Abstract
Purpose.
To delineate astrocyte-mediated inflammatory processes in glaucoma, we analyzed proteomic responses of retinal astrocytes in an experimental rat model using a cell-specific approach.
Methods.
IOP elevation was induced in rats by hypertonic saline injections into episcleral veins. Enriched samples of astrocytes were isolated through the immunomagnetic cell selection process established originally for retinal ganglion cell (RGC) sampling. Ocular hypertensive and control samples were collected by pooling from rat eyes matched for the cumulative IOP exposure. Protein expression was analyzed complementarily by quantitative two-dimensional capillary liquid chromatography and linear ion trap mass spectrometry (LC-MS/MS) followed by quantitative Western blot analysis and retinal tissue immunolabeling using specific antibodies to selected proteins.
Results.
Following validation of enriched astrocyte samples, LC-MS/MS analysis resulted in the identification of over 2000 proteins with high confidence. Bioinformatic comparison analysis of the high-throughput MS/MS data along with the findings of immunoblotting and immunohistochemistry supported distinct responses of ocular hypertensive astrocytes during the experimental paradigm, which exhibited predominantly cellular activation and immune/inflammatory responses as opposed to activation of cell death signaling in ocular hypertensive RGCs. Inflammatory responses of astrocytes in experimental glaucoma included up-regulation of a number of immune mediators/regulators linked to TNF-α/TNFR signaling, nuclear factor kappa-B (NF-κB) activation, autophagy regulation, and inflammasome assembly.
Conclusions.
These findings validate an astrocyte-specific approach to quantitatively identify proteomic alterations in experimental glaucoma, and highlight many immune mediators/regulators characteristic of the inflammatory responses of ocular hypertensive astrocytes. By dissecting the complexity of prior data obtained from whole tissue, this pioneering approach should enable astrocyte responses to be defined and new treatments targeting astrocytes to be developed.
This study introduces an astrocyte-specific approach, validates its sensitivity to quantitatively identify astrocyte responses in experimental rat glaucoma, and highlights various immune mediators/regulators characteristic of the inflammatory responses of ocular hypertensive astrocytes.
Introduction
Retinal ganglion cell (RGC) axons, somas, and synapses are specific victims of glaucomatous neurodegeneration, but glial cells, including retina and optic nerve astrocytes, survive the glaucomatous tissue stress and respond differently. By exerting both neurosupportive and detrimental effects, glial cells have key roles in determining neuronal life or death decisions in glaucoma. It has become clear over the past two decades that elucidation of RGC and glia responses are equally important for glaucoma research aiming to better understand and treat neurodegeneration.1
An unbalanced environment created by a variety of stress stimuli in glaucomatous tissues becomes a major initiator and propagator of secondary injury processes, which include neuroinflammation.1,2 Chronic activation of the glia, resident immune regulatory cells, is commonly accepted as an indicator of ongoing neuroinflammation in the glaucomatous retina and optic nerve.1 A growing number of studies analyzing gene and protein expression in these tissues support increased production of various immune mediators in human glaucoma3–5 and different animal models.6–11 Based on in vitro observations, glial immune mediators are important to establish autocrine and paracrine feedback circuits for innate immune injury, glia-T cell interactions, and antigen presentation.12 For example, TNF-α, which is a major pro-inflammatory cytokine produced increasingly by activated glial cells in glaucoma,13,14 has been linked to glial activation response, inflammatory processes, and mediation of RGC death in cell cultures.15–17
We previously used enriched samples of RGCs in proteomic analysis to illuminate different aspects of RGC responses during glaucomatous neurodegeneration.18–20 More recently, we also started to isolate enriched samples of astrocytes through a similar cell isolation technique. With the advantage of cell-specific sampling, our study aimed to determine astrocyte-mediated inflammatory processes in an experimental rat model of glaucoma. Our findings highlighted various molecules characteristic of the distinct inflammatory responses of astrocytes during the experimental paradigm. Dissection of cell-specific responses can help identify molecular pathways of glaucomatous neurodegeneration toward new treatment strategies, and better understanding of glial immune response pathways can lead us to immune modulatory treatments for neuroprotection.
Materials and Methods
Experimental Rat Model of Glaucoma
Similar to previous studies,19–21 IOP elevation was induced in 8-month-old Brown Norway rats by hypertonic saline injections into episcleral veins as originally described by Morrison et al.22 IOP was measured in awake rats twice weekly using a handheld rebound tonometer (TonoLab, Colonial Medical Supply, Franconia, NH) and monitored for up to 8 weeks.
To determine optic nerve injury, 1 μm plastic cross-sections of the optic nerves were used for imaging-based axon quantification as described in our previous studies.19–21 However, unlike previously used systematic sampling protocol, optic nerve cross-sections were imaged in their entirety as non-overlapping frames using the Zeiss/AxioVision/MosaiX-Panorama software (Carl Zeiss, Thornwood, NY). This methodological improvement allowed axon counts representing the entire surface area of optic nerve cross-sections, free from sampling bias. After image acquisition, processing and analysis of captured images were performed as described previously using the Axiovision software (Carl Zeiss).19–21 By following the same protocol, we manually traced nerve outlines on mosaics of images, and determined the size and shape parameters to exclude intervening glia, myelin debris, and highly degenerated axons to ensure accurate counts.
Cumulative IOP exposure was determined by calculating the area under the pressure-time curve in the ocular hypertensive eye, then subtracting this IOP-time integral from that in the normotensive fellow eye (expressed in units of mm Hg-days) as described previously.21 To minimize the influence of IOP variability among animals and within the same eye over time, cell-specific samples of retinal proteins were collected by pooling from rat eyes matched for the cumulative IOP exposure of 200–400 mm Hg-days. Based on optic nerve axon counts, this selection criterion corresponded to a relative axon loss value of no more than 50% (42.6 ± 7.3%). Animals with no measurable IOP exposure were excluded from further analysis.
Mass spectrometric analysis and following Western blot analysis for data validation required cell-specific protein samples collected from over 200 rats. In an adult rat retina approximately 150,000 RGCs are present and the rat retina exhibits an astrocyte-to-RGC ratio of 1:5–7. The cell selection procedure used (with cell yields of more than 75%) isolates at least 50 μg of astrocyte protein from 15 rat eyes with or without ocular hypertension. Although 50 μg or more RGC protein can be obtained by pooling from 2–3 normotensive rat eyes, the same amount of RGC protein can be obtained from 8–9 ocular hypertensive rat eyes with 50% axon loss.
All animals were handled according to the regulations of the Institutional Animal Care and Use Committee, and all procedures adhered to the tenets of the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Sampling of Enriched Astrocyte Proteins
Enriched samples of astrocytes were obtained through the immunomagnetic technique we described originally to isolate RGCs.15 Briefly, retinas were dissected from enucleated rat eyes and the neural retina was peeled gently from the retinal pigment epithelium under a microscope. Tissues were dissociated briefly in Eagles's minimum essential medium (EMEM) containing 20 U/mL papain, 1 mM L-cystein, 0.5 mM EDTA, and 0.005% DNase (Worthington, Lakewood, NJ). After rinsing in an inhibitor solution containing EMEM, 0.2% ovomucoid (US Biological, Swampscott, MA), 0.04% DNAase, and 0.1% BSA (Sigma-Aldrich, St. Louis, MO), gentle trituration through a 1 mL plastic pipette was used to yield a suspension of single cells.
The immunomagnetic technique used antibody coated magnetic beads (4 × 108 beads/ml; Invitrogen, Grand Island, NY). After blocking by incubation with 1% gamma globulin, retinal cells were incubated with a monoclonal ASTRO1 antibody specific to an astrocyte surface protein for 20 minutes (100 μg for 1 × 105 cells/ml; Abcam, Cambridge, MA). Incubation was followed by washing steps with PBS containing 0.1% BSA, and 2 mM EDTA, pH 7.4. To select astrocytes, cells then were incubated with anti-mouse IgM-coated magnetic beads (25 μl; Invitrogen) for 20 minutes at 4°C. For complementary comparative analysis, we also obtained enriched samples of RGCs using a monoclonal antibody to Thy-1.1 (100 μg/ml; Millipore/Chemicon, Billerica, MA) as described previously.15,18–20 Protein samples were prepared with a lysis buffer containing 50 mM Hepes-KOH at pH 8.0, 100 mM KCl, 2 mM EDTA, 0.10% NP-40, 2 mM dithiothreitol, 10% glycerol, and protease and phosphatase inhibitors as described previously.23
All quantitative analyses of protein expression were repeated at least three times with different sample pools. The Mann-Whitney rank sum test was used to determine the statistical significance of differences in protein expression between ocular hypertensive and control samples.
Proteomic Analysis
Cell-specific samples (10 μg) were analyzed quantitatively by two-dimensional capillary liquid chromatography and linear ion trap mass spectrometry (LC-MS/MS) as described previously.23 Briefly, trypsin-digested samples were loaded onto an analytical 2D-capillary chromatography column packed with strong cation exchange (SCX) and C18 reversed-phase (RP) resin (Phenomenex, Torrance, CA). This biphasic column was attached to an analytical RP chromatography column with an integrated, laser-pulled emitter tip. Peptides were eluted from SCX with seven-step gradients of 500 mM ammonium acetate and eluted into a linear ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) according to a linear HPLC gradient. Protein identification was performed with Sequest Sorcerer (Sage-N Research, San Jose, CA) set up to search a FASTA formatted rat protein database with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 1.2 Da. The Scaffold (Proteome Software Inc., Portland, OR) was used to validate peptide and protein identifications based on the criteria of greater than 95.0% and 99.0% probability, and at least two peptides as specified by the Prophet peptide24 and protein25 algorithms. The protein abundance was determined by normalizing the number of unique spectral counts matching to the identified protein by its predicted molecular weight.26
Similar to previous studies,3–5 we used the Ingenuity Pathways Analysis (Ingenuity Systems, Mountain View, CA) to define functional patterns and generate interaction networks of the identified proteins. Canonical pathway analysis identified the pathways from the Ingenuity Pathways Analysis library that were most significant to the data set by the right-tailed Fisher's exact test.
Quantitative Western Blot Analysis
For proteomic data validation, cell-specific samples (10 μg) were also studied by quantitative Western blot analysis as described previously.3–5 Briefly, SDS-PAGE used 7.5% or 12% gels (Bio-Rad, Hercules, CA), and separated proteins were transferred electrophoretically to a nitrocellulose membrane (Bio-Rad). Following a blocking step, membranes were probed with a primary antibody to ASTRO1 (1:100; Abcam), glial fibrillary acidic protein (GFAP; 1:1000; Abcam), Thy-1.1 (1:1000; Millipore/Chemicon), neuronal nucleus protein (NeuN; 1:1000; Millipore/Chemicon), glutamine synthetase (GS; 1:1000; Millipore/Chemicon), cellular retinaldehyde-binding protein (CRALBP; 1:2000; Abcam), retinal pigment epithelium-specific protein 65 (RPE65; 1:5000; Abcam), TNF-α (1:500; Abcam), TNF receptor-1 (TNFR1; 1:500; Abcam), cleaved caspase-8 (1:500; Cell Signaling, Danvers, MA), a subunit of nuclear factor-kappa B (NF-κB), p65 (phospho-Ser276, 1:1000; Cell Signaling), an immunity-related GTPase (IRG), IRGM (1:500; LifeSpan Biosciences, Seattle, WA), mammalian target of rapamycin (mTOR; phospho-Ser2448, 1:100; Cell Signaling), nucleotide-binding domain, leucine-rich repeat containing (NLR) family-pyrin domain containing 3 (NLRP3; 1:500; Abcam), or procaspase-1/cleaved caspase-1 (1:150; Millipore/Chemicon). The antibody dilutions used were optimum as assessed preliminarily by signal intensity, background staining, and amount of non-specific detection with varying antibody concentrations. After a second blocking step, membranes were incubated with secondary antibodies conjugated with horseradish peroxidase (1:2000; Sigma-Aldrich). Immunoreactive bands were visualized by enhanced chemiluminescence using commercial reagents (GE Healthcare, Pittsburgh, PA). Images were captured at optimum condition for proper balance between signal and background, and avoided band saturation with signal. A beta-actin antibody (1:1000; Sigma-Aldrich) was used to reprobe the stripped immunoblots for loading and transfer control.
For densitometry of immunoblots, images were acquired using a scanning device with a linear dynamic range (Typhoon 9400, GE Healthcare), image analysis used the ImageQuant software (GE Healthcare), background correction was applied to each lane by baseline subtraction, and integral values were selected as measurement parameters. Following normalization to beta-actin, the average values obtained from normotensive control samples was used to calculate the fold change in protein expression in ocular hypertensive samples.
Retinal Immunolabeling
To determine the cellular localization of selected proteins, histological sections of the rat retina were analyzed by immunohistochemistry as described previously.3,4 Immunofluorescence labeling used the same primary antibodies described above for Western blot analysis, including antibodies to TNF-α, TNFR1, cleaved caspase-8, phospho-p65, IRGM, phospho-mTOR, NLRP3, and procaspase-1/cleaved caspase-1. In addition, a specific antibody against GFAP (1:200; Santa Cruz Biotechnology, Santa Cruz, CA) was used to identify astrocytes. A mixture of Alexa Fluor 488- or 568-conjugated species-specific IgGs (1:500, Invitrogen) was used for the secondary antibody incubation. Similar to Western blot analysis, optimum antibody dilutions (1:100–500) were pre-selected based on retinal immunolabeling with minimum non-specific background staining. DAPI (Thermo Fisher Scientific) was used for nuclear counterstaining. Slides were examined by fluorescence microscopy and images were recorded by digital photomicrography (Carl Zeiss). Negative controls were performed by replacing the primary antibody with serum or using an inappropriate secondary antibody to determine species specificity. At least six histological sections obtained from three ocular hypertensive and control retinas were used for each antibody immunolabeling.
Results
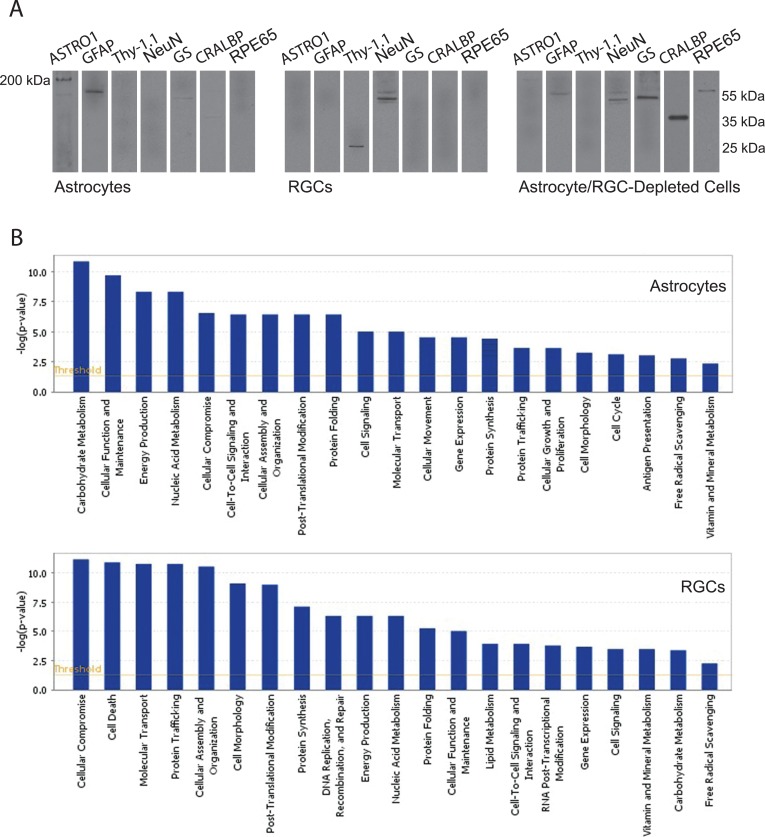
Our data validated the feasibility of astrocyte-specific sampling to detect cell-specific differences in protein expression in experimental glaucoma. We initially aimed to verify our enriched astrocyte samples by Western blot analysis using specific antibodies to different retinal cell markers. Similarly isolated RGC samples were verified previously based on retrograde fluorescence labeling,15 cell morphology and immunolabeling in culture,15 RT-PCR,20 and Western blot analysis of specific markers.18 However, to provide complementary information, we re-examined RGC samples, and also retinal protein samples obtained after depletion of astrocytes and RGCs, along with the analysis of astrocyte samples (Fig. 1A). Western blots verified GFAP and ASTRO1 expression in selected astrocytes. However, neuronal markers, including NeuN and Thy-1.1, were not detectable in these samples. Despite faint immunoreactivity for GS (a marker for Müller cells) and CRALBP (expressed in Müller cells and retinal pigment epithelium), enriched samples of astrocytes were negative for RPE65, a marker for retinal pigment epithelium. Similar to previous observations, RGC samples were positive for NeuN and Thy-1.1, but negative for ASTRO1, GFAP, GS, CRALBP, or RPE65. The minimum contamination of enriched astrocyte samples with Müller cell proteins (GS and CRALBP expressed only by developing astrocytes27) likely is due to the ASTRO1 antibody used for cell isolation, which is specific for astrocytes but also may bind weakly some Müller cells in the retina. To provide an internal control and complementary information, Figure 1A also presents Western blots of retinal protein samples obtained after depletion of astrocytes and RGCs. Note that dark immunoreactive bands for GS and CRALBP support Müller cell proteins remaining in these astrocyte/RGC-depleted samples. Altogether, the data presented in Figure 1A indicate no prominent contamination of enriched astrocyte samples with other retinal cell types and support the high purity of these samples.
Figure 1. .
Validation of enriched astrocyte samples. (A) Enriched samples of astrocytes exhibited prominent immunoreactivity for ASTRO1 and GFAP. Despite faint immunoreactive bands for GS (a marker for Müller cells) and CRALBP (expressed in Müller cells and retinal pigment epithelium), enriched samples of astrocytes were negative for RPE65, a marker for retinal pigment epithelium, as well as for neuronal markers (including NeuN and Thy-1.1). However, Western blot analysis detected neuronal markers (NeuN and Thy-1.1) in RGC samples. Also presented are Western blots of retinal protein samples obtained after depletion of astrocytes and RGCs. Note the dark immunoreactive bands for GS and CRALBP in these astrocyte/RGC-depleted samples, which support remaining Müller cell proteins. Protein samples were separated using 12% SDS-PAGE gels (except for ASTRO1 immunoblots in which 7.5% gels were used), and membranes were cut into strips to probe with different antibodies specific to target proteins. Data represent three independent sets of analyses with different samples. (B) Bioinformatics analysis of the comparative proteomic data by the Ingenuity Pathways Analysis supported distinct responses of astrocytes and RGCs in ocular hypertensive eyes. Shown are functional groups of the up-regulated proteins in ocular hypertensive astrocytes and RGCs relative to their controls.
The quantitative LC-MS/MS analysis of enriched astrocyte samples identified 2104 proteins by two peptides or more at the 0.2% peptide and 0.1% protein false discovery rates, and the MS/MS data included proteins exhibiting up-regulated or down-regulated expression in ocular hypertensive samples. Bioinformatic comparison analysis using the Ingenuity Pathways Analysis supported distinct responses of ocular hypertensive astrocytes during the experimental paradigm. To provide overall information about our high-throughput data, Figure 1B shows the functional groups of up-regulated proteins in ocular hypertensive astrocytes in comparison to ocular hypertensive RGCs. Ocular hypertensive astrocytes exhibited predominantly cellular activation and immune/inflammatory responses as opposed to the stress response and cell death signaling prominent in RGCs.
The Table lists 50 astrocyte proteins most relevant to inflammatory responses in ocular hypertensive samples. Data presented in this table include GFAP expression verifying astrocyte samples. Additional proteomic data are given in the Supplementary Table (http://www.iovs.org/content/53/7/4220/suppl/DC1). Selected proteins from this list were studied further for data validation. We ran Western blot analysis to validate increased expression and/or activation of selected proteins. As an additional effort to provide further validation for distinct responses of astrocytes during glaucomatous neurodegeneration, we also analyzed enriched samples of RGCs for these proteins and presented our astrocyte data along with this complementary data from RGC samples. In addition, we immunolabeled histological sections of the rat retina with specific antibodies to verify the cellular localization of selected proteins in astrocytes.
Table. .
Astrocyte Proteins Linked to Inflammatory Responses
|
Symbol |
Protein Name |
Protein ID |
Fold Change |
| TRADD | TNFRSF1A-associated via death domain | gi|281427214 | 2.1* |
| PEA15 | Astrocytic phosphoprotein PEA-15 | gi|61557370 | 1.8* |
| TNFAIP2 | Tumor necrosis factor alpha-induced protein 2 | gi|212549631 | 1.9* |
| RelA | Nuclear factor NF-kappa-B subunit p65 | gi|40538870 | 3.1* |
| NFKB-2 | Nuclear factor NF-kappa-B subunit p100 | gi|56605774 | 5.4* |
| IKBIP | Inhibitor of nuclear factor kappa-B kinase-interacting protein | gi|148539967 | 1.9* |
| MKK1 | Dual specificity mitogen-activated protein kinase kinase 1 | gi|13928886 | 2.2* |
| MKK3 | Mitogen activated protein kinase kinase 3 | gi|197333734 | 1.8* |
| MKK4 | Dual specificity mitogen-activated protein kinase kinase 4 | gi|71795640 | 1.5 |
| ERK2 | Mitogen-activated protein kinase 1 | gi|16758698 | 1.2 |
| ERK1 | Mitogen-activated protein kinase 3 | gi|8393331 | 3.2* |
| P38 | Mitogen-activated protein kinase 14 | gi|62461582 | 2.4* |
| ROCK2 | Rho-associated protein kinase 2 | gi|6981478 | −1.4 |
| AKT1 | RAC-alpha serine/threonine-protein kinase 1 | gi|15100164 | 2.8* |
| PDLIM1 | PDZ and LIM domain protein 1 | gi|8393153 | 1.1 |
| PDLIM7 | PDZ and LIM domain protein 7 | gi|27465579 | 1.4 |
| SGN1 | COP9 signalosome complex subunit 1 | gi|42476092 | 2.3* |
| SGN2 | COP9 signalosome complex subunit 2 | gi|23463271 | −1.2 |
| SGN3 | COP9 signalosome complex subunit 3 | gi|51948372 | 3.9* |
| SGN4 | COP9 signalosome complex subunit 4 | gi|51948518 | 0.2 |
| SGN5 | COP9 signalosome complex subunit 5 | gi|71043620 | 1.6 |
| SGN6 | COP9 signalosome complex subunit 6 | gi|157821399 | 1.2 |
| COPS7A | COP9 complex subunit 7a isoform 2 | gi|255760057 | 1.7 |
| SGN8 | COP9 signalosome complex subunit 8 | gi|61557351 | −0.7 |
| ASCC311 | Activating signal cointegrator 1 complex subunit 3-like | gi|281371480 | 4.1* |
| AP1B1 | AP-1 complex subunit beta-1 | gi|8392872 | 1.4 |
| AP1G1 | AP-1 complex subunit gamma-1 | gi|189491695 | 1.2 |
| AP1M1 | AP-1 complex subunit mu-1 | gi|112984344 | 2.2* |
| AP1S1 | AP-1 complex subunit sigma-1 | gi|205360945 | −0.9 |
| STAT1 | Signal transducer and activator of transcription 1 isoform alpha | gi|77695926 | 2.5* |
| STAT2 | Signal transducer and activator of transcription 2 | gi|58865380 | 1.6 |
| STAT3 | Signal transducer and activator of transcription 3 | gi|6981592 | 2.0* |
| STAT6 | Signal transducer and transcription activator 6 | gi|113205500 | 3.1* |
| TLR2 | Toll-like receptor 2 | gi|38454274 | 2.1* |
| TOLLIP | Toll-interacting protein | gi|157823763 | −1.6 |
| IRGQ | Immunity-related GTPase family, Q | gi|209364508 | 1.8* |
| mTOR | Serine/threonine-protein kinase mTOR | gi|9845251 | 2.1* |
| ATG3 | Ubiquitin-like-conjugating enzyme autophagy-related 3 | gi|19705511 | 1.5 |
| ATG7 | Ubiquitin-like modifier-activating enzyme autophagy-related 7 | gi|58865764 | 2.7* |
| ASC | Apoptosis-associated speck-like protein containing a CARD | gi|27229294 | 1.9* |
| CASP1 | Caspase-1 | gi|31542341 | 2.0* |
| ILF2 | Interleukin enhancer-binding factor 2 | gi|114145744 | 0.5 |
| ILF3 | Interleukin enhancer-binding factor 3 | gi|16758150 | 1.2 |
| IL-1RA | Interleukin-1 receptor antagonist protein | gi|11559964 | −1.9* |
| TGFB2 | Transforming growth factor beta-2 | gi|13592109 | 1.3 |
| TGFB3 | Transforming growth factor beta-3 | gi|6981650 | 0.3 |
| MIF | Macrophage migration inhibitory factor | gi|13591985 | 1.5 |
| GMFB | Glia maturation factor beta | gi|13624295 | 1.8* |
| GMFG | Glia maturation factor gamma | gi|30842809 | 2.2* |
| GFAP | Glial fibrillary acidic protein | gi|158186732 | 1.6 |
Protein samples of enriched astrocytes were collected by pooling from rat eyes matched for the cumulative IOP exposure and analyzed by quantitative LC-MS/MS. All listed proteins were identified with high confidence (based on at least two peptides with greater than 99.0% probability).
Significant difference in protein expression between OHT and control samples (Mann-Whitney rank sum test, P < 0.05).
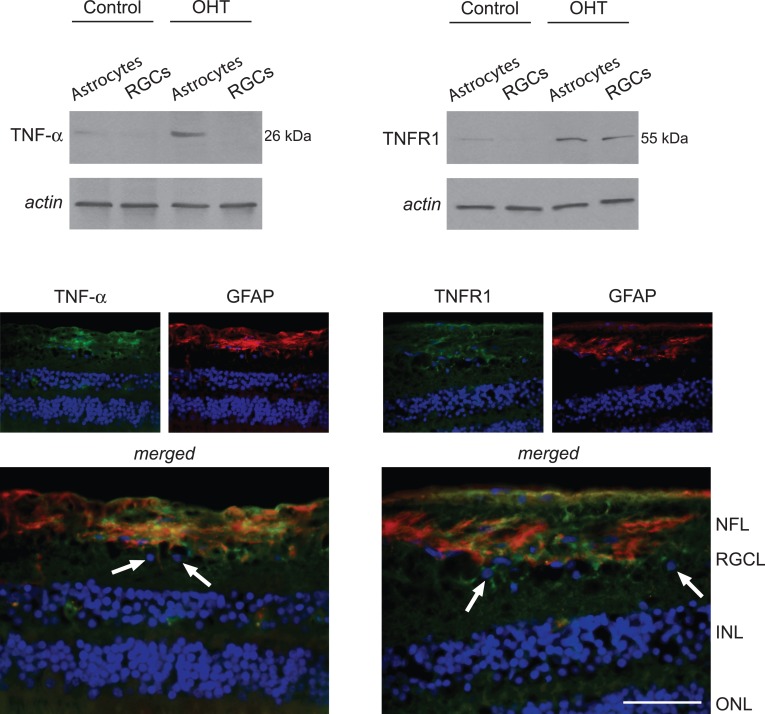
As listed in the Table, identified immune mediators/regulators in ocular hypertensive astrocyte samples included a number of downstream proteins linked to TNF-α/TNFR signaling. As an initial step for data validation, we aimed to determine whether cell-specific samples reflect previously detected differences in TNF-α and TNFR1 expression in human glaucoma.5,14 In parallel to previous observations, Western blot analysis supported increased expression of TNF-α and TNFR1 in astrocyte samples isolated from ocular hypertensive eyes relative to normotensive controls. This observation was also consistent with TNF-α immunolabeling localized predominantly to GFAP-positive astrocytes in the ocular hypertensive rat retina. However, TNFR1 immunolabeling was detectable on both GFAP-positive and GFAP-negative cells in the RGC/nerve fibers layers of the same tissues (Fig. 2).
Figure 2. .
TNF-α and TNFR1 expression in astrocytes. Western blot analysis detected TNF-α expression in astrocyte samples, not in RGCs. However, TNFR1 expression was detectable in samples of RGCs and astrocytes isolated from ocular hypertensive (OHT) rat eyes. Immunohistochemistry images show the OHT retina that indicated prominent localization of TNF-α (green) in GFAP-positive (red) astroglia. GFAP-positive cells in the RGC and NFL correspond to astrocytes, while Müller cells (located in the inner nuclear layer) and their processes through the retina may also exhibit immunolabeling for GFAP in OHT animals similar to human glaucoma. Regarding TNFR1 immunolabeling (green), in addition to GFAP-positive astroglia, some GFAP-negative cells in the RGC layer, likely corresponding to RGCs (arrows), also exhibited TNFR1 immunolabeling in the OHT rat retina. Blue corresponds to nuclear DAPI labeling. Data represent three independent sets of analyses with different samples. NFL, nerve fibers layer; RGCL, retinal ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Scale bar 100 μm.
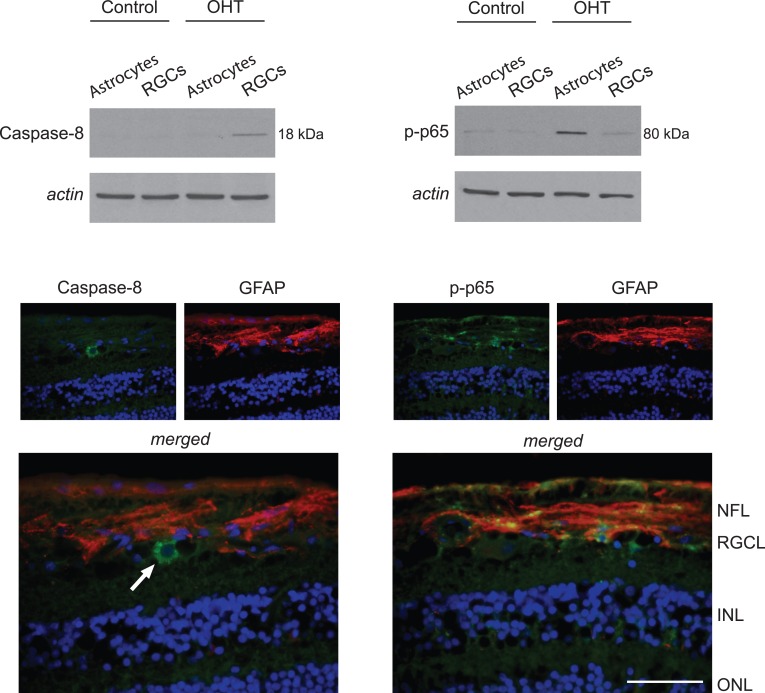
The proteins linked to TNF-α/TNFR signaling in ocular hypertensive astrocytes mainly included mediators of inflammatory processes, not mediators of apoptosis. In contrast, inflammatory proteins were not detectable by mass spectrometric analysis of ocular hypertensive RGCs. Due to prominent up-regulation of TNFR signaling in ocular hypertensive samples, the next set of validation studies determined cell-specific differences in TNFR1-mediated caspase activation by Western blot analysis and immunohistochemistry using a cleavage site-specific antibody. In support of the MS/MS data, cleaved caspase-8 (a proximal caspase in the TNFR1-mediated apoptosis pathway) was detectable in ocular hypertensive RGCs, but not in astrocytes (Fig. 3).
Figure 3. .
Components of TNF-α/TNFR signaling in OHT astrocytes. Western blot analysis using a cleavage site-specific antibody detected cleaved caspase-8 only in OHT RGCs. However, based on Western blots with a phosphorylation site-specific antibody, p65 (a subunit of NF-κB) activation was detectable most prominently in OHT astrocytes. Immunohistochemistry images show the OHT retina. No immunofluorescence labeling for cleaved caspase-8 (green) was detectable in GFAP-positive astroglia, while phospho-p65 (green) was localized predominantly to GFAP-positive (red) astrocytes. Arrow shows a GFAP-negative cell in the RGC layer, exhibiting prominent immunolabeling for cleaved caspase-8. Based on its larger, lighter, and circular nucleus, this GFAP-negative cell in the RGC layer possibly is a RGC. Blue corresponds to nuclear DAPI labeling. Data represent three independent sets of analyses with different samples. Scale bar 100 μm.
While caspase activation was not detectable in ocular hypertensive astrocytes, the opposing pathway of TNFR signaling that promotes NF-κB activation was detectable prominently. Based on the Ingenuity Pathways Analysis, the NF-κB activation pathway was among the top canonical pathways associated most significantly with our astrocyte dataset (right-tailed Fisher's exact test, P < 0.05). Western blot analysis using a phosphorylation site-specific antibody indicated increased phosphorylation of p65 (RelA) in ocular hypertensive astrocytes. Based on immunohistochemical analysis, phospho-p65 was localized predominantly to GFAP-positive astrocytes in the ocular hypertensive rat retina (Fig. 3).
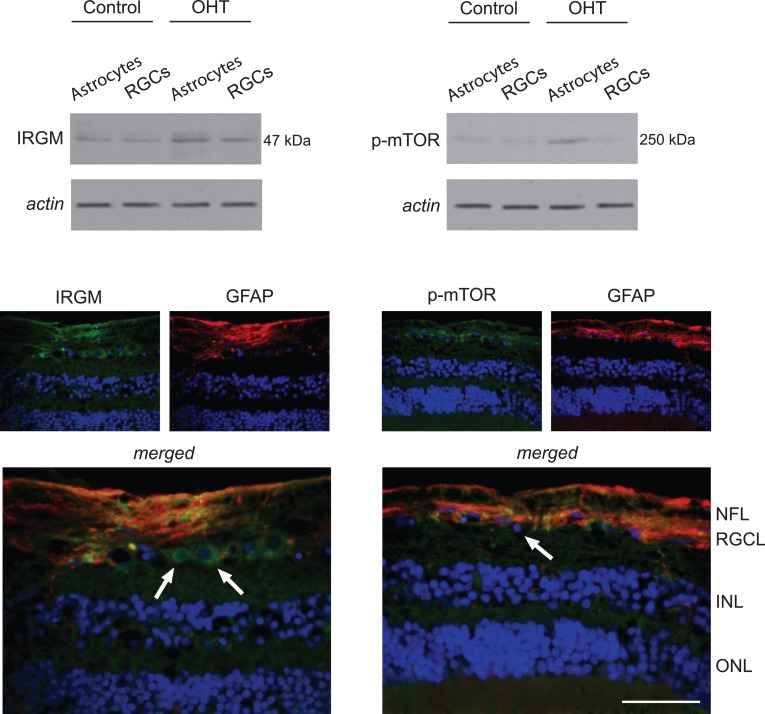
Another group of proteins up-regulated in ocular hypertensive astrocytes was related to autophagy signaling, including mTOR and autophagy-related proteins (ATGs), ATG3 and ATG7. Immunolabeling with a phosphorylation site-specific antibody also indicated phosphorylation-mediated activation of mTOR in ocular hypertensive astrocytes. However, phospho-mTOR was not prominently detectable in ocular hypertensive RGCs. An IRG linked to immunity-related autophagy, IRGM, also exhibited increased expression by Western blot analysis of ocular hypertensive astrocytes (Fig. 4).
Figure 4. .
Regulation of autophagy signaling in ocular hypertensive astrocytes. Western blot analysis detected prominent up-regulation of IRGM in OHT astrocytes. In addition, probing with a phosphorylation site-specific antibody to mTOR indicated increased immunoreactivity in these samples of OHT astrocytes. Based on immunofluorescence labeling, GFAP-positive (red) astrocytes and also some GFAP-negative cells in the RGC layer, likely corresponding to RGCs (arrows), exhibited IRGM immunolabeling (green) in the OHT rat retina. However, phospho-mTOR (green) immunolabeling was detectable predominantly in GFAP-positive astrocytes. Blue corresponds to nuclear DAPI labeling. Data represent three independent sets of analyses with different samples. Scale bar 100 μm.
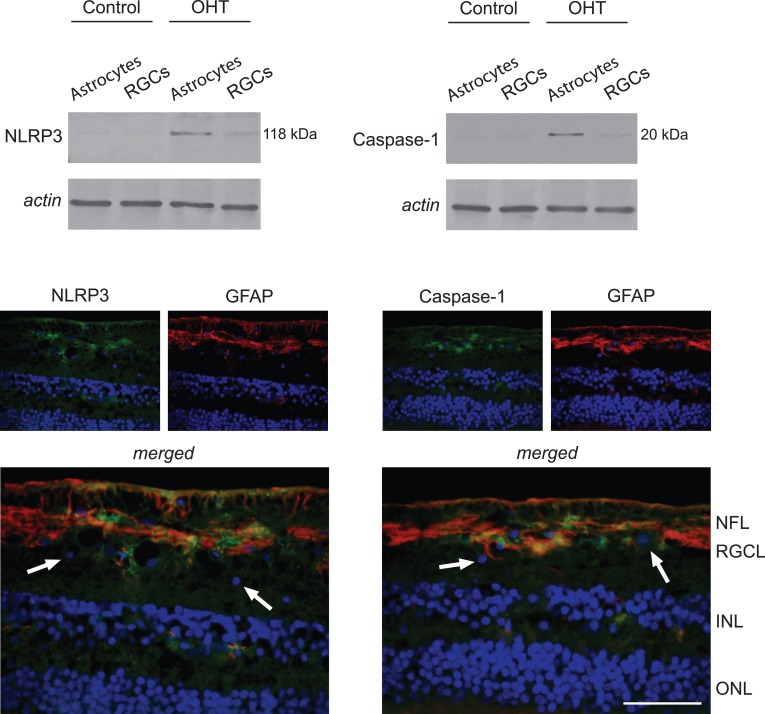
Western blot analysis also validated NLRP3 expression and caspase-1 cleavage supporting inflammasome assembly in ocular hypertensive astrocytes. In addition, immunohistochemical analysis of ocular hypertensive rat retinas indicated prominent localization of these inflammasome components in GFAP-positive astrocytes (Fig. 5).
Figure 5. .
Inflammasome components in OHT astrocytes. Western blot analysis using specific antibodies detected a subset of NLRs named NLRP3 and cleaved caspase-1 predominantly in OHT astrocytes. Immunofluorescence labeling of the OHT rat retina indicated prominent localization of NLRP3 and caspase-1 (green) in GFAP-positive (red) astrocytes. Arrows show GFAP-negative cells in the RGC layer exhibiting no detectable immunolabeling for NLRP3 or caspase-1. Blue corresponds to nuclear DAPI labeling. Data represent three independent sets of analyses with different samples. Scale bar 100 μm.
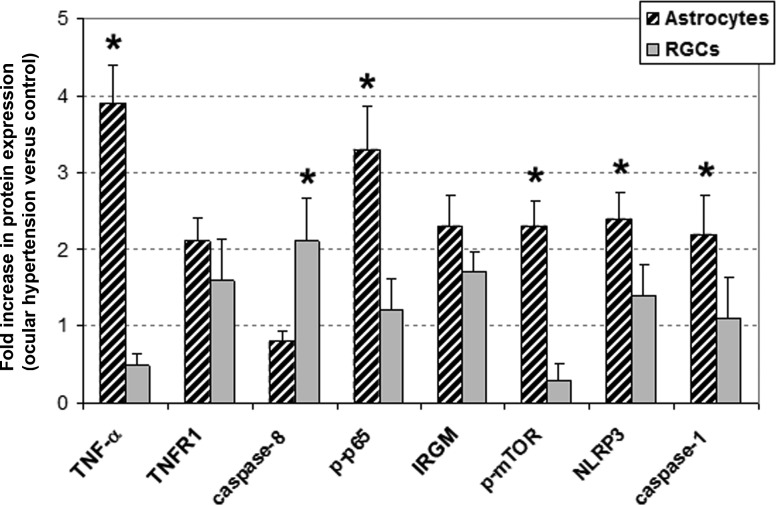
Figure 6 summarizes the protein expression data based on quantitative Western blot analysis. Figure 7 shows a simplified network of the studied proteins that characterize inflammatory responses of ocular hypertensive astrocytes.
Figure 6. .
Quantitative Western blot analysis of protein expression in cell-specific samples. Comparison of the beta-actin-normalized intensity values between OHT and control samples supported distinct responses of astrocytes and RGCs. Data represent three independent sets of analyses with different samples. Presented is the mean ± SD fold increase in protein expression, which was determined by comparing the actin-normalized densitometry values of specific immunoreactive bands in OHT samples with those of normotensive controls. Asterisk indicates statistically significant difference between astrocyte and RGC responses (Mann-Whitney rank sum test, P < 0.05).
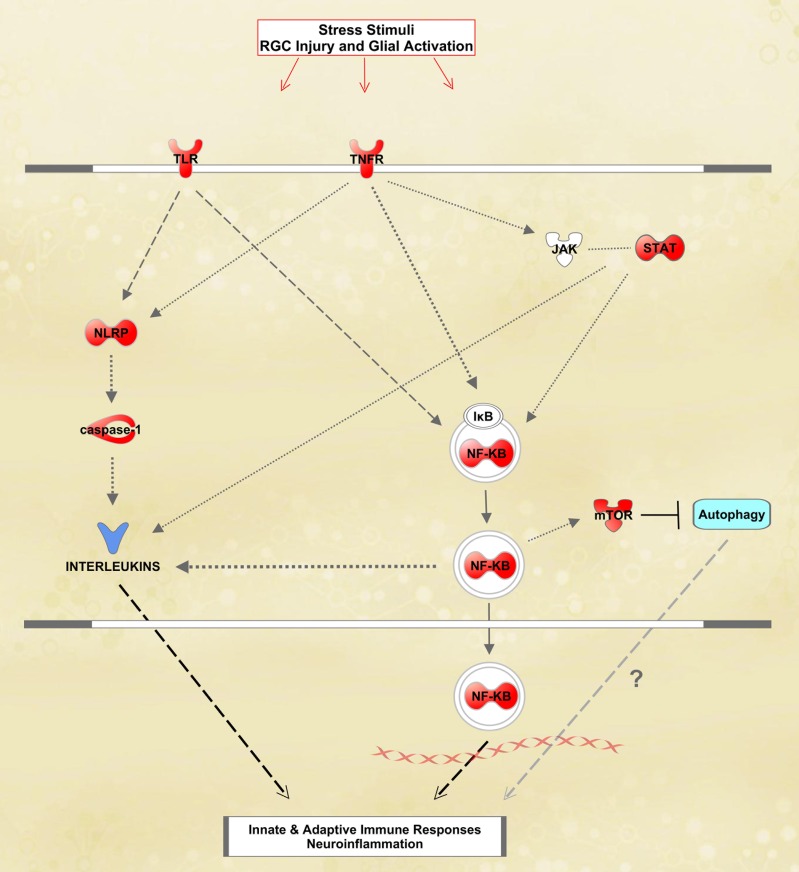
Figure 7. .
Astrocyte proteins linked to inflammatory responses in OHT samples. In this simplified network generated by The Ingenuity Pathways Analysis, the proteins shown in red exhibited significantly increased expression in OHT samples relative to normotensive controls.
Discussion
Cell-Specific Approach to Glaucomatous Neurodegeneration
Our study used a cell-specific approach to determine retinal astrocyte responses in experimental rat glaucoma. Our reproducible data supported the feasibility of cell-specific sampling, and validated the sensitivity of cell-specific analysis to characterize distinct responses of astrocytes during the experimental paradigm. Similar to our previous studies,15,16,18–20 we used a technique through which target cells are coupled immunologically to magnetic beads and then separated out from the mixed cell population using a magnetic field. This cell isolation process, which can be completed within approximately one hour, presents advantages over alternative techniques. A similar immunomagnetic technique has been used previously for rapid isolation of brain astrocytes with high purity and viability in culture.28 Cell-specific sampling allows direct analysis of primary astrocytes by avoiding phenotypic changes in tissue cultures. Due to this unique advantage, we had no intention to culture isolated astrocytes in this in vivo study, although we successfully used similarly isolated RGCs in previous in vitro studies.15,16 Despite appropriate sensitivity to determine differences in protein expression within distinct cell types, it remains unclear whether even a minimum contamination with a few other cell types may be problematic for the analysis of gene expression using these samples. In addition, sufficient sampling of astrocytes for protein analysis (and data validation by immunoblotting) may require large numbers of animals; however, rapidly emerging experimental techniques, ultrasensitive instruments, and new antibodies are expected to improve cell-specific sampling and analysis even further. There is no doubt that assessing cell-specific responses is an ideal way to determine molecular pathways involved in disease pathogenesis, since analysis of whole retina or optic nerve may reflect only the sum of opposing responses from many different cell types. By dissecting the complexity of prior data obtained from whole tissue, the cell-specific analysis enables specific cellular responses to be defined and cell-specific treatment strategies to be developed. Characterization of astrocyte responses can help design treatments to similarly enhance RGC survival in glaucoma and also enable to manipulate astrocyte responses for the gain of RGCs so that neurodestructive consequences may be interfered selectively without compromising glial neurosupportive and homeostatic functions.
Astroglial Components of TNF-α/TNFR Signaling Linked to Neuroinflammatory Responses
Analysis of enriched astrocytes detected predominantly cellular activation and immune/inflammatory responses in ocular hypertensive samples as opposed to various cell death mediators detected in RGCs isolated from the same experimental eyes. Ocular hypertensive astrocytes exhibited NF-κB activation regulating inflammatory consequences of TNF-α/TNFR signaling as discussed later below, but not caspase activation mediating TNF-α-induced apoptosis. Based on our findings, TNF-α/TNFR signaling during glaucomatous neurodegeneration may induce cell death in RGCs but mediate immune/inflammatory responses in astrocytes. This is consistent with previous in vitro findings that support a relative resistance of astrocytes, including retinal astrocytes,15 to death receptor-mediated apoptosis.29 Our present data suggested that various molecules may regulate cell-specific outcomes of TNF-α/TNFR signaling in glaucoma.
First, ocular hypertensive astrocytes exhibited up-regulated expression of a signal transducer protein involved in the multiprotein signaling complex formed after TNFR binding, namely TNFR-associated death domain protein (TRADD). This multifunctional protein not only is crucial for diverse consequences of TNFR1 signaling but also for other signaling pathways relevant to inflammatory responses.30 Second, up-regulated proteins in ocular hypertensive astrocytes included a dead domain-containing protein particularly abundant in astrocytes, named phosphoprotein enriched in astrocytes 15 (PEA-15).31 This astrocyte phosphoprotein directs cytokine outcomes toward survival and protects astrocytes from TNF-α-induced apoptosis by binding dead domain-containing proteins32,33 and controlling the activity of mitogen-activated protein kinases.34,35 Another up-regulated protein in ocular hypertensive astrocytes was TNF-α-induced protein 2, a primary response molecule involved in inflammatory processes induced by TNF-α.36,37 More studies are needed to determine the importance of identified molecules as treatment targets to modulate neurodegenerative inflammation and provide neuroprotection in glaucoma.
NF-κB Activation Regulating Neuroinflammatory Processes in Astrocytes
Our findings support that many of the astrocyte-driven pro-inflammatory processes are mediated by NF-κB. Our proteomic data supporting NF-κB activation in ocular hypertensive astrocytes included NF-κB subunits, p65 (RelA) and p100 (NF-κB2). In addition, immunolabeling with a phosphorylation site-specific antibody detected active p65. In contrast to the canonical pathway through RelA/p50 signaling, the non-canonical pathway of NF-κB signaling upon binding of a small subset of TNFR family members targets predominantly the activation of RelB/p52 complex.38,39 However, the classical pathway also feeds into the alternative pathway through up-regulation of NF-κB2 expression,40 and the alternative pathway regulates nuclear localization of RelA besides RelB.41
Phosphorylation triggers ubiquitination of inhibitor IκB molecules for proteasomal degradation. The ubiquitination machinery involved in NF-κB activation is regulated by a highly conserved protein complex designated as COP9 signalosome (CSN).42 The CSN complex interferes with ubiquitination and proteasomal degradation of the inhibitory molecule, IκB, and allows rapid activation of NF-κB.43 Our data included increased expression of different CSN subunits in ocular hypertensive astrocytes. In a fashion similar to this cytoplasmic control mechanism, NF-κB activation may also be terminated through intranuclear sequestration and degradation.44 Members of the PDZ and LIM domain proteins, also detected in astrocyte samples, have been shown to be expressed in brain astrocytes45 and implicated in nuclear regulation of NF-κB activation.46 Another protein up-regulated in ocular hypertensive astrocytes was a subunit of activating signal cointegrator 1 complex (ASCC). This protein, originally isolated as a transcriptional co-activator of nuclear receptors, is also known to stimulate the activation of NF-κB.47
Thus, various molecules appear to be involved in NF-κB activation in ocular hypertensive astrocytes. This redox-sensitive transcription factor is a master regulator of inflammatory responses and secondary injury processes in the brain,48,49 and inactivation of astroglial NF-κB reduces inflammatory environment after ischemic injury and promotes RGC survival.50 Findings of our recent study also have supported NF-κB activation in the glaucomatous human retina.5
In addition to NF-κB pathway controlling the transcription of immune mediators, the c-Jun N-terminal protein kinase/activator protein 1 (AP-1) signaling can stimulate inflammation through the activation of gene transcription.51 The AP-1, also up-regulated in ocular hypertensive astrocytes, is among the best characterized inducible DNA-binding proteins involved in a number of cellular processes linked to inflammatory responses of brain astrocytes.52
We also detected increased expression of various signal transducers and activators of transcription (STAT) in ocular hypertensive astrocytes. The janus kinase (JAK)/STAT signaling, along with the NF-κB signaling, has received a growing attention as key regulators of cytokine-mediated inflammation53,54 and promising targets for immunomodulation.55 Various components of the JAK/STAT signaling pathway recently have been indicated in the glaucomatous human retina,5 ocular hypertensive rat retina,56 and optic nerve.57
Our MS/MS data also included glia maturation factors that mediate inflammation in the central nervous system and exhibit up-regulation in neurodegenerative diseases,58 and the macrophage migration inhibitory factor (MIF), which is expressed by retinal glia59 and has an important amplifying role in cytokine-mediated inflammatory diseases.60
Cell-Specific Regulation of Autophagy
Molecular responses of ocular hypertensive astrocytes included up-regulation of IRGs that are key mediators of the host resistance to intracellular pathogens with important links to autophagy.61,62 We also detected activation of a serine/threonine protein kinase, mTOR, which is an upstream negative regulator of autophagy signaling,63,64 and ATGs, which are involved in the execution stages of autophagy.65
Autophagy is a physiological cell-autonomous defense mechanism enabling cells to digest their own cytosol, remove toxic products, and eliminate defective or surplus organelles. This cytoplasmic homeostasis pathway allows cells to survive nutrient depletion or the absence of growth factors; however, under specific conditions, it also may determine cell death.66,67 Although the autophagic pathway is activated in RGCs following optic nerve transaction68 or retinal ischemia,69 the role of autophagy in glaucomatous neurodegeneration is unknown. Many questions raised from our findings motivate further research to clarify the importance and regulation of autophagy and its crosstalk with RGC apoptosis in glaucoma. Autophagy can be induced by cytokines, including TNF-α,70,71 which also up-regulates IRGs in astrocytes,72 while NF-κB activation represses TNF-α-induced autophagy through the activation of autophagy inhibitor, mTOR.73 Since NF-κB was activated predominantly in glaucomatous astrocytes, it would be interesting to determine whether NF-κB-dependent activation of mTOR controls the balance between activator and inhibitor pathways of autophagy in astrocytes and other retinal cell types.
It also is interesting that autophagy has recently been recognized as a biological pathway broadly associated with innate and adaptive immunity.74–76 By eliminating intracellular microbial pathogens, and regulating T and B cell homeostasis, a properly functioning autophagy helps prevent inflammation and autoimmunity. However, when functioning aberrantly, it may contribute to immunogenic neurodegenerative human diseases. Not only does it act as an innate effector downstream of toll-like receptors (TLRs),77 but autophagy can also deliver danger-associated molecular patterns to endosomal TLRs78 and process cytosolic autoantigens for presentation on MHC class II molecules.79–81 In conjunction with earlier studies of increased serum autoantibodies in glaucoma, our findings bring about additional questions stimulating further research. For example, it is tempting to determine whether the autophagy-mediated route for efficient presentation of autoantigens may have a role in autoantibody generation in glaucoma. Retinal antigens identified so far, which are not limited to RGC proteins, possibly may reflect enhanced presentation of autoantigens on various cell types undergoing autophagy. With respect to chaperone-mediated autophagy,82 heat shock protein antibodies commonly present in the glaucomatous sera may particularly be relevant to autophagy-mediated immunity.
Neuroinflammatory Responses of Astrocytes Linked to Inflammasome
Our data also supported inflammasome activation in ocular hypertensive astrocytes. Upon sensing intrinsic danger signals as well as microbial components, this cytosolic multiprotein complex promotes proteolytic activation of pro-interleukins and secretion of mature cytokines.83,84 This innate mechanism mediates neuroinflammation in the brain, and its therapeutic neutralization reduces the damaging effects of post-traumatic brain inflammation.85,86 Similar to our recent study of human glaucoma,5 the present study detected various inflammasome components in experimental rat glaucoma. Up-regulated astrocyte proteins in ocular hypertensive samples included a specific NLRP adaptor for inflammasome assembly, apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC).83,84 Also detected in ocular hypertensive astrocytes was up-regulation and proteolytic activation of the inflammatory caspase, caspase-1. In addition to potassium efflux, amyloid-β, and pannexins,87 oxidative stress, evident in human glaucoma,3,88 has been implicated in inflammasome formation.89–91 Despite some controversies,92 caspase-1 activation is sensitive to alterations in the cellular redox balance.93 Emerging evidence also provides links between inflammasome and autophagic pathways.94
In summary, our findings introduced a cell-specific proteomic approach and validated its sensitivity to identify astrocyte responses in experimental rat glaucoma. Various pathways, including pro-inflammatory TNF-α/TNFR signaling, NF-κB activation, and inflammasome (along with the TLR signaling4 and complement activation3), appear to be co-players of inflammatory responses mediated by ocular hypertensive astrocytes and may represent key treatment targets for immunomodulation. Further research using focus-in strategies and functional testing are expected to generate a greater understanding of target molecules for cell-specific treatments in glaucoma.
Supplementary Material
Footnotes
Supported in part by National Eye Institute, Bethesda, Maryland (R01 EY013813 and R01 EY017131); The Robert W. Rounsavall, Jr. Family Foundation, Inc.; and an unrestricted grant to University of Louisville Department of Ophthalmology & Visual Sciences from Research to Prevent Blindness Inc., New York, New York.
Disclosure: G. Tezel, None; X. Yang, None; C. Luo, None; J. Cai, None; D.W. Powell, None
References
- 1. Tezel G. The role of glia, mitochondria, and the immune system in glaucoma. Invest Ophthalmol Vis Sci. 2009;50:1001–1012 [DOI] [PubMed] [Google Scholar]
- 2. Tezel G. The immune response in glaucoma: a perspective on the roles of oxidative stress. Exp Eye Res. 2011;93:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tezel G, Yang X, Luo C, et al. Oxidative stress and the regulation of complement activation in human glaucoma. Invest Ophthalmol Vis Sci. 2010;51:5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo C, Yang X, Kain AD, Powell DW, Kuehn MH, Tezel G. Glaucomatous tissue stress and the regulation of immune response through glial toll-like receptor signaling. Invest Ophthalmol Vis Sci. 2010;51:5697–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang X, Luo C, Cai J, et al. Neurodegenerative and inflammatory pathway components linked to TNF-alpha/TNFR1 signaling in the glaucomatous human retina. Invest Ophthalmol Vis Sci. 2011;52:8442–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steele MR, Inman DM, Calkins DJ, Horner PJ, Vetter ML. Microarray analysis of retinal gene expression in the DBA/2J model of glaucoma. Invest Ophthalmol Vis Sci. 2006;47:977–985 [DOI] [PubMed] [Google Scholar]
- 7. Yang Z, Quigley HA, Pease ME, et al. Changes in gene expression in experimental glaucoma and optic nerve transection: the equilibrium between protective and detrimental mechanisms. Invest Ophthalmol Vis Sci. 2007;48:5539–5548 [DOI] [PubMed] [Google Scholar]
- 8. Johnson EC, Jia L, Cepurna WO, Doser TA, Morrison JC. Global changes in optic nerve head gene expression after exposure to elevated intraocular pressure in a rat glaucoma model. Invest Ophthalmol Vis Sci. 2007;48:3161–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kompass KS, Agapova OA, Li W, Kaufman PL, Rasmussen CA, Hernandez MR. Bioinformatic and statistical analysis of the optic nerve head in a primate model of ocular hypertension. BMC Neurosci. 2008;9:93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang B, Harper MM, Kecova H, et al. Neuroinflammation in advanced canine glaucoma. Mol Vis. 2010;16:2092–2108 [PMC free article] [PubMed] [Google Scholar]
- 11. Panagis L, Zhao X, Ge Y, Ren L, Mittag TW, Danias J. Gene expression changes in areas of focal loss of retinal ganglion cells in the retina of DBA/2J mice. Invest Ophthalmol Vis Sci. 2010;51:2024–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tezel G, Yang X, Luo C, Peng Y, Sun SL, Sun D. Mechanisms of immune system activation in glaucoma: oxidative stress-stimulated antigen presentation by the retina and optic nerve head glia. Invest Ophthalmol Vis Sci. 2007;48:705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan X, Tezel G, Wax MB, Edward DP. Matrix metalloproteinases and tumor necrosis factor alpha in glaucomatous optic nerve head. Arch Ophthalmol. 2000;118:666–673 [DOI] [PubMed] [Google Scholar]
- 14. Tezel G, Li LY, Patil RV, Wax MB. Tumor necrosis factor-alpha and its receptor-1 in the retina of normal and glaucomatous eyes. Invest Ophthalmol Vis Sci. 2001;42:1787–1794 [PubMed] [Google Scholar]
- 15. Tezel G, Wax MB. Increased production of tumor necrosis factor-alpha by glial cells exposed to simulated ischemia or elevated hydrostatic pressure induces apoptosis in cocultured retinal ganglion cells. J Neurosci. 2000;20:8693–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tezel G, Yang X. Caspase-independent component of retinal ganglion cell death, in vitro. Invest Ophthalmol Vis Sci. 2004;45:4049–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tezel G. TNF-alpha signaling in glaucomatous neurodegeneration. Prog Brain Res. 2008;173:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tezel G. Proteomics in defining pathogenic processes involved in glaucomatous neurodegeneration. : Tombran-Tink J, Barnstable J, Shields MB, Ophthalmology Research Series, Glaucoma: Mechanisms of the Glaucomas: Disease Processes and Therapeutic Modalities. New York, NY: Humana Press Inc.; 2008:425–441 [Google Scholar]
- 19. Yang X, Luo C, Cai J, Pierce WM, Tezel G. Phosphorylation-dependent interaction with 14-3-3 in the regulation of bad trafficking in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2008;49:2483–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tezel G, Yang X, Luo C, et al. Hemoglobin expression and regulation in glaucoma: insights into retinal ganglion cell oxygenation. Invest Ophthalmol Vis Sci. 2010;51:907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tezel G, Yang X, Cai J. Proteomic identification of oxidatively modified retinal proteins in a chronic pressure-induced rat model of glaucoma. Invest Ophthalmol Vis Sci. 2005;46:3177–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morrison JC, Moore CG, Deppmeier LM, Gold BG, Meshul CK, Johnson EC. A rat model of chronic pressure-induced optic nerve damage. Exp Eye Res. 1997;64:85–96 [DOI] [PubMed] [Google Scholar]
- 23. Cummins TD, Barati MT, Coventry SC, Salyer SA, Klein JB, Powell DW. Quantitative mass spectrometry of diabetic kidney tubules identifies GRAP as a novel regulator of TGF-beta signaling. Biochim Biophys Acta. 2010;1804:653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 25. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 26. Powell DW, Weaver CM, Jennings JL, et al. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson PT, Geller SF, Lewis GP, Reese BE. Cellular retinaldehyde binding protein in developing retinal astrocytes. Exp Eye Res. 1997;64:759–766 [DOI] [PubMed] [Google Scholar]
- 28. Wright AP, Fitzgerald JJ, Colello RJ. Rapid purification of glial cells using immunomagnetic separation. J Neurosci Methods. 1997;74:37–44 [DOI] [PubMed] [Google Scholar]
- 29. Song JH, Bellail A, Tse MC, Yong VW, Hao C. Human astrocytes are resistant to Fas ligand and tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. J Neurosci. 2006;26:3299–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen NJ, Chio II, Lin WJ, et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc Natl Acad Sci U S A. 2008;105:12429–12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Araujo H, Danziger N, Cordier J, Glowinski J, Chneiweiss H. Characterization of PEA-15, a major substrate for protein kinase C in astrocytes. J Biol Chem. 1993;268:5911–5920 [PubMed] [Google Scholar]
- 32. Kitsberg D, Formstecher E, Fauquet M, et al. Knock-out of the neural death effector domain protein PEA-15 demonstrates that its expression protects astrocytes from TNFalpha-induced apoptosis. J Neurosci. 1999;19:8244–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zvalova D, Formstecher E, Fauquet M, Canton B, Chneiweiss H. Keeping TNF-induced apoptosis under control in astrocytes: PEA-15 as a ‘double key' on caspase-dependent and MAP-kinase-dependent pathways. Prog Brain Res. 2001;132:455–467 [DOI] [PubMed] [Google Scholar]
- 34. Condorelli G, Trencia A, Vigliotta G, et al. Multiple members of the mitogen-activated protein kinase family are necessary for PED/PEA-15 anti-apoptotic function. J Biol Chem. 2002;277:11013–11018 [DOI] [PubMed] [Google Scholar]
- 35. Sharif A, Canton B, Junier MP, Chneiweiss H. PEA-15 modulates TNFalpha intracellular signaling in astrocytes. Ann N Y Acad Sci. 2003;1010:43–50 [DOI] [PubMed] [Google Scholar]
- 36. Wolf FW, Sarma V, Seldin M, et al. B94, a primary response gene inducible by tumor necrosis factor-alpha, is expressed in developing hematopoietic tissues and the sperm acrosome. J Biol Chem. 1994;269:3633–3640 [PubMed] [Google Scholar]
- 37. Mookherjee N, Brown KL, Bowdish DM, et al. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J Immunol. 2006;176:2455–2464 [DOI] [PubMed] [Google Scholar]
- 38. Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105 [DOI] [PubMed] [Google Scholar]
- 39. Sarnico I, Lanzillotta A, Benarese M, et al. NF-kappaB dimers in the regulation of neuronal survival. Int Rev Neurobiol. 2009;85:351–362 [DOI] [PubMed] [Google Scholar]
- 40. Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733 [DOI] [PubMed] [Google Scholar]
- 41. Basak S, Kim H, Kearns JD, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orel L, Neumeier H, Hochrainer K, Binder BR, Schmid JA. Crosstalk between the NF-kappaB activating IKK-complex and the CSN signalosome. J Cell Mol Med. 2010;14:1555–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schweitzer K, Naumann M. Control of NF-kappaB activation by the COP9 signalosome. Biochem Soc Trans. 2010;38:156–161 [DOI] [PubMed] [Google Scholar]
- 44. Mankan AK, Lawless MW, Gray SG, Kelleher D, McManus R. NF-kappaB regulation: the nuclear response. J Cell Mol Med. 2009;13:631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iida Y, Matsuzaki T, Morishima T, et al. Localization of reversion-induced LIM protein (RIL) in the rat central nervous system. Acta Histochem Cytochem. 2009;42:9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591 [DOI] [PubMed] [Google Scholar]
- 47. Jung DJ, Sung HS, Goo YW, et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol Cell Biol. 2002;22:5203–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362 [DOI] [PubMed] [Google Scholar]
- 49. Harari OA, Liao JK. NF-kappaB and innate immunity in ischemic stroke. Ann N Y Acad Sci. 2010;1207:32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dvoriantchikova G, Barakat D, Brambilla R, et al. Inactivation of astroglial NF-kappa B promotes survival of retinal neurons following ischemic injury. Eur J Neurosci. 2009;30:175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu ZG. Molecular mechanism of TNF signaling and beyond. Cell Res. 2005;15:24–27 [DOI] [PubMed] [Google Scholar]
- 52. Hua LL, Zhao ML, Cosenza M, et al. Role of mitogen-activated protein kinases in inducible nitric oxide synthase and TNFalpha expression in human fetal astrocytes. J Neuroimmunol. 2002;126:180–189 [DOI] [PubMed] [Google Scholar]
- 53. Guo D, Dunbar JD, Yang CH, Pfeffer LM, Donner DB. Induction of Jak/STAT signaling by activation of the type 1 TNF receptor. J Immunol. 1998;160:2742–2750 [PubMed] [Google Scholar]
- 54. Wang Y, Wu TR, Cai S, Welte T, Chin YE. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol Cell Biol. 2000;20:4505–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaminska B, Swiatek-Machado K. Targeting signaling pathways with small molecules to treat autoimmune disorders. Expert Rev Clin Immunol. 2008;4:93–112 [DOI] [PubMed] [Google Scholar]
- 56. Wang DY, Ray A, Rodgers K, et al. Global gene expression changes in rat retinal ganglion cells in experimental glaucoma. Invest Ophthalmol Vis Sci. 2010;51:4084–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Johnson EC, Doser TA, Cepurna WO, et al. Cell proliferation and interleukin-6-type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest Ophthalmol Vis Sci. 2011;52:504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zaheer S, Thangavel R, Sahu SK, Zaheer A. Augmented expression of glia maturation factor in Alzheimer's disease. Neuroscience. 2011;194:227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matsuda A, Tagawa Y, Yoshida K, Matsuda H, Nishihira J. Expression of macrophage migration inhibitory factor in rat retina and its immunohistochemical localization. J Neuroimmunol. 1997;77:85–90 [DOI] [PubMed] [Google Scholar]
- 60. Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441 [DOI] [PubMed] [Google Scholar]
- 62. Bekpen C, Xavier RJ, Eichler EE. Human IRGM gene “to be or not to be.”. Semin Immunopathol. 2010;32:437–444 [DOI] [PubMed] [Google Scholar]
- 63. Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Levine B, Yuan J. Autophagy in cell death: an innocent convict?. J Clin Invest. 2005;115:2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752 [DOI] [PubMed] [Google Scholar]
- 67. Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol. 2011;21:387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim SH, Munemasa Y, Kwong JM, et al. Activation of autophagy in retinal ganglion cells. J Neurosci Res. 2008;86:2943–2951 [DOI] [PubMed] [Google Scholar]
- 69. Piras A, Gianetto D, Conte D, Bosone A, Vercelli A. Activation of autophagy in a rat model of retinal ischemia following high intraocular pressure. PLoS One. 2011;6:e22514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jia G, Cheng G, Gangahar DM, Agrawal DK. Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol Cell Biol. 2006;84:448–454 [DOI] [PubMed] [Google Scholar]
- 71. Keller CW, Fokken C, Turville SG, et al. TNF-alpha induces macroautophagy and regulates MHC class II expression in human skeletal muscle cells. J Biol Chem. 2011;286:3970–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yamada K, Akimoto H, Ogawa Y, Kinumi T, Kamagata Y, Ohmiya Y. Upregulation of immunity-related GTPase (IRG) proteins by TNF-alpha in murine astrocytes. Biochem Biophys Res Commun. 2009;382:434–439 [DOI] [PubMed] [Google Scholar]
- 73. Djavaheri-Mergny M, Amelotti M, Mathieu J, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281:30373–30382 [DOI] [PubMed] [Google Scholar]
- 74. Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schmid D, Münz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Deretic V. Multiple regulatory and effector roles of autophagy in immunity. Curr Opin Immunol. 2009;21:53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257 [DOI] [PubMed] [Google Scholar]
- 78. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401 [DOI] [PubMed] [Google Scholar]
- 79. Dengjel J, Schoor O, Fischer R, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005;102:7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Crotzer VL, Blum JS. Autophagy and its role in MHC-mediated antigen presentation. J Immunol. 2009;182:3335–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235 [DOI] [PubMed] [Google Scholar]
- 83. Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426 [DOI] [PubMed] [Google Scholar]
- 84. Ogura Y, Sutterwala FS, Flavell RA. The inflammasome: first line of the immune response to cell stress. Cell. 2006;126:659–662 [DOI] [PubMed] [Google Scholar]
- 85. de Rivero Vaccari JP, Lotocki G, Alonso OF, Bramlett HM, Dietrich WD, Keane RW. Therapeutic neutralization of the NLRP1 inflammasome reduces the innate immune response and improves histopathology after traumatic brain injury. J Cereb Blood Flow Metab. 2009;29:1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jha S, Srivastava SY, Brickey WJ, et al. The inflammasome sensor, NLRP3, regulates CNS inflammation and demyelination via caspase-1 and interleukin-18. J Neurosci. 2010;30:15811–15820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Silverman WR, de Rivero Vaccari JP, Locovei S, et al. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. J Biol Chem. 2009;284:18143–18151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tezel G, Luo C, Yang X. Accelerated aging in glaucoma: immunohistochemical assessment of advanced glycation end products in the human retina and optic nerve head. Invest Ophthalmol Vis Sci. 2007;48:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Martinon F. Signaling by ROS drives inflammasome activation. Eur J Immunol. 2010;40:616–619 [DOI] [PubMed] [Google Scholar]
- 90. Tassi S, Carta S, Delfino L, et al. Altered redox state of monocytes from cryopyrin-associated periodic syndromes causes accelerated IL-1beta secretion. Proc Natl Acad Sci U S A. 2010;107:9789–9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140 [DOI] [PubMed] [Google Scholar]
- 92. Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat Immunol. 2008;9:866–872 [DOI] [PubMed] [Google Scholar]
- 93. Chakraborty S, Kaushik DK, Gupta M, Basu A. Inflammasome signaling at the heart of central nervous system pathology. J Neurosci Res. 2010;88:1615–1631 [DOI] [PubMed] [Google Scholar]
- 94. Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.