Abstract
ALKBH2 is a direct DNA repair dioxygenase guarding mammalian genome against N1-methyladenine, N3-methylcytosine, and 1,N6-ethenoadenine damage. A prerequisite for repair is to identify these lesions in the genome. Here we present crystal structures of ALKBH2 bound to different duplex DNAs. Together with computational and biochemical analyses, our results suggest that DNA interrogation by ALKBH2 displays two novel features: i) ALKBH2 probes base-pair stability and detects base pairs with reduced stability; ii) ALKBH2 does not have nor need a “damage-checking site”, which is critical for preventing spurious base-cleavage for several glycosylases. The demethylation mechanism of ALKBH2 insures that only cognate lesions are oxidized and reversed to normal bases, and that a flipped, non-substrate base remains intact in the active site. Overall, the combination of duplex interrogation and oxidation chemistry allows ALKBH2 to detect and process diverse lesions efficiently and correctly.
Genomic DNA can be damaged by alkylating agents from both endogenous and exogenous sources, leading to cytotoxicity and carcinogenic mutations1. Three mechanisms exist to battle cellular methylation/alkylation damage: DNA glycosylases, the suicidal O6-methylguanine-DNA methyltransferases, and direct oxidative repair mediated by the ALKBH2 type iron(II)/2-ketoglutarate(2KG)-dependent dioxygenases2–3. ALKBH2 is one of the first human homologues identified within the AlkB family of DNA/RNA repair dioxygenases to exhibit demethylation activity against the cytotoxic N1-methyladenine (1-meA) and N3-methylcytosine (3-meC) DNA adducts4–5; knockout experiments in mice have further established mALKBH2 as the house-keeping enzyme guarding the mammalian genome against such lesions6–7. Consistent with these observations, ALKBH2 has been shown to play an efficient role in pediatric brain tumors during chemotherapy treatment, and the combination of ALKBH2 knockdown and cisplatin chemotherapy could potentially improve the efficacy of treatment of H1299 lung cancer cells8–9. Mechanistically, ALKBH2 has been reported to interact with proliferating cell nuclear antigen through its N-terminal sequence10; and previous crystal structures of ALKBH2/dsDNA complexes have provided the molecular basis for 1-meA recognition by ALKBH211.
Regardless of the differences in repair mechanisms of the aforementioned DNA repair proteins, a major challenge that all repair proteins face is how to single out their cognate damage in the vast background of normal DNA bases12–13. Human N3-methyladenine DNA glycosylase AAG gains selective recognition of its substrates through combination of the shape, aromaticity and hydrogen-bonding characteristics of the damaged bases14, and kinetic experiments have also revealed a two-step lesion-binding process of AAG, including an initial recognition step and a stable extrahelical-complex stage15–16; E. coli glycosylase AlkA invasively interrogates the minor groove of DNA while probing for damage and induces a sharp DNA bend upon nucleotide-flipping17–18; bacterial 8-oxoguanine (oxoG) DNA glycosylase MutM can interrogate undamaged DNA in an intrahelical fashion and also detect the presence of oxoG even at a very early stage of encounter19–20; the human oxoG repair protein, hOGG1, discriminates guanine from oxoG (which is bound in the active site) in an alternative damage-checking site, thus representing a “gate-keeping” strategy to prevent incorrect cleavage of normal bases21; and uracil DNA glycosylase (UDG) utilizes the spontaneous thermal opening of U:A pairs as the initial step of its “passive” base-flipping process22–24. Despite different strategies employed by these base-excision glycosylases, an invariant goal is to efficiently discriminate cognate substrates from their undamaged counterparts and prevent entry of normal DNA bases to the active site so that they will not be mistakenly cleaved25–26. In contrast, little is known about how most direct repair proteins, for example ALKBH24–7, search for the structurally diverse base lesions in the genome and whether or not ALKBH2 also uses a gate-keeping strategy for efficient lesion discrimination.
RESULTS
Duplex interrogation by ALKBH2 to differentiate base pair stability
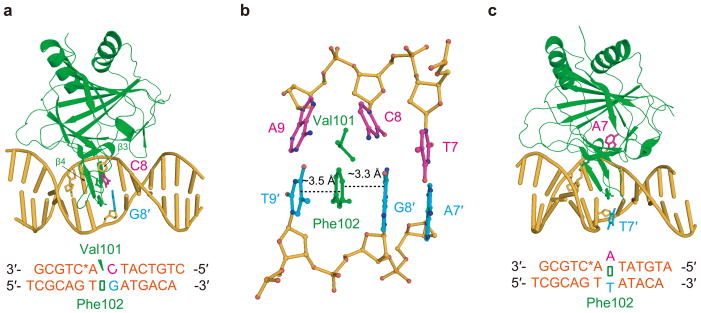
Using a previously developed disulfide cross-linking method11,27, we first solved complex structures of ALKBH2-dsDNA containing a central C:G or A:T pair (referred to hereafter as the CG and AT structures, respectively) (Fig. 1; Table 1). One prominent feature of the CG structure is that the central C:G pair is severely distorted by ALKBH2 while remaining fully intrahelical (Fig. 1a). The side chain of Val101 wedges into C8 and A9, disrupting the stacking of C8 to A9 and significantly buckling the C8 base; the finger residue Phe102 intercalates between G8′ and T9′, leading to a continuous four base-pair-wide stack in the complementary strand from A7′ to T9′ (Fig. 1b). As a result, while the flanking pairs of T7:A7′ and A9:T9′ remain hydrogen-bonded with the duplex structure outside of this area not much affected, the central target pair C8:G8′ is weakened and the stacking of C8 with neighboring bases is essentially abolished. Activity assays confirmed the important roles of the Val101 and Phe102 residues (which are in the β3-β4 hairpin): both ALKBH2 Val101Ala and Phe102Ala mutations cause reduced repair activity towards 1-meA and 3-meC; introducing a Val101Gly Phe102Ala double mutation to the protein almost eliminated its enzymatic activity (Supplementary Fig. 1a–c)28–29. Since the active site of ALKBH2 in the CG structure is not occupied by a DNA base, the substrate recognition lid comprised of the β6–β7 hairpin motif adopts an “open” conformation (Supplementary Fig. 2a).
Figure 1.
Base pairs with different stability are discernible by ALKBH2. (a) Cartoon of the CG structure. (b) Local view showing the interrogation of the target C8:G8′ pair by ALKBH2, with residues Val101 and Phe102 highlighted. (c) Overall view of the AT structure. ALKBH2 is shown in green, DNA in yellow-orange, DNA bases from the upper strand in light magenta and those from the bottom strand in cyan.
Table 1.
Data collection and refinement statistics
| CG structure | AT structure | CI structure | (Mn/2KG) εA structure | 3-meC structure | (Mn/2KG) 3-meC structure | (Mn/2KG) CG structure | (Mn/2KG) CG-DNA2 | |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | P1211 | P41212 | P212121 | P6522 | P6522 | P6522 | P1211 | P1211 |
| Cell dimensions | ||||||||
| a, b, c (Å) | 41.2, 61.0, 65.2 | 74.9, 74.9, 169.6 | 55.9, 65.0, 167.9 | 78.6, 78.6, 228.9 | 78.1, 78.1, 230.0 | 77.8, 77.8, 229.0 | 46.0, 60.5, 65.5 | 46.0, 60.6, 65.6 |
| α, β γ (°) | 90, 89.2, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 | 90, 90,120 | 90, 90, 120 | 90, 90.9, 90 | 90, 90.5, 90 |
| Resolution* (Å) | 50 – 1.58 (1.65 – 1.58) | 50 – 3.06 (3.17 – 3.06) | 50 – 2.60 (2.69 – 2.60) | 50 – 2.78 (2.88 – 2.78) | 50 – 2.25 (2.33 – 2.25) | 50 – 2.50 (2.59 – 2.50) | 50 – 1.49 (1.54 – 1.49) | 50 – 1.55 (1.61 – 1.55) |
| Rmerge | 0.03 (0.408) | 0.07 (0.722) | 0.07 (0.596) | 0.07 (0.829) | 0.07 (0.821) | 0.06 (0.872) | 0.06 (0.308) | 0.04 (0.666) |
| I/σI | 20.8 (2.2) | 17.1 (2.2) | 19.8 (2.1) | 30.7 (3.4) | 39.7 (3.9) | 35.1 (4.2) | 35.7 (6.0) | 30.2 (2.0) |
| Completeness (%) | 94.3 (93.4) | 99.6 (98.6) | 94.9 (96.5) | 99.9 (100) | 99.7 (98.4) | 99.3 (98.2) | 96.2 (89.8) | 99.7 (99.5) |
| Redundancy | 2.0 (2.0) | 5.5 (6.0) | 4.9 (5.2) | 13.2 (13.0) | 17.8 (16.9) | 15.5 (15.3) | 7.7 (7.7) | 7.3 (6.4) |
| Refinement | ||||||||
| Resolution (Å) | 20 – 1.62 (1.66 – 1.62) | 50 – 3.06 (3.14 – 3.06) | 20 – 2.60 (2.67 – 2.60) | 20 – 2.78 (2.85 – 2.78) | 20 – 2.25 (2.31 – 2.25) | 50 – 2.50 (2.57 – 2.50) | 20 – 1.60 (1.64 – 1.60) | 20 – 1.70 (1.74 – 1.70) |
| No. reflections | 43430 | 9236 | 17484 | 10604 | 19430 | 14048 | 43675 | 37531 |
| Rwork/Rfree | 20.1/22.3 | 25.3/28.9 | 22.1/28.9 | 22.8/25.9 | 22.4/25.6 | 22.7/25.9 | 17.5/20.2 | 19.4/23.4 |
| No. atoms | ||||||||
| Protein | 1695 | 1614 | 3210 | 1626 | 1636 | 1636 | 1727 | 1703 |
| DNA | 566 | 527 | 1052 | 527 | 526 | 526 | 566 | 566 |
| Ligands/water | 318 | 4 | 26 | 27 | 75 | 59 | 364 | 361 |
| B-factors | ||||||||
| Protein | 16.8 | 45.6 | 33.5 | 48.9 | 28.0 | 31.2 | 22.8 | 28.2 |
| DNA | 23.8 | 84.5 | 33.8 | 45.4 | 28.7 | 30.8 | 27.8 | 42.1 |
| Ligands/water | 28.0 | 63.7 | 38.9 | 52.8 | 27.7 | 34.4 | 33.4 | 41.2 |
| R.m.s deviations | ||||||||
| Bond lengths (Å) | 0.006 | 0.007 | 0.014 | 0.007 | 0.011 | 0.006 | 0.008 | 0.010 |
| Bond angles (°) | 1.191 | 1.255 | 1.718 | 1.359 | 1.541 | 1.112 | 1.464 | 1.479 |
Values in parentheses are for highest-resolution shell. Each structure was solved using one crystal.
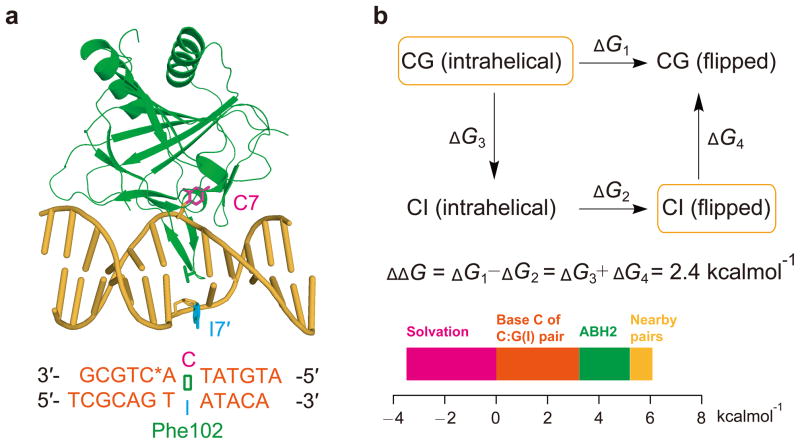
To our surprise, the AT structure revealed that the central A:T pair is broken and A7 is flipped into the active site of ALKBH2 (Fig. 1c). It is unlikely that ALKBH2 disrupts every normal base pair it encounters in order to target a lesion; however, this cross-linking-stabilized structure implies that base pairs with different stabilities are discernible by ALKBH2 when it binds duplex DNA. To make a fair comparison to the CG structure, we crystallized an ALKBH2-dsDNA complex containing a central C:I (inosine) pair (CI structure), which resembles C:G but possesses one fewer hydrogen bond. In the resulting structure, instead of remaining intrahelical as observed in the CG structure, C7 is flipped by ALKBH2 (Fig. 2a).
Figure 2.
Contributions of ALKBH2 to base-flipping in duplex DNA. (a) Cartoon view of the CI structure. The same color coding in Fig. 1 is used. (b) Computational analysis of free energy difference for ALKBH2 to break a C:G or C:I pair. States observed in crystal structures are in the orange boxes and contributions to the free energy difference ΔΔG are plotted.
Contribution of ALKBH2 β3–β4 hairpin in stability interrogation
To evaluate the relative free energies for the protein to flip a cytosine base out of C:G and C:I pairs, we performed “alchemical” molecular dynamics simulations as described in the Supplementary Information30. Two hypothetical states—one with an intra-helical C:I pair being interrogated and the other with a broken C:G pair—were constructed by computationally mutating the CG and CI crystal structures (Supplementary Fig. 3). The simulation indicates that it is ~ 2.4 kcalmol−1 less favorable to flip a cytosine from a C:G than a C:I pair (Fig. 2b). This calculated value can be further decomposed to understand its physical basis (Supplementary Table 1). The solvation energy of the 2-amino group of guanine (~ −3.6 kcalmol−1) and the penalty of decreased hydrogen bonding capacity (~ 3.3 kcalmol−1) almost balance. As a result, the net contribution comes mainly from the protein (~ 2.1 kcalmol−1). More specifically, both Gln100 and Phe102 prefer the intra-helical C:G pair to an intra-helical C:I state through either electrostatic or van der Waals interactions (Supplementary Fig. 3e,f). Thus, our computational analysis suggests that ALKBH2 applies a moderate mean force to DNA, which can be used to distinguish the stability of different base pairs.
We next performed fluorescence studies using 2-amino purine (2AP) to further investigate how ALKBH2 senses base pair stability in duplex DNA. When 2AP:T was incorporated into the flanking base pairs of the central 1-meA:T pair, the addition of ALKBH2 enhanced fluorescence of 2AP, suggesting significant conformational change of DNA upon formation of the specific ALKBH2/1-meA complex (Supplementary Fig. 1d). Furthermore, a dsDNA probe with a 2AP:T pair adjacent to an A:C pair also showed increased fluorescence upon protein binding. Thus, a non-cognate mismatched base pair could also be readily detected by ALKBH2 to trigger a substantial conformational change. With the Phe102Ala mutant protein, reduced fluorescence increases were observed to both 1-meA:T- and A:C-containing DNAs, and no fluorescence enhancement took place with the Val101Gly Phe102Ala double mutant ALKBH2, as expected (Supplementary Fig. 1e).
ALKBH2 detect weakened base pairs to locate its substrates
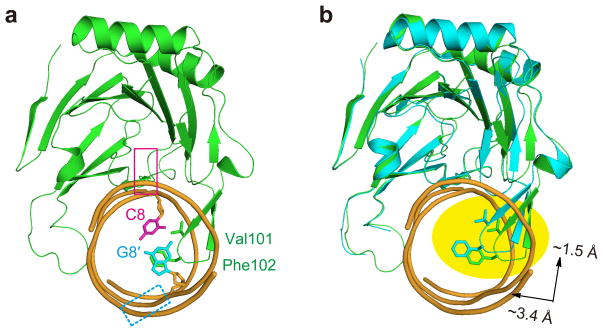
ALKBH2 is a promiscuous repair protein whose substrates include both purines (1-meA and 1,N6-ethenoadenine [or εA]) and pyrimidines (3-meC and 3-meT). While structurally diverse, a common feature shared by these N-alkylated lesions is the disrupted Watson-Crick interface, which leads to significantly weakened base pairs compared to their undamaged Watson-Crick counterparts31–32. A superposition of the CG structure with the 1-meA structure (PDB code 3BTY) (or any other ALKBH2/dsDNA structure with a flipped base) reveals how the interrogation mechanism of ALKBH2 could facilitate detection of these lesions hidden in different contexts. The β3–β4 hairpin motif inserts itself much deeper into the duplex in the 1-meA structure (Fig. 3). This motif also shifts up significantly towards the lesion-containing strand in the 1-meA structure as compared to the CG structure (Fig. 3b), which allows the finger residue Phe102 to fully occupy the vacant space created by base-flipping in the lesion-specific complex. On the other hand, prior to the location of a damaged base, the shallow insertion of the hairpin tip, observed in the CG interrogation complex, would exert force on the target base pair. In the case of a weakened base pair that often harbors DNA damage, interrogation by the hydrophobic hairpin tip enables ALKBH2 to readily detect the lesioned base and initiate base-flipping. The exact identity of the lesioned base does not affect the probing mechanism used by ALKBH2. In contrast, the class of glycosylases that have been suggested to use active interrogation (for instance AlkA and MutM) recognizes unique characteristics of their cognate lesions17,20; whether or not such indiscriminate stability-interrogation can also be utilized by other DNA glycosylases to directly probe an intra-helical base pair remains to be determined.
Figure 3.
ALKBH2 probes the stability of a base pair to detect DNA damages. (a) Side view of the CG structure. The approximate location of a flipped base is indicated with a magenta box and that of the orphaned base (which could have multiple conformations) is indicated with a dashed cyan box. (b) Overlay of the 1-meA structure (3BTY) and the CG structure. A clear shift of the hairpin loop is highlighted. The protein portion of 3BTY is shown in cyan and the DNA part is omitted for clarity purpose.
Unlike many glycosylases, ALKBH2 does not bend or induce significant global distortion to duplex DNA, in the process of both base interrogation and damaged base recognition in the active site. It utilizes several structural motifs, especially the finger bearing β3-β4 hairpin and the long loop (between β11 and β12), to clamp a dsDNA from two sides (Fig. 3a), which also represent the major elements involved in DNA duplex interrogation. A few other base-modification enzymes do not cause significant disruption to the duplex structure either33. In fact, these duplex-interacting elements are unique for ALKBH2 among the other human homologue proteins: swapping the hairpin sequence of ALKBH2 with the corresponding sequence from ABH3, a single-stranded nucleic acid demethylase, leads to switched preference of ALKBH2 towards ssDNA28–29.
Crystal structures of ALKBH2 bound to εA- and 3-meC-containing DNA
Once ALKBH2 locates a weakened base pair in the genome, it then needs to recognize the potential base lesion. To investigate how ALKBH2 accommodates various bases, we also crystallized ALKBH2-dsDNA complexes containing εA and 3-meC, respectively (εA and 3-meC structure) (Supplementary Fig. 2b). ALKBH2 was shown to use at least five active site residues for 1-meA recognition11, but base recognition for εA or 3-meC is less tight (Supplementary Fig. 2c). ALKBH2, like its E. coli homologue protein AlkB, can well accommodate the flipped lesion, despite differing in hydrogen-bonding capacities and base-dimensions, in its active site to allow efficient catalysis (Supplementary Fig. 2d,e)11,34–36.
εA was previously found to form base pairs with thymine and guanine in B-form DNA32,37, while 3-meC is not able to pair with an opposite guanine base31. Because the stability of εA:T was thus expected to be much closer to C:G, we focused our quantitative analysis on the εA:T case. To evaluate the energetics for ALKBH2 to flip an εA lesion out of a weak εA:T pair, we again used alchemical molecular dynamics simulations (Supplementary Fig. 4). These simulations were based on the εA and the CG structures as discussed in the Supplementary Information. According to the calculations, flipping an εA lesion from the εA:T pair is ~3.5 kcalmol−1 more favorable than flipping a cytosine from a C:G pair (Supplementary Table 1); therefore, the εA:T pair is calculated to be ~1.1 kcalmol−1 weaker than the C:I pair discussed above. Overall, the contributions of individual parts of the system are larger in magnitude than in the CI to CG case, and the difference in stability arises from a balance of many compensating interactions.
ALKBH2 does not have nor need a “damage-checking” site
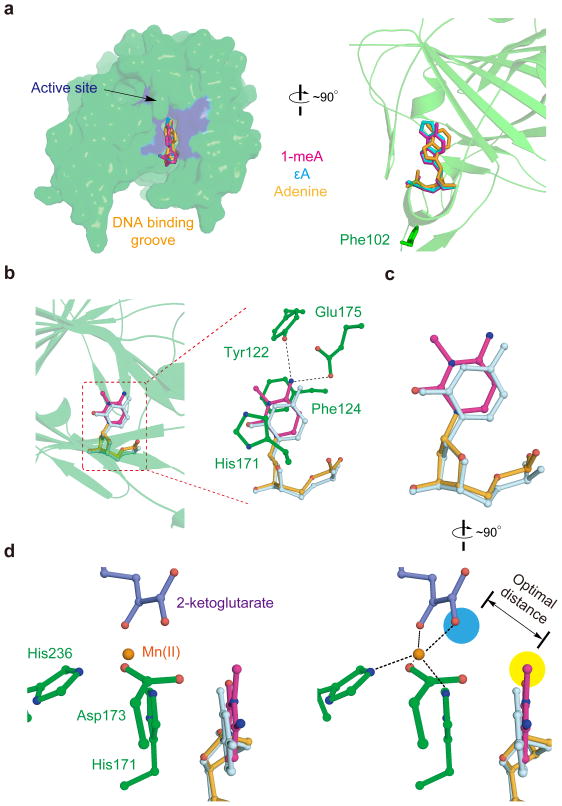
Having the complex structures with each of the five bases (1-meA, εA, adenine, 3-meC and cytosine) flipped by ALKBH2, the location of an undamaged base versus its derived damage can be compared. An overlay using 1-meA, εA, and AT structures reveals that all these bases reside at a similar position in the active site of ALKBH2 (Fig. 4a); moreover, 3-meC and cytosine are also found to be located at the identical spot (Figs. 4b,c). Such observations are in sharp contrast to the previous knowledge with regard to DNA glycosylases: to prevent accidental glycosidic bond cleavages that generate potential DNA breaks, DNA glycosylases employ different and strict strategies to insure that a normal base cannot be flipped into the catalytic site to initiate glycosylation25–26; ALKBH2 must accommodate and accurately position the aberrant alkyl group on different base lesions in the active site in order to perform the oxidative dealkylation. Even if bases from weakened pairs (substrates or not) are flipped to the same position in the active site, only the damaged bases possessing the alkyl adduct can be oxidatively modified due to the proximity of the extra alkyl portion to the catalytic metal site (Fig. 4d). Therefore, flipping a non-substrate base into the enzymatic pocket of ALKBH2 does not run a risk of unwanted modification to the base (Supplementary Fig. 5a,b), and a damage checking site, which has been shown to be critical for DNA glycosylases to function properly, is not necessary for ALKBH2. Similarly, the recently discovered TET family proteins, which use the same oxidation mechanism to convert 5-methyl cytosine (5-meC) to 5-hydroxymethyl cytosine in genomic DNA38, should oxidatively modify only 5-meC but not cytosine even if a cytosine base is accidently flipped, assuming the TET family proteins also flip 5-meC out of the duplex for oxidation.
Figure 4.
ALKBH2 does not have nor need a damage-checking site; its oxidation chemistry insures that non-substrate bases are not modified. (a) Superposition of the AT structure, εA structure, and 3BTY to show that the three flipped bases, damaged or not, are bound to the same site of ALKBH2. The view on the left panel is of the same angle as in Fig. 3; a 90° clockwise rotation of the left panel gives the view on the right. (b) Overlay of the CI and 3-meC structures using the protein part of the complex. A zoom-in view on the right shows the four active site residues that interact with the 3-meC base (Tyr122 and Glu175 form hydrogen-bonds to N4 of 3-meC; Phe124 and His171 stack against 3-meC). (c) Final positions of 3-meC (light magenta) and cytosine (pale cyan) in the active site of ALKBH2 are the same. The identical view as in b is shown, and all protein residues are omitted for clarity purpose. (d) Stereo view (which is a 90° clockwise rotation of c) of the metal site of the 3-meC structure. The aberrant methyl group (highlighted with yellow background color) is precisely positioned by ALKBH2 for efficient oxidation. The approximate location of the putative iron(IV)-oxo species, after 2-ketoglutarate is converted to succinate, is colored in blue.
DISCUSSION
DNA damage constitutes only a very small percentage of human genome, yet the lesions must be promptly repaired to avoid cytotoxic and/or mutagenic consequences. To search for these sporadic sites, repair proteins have evolved different strategies to assure their efficiencies in damage location. We report here that the direct repair dioxygenase ALKBH2 interrogates DNA using mainly a hydrophobic hairpin motif, which is partially inserted into the DNA duplex. This interrogation mechanism enables ALKBH2 to probe base pair stability irrespective of the base pair identity, thus narrowing down its structurally-diverse cognate lesions to be within the weakened base pairs (Supplementary Fig. 5c).
Invasive interrogation of undamaged DNA has previously been shown in the case of MutM19. hOGG1 could also break a G:C base pair when it is bound to undamaged DNA21. Although results reported in this study suggest that ALKBH2 can discern base pairs with different stabilities, we do not think that ALKBH2 flips every base it encounters during the damage-searching process. Both sliding (in which protein diffuses along DNA and makes continuous contact) and hopping (in which protein undergoes microscopic dissociation and re-association) have been suggested to be effective mechanisms for DNA repair proteins to locate lesions in the genome26,39–42; for example, the time scale of hopping and sliding of UDG has been established and studied43. Recent kinetic experiments have shown that lesion-searching is a multistep process which could involve a nonspecific “search complex”, an “interrogation complex” capable of base discrimination, and eventually a “substrate-recognition complex” featuring a fully flipped base15–16,23,44. As for ALKBH2, duplex distortion occurs locally in both the CG structure and structures with a flipped substrate (Fig. 1a and Supplementary Fig. 2c). It is tempting to speculate that such minimal disturbance would reduce the energetic cost for short-range sliding. Further biophysical and biochemical studies can help to provide mechanistic insights and kinetic characteristics of the nature of ALKBH2-mediated damage-searching process. It is interesting to note that ALKBH3, a homologue of ALKBH2, relies on partner proteins to open DNA duplex for damage-searching, presenting an intriguing example of DNA damage-searching and repair.45–46
An immediate consequence of the non-selective lesion-detection process is that ALKBH2 may flip into its active site a non-cognate lesion, or even a normal base from a mismatched base pair. Our structures show that ALKBH2 does not have nor need a damage-checking site, which is demonstrated to be critical for assuring correct enzymatic activity in the cases of the MutM and UDG DNA glycosylases. Whether or not other DNA glycosylases also possess such a damage-checking site requires further investigation. For ALKBH2, a flipped base is always at the same position in the active site; then the oxidative demethylation chemistry of ALKBH2 must ensure that a non-cognate base does not get modified in its active site. Previous studies on both model compounds and iron(II)/2KG dioxygenases (including a density functional theory study of AlkB) have yielded rich knowledge on dioxygen activation in the context of the conserved 2-His-1-Asp/Glu motif (metal-chelating ligand of AlkB and ALKBH2)47–50. The spatial relationship of the modified base to the putative, catalytic iron(IV)-oxo species has been shown to be essential to allow efficient catalysis50. Thus, when a non-cognate base is flipped into the active site, due to the absence of a geometrically optimal chemical group, the flipped base should not be oxidized by ALKBH2 (Supplementary Fig. 5a,b).
On the other hand, when ALKBH2 detects weakened base pairs that does not contain cognate lesions (for example base pairs with other types of lesions or mismatched pairs), it remains to be seen if ALKBH2 carries signaling functions, as demonstrated by the ATL protein, and perhaps the AlkD protein (Supplementary Fig. 5c)51–52. Overall, the combination of DNA interrogation, non-discriminative base-flipping, and oxidation chemistry by the dioxygenase ALKBH2 can improve the efficiency of DNA repair by increasing the odds of finding hot spots with structurally diverse base lesions while not endangering the integrity of the genome by accidental and unwanted DNA modification.
ONLINE METHODS
Expression and purification of ALKBH2-ΔN55
The ALKBH2-ΔN55 gene was cloned between the NdeI and HindIII sites of a pET28a+ vector (Novagen). This plasmid was transformed into BL21 (DE3) E. coli cells (Stratagene) for protein over-expression. The protein was purified using Ni-NTA chromatography (GE Healthcare) with a binding buffer of 50 mM sodium phosphate (pH 8.0), 300 mM NaCl and 50 mM imidazole and an elution buffer of 50 mM sodium phosphate (pH 8.0), 300 mM NaCl and 400 mM imidazole. The N-terminal His-tag was removed by an overnight thrombin digestion at 4 °C. About 4–6 mg of protein can be obtained from 1 L of bacterial cells.
Oligonucleotide synthesis
Oligonucleotides containing modified bases were prepared using solid-phase synthesis53. Phosphoramidites were all purchased from Glen Research Corporation and ChemGenes Corporation. All synthetic oligonucleotides were purified with denaturing PAGE and verified with MALDI-TOF mass spectrometry.
Cross-linking and crystallization of the ALKBH2-dsDNA complexes
Cross-linking reactions between ALKBH2 and dsDNA were performed as the decribed below: to set up distal cross-linking, a G169C mutation was introduced as determined previously; to avoid non-specific disulfide cross-linking, three additional mutations, C67S, C165S and C192S, were introduced into ALKBH2-ΔN55 G169C. This mutant protein was used to cross-link with different synthetic oligonucleotides: 5′-CTGTCATCAC*TGCG -3′ paired with 5′-TCGCAGTGATGACA -3′ for the CG structure, 5′-ATGTATAAC*TGCG -3′ with 5′-TCGCAGTTATACA -3′ for the AT structure, 5′-ATGTATCAC*TGCG -3′ paired with 5′-TCGCAGTIATACA -3′ for the CI structure, 5′-CTGTCT(3-meC)AC*TGCG -3′ paired with 5′-TCGCAGTGAGACA -3′ for the 3-meC structure and 5′-CTGTCA(εA)ACTGCG -3′ paired with 5′-TCGCAGTTAGACA –3′ for the εA structure. The cross-linking reactions were performed at 4 °C for 16 h and the covalently-linked ALKBH2-dsDNA complexes were purified using Mono-Q anion exchange chromatography, which were then buffer-exchanged to 100 mM NaCl, 10 mM Tris-HCl (pH 8.0). The complexes were then concentrated to 5 mg/mL for crystallization trials. Rod-shaped crystals grew in 1–4 weeks at 4 °C, in hanging drops containing 1 μL of complex solution and 1 μL of reservoir solution of 100 mM NaCl, 50 mM MgCl2 and 100 mM cacodylate (pH 6.5) and 8% 8K PEG. Cofactor-containing ALKBH2-DNA structures were obtained by soaking apo-crystals in well solutions containing MnCl2 (~2.5 mM) and 2-KG (~5 mM) for 2–4 h to give (Mn/2KG)ALKBH2-DNA structures.
Experimental details of activity assays
A 100 μL reaction mixture containing 50 mM Tris (pH 8.0), 2 mM ascorbic acid, 1 mM 2-ketoglutarate, 0.28 mM (NH4)2Fe(SO4)2 and 1 nmol dsDNA or ssDNA substrate was incubated at 4 °C for ~60 min in the presence of 0.5 nmol wild type ALKBH2 protein and different ALKBH2 mutants (F102A and V101G F102A double mutant protein). The reaction was terminated by adding EDTA to 5 mM to quench the enzymatic activity. The reaction mixture was run on a DNAPac PA200 analytical column (4×250 mm, Dionex) connected to a Dionex HPLC system, with a flow rate of 1 ml/min at room temperature. Buffer A contains 50 mM sodium citrate (pH = ~5.3) and buffer B contains 50 mM sodium citrate (pH = ~5.3), 1 M NaCl. The red lines in Supplementary Fig. 1a–b represent the exact gradient of the HPLC program. Each experiment was performed in triplicate.
Experimental details of the 2AP fluorescence studies
For a typical fluorescence study, 1 μM 2AP-containg dsDNA was prepared in a buffer containing 50 mM NaCl, 10 mM Tris-HCl (pH = 8.0) and 1 mM DTT. ALKBH2 stock (~0.12 mM) was also prepared in the same buffer, and the appropriate amount (from 0.25 equiv to up to 1.25 equiv of dsDNA) was added to the same dsDNA solution (~120 μL). Fluorescence traces (fluorescence from ALKBH2 has been subtracted) when increasing amount of stock ALKBH2 protein was added to the dsDNA-containing solution were shown in Supplementary Fig. 1d–f. Fold of fluorescence change (the fluorescence when 1 equiv of ALKBH2 was added to DNA divided by that of free DNA) is plotted. Emission spectra (340–420 nm) were recorded at an excitation wavelength λex = 320 nm with excitation and emission bandwidths of 5 nm. All fluorescence measurements (performed at least in triplicate) were acquired at 4 °C, using a Varian Cary Bio 300 UV-Vis Spectrophotometer.
Structure determination and refinement
All X-ray data were integrated and scaled using HKL2000 program suite54, and converted to structure factors with the CCP4 program. The ALKBH2-DNA complex structure was phased by molecular replacement (with Phaser)55, using the previously published ALKBH2 structure as a search model. The model was built by using COOT56, and refinement was carried out with the program REFMAC5 from the CCP4 suite57. Data collection and refinement parameters for all structures are given in Table 1. Molecular graphics figures were prepared with PyMOL58.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health (GM071440 to C.H.), Hundred Talent Program of the Chinese Academy of Sciences (to C.-G.Y.), the State Key Development Program of Basic Research of China (2009CB918502 to C.-G.Y.), the special grant for Stem Cell and Regenerative Medicine (XDA01040305 to C.-G.Y.), National Science Foundation (MCB-0547854 to A.R.D.), and Beamlines 23ID-B(General Medicine and Cancer Institutes Collaborative Access Team [GM/CA-CAT]), 21ID-D (Life Sciences Collaborative Access Team [LS-CAT]) and 24ID-E (The Northeastern Collaborative Access Team [NE-CAT]) at the Advanced Photon Source at Argonne National Laboratory, and Shanghai Synchrotron Radiation Facility (BL17U1); National Institutes of Health and the United States Department of Energy.
Footnotes
ACCESSION CODES.
Protein Data Bank: Coordinates and structure factors for the ALKBH2-dsDNA complex have been deposited under accession codes 3RZG [CG structure], 3RZM [AT structure], 3RZL [CI structure], 3RZK[(Mn/2KG) εA structure], 3RZH [3-meC structure], 3RZJ [(Mn/2KG) 3-meC structure], 3S57[(Mn/2KG) CG structure], and 3S5A [(Mn/2KG) CG-DNA2 structure].
Note: Supplementary information is available at the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
C.Y., C.-G.Y. and C.H. designed the experiments. Experiments were performed by C.Y., B.C., W.Z., G.J., L.Z. and C.L.; computational analyses were performed by B.Q. and A.R.D. C.Y. and C.H. wrote the paper and B.Q., A.R.D. and C.-G.Y. contributed to editing the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 2.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: Recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Yi C, Yang CG, He C. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc Chem Res. 2009;42:530–541. doi: 10.1021/ar800178j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan T, et al. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci U S A. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aas PA, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 6.Ringvoll J, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringvoll J, et al. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res. 2008;68:4142–4149. doi: 10.1158/0008-5472.CAN-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cetica V, et al. Pediatric brain tumors: mutations of two dioxygenases (hABH2 and hABH3) that directly repair alkylation damage. J Neurooncol. 2009;94:195–201. doi: 10.1007/s11060-009-9837-0. [DOI] [PubMed] [Google Scholar]
- 9.Wu SS, et al. Down-regulation of ALKBH2 increases cisplatin sensitivity in H1299 lung cancer cells. Acta Pharmacol Sin. 2011;32:393–398. doi: 10.1038/aps.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilljam KM, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol. 2009;186:645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang CG, et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Yang CG, Garcia K, He C. Damage detection and base flipping in direct DNA alkylation repair. Chembiochem. 2009;10:417–423. doi: 10.1002/cbic.200800580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci U S A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe AE, O’Brien PJ. Kinetic mechanism for the flipping and excision of 1,N(6)-ethenoadenine by human alkyladenine DNA glycosylase. Biochemistry. 2009;48:11357–11369. doi: 10.1021/bi9015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendershot JM, Wolfe AE, O’Brien PJ. Substitution of active site tyrosines with tryptophan alters the free energy for nucleotide flipping by human alkyladenine DNA glycosylase. Biochemistry. 2011;50:1864–1874. doi: 10.1021/bi101856a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman BR, Lee S, Wang S, Verdine GL. Structure of Escherichia coli AlkA in complex with undamaged DNA. J Biol Chem. 2010;285:35783–35791. doi: 10.1074/jbc.M110.155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y, et al. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 22.Slupphaug G, et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 23.Cao CY, Jiang YL, Stivers JT, Song FH. Dynamic opening of DNA during the enzymatic search for a damaged base. Nature Structural & Molecular Biology. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 24.Parker JB, et al. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–U432. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 28.Monsen VT, SO, Aas PA, Westbye MP, Sousa MM, Slupphaug G, Krokan HE. Divergent {beta}-hairpins determine double-strand versus single-strand substrate recognition of human AlkB-homologues 2 and 3. Nucleic Acids Research. 2010 doi: 10.1093/nar/gkq518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B, Liu H, Sun X, Yang CG. Mechanistic insight into the recognition of single-stranded and double-stranded DNA substrates by ABH2 and ABH3. Mol Biosyst. 2010;6:2143–2149. doi: 10.1039/c005148a. [DOI] [PubMed] [Google Scholar]
- 30.Ma A, Hu J, Karplus M, Dinner AR. Implications of alternative substrate binding modes for catalysis by uracil-DNA glycosylase: an apparent discrepancy resolved. Biochemistry. 2006;45:13687–13696. doi: 10.1021/bi061061y. [DOI] [PubMed] [Google Scholar]
- 31.Lu L, Yi C, Jian X, Zheng G, He C. Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system. Nucleic Acids Res. 2010;38:4415–4425. doi: 10.1093/nar/gkq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman BR, Lee S, Wang S, Verdine GL. Structure of the E. coli DNA glycosylase AlkA bound to the ends of duplex DNA: a system for the structure determination of lesion-containing DNA. Structure. 2008;16:1166–1174. doi: 10.1016/j.str.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu B, et al. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 35.Yi C, et al. Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature. 2010;468:330–333. doi: 10.1038/nature09497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu B, Hunt JF. Enzymological and structural studies of the mechanism of promiscuous substrate recognition by the oxidative DNA repair enzyme AlkB. Proc Natl Acad Sci U S A. 2009;106:14315–14320. doi: 10.1073/pnas.0812938106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard GA, et al. Guanine-1,N6-ethenoadenine base pairs in the crystal structure of d(CGCGAATT(epsilon dA)GCG) Biochemistry. 1994;33:4755–4761. doi: 10.1021/bi00182a002. [DOI] [PubMed] [Google Scholar]
- 38.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blainey PC, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nature Structural & Molecular Biology. 2009;16:1224–U1234. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin YH, et al. Using the Bias from Flow to Elucidate Single DNA Repair Protein Sliding and Interactions with DNA. Biophysical Journal. 2009;96:1911–1917. doi: 10.1016/j.bpj.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hedglin M, O’Brien PJ. Hopping Enables a DNA Repair Glycosylase To Search Both Strands and Bypass a Bound Protein. Acs Chem Biol. 2010;5:427–436. doi: 10.1021/cb1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schonhoft JD, Stivers JT. Timing facilitated site transfer of an enzyme on DNA. Nat Chem Biol. 2012;8:205–210. doi: 10.1038/nchembio.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y, Friedman JI, Stivers JT. Cosolute paramagnetic relaxation enhancements detect transient conformations of human uracil DNA glycosylase (hUNG) Biochemistry. 2011;50:10724–10731. doi: 10.1021/bi201572g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dango S, et al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell. 2011;44:373–384. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundheim O, et al. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krebs C, et al. Rapid freeze-quench 57Fe Mossbauer spectroscopy: monitoring changes of an iron-containing active site during a biochemical reaction. Inorg Chem. 2005;44:742–757. doi: 10.1021/ic048523l. [DOI] [PubMed] [Google Scholar]
- 48.Schofield CJ, Zhang ZH. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr Opin Struc Biol. 1999;9:722–731. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 49.Costas M, Mehn MP, Jensen MP, Que L. Dioxygen Activation at Mononuclear Nonheme Iron Active Sites: Enzymes, Models, and Intermediates. Chemical Reviews. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, Llano J, Gauld JW. A DFT study of nucleobase dealkylation by the DNA repair enzyme AlkB. J Phys Chem B. 2009;113:4887–4898. doi: 10.1021/jp810715t. [DOI] [PubMed] [Google Scholar]
- 51.Tubbs JL, et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mishina Y, Chen LX, He C. Preparation and characterization of the native iron(II)-containing DNA repair AlkB protein directly from Escherichia coli. J Am Chem Soc. 2004;126:16930–16936. doi: 10.1021/ja045066z. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.The PyMOL Molecular Graphics System, V. S., LLC. 2002 http://www.pymol.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.