SUMMARY
Human bone marrow mesenchymal stem/stromal cells (MSCs) are multipotent progenitor cells with multilineage differentiation potentials including osteogenesis and adipogenesis. While significant progress has been made in understanding transcriptional controls of MSC fate, little is known about how MSC differentiation is epigenetically regulated. Here we show that the histone demethylases KDM4B and KDM6B play critical roles in osteogenic commitment of MSCs by removing H3K9me3 and H3K27me3. Depletion of KDM4B or KDM6B significantly reduced osteogenic differentiation and increased adipogenic differentiation. Mechanistically, while KDM6B controlled HOX expression by removing H3K27me3, KDM4B promoted DLX expression by removing H3K9me3. Importantly, H3K27me3- and H3K9me3-positive MSCs of bone marrow were significantly elevated in ovariectomized and aging mice in which adipogenesis was highly active. Since histone demethylases are chemically modifiable, KDM4B and KDM6B may present as novel therapeutic targets for controlling MSC fate choices, and lead to clues for new treatment in metabolic bone diseases such as osteoporosis.
INTRODUCTION
Human bone marrow mesenchymal/stromal stem cells (MSCs) are multipotent progenitor cells with self-renewal capabilities and multilineage differentiation potentials including osteogenesis, chondrogenesis and adipogenesis (Dominici et al., 2006; Bianco et al., 2008; Caplan, 2007; Levi and Longaker, 2011; Teitelbaum, 2010). MSCs hold significant promise for regenerative therapies due to their convenient isolation, lack of immunogenicity, as well as their ability to transdifferentiate and to create a tissue microenvironment favorable for tissue repair (Kolf et al., 2007; Phinney and Prockop, 2007; Undale et al., 2009). Their therapeutic utility hinges upon the understanding of molecular mechanisms that regulate their differentiation (Tang et al., 2009; Sacchetti et al., 2007; Medici et al., 2010; Kronenberg, 2010; Huebsch et al., 2010). Normal bone homeostasis relies on the inverse relation between adipogenesis and osteogenesis in MSCs. As observed in both healthy and diseased bone, inhibition of adipogenesis may enhance bone growth and repair (McCauley, 2010; Kawai and Rosen, 2010). Thus, it is crucial to gain an understanding of how the MSC lineage is regulated. Growing evidence suggest that the final cell fate decision of MSCs relies on an orchestrated activation of lineage specific genes and repression of genes promoting cell stemness or commitment to other lineages (Méndez-Ferrer et al., 2010; Takada et al., 2009; Wei et al., 2011). At present, studies have been focused on identifying a number of extrinsic regulators and their downstream transcription factors that control cell fate commitment (Lian et al., 2006; Karsenty et al., 2009; Phimphilai et al., 2006). For example, bone morphogenic proteins (BMP) act as major osteogenic inducers and have been utilized for promoting bone regeneration and repair (Chen et al., 2004; Medici et al., 2010). While significant progress has been made in understanding of molecular regulation of MSC differentiation, very little is known with regard to the epigenetic events that control MSC commitment to bone, adipose tissue and cartilage.
Cell fate is determined by DNA-binding transcription factors that are regulated more specifically at the epigenetic level, where a specific chromatin configuration is conferred to control the expression of key transcription factors for differentiation (Jenuwein and Allis, 2001; Shi, 2007; Agger et al., 2008; Stein et al., 2010). Progressive restriction of differentiation potential in MSCs relies on orchestrated activation of repression of lineage specific genes (Lian et al., 2006; Kronenberg, 2010). While epigenetic regulatory mechanisms that govern adipogenic and osteogenic specific genes remain largely elusive, they are the crucial missing links in understanding extracellular signaling and lineage-specific decisions during MSC differentiation. One of the central chromatin modifications, histone lysine methylation at the N-terminal tails, controls the access to regulatory regions on the DNA by altering chromatin compaction (Jenuwein and Allis, 2001). Upon differentiation of stem cells, the relative levels of epigenetic marks change to reflect the activation and repression of genes, guiding development towards one cell lineage. Modified histone domains have thus become epigenetic signatures that either mark for gene activation, such as acetylated histone 3 Lysine 9 (H3K9) and trimethylated Histone 3 Lysine 4 (H3K4), or mark for gene repression, such as trimethylated H3K9 and trimethylated H3K27 (Shi, 2007; Agger et al., 2008). Recently, histone demethylases have been discovered to possess the ability of removing methyl groups from various methylated histone lysine residues such as K4, K9, K27, K36 and K79, thereby enabling the dynamic and reversible regulation of transcription (Klose et al., 2006; Shi and Whetstine, 2007; Agger et al., 2008). As such, growing evidence suggest that histone demethylases may be associated with cell fate choices and cell differentiation (Shi, 2007; Sen et al. 2008; Lan et al., 2007; Agger et al., 2007; Agger et al., 2009).
Recently, we have identified a demethylase complex regulating the function of MSCs isolated from craniofacial tissues (Fan et al., 2009). To further explore the role of demethylases in MSC cell fate determination, we systemically profiled the expression of histone demethylases in BMP-stimulated MSCs, as BMPs are potent inducers of osteogenic differentiation (Chen et al., 2004). We found that BMP4/7 heterodimer rapidly induced the expression of two histone demethylases, KDM4B (also known as JMJD2B; Fodor et al., 2006; Cloos et al., 2006; Klose et al., 2006; Whetstine et al., 2006) and KDM6B (also known as JMJD3; Lan et al. 2007; Agger et al., 2007), through Smad signaling. The knock-down of either KDM4B or KDM6B impaired osteogenic commitment of MSCs in vitro and in vivo whereas adipogenic lineage commitment of MSCs was enhanced. Mechanistically, while KDM6B was required for the expression of HOX genes by removing histone 3 K27 trimethylation (H3K27me3), KDM4B regulated the expression of DLX genes by removing histone 3 K9 trimethylation (H3K9me3). Intriguingly, we found that H3K27me3 and H3K9me3 marks were significantly increased in bone marrow MSCs of ovariectomized mice or aging mice in which adipogenesis was significantly elevated.
RESULTS
Induction of KDM4B and KDM6B by BMP-4/7 through Smad in MSCs
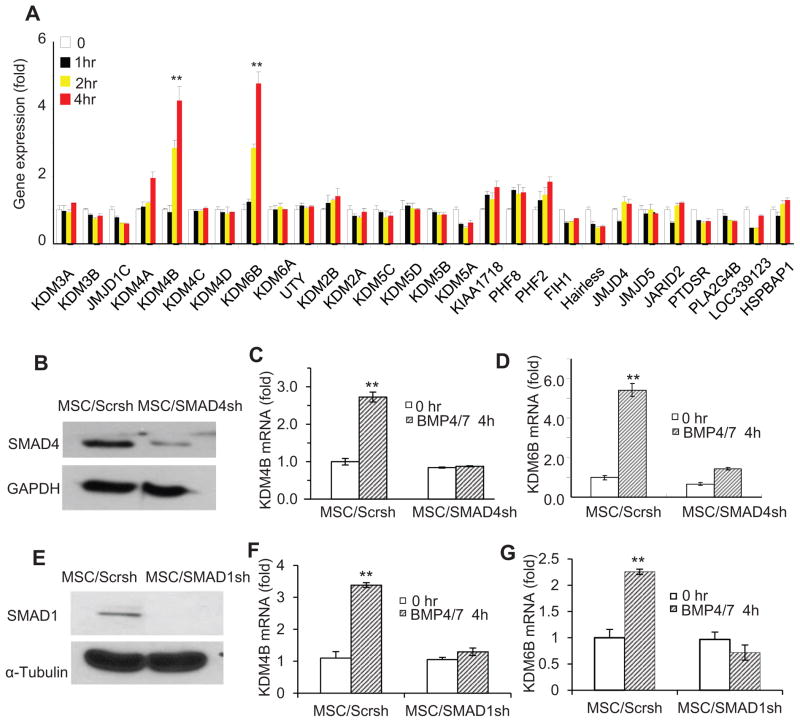
BMP-4/7 are potent inducers of osteogenic differentiation of MSCs, offering a great promise for bone regeneration and repair. To investigate potential roles of histone demethylases in osteogenic differentiation of MSCs, we profiled expression of 28 histone demethylases in BMP-4/7-induced MSCs from human bone marrow. Human MSCs used in our studies highly expressed CD73, CD90, CD105 and CD146, and were negative for CD34, CD45 and HLA (Fig. S1). Real-time RT-PCR revealed that KDM4B and KDM6B were most strongly induced by BMP4/7 (Fig. 1A). KDM4B and KDM6B are histone demethylases that remove H3K9me3 and H3K27me3, respectively (Fodor et al., 2006; Cloos et al., 2006; Klose et al., 2006; Whetstine et al., 2006; Lan et al. 2007; Agger et al., 2007). Both H3K9me3 and H3K27me3 are associated with gene silencing. Previously, KDM6B was identified as an early response gene induced by LPS through NF-κB signaling in macrophages (De Santa et al., 2007). Since BMPs mainly induce gene expression through SMAD signaling (Feng and Derynck, 2005; Attisano and Wrana, 2002; Massagué et al., 2005), we examined whether the inductions of KDM4B and KDM6B was dependent on SMAD signaling. Lentiviruses expressing SMAD4 shRNA were generated to knock-down SMAD4. Western blot analysis showed that more than 85% SMAD4 was depleted in MSCs expressing SMAD4 shRNA (MSC/SMAD4sh) compared with MSCs expressing scramble shRNA (MSC/Scrsh) (Fig. 1B). Real-time RT-PCR revealed that knock-down of SMAD4 significantly reduced the expression of KDM4B and KDM6B in MSCs induced by BMP4/7 (Fig. 1C and D). Furthermore, the knock-down of SMAD1, a key factor associated with BMP signaling, also inhibited the expression of KDM4B and KDM6B induced by BMP4/7 (Fig. 1E–G).
Figure 1. KDM4B and KDM6B were induced by BMP/SMAD signaling in MSCs.
(A) BMP4/7-induced histone demethylases in MSC by gene expression profiling. The mRNA expression of histone demethylases was assessed by Real-time RT-PCR. (B) The knock-down of SMAD4 by shRNA. SMAD4 expression was examined by Western Blot analysis. (C) The knock-down of SMAD4 inhibited KDM4B in MSCs. KDM4B mRNA was examined by Real-time RT-PCR. (D) The knock-down of SMAD4 inhibited KDM6B expression in MSCs. KDM6B mRNA levels were assessed by Real-time RT-PCR. ** P < 0.01. (E) The knock-down of SMAD1 by siRNA. (F) The knock-down of SMAD1 inhibited KDM4B in MSCs. (G) The knock-down of SMAD1 inhibited KDM6B expression in MSCs. ** P < 0.01. See also Figure S1.
KDM4B and KDM6B are required for osteogenic differentiation of MSCs
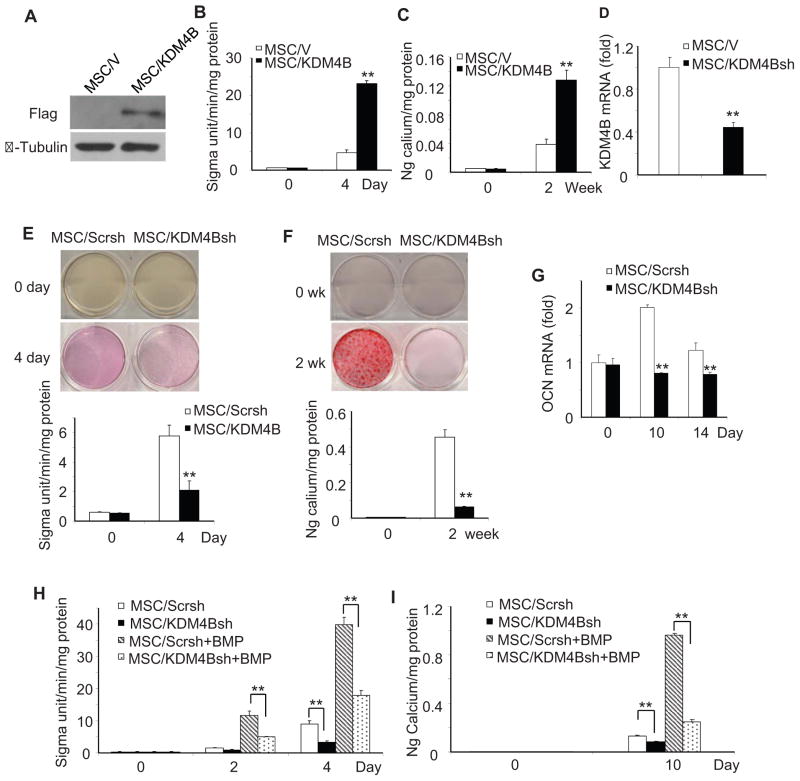
Since KDM4B and KDM6B were induced by BMP4/7, we explored whether they played a role in MSC cell fate determination. MSCs can be differentiated into osteoblasts, chondrocytes and adipocytes in vitro depending on the culture conditions. When stimulated by culture medium containing dexamethasone, β-glycerophosphate, and inorganic phosphate, MSCs can be induced to undergo osteogenic differentiation in vitro (Gronthos et al., 1994; Fan et al., 2009). We over-expressed KDM6B in human MSCs (Fig. 2A). After osteogenic induction, we found that over-expression of KDM6B significantly enhanced alkaline phosphatase (ALP) activity, an early marker of osteoblast differentiation (Fig. 2B). Consistently, over-expression of KDM6B enhanced mineralization in vitro, as assessed by Alizarin Red staining (Fig. 2C). Similarly, we found that over-expression of KDM4B also enhanced ALP activity and mineralization in MSCs (Fig. 3A–C).
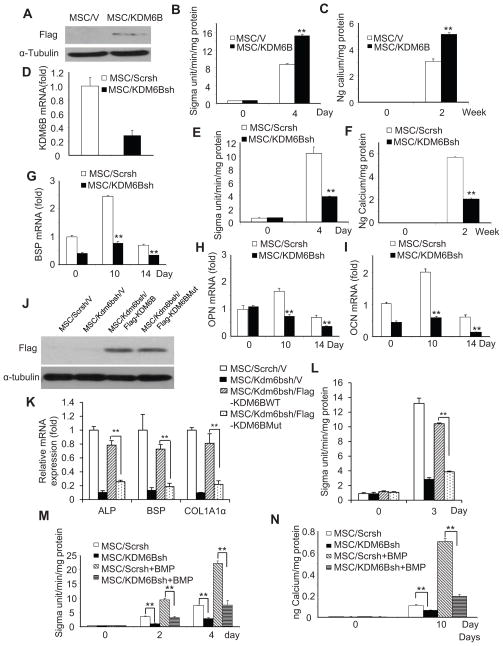
Figure 2. KDM6B is required for osteogenic differentiation of MSCs.
(A) Over-expression of KDM6B in MSCs. (B) Over-expression of KDM6B enhanced ALP activities in MSC differentiation. Values are mean ± s.d for triplicate samples from a representative experiment. **P < 0.01. (C) The over-expression of KDM6B enhanced mineralization in MSC differentiation. Mineralization was stained with Alizarin Red and qualitatively measured. **P < 0.01. (D) The knock-down of KDM6B by shRNA. MSC/KDM6Bsh, MSCs expressing KDM6B shRNA; MSC/Scrsh, MSCs expressing scramble shRNA. (E) The knock-down of KDM6B inhibited ALP activities in MSCs. Values are mean ± s.d for triplicate samples from a representative experiment. **P < 0.01. (F) The knock-down of KDM6B inhibited mineralization in MSCs. **P < 0.01. (G) The knock-down of KDM6B inhibited BSP expression in MSCs as determined by Real-time RT-PCR. **P < 0.01. (H) The knock-down of KDM6B inhibited OPN expression in MSCs as determined by Real-time RT-PCR. **P < 0.01. (I) The knock-down of KDM6B inhibited OCN expression in MSCs as determined by Real-time RT-PCR. **P < 0.01. (J) The restoration of KDM6B and mutant KDM6B in Kdm6b knock-down MSCs by Western blot analysis. (K) The catalytically mutant KDM6B could not restore the expression of osteoblast marker genes in MSCs. (L) The catalytically mutant KDM6B could not restore ALP activities in MSCs. (M) The knock-down of KDM6B inhibited BMP-enhanced ALP activities in MSCs. **P < 0.01. (N) The knock-down of KDM6B inhibited BMP-enhanced mineralization in MSCs. **P < 0.01. See also Figure S2
Figure 3. KDM4B is required for osteogenic differentiation of MSCs.
(A) Over-expression of KDM4B in MSCs. MSC/KDM6B, MSCs expressing KDM6B; MSC/V, MSCs expressing empty vector. (B) Over-expression of KDM4B enhanced ALP activities in MSC differentiation. Values are mean ± s.d for triplicate samples from a representative experiment. **P < 0.01. (C) Over-expression of KDM4B enhanced mineralization in MSC differentiation. **P < 0.01. (D) The knock-down of KDM4B by shRNA. MSC/KDM4Bsh, MSCs expressing KDM4B shRNA; MSC/Scrsh, MSCs expressing scramble shRNA. (E) The knock-down of KDM4B inhibited ALP activities in MSCs. Values are mean ± s.d for triplicate samples from a representative experiment. **P < 0.01. (F) Knock-down of KDM4B inhibited mineralization in MSCs. **P < 0.01. (G) Knock-down of KDM4B inhibited OCN expression in MSCs as determined by Real-time RT-PCR. **P < 0.01. (H) Knock-down of KDM4B inhibited BMP-enhanced ALP activities in MSCs. **P < 0.01. (I) Knock-down of KDM4B inhibited BMP-enhanced mineralization in MSCs. **P < 0.01. See also Figure S3
To examine whether KDM4B and KDM6B played important roles in osteogenic differentiation of MSCs, we generated lentiviruses expressing KDM4B or KDM6B shRNA. The knock-down of KDM6B in MSCs was confirmed using Real-time RT-PCR (Fig. 2D). After treating MSCs with osteogenic inducing media for 4–5 days, ALP activity of differentiating MSCs was significantly suppressed by knocking down KDM6B (Fig. 2E). Moreover, KDM6B knock-down also reduced formation of mineralized nodules after prolonged treatment with inducing media for 2 weeks (Fig. 2F). To confirm that KDM6B depletion inhibited osteogenic differentiation in MSCs, we assessed the mRNA expression of several osteogenic markers 0, 10, and 14 days after induction. The knock-down of KDM6B significantly inhibited the expression of bone sialoprotein (BSP), osteopontin (OPN) and osteocalcin (OCN) (Fig. 2G–I). Moreover, to rule out off-target effects, we made another vector targeting a different region of KDM6B and obtained similar results (Fig. S2A–C). We found that the knock-down of Kdm6b in mouse MSCs also inhibited osteogenic differentiation. In mouse MSC/Kdm6bsh, the over-expression of human KDM6B, which was resistant to mouse Kdm6sh shRNA, restored osteogenic differentiation of MSCs (Fig. S2D–G). Elegant studies by Miller et al (2010a, 2010b, 2008) showed that KDM6B may function independently from its demethylase activity to achieve chromatin remodeling of its target genes in T cell commitment. To test this possibility, we generated a catalytically-dead KDM6B mutant in which histidin 1390 was changed to alanine, disrupting the non-heme center for its demethylase activity. However, the over-expression of KDM6B, but not the catalytically mutant KDM6B in mouse MSC/Kdm6sh rescued osteogenic differentiation of MSCs (Fig. 2J–L), indicating that KDM6B demethylase activity is required for MSC differentiation. It is well known that addition of exogenous BMPs enhances osteogenic differentiation in MSCs. More importantly, since KDM6B was induced by BMP4, we examined whether the depletion of KDM6B would affect BMP4/7 enhancement. As shown in Fig. 2M and N, the knock-down of KDM6B significantly suppressed osteogenic differentiation of MSC induced by BMP4/7. Consistently, the knock-down of KDM6B inhibited the expression of RUNX2 and OSX induced by BMP4/7 (Fig. S2H and I).
We also utilized shRNA to knock-down KDM4B in MSCs. The depletion of KDM4B inhibited ALP activity, mineralization and expression of extracellular matrix proteins in MSCs stimulated with osteogenic-inducing media (Fig. 3D–G). To rule out off-target effects, we made another vector targeting different region of KDM4B and obtained similar results (Fig. S3A–C). The restoration of KDM4B in MSC/KDM4Bsh cells, which was resistant to KDM4B shRNA, also reinstated osteogenic differentiation of MSCs (Fig. S3D–F). The knock-down of KDM4B also inhibited osteogenic differentiation of MSCs enhanced by BMP4/7 (Fig. 3H and I).
KDM4B and KDM6B are required for MSC-mediated bone formation in vivo
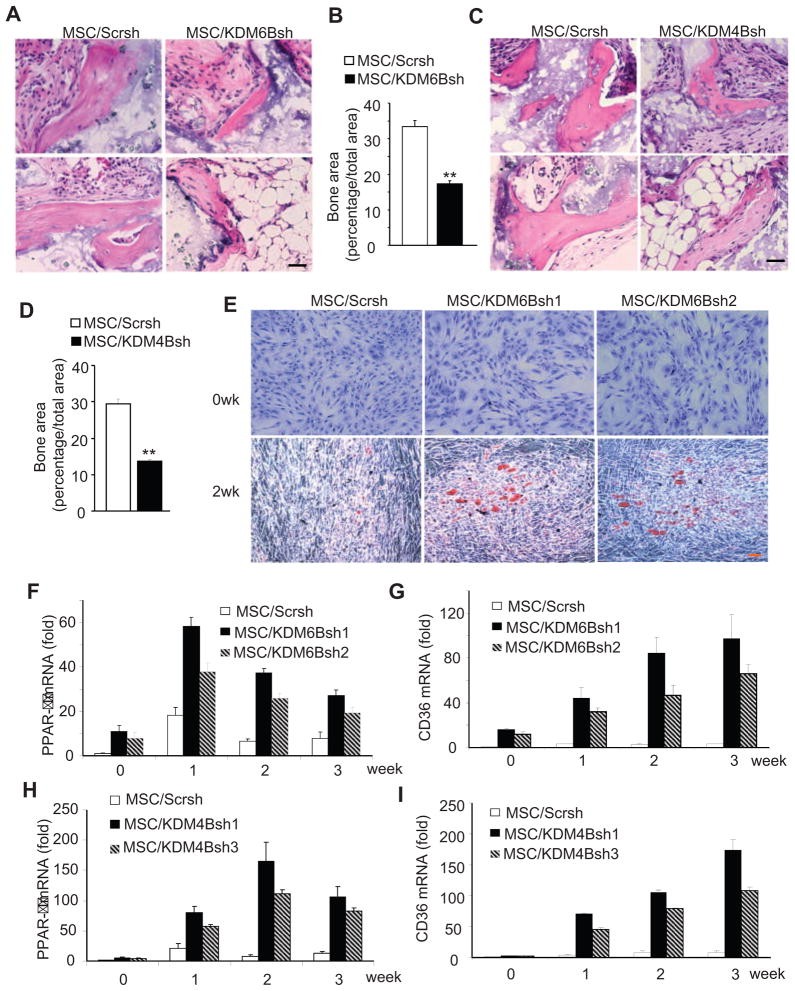
To verify our in vitro findings, we examined whether the knock-down of KDM4B and KDM6B affected MSC-mediated bone formation in vivo. MSC/KDM4Bsh, MSC/KDM6Bsh and MSC/Scrsh were mixed with hydroxyapatite/tricalcium phosphate (HA/TCP) carriers, and then transplanted subcutaneously into the dorsal side of 10-week old nude mice. After 8 weeks, transplants were harvested and prepared for histological analysis. H&E staining showed that MSC/KDM4Bsh and MSC/KDM6Bsh cells formed less bone tissues than MSC/Scrsh cells (Fig. 4A and C). Quantitative measurement of mineralized tissue areas revealed a greater than 50% decrease in bone formation by MSC/KDM4Bsh or MSC/KDM6Bsh cells compared with MSC/Scrsh cells (Fig. 4B and D). Interestingly, although it was difficult to perform qualitative measurement, we noted that adipose tissue formation in some regions was increased in MSC/KDM4B or MSC/KDM6B transplants as compared with MSC/Scrch transplants (Fig. 4A and C, bottom right panel), suggesting that KDM4B and KDM6B may inhibit adipogenic differentiation of MSCs.
Figure 4. KDM4B and KDM6B are required for MSC-mediated bone formation in vivo.
(A) Knock-down of KDM6B reduced MSC-mediated bone formation in vivo. Scale bar, 100 μm. (B) Qualitative measurement of bone formation by MSC/KDM6Bsh and MSC/V in vivo. Values are mean ± s.d., n = 5. **P < 0.01. (C) Knock-down of KDM4B reduced MSC-mediated bone formation in vivo. Scale bar, 100 μm. (D) Qualitative measurement of bone formation by MSC/KDM4Bsh and MSC/V in vivo. Values are mean ± s.d., n = 5. **P < 0.01. (E) Knockdown of KDM6B promoted adipogenic differentiation of MSCs. MSC/Scrsh, MSCs expressing scramble shRNA; MSC/KDM6Bsh1, MSCs expressing KDM6B shRNA1; MSC/KDM6Bsh2, MSCs expressing KDM6B shRNA2. (F) Knock-down of KDM6B promoted PPAR-γ expression as determined by Real-time RT-PCR. (G) Knock-down of KDM6B promoted CD36 expressing as determined by Real-time RT-PCR. (H) Knock-down of KDM4B promoted PPAR-γ expression as determined by Real-time RT-PCR. MSC/Scrsh, MSCs expressing scramble shRNA; MSC/KDM4Bsh1, MSCs expressing KDM4B shRNA1; MSC/KDM4Bsh2, MSCs expressing KDM4B shRNA2. (I) Knock-down of KDM4B promoted CD36 expressing as determined by Real-time RT-PCR.
Inhibition of adipocyte lineage commitment of MSCs by KDM4B and KDM6B
As normal bone homeostasis relies on the dynamic balance between osteogenesis and adipogenesis in differentiating MSCs, an osteogenic lineage commitment could be coupled with an inhibition of adipogenic differentiation. To explore whether KDM4B and KDM6B regulated adipogenic differentiation of MSCs, we assessed phenotypical changes of MSC/KDM4Bsh and MSC/KDM6Bsh cultured with adipogenic inducing medium. Oil-red-O staining revealed significant enhancement of adipogenesis in MSC/KDM6B cells as compared with MSC/Scrsh cells (Fig. 4E). To examine if the early onset of adipogenesis was influenced by KDM6B, we quantified the mRNA levels of the master adipogenic transcription factor PPAR-γ. As shown in Fig 4F, the depletion of KDM6B significantly enhanced PPAR-γ expression. CD36, also known as fatty acid translocase, is highly expressed in adipocytes and is important in fatty acid uptake and lipid accumulation. Its expression is induced by PPAR-γ (Qiao et al., 2008). The depletion of KDM6B significantly increased CD36 expression over a longer period (Fig. 4G). Consistently, we found that the knock-down of KDM4B also enhanced adipogenic differentiation of MSCs (Fig. 4H and I).
KDM4B and KDM6B target distinct transcription factors to induce MSC cell lineage commitment through H3K9me3 and H3K27me3
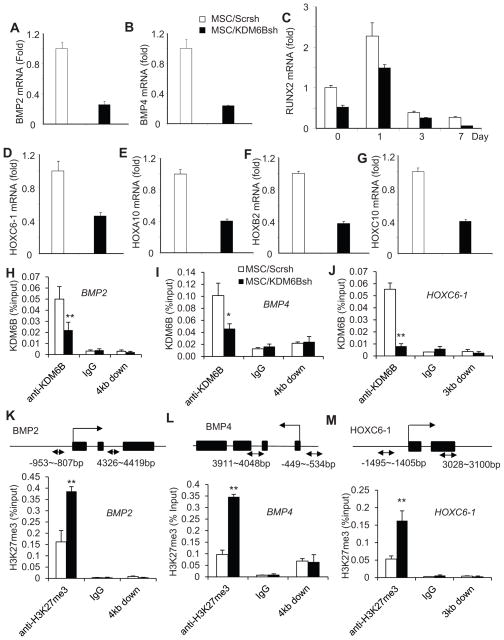
Previously, it was shown that KDM6B regulates the expression of HOX and BMP genes in embryonic stem cell differentiation through the removal of H3K27me3 (Lan et al. 2007; Agger et al., 2007). Since HOX and BMP genes are implicated in osteogenic differentiation, we tested whether the depletion of KDM6B affected the endogenous expression of BMP and HOX genes. Interestingly, in MSC/KDM6Bsh cells, the basal levels of endogenous BMP2 and BMP4 were significantly suppressed (Fig. 5A and B). Consistently, the induction of RUNX2 was inhibited upon induction of osteogenic differentiation (Fig. 5C). Homeobox (HOX) genes encode a large family of homeodomain-containing transcription factors that are implicated in anterior-posterior axis patterning in embryonic development (Wacker et al., 2004; Lan et al. 2007; Agger et al., 2007). Moreover, HOX transcription factors were found to activate RUNX2 transcription and can also mediate osteoblastogenesis in a RUNX2-independent manner (Hassan et al., 2007). Real-time RT-PCR revealed that the depletion of KDM6B significantly inhibited the expressions of HOXC6-1, HOXA10, HOXB2 and HOXC10 (Fig. 5D–G), implying that these HOX transcription factors may be regulated by KDM6B, thereby playing roles in promoting the osteogenic differentiation of MSCs. To further examine how KDM6B globally controlled MSC differentiation, we also performed microarray analysis on MSC/Scrsh and MSC/KDM6Bsh. KDM6B knockdown significantly inhibited the expression of 1867 genes at the basal level and 638 genes induced by BMP4/7, including BMP2 and BMP4 as well as members of HOX family genes (Fig. S4). A gene ontology (GO) analysis revealed that KDM6B controls the pathways associated with skeletal system development, development process, cell morphogenesis, and extracellular matrix organization (Table S1–3).
Figure 5. KDM6B is required for BMP and HOX gene expression in MSCs by removal of H3K27me3 marks.
(A and B) Knock-down of KDM6B inhibited the basal levels of BMP2 and BMP4 expression as determined by Real-time RT-PCR. (C) Knock-down of KDM6B inhibited RUNX2 expression during MSC differentiation. (D, E, F and G) Knock-down of KDM6B inhibited the basal levels of HOX gene expression in MSCs as assessed Real-time RT-PCR. (H, I and J) Knock-down of KDM6B reduced KDM6B binding to the promoters of BMP2, BMP4 and HOXC6-1 in MSCs. The promoters of BMP2, BMP4 and HOXC6-1 were ChIP-ed with anti-KDM6B antibodies or IgG control. The 3 to 4 kb downstream of the transcriptional start site was also ChIPed with anti-KDM6B. (K, L and M) Knock-down of KDM6B increased H3K27me3 levels at the promoters of BMP2, BMP4 and HOXC6-1. **P < 0.01. See also Figure S4 and S5 and Table S1–3.
In order to examine if KDM6B regulated MSC differentiation by demethylating H3K27me3, we performed ChIP assay to assess the changes in histone methylation status at the promoter regions of those master differentiation genes. Indeed, we found that KDM6B bound to the promoter regions of BMP2 and BMP4 (Fig. 5H and I). Since our gain- and loss-of-function analysis revealed that HOXC6-1 played a functional role in osteogenic differentiation of MSCs (Fig. S5A–I), we also examined the epigenetic status of the HOXC6-1 promoter. KDM6B was also found to be present on the HOXC6-1 promoter (Fig. 5J). KDM6B occupancy on the promoters of BMP2, BMP4, and HOXC6-1 was reduced in MSC/KDM6B cells (Fig. 5H–J). Consistently, decreased binding of KDM6B at the promoter regions was associated with increased occurrence of its substrate, H3K27me3, which is a hallmark of gene silencing (Fig. 5K–M). As a control, we could not detect KDM6B occupancy 3 to 4kb downstream of the transcription start sites, and the knock-down of KDM6B did not affect H3K27me3 on that region. Importantly, we found that knock-down of KDM6B decreased H3 acetylation and the recruitment of RNA polymerase II on the promoter of BMP2 and HOXC6-1 (Fig. S5J–M), further suggesting that KDM6B is required for osteogenic differentiation of MSCs. However, despite our repeated efforts, we could not detect KDM6B on the RUNX2 and OSX promoter, suggesting that KDM6B does not directly regulate the RUNX2 and OSX expression.
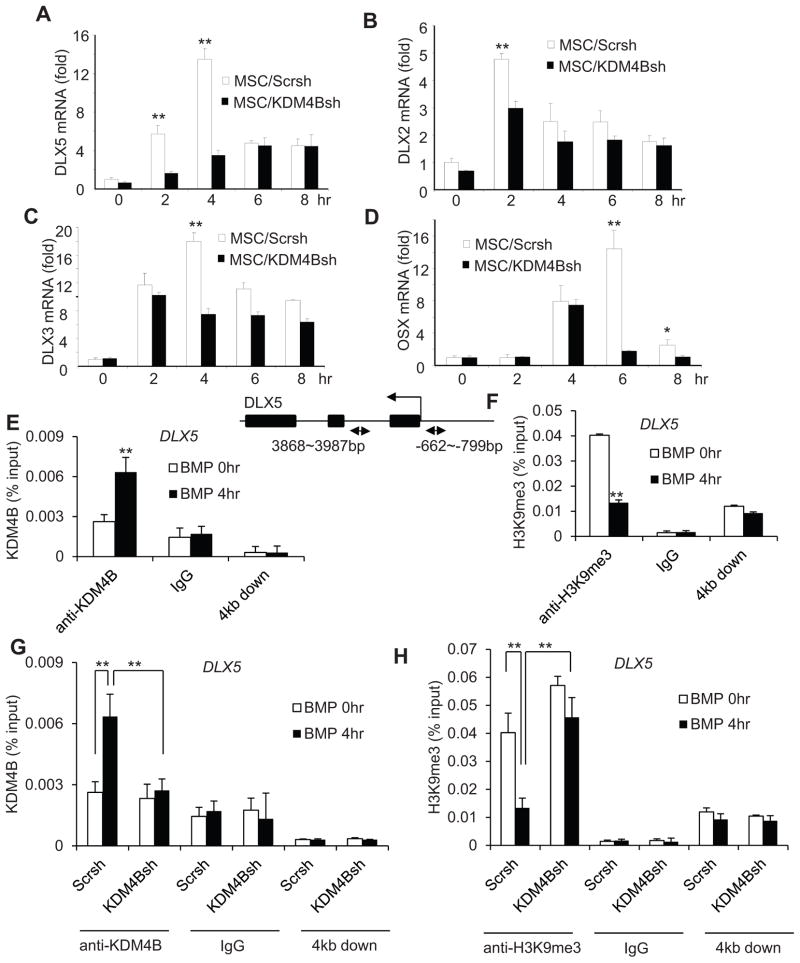
To explore the molecular mechanisms underlying the regulation of osteogenic differentiation by KDM4B, we performed Real-Time RT-PCR analysis to assess the expression of key genes associated with osteogenic differentiation of MSCs. In contrast to KDM6B, we found that the depletion of KDM4B did not affect the basal levels of HOX genes (Fig. S6A and B). Interestingly, we observed that the depletion of KDM4B significantly reduced DLX5 expression that was induced by BMP4/7 (Fig. 6A). The depletion of KDM4B also significantly repressed expression of DLX2 and DLX3 induced by BMP4/7 (Fig. 6B and C). Additionally, we found that KDM4B depletion also inhibited OSX expression, although OSX induction was slower than that that of DLX5 (Fig. 6D). We also examined how KDM4B globally controlled gene expression profile during MSC differentiation using microarray analysis. KDM4B knockdown inhibited the expression of 2448 genes at the basal level and 643 genes induced by BMP4/7. Unlike the results from KDM6B knockdown, the DLX family genes, including DLX2, DLX3, DLX5 and DLX6, were mainly dependent on KDM4B (Fig. S7). The GO analysis also indicated that KDM4B globally controls the expression of genes associated with skeletal development and osteoblast differentiation (Table S4–6). Of note, we found that there were some overlaps between KDM4B- and KDM6B-depenent genes. The demethylation of both H3K27me3 and H3K9me3 might be required for their transcription. However, it also could be due to secondary effects because KDM4B or KDM6B knock-down impaired MSC differentiation through master regulators and subsequently affected common genes associated with osteogenesis.
Figure 6. KDM4B is required for DLX gene expression in MSCs by removal of H3K9me3 marks.
(A, B and C) Knock-down of KDM4B inhibited the expression of DLX5, DLX2 and DLX3 induced by BMP4/7, as determined by Real-time RT-PCR. **P < 0.01. (D) The knock-down of KDM4B inhibited OSX expression induced by BMP4/7. (E) BMP4/7 induced KDM4B binding to the DLX5 promoter. MSCs were treated with BMP4/7 for 4 hr and the DLX5 promoter was ChIP-ed with anti-KDM4B or IgG control. The 4 kb downstream of the transcriptional start site was also ChIPed with anti-KDM4B. (F) BMP4/7 promoted the removal of H3K9me3 in the DLX5 promoter. (G) Knock-down of KDM4B inhibited the recruitment of KDM4B to the DLX5 promoter. (H) Knock-down of KDM4B abolished BMP-induced removal of H3K9me3 marks at the DLX5 promoter. **P < 0.01. See also Figure S6 and S7 and Table S1–3.
DLX family genes play critical roles in osteoblast differentiation (Hassan et al., 2004). Importantly, DLX5 has been found to control OSX expression (Lee et al., 2003). Since DLX5 was dramatically induced by BMP4/7, we examined whether KDM4B directly regulated DLX5 expression by removing H3K9me3 silencing marks using ChIP assays. While KDM4B was induced to bind to the DLX5 promoter (Fig. 6E), H3K9me3 levels at the DLX5 promoter were significantly reduced in MSCs following BMP4/7 treatment, (Fig. 6F). Moreover, the knockdown of KDM4B reduced KDM4B binding to the DLX5 promoter induced by BMP4/7 (Fig. 6G). Consistently, H3K9me3 levels on the DLX5 promoter were not reduced in MSC/KDM4Bsh cells as compared with MSC/Scrsh cells upon BMP4/7 treatment (Fig. 6H). As a control, KDM4B was not present 4kb downstream of the transcription start site, and the knock-down of KDM4B did not affect H3K9me3 on that region (Fig. 6G and H). Since KDM4 family demethylases might help to remove H3K36me3 marks, we also examined the levels of H3K36me3 on the DLX5 promoter. ChIP assays showed that KDM4B knock-down did not affect H3K36me3 levels on the DLX5 promoter (Fig. S6C). Importantly, knock-down of KDM4B decreased H3 acetylation and the recruitment of RNA polymerase II on the DLX5 promoter, further implicating that KDM4B is required for DLX5 gene transcription (Fig. S6D and E).
H3K27me3 and H3K9me3 are elevated in MSCs of osteoporotic bone marrow
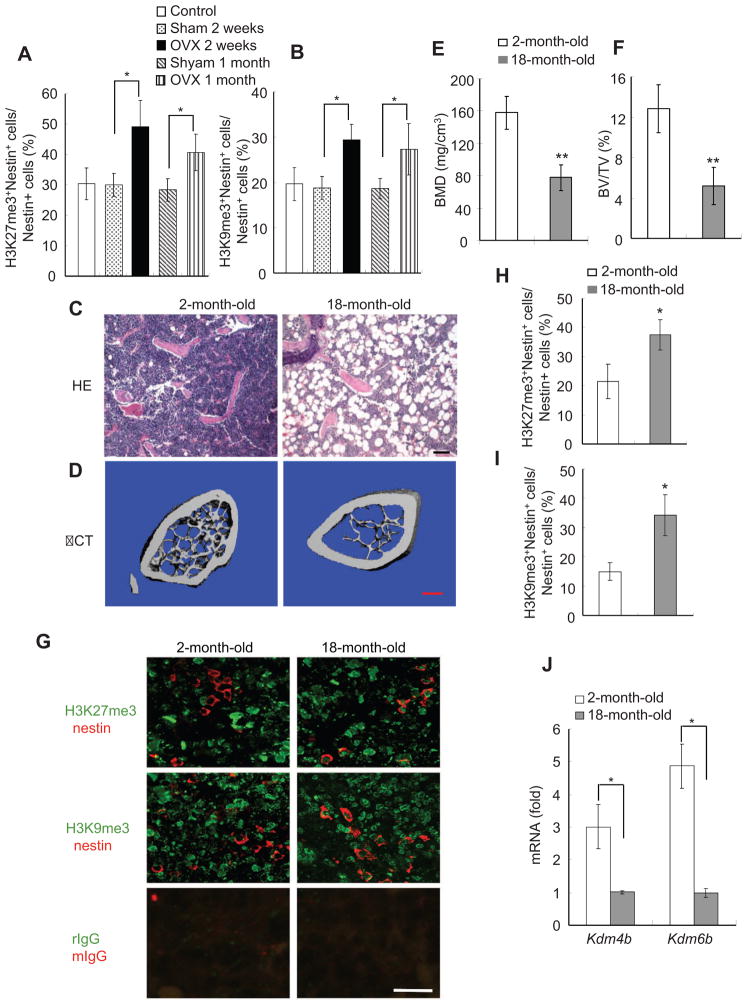
MSCs in bone marrow can differentiate into either osteoblasts or adipocytes, and there is an inverse relationship between osteogenesis and adipogenesis in MSC differentiation (McCauley, 2010; Takada et al., 2007; Takada et al., 2009; Kawai and Rosen, 2010). Excessive adipogenic differentiation has been observed in bone marrow of osteoporotic postmenopausal women (Meunier et al., 1971; Justesen et al., 2001). Our results suggest that the methylation status of H3K9me3 and H3K27me3 may be a critical epigenetic factor that determines MSC fate in bone marrow. Previously, we showed that the adipogenesis was significantly increased in the bone marrow of mice with osteoporosis induced by ovariectomy (OVX) (Chang et al., 2009). Based on our in vitro studies, it is possible that H3K9me3 and H3K27me3 might be abnormally elevated in bone marrow MSCs of OVX-ed mice. To test this possibility in vivo, we performed immunofluorescence double labeling to examine the status of H3K9me3 and H3K27me3 in bone marrow MSCs of OVX-ed mice using anti-nestin antibodies and anti-H3K9me3 or anti-H3K27me3 antibodies. Very recently, elegant studies by Mendez-Ferrer et al (2010) showed that mouse MSCs expressed nestin which can be utilized to directly detect MSCs in mouse bone marrow. The total number of cells positive for both nestin and H3K9me3 or H3K27me3 were counted and compared with the total number of nestin-positive cells. Double immunofluorescence labeling revealed that the percentage of MSCs positive for both nestin and H3K9me3 or H3K27me3 was significantly higher in ovariectomized mice than in sham-operated mice (Fig. 7A–B).
Figure 7. H3K27me3 and H3K9me3 in MSCs are significantly increased in osteoporotic or aging bone marrow of mice.
(A) The percentage of H3K27me3-positive MSCs was increased in bone marrow after OVX. The total number of double nestin- and H3K27me3-positive MSCs from 5 different fields were counted and compared with the total number of nestin-positive MSCs. n = 6; *P<0.05. (B) The percentage of H3K9me3-positive MSCs was increased in bone marrow after OVX. n= 6, *P<0.05. (C) Trabecular bones were decreased and adipose tissues were increased in ageing mice by histological analysis. Scale bar, 100 μm. (D) Representative μCT image of bone loss in aging mice. Scale bar, 0.4 mm. (E) BMD was reduced in ageing mice. n = 8, *P<0.05. (F) Bone volume was reduced in aging mice. *P<0.05. (G) Immunofluorescence double staining of nestin- and H3K27me3- or H3K9me3-positive MSCs in bone marrow from 2- and 18-month-old mice. Nestin was stained red; and H3K27me3 or H3K9me3 was stained green. Yellow arrow, MSCs double positive for nestin and H3K27me3 or H3K9me3; white arrow, nestin-positive and H3K27me3- or H3K9me3-negative MSCs. rIgG, rabbit IgG; mIgG, mouse IgG. Scale bar, 25 μm. (H) The percentage of H3K27me3-positive MSCs was higher in aging mice. *P<0.05. (I) The percentage of H3K9me3-positive MSCs was higher in aging mice. *P<0.05. Scale bar, 25 μm. (J) The expressions of Kdm4b and Kdm6b were reduced in MSCs from aging mice. n = 3, *P<0.05.
In aging mice, adipogenesis is increased in bone marrow whereas osteoblastogenesis is reduced (Muruganandan et al., 2009). To further confirm that H3K9me3 and H3K27me3 are associated with MSC fate commitments, we also examined their status in bone marrow MSCs of aging mice. Compared with younger, 2-month-old mice, we observed that adipose tissues were significantly increased in the bone marrows of 18-month-old mice (Fig. 7C). μCT showed that the trabecular bone was significantly reduced in 18-month-old mice as compared with 2-month-old mice (Fig. 7D–F). Immnofluorescence double labeling revealed that the percentage of MSCs positive for both nestin and H3K9me3 or H3K27me3 was significantly higher in 18 month-old mice than in 2 month-old mice (Fig. 7G–I). Despite repeated efforts, we could not co-stain nestin with KDM4B and KDM6B in bone marrow using commercially-available antibodies. Thus, we isolated MSC from both young and old mice. Real-time RT-PCR revealed that the expression of KDM4B and KDM6B was significantly reduced in MSCs from old mice compared to those from young mice (Fig. 7J).
DISCUSSION
MSCs have multiple differentiation potentials and can be induced to differentiate into osteoblasts or adipocytes. However, both lineages are mutually exclusive (McCauley, 2010). The activation of RUNX2, OSX and DLX5 has been found to inhibit adipogenesis. Conversely, forced expression of PPAR-γ inhibits osteogenic differentiation of MSCs (Takada et al., 2007). The commitment of MSCs to an osteogenic lineage requires coordinated inhibition of differentiation toward other lineages, including the adipogenic lineage. The activation of multiple transcription factors has been identified to be associated with MSC differentiation (Lian et al., 2006; Caplan, 2007; Levi and Longaker, 2011). To differentiate into osteoblastic or adipogenic lineages, in theory, the chromatin of those genes needs to be reprogrammed or modified. In this study, using gene expression profiling, we identified that KDM4B and KDM6B played a critical role in MSC cell fate commitment by removing H3K9me3 and H3K27me3 on different sets of lineage specific genes. The HOX and DLX genes that play essential roles in MSC osteogenic differentiation were found to be regulated by KDM6B and KDM4B through the removal of H3K27me3 and H3K9me3 respectively. More importantly, given the inverse relationship between osteogenic and adipogenic lineage commitment, we found that H3K9me3 and H3K27me3 marks were abnormally elevated in MSCs of osteoporotic bone marrow in vivo. Our results provide the first demonstration that histone demethylation controls the imbalance between osteogenesis and adipogenesis in metabolic bone diseases.
BMPs are potent inducers of osteogenic differentiation of MSC that stimulate master transcription factors (Chen et al., 2004). In this study, we provide novel insights into BMP-stimulated MSC differentiation. The H3K27me3 gene silencing function is essential for maintaining the balance between embryonic stem cell (ESC) self-renewal and differentiation (Lan et al. 2007; Agger et al., 2007). Decrease of H3K27me3 marks is associated with embryonic development and ESC differentiation. It is possible that chromatins of master differentiation genes in MSCs are enriched with repressive H3K27me3 marks prior to lineage commitment. Recently, Wei et al (2011) found that the H3K27 methyltransferase EZH2 played an important role in the inhibition of MSC differentiation. Interestingly, they found that EZH2 directly binds to the promoter of RUNX2. The inactivation of EZH2 by CDK1-dependent phosphorylation reduced the levels of H3K27me3 and promoted osteogenic differentiation of MSC by inducing Runx2. Although the knock-down of KDM6B inhibited RUNX2 expression, we found that KDM6B did not directly bind to the promoter of RUNX2. As in the case of ESCs, we found that KDM6B directly regulated the expression of BMP and HOX genes by removing H3K27me3 marks. Therefore, it is likely that KDM6B indirectly controls RUNX2 expression through BMPs and HOX. Given the fact that KDM6B is also induced by BMPs, our results suggest a possible feedback system and that MSC fate commitment can be regulated at multiple levels by modulating the levels of H3K27me3.
KDM4B has been characterized as being capable of selectively reducing chromosomal H3K9me3 marks to H3K9me1 with the H3K9me2 state remaining unaltered (Fodor et al., 2006; Cloos et al., 2006; Klose et al., 2006; Whetstine et al., 2006). H3K9me3 marks are generally associated with gene repression. The majority of H3K9me3 marks are found in inactive regions of the genome, such as constitutive heterochromatin. Whereas in some reports these marks are enriched in actively transcribed regions of the genome, they are never at the promoter regions of expressed genes (Barski et al., 2007; McEwen and Ferguson-Smith, 2010). Depletion of KDM4B homolog in C. elegans led to a global increase in H3K9me3 levels and activated p53-mediated apoptosis (Whetstine et al., 2006). However, we found that the knock-down of KMD4B did not affect MSC apoptosis. Unlike KDM6B, KDM4B knock-down mainly inhibited the expression of DLX genes. Our results suggest that KDM4B and KDM6B may target different transcription factors by removing H3K9me3 or H3K27me27 marks, respectively. DLX family transcription factors have been found to play a critical role in the induction of RUNX2 and OSX (Acampora et al., 1999; Lee et al., 2003; Hassan et al., 2004). Although knock-down of KDM4B inhibited OSX expression, our ChIP assay was unable to detect KDM4B binding to the OSX promoter, thereby implying an indirect regulation of OSX expression through DLX.
Osteoporosis is one of the most common bone metabolic diseases that are associated with a shift in MSC lineage commitment, and bone loss is accompanied by increased adipose tissues in the bone marrow cavity. Our immunostaining revealed a significant higher percentage of H3K27me3- and H3K9me3-positive MSCs in bone marrow of OVX-ed mice or aging mice. These results suggest that increased H3K27me3 and H3K9me3 levels might play an important role in the development of osteoporosis by influencing determination of MSC fate choices in bone marrow. As a chemically modifiable enzyme, KDM4B and KDM6B could be activated or deactivated to regulate specific lineage decisions of MSCs, thereby holding promising potentials as a therapeutic target for stem cell-mediated regenerative medicine as well as the treatment of human metabolic bone diseases such as osteoporosis.
Experimental Procedures
Cell Culture and Viral Infection
Primary human and mouse MSCs were purchased from Texas A&M Health Science Center College of Medicine Institute for Regenerative Medicine at Scott & White Hospital. Viral packaging and infection was prepared as described previously (Fan et al., 2009; Park et al., 2006). The target sequences for shRNA were: KDM6Bsh1, 5′-GCAGTCGGAAACCGTTCTT-3′; KDM6Bsh2, 5′-GTGGGAACTGAAATGGTAT-3′; KDM4Bsh1, CCTGCCTCTAGGTTCATAA; and KDM4Bsh3, GCTACGAAGTGAACTTCGA.
ALP, Alizarin Red Staining and Oil-Red Staining
To induce MSC differentiation, MSCs were grown in mineralization-inducing media containing 100 μM/ml ascorbic acid, 2 mM β-glycerophosphate and 10 nM dexamethasone. ALP activity assay and Alizarin Red staining were performed as described previously (Chang et al., 2009). To induce adipogenic differentiation, MSCs were grown in adipogenic-inducing media containing 0.5 mM isobutylmethylxanthine, 0.5 μM hydrocortisone, and 60 μM indomethacin (Sigma-Aldrich). Media were changed every 3 days. After 3 weeks of culture in vitro; Oil-Red-O staining was performed to detect the lipid droplets using an OIL-RED-O STAIN KIT according to the manufacturer’s instruction (DBS, Pleasanton, CA, USA).
Real-time Reverse Transcriptase-Polymerase Chain Reaction (Real-time RT-PCR)
Total RNA was isolated from MSCs using Trizol reagents (Invitrogen). 2-μg aliquots of RNAs were synthesized using random hexamers and reverse transcriptase according to the manufacturer’s protocol (Invitrogen). The real-time PCR reactions were performed using the QuantiTect SYBR Green PCR kit (Qiagen) and the Icycler iQ Multi-color Real-time PCR Detection System. The primers for 18S rRNA are: forward, 5′-CGGCTACCAC ATCCAAGGAA-3′; reverse, 5′-GCTGGAATTACCGCGGCT-3′. The primers for OPN are: forward, 5′-ATGATGGCCGAGGTGATAGT-3′; reverse, 5′-ACCATTCAACTCCTCGCTTT-3′. The primers for OCN are: forward, 5′-AGCAAAGGTGCAGCCTTTGT-3′; reverse, 5′-GCGCCTGGGTCTCTTCACT-3′. The primers for ALP are: forward, 5′-GACCTCCTCGGAAGACACTC-3′; reverse: 5′-TGAAGGGCTTCTTGTCTGTG-3′. The primers for BSP are: forward, 5′-CAGGCCACGATATTATCTTTACA; reverse: 5′-CTCCTCTTCTTCCTCCTCCTC-3′. The primers for HOXC6-1 are: forward, 5′-CGGATAAGGGAGTCGAGTAGATCCG-3′; reverse, 5′-TGAGGCATTTCTCGACCTATGG-3′. The primers for DLX5 are: forward, CGCTAGCTCCTACCACCAGT; reverse, GGCTCGGTCACTTCTTTCTC. The primers for DLX2 are: forward, CCTGAGAAGGAGGACCTTGA; Reverse, TTCCGGACTTTCTTTGGCT. The primers for KDM6B are: forward, 5′-CTCAACTTGGGCCTCTTCTC-3″; reverse, 5′-GCCTGTCAGATCCCAGTTCT-3″. The primers for KDM4B are: forward, 5′-CGGGTTCTATCTTTGTTTCTCTCACCCG-3′; reverse, 5′-AAGGAAGCCTCTGGAACACCTG-3′. The primers for PPARγ are: forward, CGAGACCAACAGCTTCTCCTTCTCG; reverse, TTTCAGAAATGCCTTGCAGTGGT. The primers for CD36 are: forward, CGATTAACATAAGTAAAGTTGCCATA; reverse, CGCAGTGACTTTCCCAATAGGAC. The primers for histone demethylase profiling are provided in supplemental data.
Microarray Procedure and Data Analysis
MSCs were treated with 150ng/ml BMP4/7 for 0, 4 and 24hrs respectively. The extraction of total RNA was performed with miRNeasy kit (Qiagen) per manufacturers’ instructions. 1μg of RNA from each sample was hybridized to Affymetrix U133 Plus 2.0 Arrays at the UCLA DNA Microarray Facility. Differential gene expression analysis was performed based on the generated metadata txt file. >1.6 fold change in expression was considered significant. To investigate enriched biological functions associated with KDM4B/KDM6B-dependent genes, GO analysis was performed using the online Database for Annotation, Visualization and Integration Discovery (DAVID) Bioinformatics Resources v6.7. P values were calculated based on hypergeometric distribution. Only significantly overexpressed (p<0.05) GO terms were included. Heatmap was generated via PermutMatrix v1.9.3 (Caraux and Pinloche, 2005) to reflect fold change relative to median expression of each gene in Log2 ratio. Microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE36970.
ChIP Assays
The assay was performed using a ChIP assay kit (Upstate, USA) according to the manufacturer’s protocol as described previously (Fan et al., 2009). Data are expressed as the percentage of input DNA. Antibodies for CHIP assays were purchased from the following commercial sources: polyclonal anti-KDM6B (Abgent, San Diego, CA); polyclonal anti-KDM4B (Millipore); polyclonal anti-H3K27me3 (Millipore); Polyclonal anti-H3K9me3 (Abcam). The primers for BMP2 are: forward, 5′-TGGATCCCACGTCTATGCTA-3′; reverse, 5′-TCAGGGTCTGGCCTCTTATT-3′; 4kb downstream for BMP2: forward, 5′-CAATCATAAGAATTACCTGTTGGG-3′; reverse, 5′-TGGTCGCATTATACTCATATTGG-3′. The primers for BMP4 are: forward: 5′-CAGTTTATGGAAGGCCACCT -3′; reverse, 5′-GTAACTGCTGCCCAAACTGA-3′; 4kb downstream for BMP4: forward, 5′-TCTCTCCGGAATCGTCTCTC-3′; reverse, 5′-AGAAGGTGGTCTGGGAGCTA-3′. The primers for HOXC6-1 are: forward, 5′-CGTGATGGCATTTTCAGCCTATATCACG-3′; reverse, 5′-GAAACAGCGGAAAGTGTGCAG-3′; 3 kb downstream for HOXC6-1: forward, 5′-CATTTATCAACCCTGGCCTT-3′; reverse, 5′-GACCCTCCTCAACACCTCTC-3′. The primers for DLX5 are: forward, 5′-GGATTTCGGGAGTTTCCATT-3′; reverse, 5′-CGGAGTCGCCAGAAATAAAG-3′; 4kb downstream for DLX5: forward, 5′-CCAAGAGCTGCAGTCAGATT-3′; reverse, 5′-CCGTCCTACTCCCTTCACTC-3′.
Transplantation, Immunostaining and μCT Analysis of Mice
The animal experiments were performed in accordance with the approved animal protocol by The University of California Los Angeles. Transplantation in nude mice was performed as previously described (Fan et al., 2009). For quantification of mineralized tissue, 3 pictures of each sample were taken randomly (Olympus 10x NA 0.25, 40x NA 0.6). SPOT 4.0 software (Diagnostic Instruments) was used to measure the area of mineralized tissue versus total area (Chang et al., 2009).
For immunostaining in ovaiectomized mice, we utilized the sections of femurs from previous studies (Chang et al., 2009). For aging studies, femurs were harvested from 2-month-old and 18-month-old mice and fixed in 10% neutral buffered formalin for 24 hrs. μCT scanning was performed as described previously (Chang et al., 2009). After scanning, the specimens were decalcified and sectioned. For immunofluorescence double staining, antigen retrieval was performed as described previously (Chang et al., 2009). The sections were incubated with monoclonal anti-nestin (Lifespan BioSciences, 1:500) and polyclonal anti-H3K9me3 (Abcam, 1:500) or polyclonal anti-H3K27me3 (Millipore, 1:500) at 4°C overnight. After washing with PBS solution for 3 times, the sections were incubated with appropriate Cy2 (red) and Cy3 (green) conjugated secondary antibodies (Jackson ImmunoResearch, 1:100) in darkness at room temperature for 1 hour. The sections were imaged with a confocal laser scanning microscope (Leica TCS-SP inverted), and the Leica Confocal Software (LCS version 2.61 Build 1537).
Supplementary Material
Highlights.
Histone demethmylase KDM4B promotes osteogenic differentiation of human MSCs
Histone demethmylase KDM6B promotes osteogenic differentiation of human MSCs
H3K27me3- and H3K9me3-positive MSCs of bone marrow were elevated in osteoporosis
KDM4B and KDM6B may present as novel therapeutic targets in metabolic bone diseases
Acknowledgments
We thank Dr. Steve Smale for critical reading the manuscript and suggestions and Dr. Tori Xiao for technical supports. We thank Dr. Paul Khavari for providing the catalytically mutant KDM6B construct. Confocal laser scanning microscopy was performed at CNSI Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA, supported with funding from NIH-NCRR shared resources grant (CJX1-443835-WS-29646) and NSF Major Research Instrumentation grant (CHE-0722519). This study was support by National Institute of Dental and Craniofacial Research grants DE019412 and DE16513.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved inHOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–68. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–6. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–7. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AI. Adult mesenchymalstem cells for tissue engineering versus regenerative medicine. J Cell Physiol. 2007;213:341–7. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- Caraux G, Pinloche S. Permutmatrix: A Graphical Environment to Arrange Gene Expression Profiles in Optimal Linear Order. Bioinformatics. 2005;21:1280–81. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- Chang J, Wang Z, Tang E, Fan Z, McCauley L, Franceschi R, Guan K, Krebsbach PH, Wang CY. Inhibition of osteoblastic bone formation by nuclear factor-kappaB. Nat Med. 2009;15:682–9. doi: 10.1038/nm.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–11. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–94. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Fan Z, Yamaza T, Lee JS, Yu J, Wang S, Fan G, Shi S, Wang CY. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol. 2009;11:1002–9. doi: 10.1038/ncb1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. AnnuRev Cell Dev Biol. 2005;21:659–93. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–62. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronthos S, Graves SE, Ohta S, Simmons PJ. The STRO-1+ fraction of adult human bone marrow contains the osteogenic precursors. Blood. 1994;84:4164–73. [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: Temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–52. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera -Feliciano J, Mooney DJ. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–26. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Justesen J, Stenderup K, Ebbesen EN, Mosekilde L, Steiniche T, Kassem M. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology. 2001;2:165–171. doi: 10.1023/a:1011513223894. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–48. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- Kawai M, Rosen CJ. PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nat Rev Endocrinol. 2010;6:629–36. doi: 10.1038/nrendo.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Kolf CM, Cho E, Tuan RS. Mesenchymal Stromal Cells. Biology of Adult Mesenchymal Stem Cells: Regulation of Niche, Self-Renewal And Differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg HM. Gs signaling in osteoblasts and hematopoietic stem cells. Ann N Y Acad Sci. 2010;1192:327–9. doi: 10.1111/j.1749-6632.2009.05251.x. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–94. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by DLX5 but is independent of Runx2. Biochem Biophys Res Commun. 2003;309:689–694. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- Levi B, Longaker MT. Concise review: adipose-derived stromal cells for skeletal regenerative medicine. Stem Cells. 2011;29:576–82. doi: 10.1002/stem.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- McCauley LK. c-Maf and you won’t see fat. J Clin Invest. 2010;120:3440–2. doi: 10.1172/JCI44786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen KR, Ferguson-Smith AC. Distinguishing epigenetic marks of developmental and imprinting regulation. Epigenetics & Chromatin. 2010;3:2. doi: 10.1186/1756-8935-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici D, Shore EM, Lounev VY, Kaplan FS, Kalluri R, Olsen BR. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16:1400–6. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–34. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier P, Aaron J, Edouard C, Vignon G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. Aquantitative study of 84 iliac bone biopsies. Clin Orthop Relat Res. 1971;80:147–154. doi: 10.1097/00003086-197110000-00021. [DOI] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010a;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Weinmann AS. Molecular mechanisms by which T-bet regulates T-helper cell commitment. Immunol Rev. 2010b;238:233–146. doi: 10.1111/j.1600-065X.2010.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4 methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruganandan S, Roman AA, Sinal CJ. Adipocyte differentiation of bone marrow- derived mesenchymal stem cells: cross talk with the osteoblastogenic program. Cell Mol Life Sci. 2009;66:236–253. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BK, Zhang H, Zeng Q, Dai J, Keller ET, Giordano T, Gu K, Shah V, Pei L, Zarbo RJ, McCauley L, Shi S, Chen S, Wang CY. NF-κB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nature Medicine. Nature Medicine. 2006;13:62–69. doi: 10.1038/nm1519. [DOI] [PubMed] [Google Scholar]
- Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cell. 2007;25:2896–902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- Phimphilai M, Zhao Z, Boules H, Roca H, Franceschi RT. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J Bone Miner Res. 2006;21:637–44. doi: 10.1359/JBMR.060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Zou C, Shao P, Schaack J, Johnson PF, Shao J. Transcriptional regulation of fatty acid translocase/CD36 expression by CCAAT/enhancer-binding protein alpha. J Biol Chem. 2008;283:8788–95. doi: 10.1074/jbc.M800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, Bianco P. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–36. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–70. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–33. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Imbalzano AN, Montecino M, Zaidi SK, Lian JB, Nickerson JA, Stein JL. Architectural genetic and epigenetic control of regulatory networks: compartmentalizing machinery for transcription and chromatinremodeling in nuclear microenvironments. Crit Rev Eukaryot Gene Expr. 2010;20:149–55. doi: 10.1615/critreveukargeneexpr.v20.i2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada I, Kouzmenko AP, Kato S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol. 2009;5:442–7. doi: 10.1038/nrrheum.2009.137. [DOI] [PubMed] [Google Scholar]
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, Takezawa S, Yogiashi Y, Kitagawa H, Yamada G, Takada S, Minami Y, Shibuya H, Matsumoto K, Kato S. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–85. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z, Zhao L, Nagy TR, Peng X, Hu J, Feng X, Van Hul W, Wan M, Cao X. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757–65. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL. Stem cells and osteoporosis therapy. Cell Stem Cell. 2010;7:553–4. doi: 10.1016/j.stem.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Undale AH, Westendorf JJ, Yaszemski MJ, Khosla S. Mesenchymal stem cells for bone repair and metabolic bone diseases. Mayo Clin Proc. 2009;84:893–902. doi: 10.4065/84.10.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker SA, McNulty CL, Durston AJ. The initiation of Hox gene expression in Xenopus laevis is controlled by Brachyury and BMP-4. Dev Biol. 2004;266:123–137. doi: 10.1016/j.ydbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of humanmesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.