesterol levels: the HERITAGE family study. Hum Biol. 2006;78:317–27. doi: 10.1353/hub.2006.0043. [DOI] [PubMed] [Google Scholar]
- Roberts CG, Shen H, Mitchell BD, Damcott CM, Shuldiner AR, Rodriguez A. Variants in scavenger receptor class B type I gene are associated with HDL cholesterol levels in younger women. Hum Hered. 2007;64:107–13. doi: 10.1159/000101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton S, Osgood D, Donoghue M, Corella D, Pocovi M, Cenarro A, Mozas P, Keilty J, Squazzo S, Woolf EA, Ordovas JM. Association of polymorphisms at the SR-BI gene locus with plasma lipid levels and body mass index in a white population. Arterioscler Thromb Vasc Biol. 1999;19:1734–43. doi: 10.1161/01.ATV.19.7.1734. [DOI] [PubMed] [Google Scholar]
- Morabia A, Ross BM, Costanza MC, Cayanis E, Flaherty MS, Alvin GB, Das K, James R, Yang AS, Evagrafov O, Gilliam TC. Population-based study of SR-BI genetic variation and lipid profile. Atherosclerosis. 2004;175:159–68. doi: 10.1016/j.atherosclerosis.2004.03.014. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Lewitzky S, Reeves C, Permutt A, Glaser B, Groop LC, Lehner T, Meyer JM. Polymorphisms of the HDL receptor gene associated with HDL cholesterol levels in diabetic kindred from three populations. Hum Hered. 2003;55:163–70. doi: 10.1159/000073986. [DOI] [PubMed] [Google Scholar]
- Osgood D, Corella D, Demissie S, Cupples LA, Wilson PWF, Meigs JB, Schaefer EJ, Coltell O, Ordovas JM. Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: the framingham study. J Clin Endocrinol Metab. 2003;88:2869–79. doi: 10.1210/jc.2002-021664. [DOI] [PubMed] [Google Scholar]
- Hong SH, Kim YR, Yoon YM, Min WK, Chun SI, Kim JQ. Association between HaeIII polymorphism of scavenger receptor class B type I gene and plasma HDL-cholesterol concentration. Ann Clin Biochem. 2002;39:478–81. doi: 10.1258/000456302320314485. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Qiming F, Dezhai Y, Shuquan L, Weixiong L, Shangling P, Hai W, Yongzhong Y, Feng H, Shuming Q. Comparison of demography, diet, lifestyle, and serum lipid levels between the Guangxi Bai Ku Yao and Han populations. J Lipid Res. 2007;48:2673–81. doi: 10.1194/jlr.M700335-JLR200. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Dezhai Y, Shuquan L, Yuming C, Hanjun Y, Qiming F, Shangling P, Weixiong L, Jing T, Yiyang L. Hyperlipidaemia and its risk factors in the Guangxi Bai Ku Yao and Han populations. Public Health Nutr. 2009;12:816–24. doi: 10.1017/S1368980008003273. [DOI] [PubMed] [Google Scholar]
- People's Republic of China-United States Cardiovascular and Cardiopulmonary Epidemiology Research Group. An epidemiological study of cardiovascular and cardiopulmonary disease risk factors in four populations in the People's Republic of China. Baseline report from the P.R.C.-U.S.A. Collaborative Study. Circulation. 1992;85:1083–96. doi: 10.1161/01.cir.85.3.1083. [DOI] [PubMed] [Google Scholar]
- Wu DF, Yin RX, Aung LH, Hu XJ, Cao XL, Miao L, Li Q, Yan TT, Wu JZ, Pan SL. Polymorphism of rs1044925 in the acyl-CoA:cholesterol acyltransferase-1 gene and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:139. doi: 10.1186/1476-511X-9-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruixing Y, Weixiong L, Hanjun Y, Dezhai Y, Shuquan L, Shangling P, Qiming F, Jinzhen W, Jianting G, Yaju D. Diet, lifestyle, and blood pressure of the middle-aged and elderly in the Guangxi Bai Ku Yao and Han populations. Am J Hypertens. 2008;21:382–7. doi: 10.1038/ajh.2008.1. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Lehner T, Reeves C, Moliterno DJ, Newby LK, Rogers WJ, Topol EJ. Association of genetic variants in the HDL receptor, SR-B1, with abnormal lipids in women with coronary artery disease. J Med Genet. 2003;40:453–8. doi: 10.1136/jmg.40.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai ES, Adiconis X, Ordovas JM, Carmena-Ramon R, Real J, Corella D, Ascaso J, Carmena R. Polymorphisms at the SRBI locus are associated with lipoprotein levels in subjects with heterozygous familial hypercholesterolemia. Clin Genet. 2003;63:53–8. doi: 10.1034/j.1399-0004.2003.630108.x. [DOI] [PubMed] [Google Scholar]
- Hsu LA, Ko YL, Wu S, Teng MS, Peng TY, Chen CF, Chen CF, Lee YS. Association between a novel 11-base pair deletion mutation in the promoter region of the scavenger receptor class B type I gene and plasma HDL cholesterol levels in Taiwanese Chinese. Arterioscler Thromb Vasc Biol. 2003;23:1869–74. doi: 10.1161/01.ATV.0000082525.84814.A9. [DOI] [PubMed] [Google Scholar]
- West M, Greason E, Kolmakova A, Jahangiri A, Asztalos B, Pollin TI, Rodriguez A. Scavenger receptor class B type I protein as an independent predictor of high-density lipoprotein cholesterol levels in subjects with hyperalphalipoproteinemia. J Clin Endocrinol Metab. 2009;94:1451–57. doi: 10.1210/jc.2008-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantineau J, Greason E, West M, Filbin M, Kieft JS, Carletti MZ, Christenson LK, Rodriguez A. A synonymous variant in scavenger receptor, class B, type I gene is associated with lower SR-BI protein expression and function. Atherosclerosis. 2010;210:177–82. doi: 10.1016/j.atherosclerosis.2009.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Ma X, Zhang D, Yu S. Effect of maize embryo on delaying aging. Food Sci. 2002;23:95–97. [Google Scholar]
- Lairon D. Dietary fibres: effects on lipid metabolism and mechanisms of action. Eur J Clin Nutr. 1996;50:125–33. [PubMed] [Google Scholar]
- Jenkins DJ, Kendall CW, Axelsen M, Augustin LS, Vuksan V. Viscous and nonviscous fibres, nonabsorbable and low glycaemic index carbohydrates, blood lipids and coronary heart disease. Curr Opin Lipidol. 2000;11:49–56. doi: 10.1097/00041433-200002000-00008. [DOI] [PubMed] [Google Scholar]
- Weggemans RM, Trautwein EA. Relation between soy-associated isoflavones and LDL and HDL cholesterol concentrations in humans: a meta-analysis. Eur J Clin Nutr. 2003;57:940–6. doi: 10.1038/sj.ejcn.1601628. [DOI] [PubMed] [Google Scholar]
- Zhan S, Ho SC. Meta-analysis of the effects of soy protein containing isoflavones on the lipid profile. Am J Clin Nutr. 2005;81:397–408. doi: 10.1093/ajcn.81.2.397. [DOI] [PubMed] [Google Scholar]
- Tomotake H, Shimaoka I, Kayashita J, Yokoyama F, Nakajoh M, Kato N. Stronger suppression of plasma cholesterol and enhancement of the fecal excretion of steroids by a buckwheat protein product than by a soy protein isolate in rats fed on a cholesterol-free diet. Biosci Biotechnol Biochem. 2001;65:1412–4. doi: 10.1271/bbb.65.1412. [DOI] [PubMed] [Google Scholar]
- Kayashita J, Shimaoka I, Nakajoh M, Yamazaki M, Kato N. Consumption of buckwheat protein lowers plasma cholesterol and raises fecal neutral sterols in cholesterol-Fed rats because of its low digestibility. J Nutr. 1997;127:1395–400. doi: 10.1093/jn/127.7.1395. [DOI] [PubMed] [Google Scholar]
- Ludvik BH, Mahdjoobian K, Waldhaeusl W, Hofer A, Prager R, Kautzky-Willer A, Pacini G. The effect of Ipomoea batatas (Caiapo) on glucose metabolism and serum cholesterol in patients with type 2 diabetes: a randomized study. Diabetes Care. 2002;25:239–40. doi: 10.2337/diacare.25.1.239. [DOI] [PubMed] [Google Scholar]
- Adaramoye OA, Achem J, Akintayo OO, Fafunso MA. Hypolipidemic effect of Telfairia occidentalis (fluted pumpkin) in rats fed a cholesterol-rich diet. J Med Food. 2007;10:330–6. doi: 10.1089/jmf.2006.213. [DOI] [PubMed] [Google Scholar]
- Ross SA, Mehmedic Z, Murphy TP, Elsohly MA. GC-MS analysis of the total delta9-THC content of both drug- and fiber-type cannabis seeds. J Anal Toxicol. 2000;24:715–7. doi: 10.1093/jat/24.8.715. [DOI] [PubMed] [Google Scholar]
- Schwab US, Callaway JC, Erkkila AT, Gynther J, Uusitupa MI, Jarvinen T. Effects of hempseed and flaxseed oils on the profile of serum lipids, serum total and lipoprotein lipid concentrations and haemostatic factors. Eur J Nutr. 2006;45:470–7. doi: 10.1007/s00394-006-0621-z. [DOI] [PubMed] [Google Scholar]