Abstract
Next Generation Sequencing is highly resource intensive. NGS Tasks related to data processing, management and analysis require high-end computing servers or even clusters. Additionally, processing NGS experiments requires suitable storage space and significant manual interaction. At The Ohio State University's Biomedical Informatics Shared Resource, we designed and implemented a scalable architecture to address the challenges associated with the resource intensive nature of NGS secondary analysis built around Illumina Genome Analyzer II sequencers and Illumina’s Gerald data processing pipeline. The software infrastructure includes a distributed computing platform consisting of a LIMS called QUEST (http://bisr.osumc.edu), an Automation Server, a computer cluster for processing NGS pipelines, and a network attached storage device expandable up to 40TB. The system has been architected to scale to multiple sequencers without requiring additional computing or labor resources. This platform provides demonstrates how to manage and automate NGS experiments in an institutional or core facility setting.
Introduction
During the past three years, the Next Generation Sequencing (NGS) technology has been widely adopted in biomedical research and is revolutionizing many research areas since it enables the researchers to directly examine the genome at single base resolution (1–4). However, the processing and analysis of NGS data presents many new computational challenges (5). Specifically, from the computational point of view, NGS is highly resource intensive in the following ways:
-
NGS data processing is computationally intensive
NGS requires dedicated high-end computer servers for long-running data processing pipelines to convert raw data formats (i.e. intensity files) into sequence data and map the massive amount of sequences to the reference genome as well as converting the results to useful output formats such as BAM and SAM for further processing or WIG and BED files for visualization in tools such as the UCSC Genome Browser. The typical minimum hardware specification for NGS data processing is at least 8 cores in CPU and 48 GB of memory on a 64-bit Linux server. These minimum requirements far surpass the computing capabilities of a typically workstation. Additionally, storage requirements for NGS data are quite significant based on archiving policy (e.g., which files to keep, for how long, etc). For instance, for a Illumina Genome Analyzer II (GAII), the average results from a single flow cell (ie, a single run) can range anywhere from 200GB to 500 GB.
-
NGS data processing is labor intensive
The human effort involved in maintaining an NGS operation is considerable after taking account of the staff required to manually run data processing pipelines, manage NGS result files, administer processing and ftp servers (for data dissemination), set up sequencing runs, adjust configuration files and processing parameters. In addition, numerous miscellaneous programming and scripting are needed to maintain and automate mundane tasks encountered in NGS processing.
Our goal is to mitigate, if not completely eliminate, the above hurdles, as well as accommodate the anticipated advances in NGS high throughput data generation. To achieve this goal we designed and implemented an automation pipeline based on our previous work on NGS data processing pipeline (6) and data management system (7).
Methods
With the fast progress in the NGS technology, the problem of managing and processing NGS data becomes a major hurdle for many sequencing cores and labs and will be an even more widespread issue once the third generation sequencers are widely available to a large number of small labs. Currently several commercial systems are available for dealing with this issue but the prices for such LIMS systems are usually too high to be adopted by a small lab or sequencing core. Our system thus provides an exampler solution for this problem with the described features and architecture. In addition, our solution avoided the use of individual computing servers. Instead we take advantage of the computer cluster in the publicly accessible Supercomputer Center. This approach is highly scalable once we want to obtain additional sequencers.
It is our goal to eventually release our system with a set of open source codes and modules. The users can then pick the relevant modules to fit their own pipeline and processing/dissemination needs. In addition, we are also working on developing a publicly accessible web portal hosted on the their own data for processing and analysis.
System architecture
Our system incorporated the following features:
A configuration manager for setting up NGS experiments that tracks metadata related to accounts/labs, users, samples, projects, experiments, genomes, flow cells, and lanes. The details of an NGS experiment are then captured as a structured configuration file which is parsed and executed by our Automation Server.
An automation server which executes the instructions in a configuration file including bundling NGS raw data sets, transferring data to and from a compute cluster (supercomputer) setting up a series of jobs or a pipeline, reporting progress and status updates to the QUEST LIMS, and copying results to a backup location and ftp server if applicable.
A computer cluster with the required software programs, genomes, and pipelines installed to process NGS raw data into a suitable result format including mapping the short reads to the reference genomes.
Integration with QUEST, a Laboratory Information Management System (LIMS) system for cataloguing NGS runs and the resulting output files for easy lookup and retrieval.
Notification and logging system for reporting pipeline and data transfer progress, any data processing errors, and automatic emails sent to end-users when pipeline execution and data transfer is completed.
FTP server for external users to download their NGS results.
Pluggable architecture to accommodate additional NGS sequencers from different manufacturers, computing clusters, and/or data portals.
Loosely coupled automation server and compute cluster to simplify pipeline modifications.
System workflow
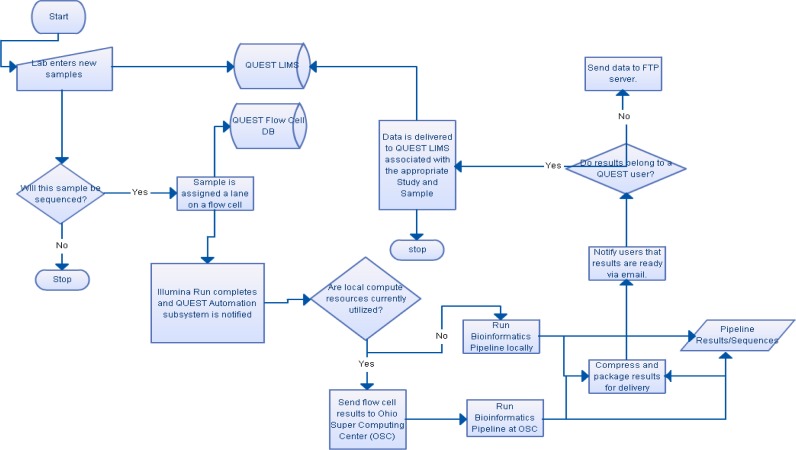
The workflow for the NGS run is shown in Figure 2 and we describe the workflow below in details. The workflow begins when a user enters a sample (on a study) in QUEST. Once a sample is entered, it will be assigned by the sequencing core facility to a lane corresponding to the physical process of preparing a sample on a lane of an Illumina flow cell. This association is made once the sequencing core manager configures the flow cell via the Configuration Manager in QUEST. Once the sequencing core facility has completed the GAII run, it indicates that the run is completed with notifies the Biomedical Informatics Shared Resource (BISR) that the run’s raw data files are ready for processing. The BISR analyst will then look over the auto-generated configuration files and then launch the automated pipeline. This is done by selecting the “Execute this configuration” button as shown in Figure 3.
Figure 2:
The NGS data processing and automation pipeline.
Figure 3.
Execution of the Configuration file.
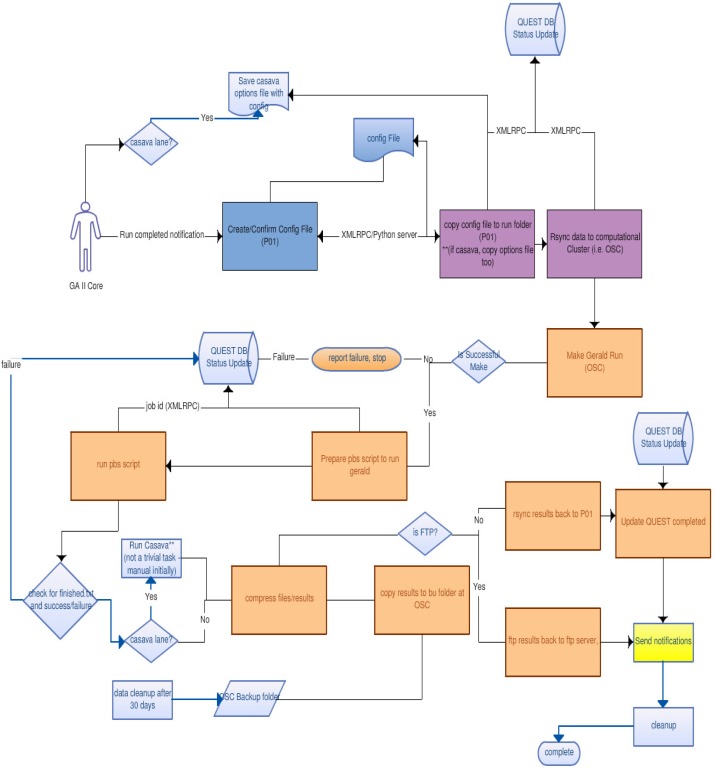
Once the NGS automation pipeline has been launched with the given configuration, QUEST makes an XML-RPC call to a Python server listening on a registered port. QUEST sends the Python server information about the run such as run id, run folder name, run location, and the configuration files generated with the Configuration Manager. The configuration server resides on a Linux machine that has access to the storage device the GAII instrument writes the raw data files,
The python server processes the run in the following basic steps:
Transfer data to the compute cluster at the Ohio Supercomputer Center (OSC).
Generate PBS scripts to make and run the Gerald pipeline. PBS scripts are used for the scheduling task on OSC computer cluster. The scripts will automatically specify the number of nodes needed and the expected CPU time of execution.
Update QUEST with status updates about the run. (report errors if encountered)
When a sequencing run is completed, transfer data to a sftp server and QUEST (Users can either ftp results or use QUEST to download http style).
Updates QUEST that run is completed which automatically sends notification to the end users via e-mail including the information such as sftp server directory and temporary password.
Key technical challenges and solutions
The system we presented here involves many different components including interfacing with instruments, heterogeneous computing facilities (i.e., computer on instrument, local data processing server, remote computer cluster), database, graphical user interface and both proprietary and public softwares. These heterogeneous hardware and software components cause many challenges for the system development. Below is a list of key technical challenges encountered during the implementation and our solutions.
Since NGS result files are very large, sessions often timeout during downloads. To keep sessions alive, file downloads are streamed to the client. Currently our rates range from 1 Mbs to 5 Mbs on a typical day. This part of our architecture is currently being upgraded due to limitations in the current hardware and network. We are looking at ways to speed up transfer rates to around 15–20 Mbs.
In order to transfer files to and from the compute cluster, the python scripts make rsync calls. This is done in the server on a separate thread (i.e. an asynchronous call since rsync can take a long time.) and uses both ‘–arv’ and ‘–e ssh’ options. These options can slow down an rsync, but since this step is not a bottleneck for us and we generated an ssh key, we are content for now using this approach.
A specific Gerald pipeline run must first be generated using make. Therefore we enforce that our python server generates a PBS file dynamically that takes the name of the newly created Gerald folder before Gerald can be ran. It does that by parsing the Gerald make command output and passing the folder name as a variable to the new PBS script. This is all done using Python on the compute cluster.
The Automation pipeline is distributed at four different points: The GAII raw data folder and the Python server, the QUEST LIMS, the compute cluster, and finally the data portal. To integrate this distributed architecture, messages are sent and received using XML-RPC with the Cook Computing libraries [8].
QUEST – the LIMS system interfacing with the sequencer
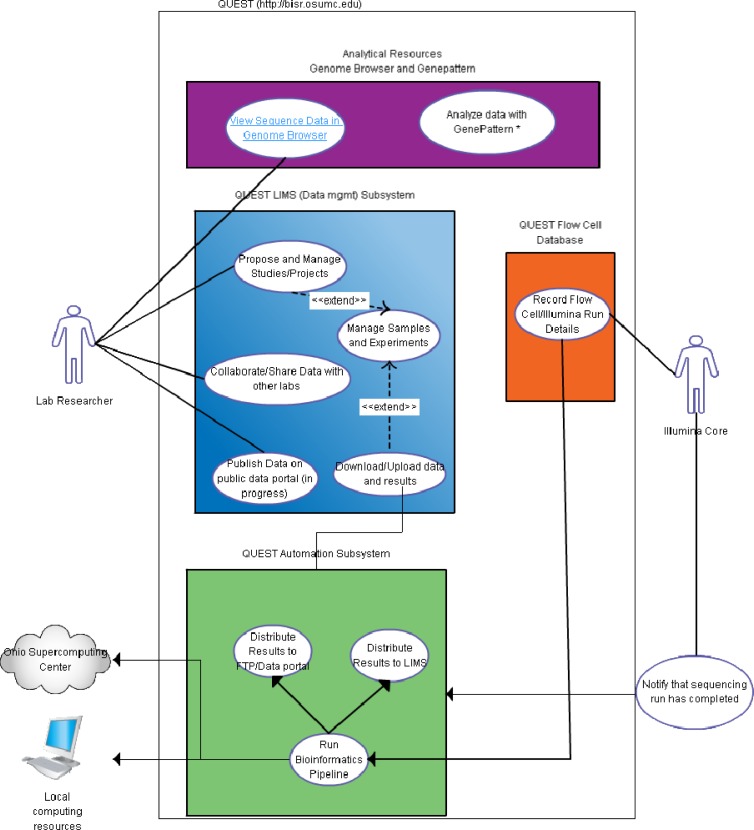
The features listed above were added to an existing LIMS system, called QUEST (https://bisr.osumc.edu), which enables users and labs, to track results in studies, samples, and experiments. Figure 1 shows the two main actor groups: A researcher or lab and the NGS sequencing core. Also shown in Figure 1 are the various use-cases and an overview of the QUEST system.
Figure 1.
An overview of the QUEST system and its use Cases.
For the lab researchers, this system enables pre-experiment planning by submitting information sample, lab and experimental protocol information. Such information can be deposited to the database even before the samples are generated. This greatly facilitated the planning for the sequencing core manager. Once the experiments are finished and data are available, the researchers can download the data, share the data (e.g., to bioinformatics collaborators for advanced analysis), visualize the data (e.g., on UCSC Genome Browser or Integrate Genome Viewer), and publish the data to public once the paper is accepted.
For the sequencing core manager, the system allows the tracking of the sample and managing of the metadata over the entire experiment and sequencing process.
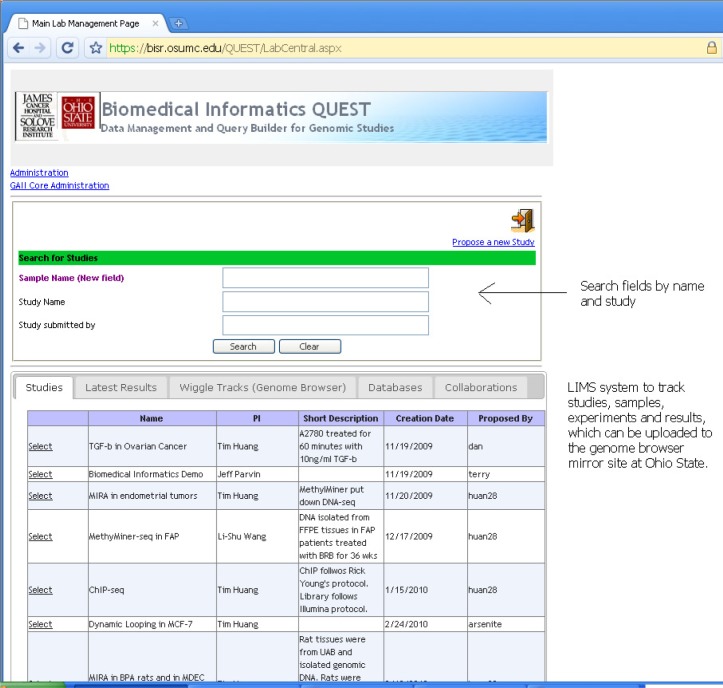
Figures 4 and 5 are the primary starting points for both the LIMS portion and the NGS Configuration section. Figure 4 shows the LabCentral screen where researchers can search through existing studies and samples to access specific results. Here each study is defined as a project with a focused research topic. Thus multiple datasets can be included in one study and one dataset can be involved in many studies. In addition, each lab can have as many different studies as they want. This flexibility greatly facilitates collaborations and also maximizes the usage of the data. For instance, a ChIP-seq dataset for RNA Polymerase II (PolII) on MCF7 breast cancer cell lines can be used in a study for cancer epigenetics by combining with other ChIP-seq data for histone markers. It can also be included in a study for investigating PolII dynamics.
Figure 4.
Main page for listing the studies.
Figure 5.
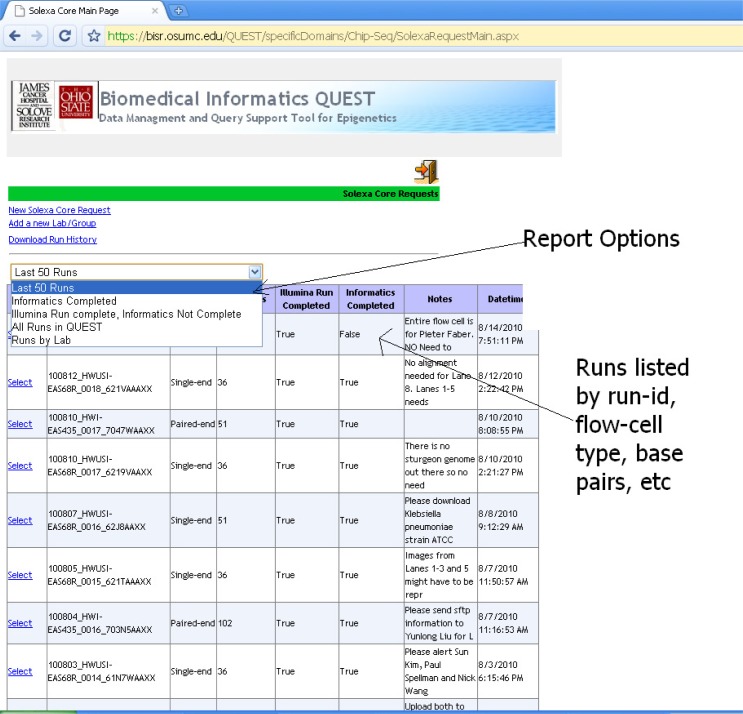
NGS/GAII run history and comments/analyzing instructions.
From the LabCentral page, users can also upload results to the UCSC Genome Browser (via an OSU mirror installation) and share results with other labs through the Collaboration panel. The database panel shown in Figure 2 also enables researchers to integrate high throughput data besides NGS data such as microarray results.
Figure 5 shows the new feature of the Configuration Management screen for configuring Illumina GAII runs that associates flow cell and lane information with sample meta-data. The sequencing core manager can list sequencing runs filtering and displaying by a variety of options and can link to a particular run. All sequencing run configurations, results, processing status, and associated meta-data are archived for easy retrieval. The processing status is also visible to the lab researchers so that they can track the progress of their sequencing experiments.
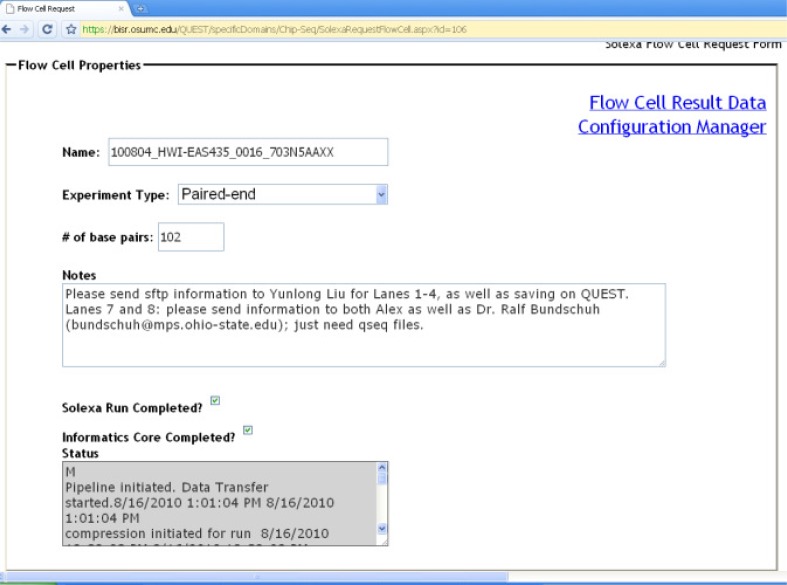
Before the sequencing experiments, sequencing runs needs to be configured by the sequencing core manager or operator. The configurations are comprised of two primary panels. One is for flow-cell properties (Figure 6) and the other is for lane properties (Figure 7). Each panel is configurable and its settings persisted between sessions. In GAII, there are eight lanes per flow cell and a sample can be sequenced in either one lane or multiple lanes. This can be easily expanded to accommodate more lanes used in the updated Illumina HiSeq2000. The logging information such as the time for the sequencing run and processing is shown in the bottom window in Figure 6.
Figure 6.
Flow cell properties header.
Figure 7.
An example of lane properties panel for a flowcell (showing first 4 lanes).
The Lane Properties panel stores information about the study, sample, genome, assay, organism, and group information (figure 7) for each of eight lanes on a flow cell. By choosing the organism associate with each sample, the operator also specifies the reference genome to which the short reads will be mapped.
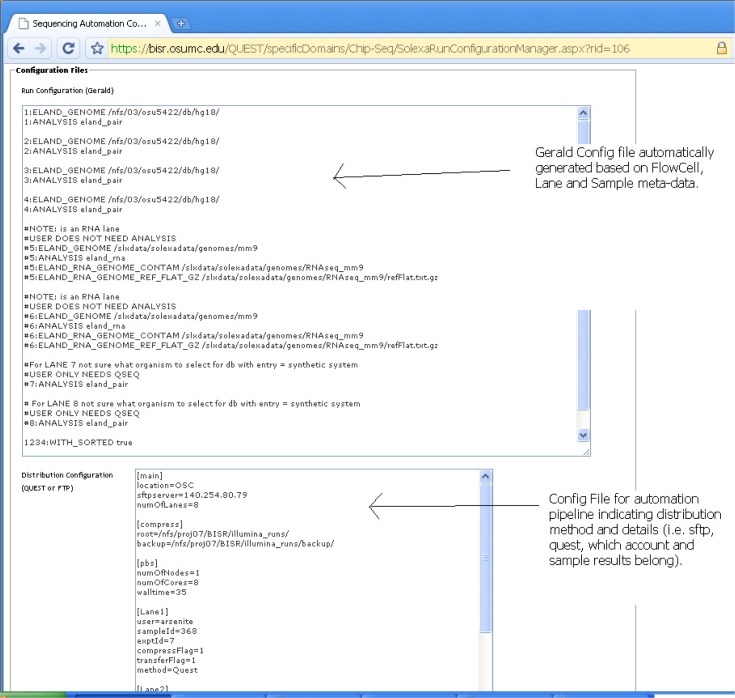
From the information on the two aforementioned panels, QUEST generates two configuration files displayed on a Configuration Manager (Figure 8). One configuration is for running Illumina’s Gerald pipeline and the other is for creating a custom configuration file for packaging and distributing results to the client.
Figure 8.
Configuration Manager.
The Gerald pipeline is Illumina’s proprietary data process pipeline which handles tasks including mapping short reads to the reference genomes using the ELAND algorithm. In this configuration, the users can specify many parameters including the mapping parameters for ELAND (e.g., allowed number of mismatches, expected sequence length, allowed repeated mapping instances), the file paths (e.g., the folder for the mapping results) and the computing parameters (e.g., the number of CPUs to be used) for parallel computing. The second configuration file specifies the automation parameters and gets information from the previous lane and flow cell configuration panels. The automation parameters allow the users to command the system on the type of processing needed and the depositary location for the results files as well as miscellaneous requests such as if the results need to be copied to an FTP server for dissemination.
The Both configuration files are saved (and can be overwritten) and sent to the compute cluster, along with the raw data files, as one of the initial steps of the automation pipeline. These configuration files are also reusable. The operator can load saved files and edit them as necessary before a new sequencing experiment.
Results
The Configuration Manager has been deployed in a production capacity since December 2009 and has configured and catalogued results from over 75 Illumina GAII flow cells for two GAII sequencers to January 2011. The automation pipeline has been in production since May 2010. Over 700 samples have been processed and we have over 5,000 result files and 4 TB of processed data have been stored till early 2011.
Conclusion and discussion
In this paper we present a scalable automation pipeline for managing and processing the massive amount of sequencing data generated by the NGS sequencers. Our system is extremely configurable. The compute cluster, and data portal can be changed with a simple modification to a configuration file. To add a new pipeline, all that is needed is a new python module, or handler, to generate the required PBS file for the compute cluster. We anticipate adding new modules as additional NGS technologies are acquired, such as ABI SOLiD, or as we adopt additional algorithms for analysis. There is actually no change to the architecture when new sequencers are added that already have existing python handlers – such as GAII; only an entry in a configuration file denoting the name and location of the new sequencer is needed.
References
- 1.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008 Jul;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009 Jan;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park PJ. Epigenetics meets next-generation sequencing. Epigenetics. 2008 Nov;3(6):318–21. doi: 10.4161/epi.3.6.7249. [DOI] [PubMed] [Google Scholar]
- 4.Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nature biotechnology. 2009 May;27(5):455–7. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nature methods. 2009 Nov;6(11 Suppl):S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozer HG, Bozdag D, Camerlengo T, et al. IEEE Press; 2009. A Comprehensive Analysis Workflow for Genome-Wide Screening Data from ChIP-Sequencing Experiments; pp. 320–330. BICoB: [Google Scholar]
- 7.Camerlengo T, Ozer HG, Teng M, et al. Enabling Data Analysis on High-throughput Data in Large Data Depository Using Web-based Analysis Platform – A Case Study on Integrating QUEST with GenePattern in Epigenetics Research. IEEE International Conference on Bioinformatics in Biomedicine; Washington, D.C.. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]