Abstract
Druggability assessment of a target protein has emerged in recent years as an important concept in hit-to-lead optimization. A reliable and physically relevant measure of druggability would allow informed decisions on the risk of investing in a particular target. Here, we define “druggability” as a quantitative estimate of binding sites and affinities for a potential drug acting on a specific protein target. In the present study, we describe a new methodology that successfully predicts the druggability and maximal binding affinity for a series of challenging targets, including those that function through allosteric mechanisms. Two distinguishing features of the methodology are (i) simulation of the binding dynamics of a diversity of probe molecules selected on the basis of an analysis of approved drugs and (ii) identification of druggable sites and estimation of corresponding binding affinities on the basis of an evaluation of the geometry and energetics of bound probe clusters. The use of the methodology for a variety of targets such as murine double mutant-2, protein tyrosine phosphatase 1B (PTP1B), lymphocyte function-associated antigen 1, vertebrate kinesin-5 (Eg5), and p38 mitogen-activated protein kinase provides examples for which the method correctly captures the location and binding affinities of known drugs. It also provides insights into novel druggable sites and the target’s structural changes that would accommodate, if not promote and stabilize, drug binding. Notably, the ability to identify high affinity spots even in challenging cases such as PTP1B or Eg5 shows promise as a rational tool for assessing the druggability of protein targets and identifying allosteric or novel sites for drug binding.
Introduction
Recent genome-wide analyses suggest that 10% of the human genome is druggable, and among druggable proteins about half correspond to disease-causing genes.1 Assessing the druggability of the target protein at a relatively early stage in the drug discovery process is now becoming common practice, with the realization that the nondruggability of a target is a major obstacle in advancing a small molecule from hit to lead.2,3 When evaluating druggability, one often wants to determine the likelihood of finding an inhibitor for the protein of interest and make a quantitative estimate of the inhibitor molecular size and affinity to help assess the risk of specializing on those targets.
Two experimental methods have been particularly useful in making such assessments: (i) NMR screening of libraries of small molecules against target proteins4 to identify binding sites and corresponding achievable affinities and (ii) multiple-solvent crystal structure (MSCS) determination,5 where a target protein structure is resolved in complex with small organic molecules used as a probe to infer potential druggable sites. Both approaches are based on the premise that probe binding sites and frequencies correlate with drug-binding sites and affinities. Hajduk and co-workers demonstrated that sites that bind a relatively large fraction of fragments (e.g., hit rates of 0.2% or higher for thousands of screened small molecules) indeed coincide with known high affinity (KD < 300 nM) sites. On the basis of these observations, they proposed a metric, the druggability index, for quantifying the druggability of target proteins. The druggability index is obtained by optimal assignment of linear and logarithmic weights to structure-based binding site descriptors to match the hit rates observed in NMR-based fragment screening.4 The resulting model was shown to distinguish the correct binding pocket as the most druggable site in 71% of test proteins.
The physical basis of experimentally observed druggability behavior has been further established with the help of theoretical and computational studies performed in recent years.6−8 Cheng et al. proposed a simpler model that involved two dominant descriptors, nonpolar surface area and pocket curvature, to estimate the maximal binding affinity achievable by a drug-like molecule, which successfully explained the behavior of a series of targets.6 Recent in silico screening of a library of fragment-like molecules and organic probe molecules against known binding sites also showed that computations successfully distinguish druggable and nondruggable targets.9 In particular, the FTMap approach based on fast Fourier transform correlation methods, combined with clustering methods and atomic force fields, was found to yield results in good agreement with MSCS experiments,10,11 in support of the utility of computations for identifying druggable sites.
Following significant progress in the field, attention has been drawn to the impact of protein flexibility in binding site identification and druggability calculations. It has become clear that experimental data are practically irreproducible when significant dynamics and conformational changes occur in binding sites.12,13 Examination of the conformational space accessible to Bcl-xL and the β-adrenergic receptor by molecular dynamics (MD) simulations has exemplified the implications of protein dynamics. Simulations of Bcl-xL revealed that the protein undergoes a change from a seemingly nondruggable conformation to a druggable one, yielding inhibitor-binding affinities more consistent with experimental data. Similarly, Ivetac and McCammon used MD simulations to generate an ensemble of β-adrenergic receptor structures for FTMap calculations to identify potential allosteric and druggable pockets,13 which could not be identified by calculations based on the crystal structure of the protein alone. Many other studies point to the significance of considering protein dynamics, albeit at low resolution (e.g., coarse-grained normal-mode analysis), in computational predictions of inhibitor-binding mechanisms.14−21 On the other hand, the need for protein conformational sampling remains a debated issue when the proteins exhibit changes limited to side-chain rearrangements in their binding site.22−24
Recently, methods based on MD simulations in water and organic molecule mixtures were introduced for binding site identification.25−27 Guvench and MacKerell simulated the dynamics of target proteins in propane, benzene, and a water mixture to generate a map of protein binding preferences.25,26 Results were evaluated in a qualitative manner by visualization of probe binding probability maps. Seco and co-workers, on the other hand, simulated proteins in a mixed solvent box of water and isopropanol.27 On the basis of previous observations that small organic molecules tend to bind druggable sites,5,28,29 they also developed a method to convert isopropanol binding propensities into achievable binding affinities of drug-like molecules. The exclusive use of a single probe that contains hydrophobic and polar groups (or other purely hydrophobic probes) limits the applicability of these methods. Additionally, the atomic contributions to binding free energy require careful evaluation to avoid redundant inclusion of interdependent interactions. The inclusion of all of the four heavy atoms of isopropanol molecules in contact with the protein as separate entities, for example, led to an overestimation of binding affinities, which were then rescaled by applying a correction factor.27
In the present study, we propose a novel methodology using a probe set with diverse physicochemical properties (Table 1) and a binding free energy estimation methodology with simplified assumptions. We developed an automated algorithm for analyzing MD trajectories of target molecules generated in the presence of diverse probe molecules to make druggability assessments. Thorough examination and comparative analysis of the results for five test proteins’ (a total of six different cases) probe-binding dynamics in the presence of two different probe/water compositions (shortly referred to as isopropanol-only and probe-mixture, both in an aqueous medium; presented in Table 2) lends support to the utility of the methodology. We note in particular the accurate prediction of experimentally observed binding sites and affinities for challenging targets such as protein tyrosine phosphatase 1B (PTP1B) and Eg5 kinesin.
Table 1. Number of Occurrences of Small Organic Fragments As Substructures in Drug Moleculesa.
| fragment name | approvedb | all drugsc |
|---|---|---|
| isobutane | 1022 (76%)d | 2697 (42%) |
| isopropanol | 768 (57%) | 3559 (55%) |
| isopropylamine (IPAM) | 337 (25%) | 1483 (23%) |
| acetic acid | 284 (21%) | 1862 (29%) |
| acetamide | 280 (21%) | 1722 (27%) |
| acetone | 239 (17%) | 691 (11%) |
| urea | 61 (5%) | 211 (3%) |
| DMSO | 37 (2%) | 150 (2%) |
Fragments used as probe molecules are highlighted in boldface.
A total of 1341 approved small-molecule drugs are included, with a molecular weight less than 800 Da.
A total of 6435 approved or experimental drugs are included, with a molecular weight less than 800 Da.
Percentage of drugs that contain the fragment as substructures.
Table 2. Comparison of Predicted Druggable Sites and Binding Affinities to Those Observed in Experiments.
| experimental data |
computational data |
||||
|---|---|---|---|---|---|
| target | binding site | best Kda | iso-propanolb | probe mixturec | protein hotspotse |
| MDM2 | p53 | 0.6 nM83 | 0.4–1.0 nM | 0.3–2.0 nM | L54, I61, M62, Y67, V93, H96 |
| 2nd site | na | 0.1 μM | nd | F86, E95, K98 | |
| MDM2 NMR | p53 | 0.6 nM84 | 0.1–1 nM | L54, I61, M62, Y67, V93, H96 | |
| PTP1B | pTyr | 2.2 nM41 | nd | 0.3–0.9 nM | R24, Y46, R254, K120, R221, S216 |
| allostericd | 8 μM43 | 2.8 μM | 9.5–18 μM | L192, F196, F280 | |
| IRK interface | na | nd | 43 nM | E4, F7, E8, D11, L272, E276 | |
| 4th site | na | 4 nM | 200 nM | V113, M114, L119, A122, R112, R156, H175 | |
| LFA-1 | induced | 18.3 nM47 | 0.5–0.8 nM | 0.03–0.5 nM | L132, Y166, V233, I235, I259, Y257 |
| Eg5 | allostericd | 0.2 nM51 | 27 nM | 0.3 nM | E116, I136, P137, Y211, L214 |
| tubulin site | na | 2 nM | 0.2 nM | I272, L293, S348, Y352 | |
| 3rd site | na | 600 nM | 47 nM | Q20, M70, V85 | |
| p38 | ATP | 0.05 nM | 1–2 nM | 0.01–0.12 nM | A51, D71, L74, L75, L108, M109 |
| MK2 site | na | 2–3 nM | 2–3 nM | T218, R220, L222, L238, V273 | |
| MAPK insert | na | 13–90 nM | 5–210 nM | I229 L232 Y258, I259 | |
Kd or Ki of the most potent inhibitor.
Predictions based on isopropanol–water environment.
Predictions based on probe mixture–water environment with 10% of isopropanols replaced with acetamide, acetate, and IPAM.
IC50 values of best inhibitors known to date.
Hotspot residues found here to interact with high-affinity probe interaction spots (LE > 0.4 kcal/mol) at the binding site. nd, not determined; na, not available.
Results and Discussion
Probe Molecules
Small organic molecules were selected as probes on the basis of the frequency of occurrence of their functional fragments, or substructures, in FDA-approved and/or experimental drug molecules listed in DrugBank.30 Isobutane, isopropylamine (IPAM), acetic acid, and acetamide, as well as isopropanol groups, were found each to take part in at least 20% of drug molecules (Table 1). Among these, we have selected acetamide (representative of polar molecules) as well acetic acid and IPAM (representative of molecules that are charged at physiological pH) as probes, to avoid the potential problem of aggregation observed in the simulations of aliphatic groups, e.g., isobutane.25 For detailed structure and energy parameters of probe molecules, see the Supporting Information (SI) text and Table S1.
Overview of the Method of Approach
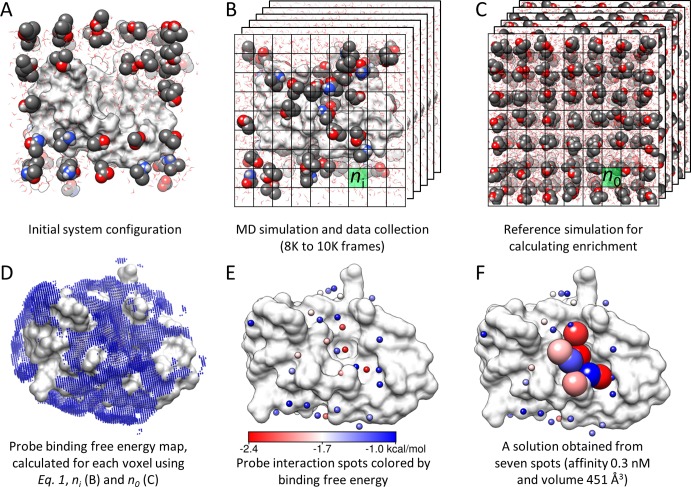
Explicit inclusions of both desolvation effects and the coupled dynamics of water/probe molecules and the protein are key features of MD-based druggability simulations.25,27 We describe our methodology in detail in the Methods section and the SI. We outline here the main steps, illustrated in Figure 1. We simulated the target proteins in probe-mixture/water at a fixed ratio of 20 water molecules per probe molecule (Figure 1A). This roughly corresponds to 20% probes by volume or a ∼2.3 M concentration of probes and is a reasonable ratio to saturate druggable sites and detect probe binding events.25,27 Assuming that probe molecules reach Boltzmann distribution within nanoseconds, runs of 32–40 ns provide a reference state for estimating the binding free energy. Using a grid-based approach27 and the inverse Boltzmann relation, the binding free energy at voxel i is
| 1 |
Here, ni/n0 is the ratio of the observed density of probes (ni in Figure 1B) to the expected density (or the reference state; n0 in Figure 1C), also referred to as enrichment, R is the gas constant, and T is the absolute temperature (K). It should be noted that despite its broad use in docking and folding studies, the physical basis of inverse Boltzmann relation for relative free energy calculations is debated.31 Concerns usually arise in cases where interacting particles are not independent, such as covalently linked amino acids in a protein.32 However, situations where it is clearly applicable for relative free energy calculations include equilibrium systems of independent particles.31 Thus, we accounted for binding events by considering probe molecules as whole independent particles. The resulting relative binding free energy map (Figure 1D) is refined to identify interaction spots (Figure 1E), each representing a probe molecule. Interaction spots within close proximity are clustered into distinct binding sites. Maximal achievable binding affinity is calculated for those sites composed of seven or more spots, by merging seven or eight of them (28 or 32 heavy atoms) located within 5.5 to 6.5 Å of each other in a deterministic way (see the Methods section). The location of such interaction spots is proposed to be a potential druggable site provided that the corresponding maximal affinity is 10 μM (with a binding free energy of −6.86 kcal/mol or stronger). Figure 1F shows such a druggable site indicated with large, color-coded interaction spots. Maximal affinity predictions for all druggable sites were compared with experimentally determined affinity data, as summarized in Table 2. See the Methods section and SI for more details.
Figure 1.
Overview of methodology. (A) The druggability simulation box is prepared by immersing the target protein in a box of water and probe molecules. (B) After the superposition of frames onto the X-ray structure using Cα atom positions, a grid representation is used to measure the probe density (ni). (C) A protein-free system is simulated to calculate the expected probe density (n0) used in eq 1. (D) The binding free energy for each voxel is calculated using eq 1. Note that only the outer layer (weaker) interactions are visible in the map. (E) Interaction spots (small spheres) are identified by removing the voxels that overlap with the lower energy voxels. The energy scale in this panel holds for panels D and F as well. (F) Proximal spots are merged to predict maximal affinity. Interaction spots that are in a druggable site are shown as larger spheres color-coded by the corresponding interaction energies with the target. Molecular graphics in this study are generated using Chimera.79
Target Proteins
We selected five test proteins: (i, ii) murine double mutant-2 (MDM2; truncated N-terminal domain and solution structure of N-terminal domain in which C- and N-terminal tails are present), (iii) PTP1B, (iv) lymphocyte function-associated antigen 1 (LFA-1), (v) kinesin Eg5, and (vi) p38 mitogen-activated protein (MAP) kinase (MAPK). These targets offer a set of binding sites with diverse shapes and physicochemical and dynamic properties. Druggability simulations were performed with two types of solvent mixtures: (i) isopropanol-only and (ii) an isopropanol, acetamide, acetate, and IPAM mixture, both in water, shortly referred to as “probe-mixture/water”, with varying mole fractions of probe molecules. See Table 2 for the description of different runs for the six cases, summing up to a cumulative simulation time of 1.3 μs. In the following, we present detailed results for each case.
MDM2 is a negative feedback regulator of the p53 tumor suppressor33 and features a protein–protein interaction site. Due to its small size and the availability of extensive experimental data,34 we used the 109-residue truncated N-terminal domain of MDM2 for method development and optimization. We used as input the structure resolved by Kussie et al.35 We performed 11 MD runs with different probe compositions and input parameters (Table S2), summing up to a total of 0.4 μs run time.
All druggability simulations invariably yielded the p53 interaction site of MDM2 as the most druggable site, with maximal affinities being in the range 0.3 to 3 nM. Figure 2A displays the interaction spots (spheres) distinguished by their high binding energy in probe mixture simulations, color-coded by their interaction strengths with MDM2. Seven of them, shown as larger spheres, are clustered together in space, indicating a druggable site. Panel B compares the positions of the interaction spots with those of the p53 side chains at the interface of the known MDM2-p53 complex. Notably, the interaction spots that make the largest contribution to binding free energy (colored red) overlap with the side-chains of p53 residues F19, W23, and L26, which are mimicked by many MDM2 inhibitors.36 MDM2 residues that are in contact with the probes at this high affinity site are L54, I61, M62, Y67, V93, and H96, consistent with the hotspots previously identified by computational alanine scanning methodology.37
Figure 2.

p53 binding pocket confirmed as the single most druggable site of MDM2. (A) The MDM2 structure35 (1YCR) in ribbon representation and interaction spots from probe mixture simulation 1–5 (Table S2) as spheres are shown. Coloring is based on binding free energy. The p53 pocket, identified as a druggable binding site, is indicated by the large spheres. Some interaction spots with a binding free energy greater than −1.5 kcal/mol are not displayed for the clarity of the figure. Coordinates of interaction spots and the protein are provided as Supporting Information. (B) A closeup view of the p53 binding pocket. p53 is colored yellow, and its labels are italicized.35 The interaction spots contributing to the predicted maximal affinity are shown. We note their overlap with the p53 hotspot residues F19, W23, and L26.
The maximal binding affinity predicted for MDM2 exhibited minor dependence on probe composition. Isopropanol/water simulations (runs 1–1 to 1–3 in Table S2) yielded a maximal achievable affinity of 0.4–1.0 nM for this site (Figure 2); simulations in a mixture of isopropanol (70% or 40%), acetamide (10% or 20%), acetate (10% or 20%), and IPAM (10% or 20%) in water (runs 1–4 to 1–6) led to 0.3–2.0 nM. These values are in excellent agreement with experiments: the affinity of the best known MDM2 inhibitor is 0.6 nM.38
We note that isopropanol molecules contributed by 80% or more to the predicted maximal binding affinity in the runs performed with the probe mixture. The maximal affinity evaluated in earlier work27 with the same probe was 0.02 nM. This affinity, about 1 order of magnitude higher than current predictions (and experimental data), has been verified by independent simulations (run 1–9, Table S2) to result from the previous assignment of partial charges, which led to an underestimation of the polarization of isopropanol. This, in turn, gave rise to more favorable interactions with the hydrophobic binding pocket of MDM2. We note that a second binding pocket was observed in earlier work,27 formed by the rearrangements of F86, E95, K98, and Met102 side chains. The corresponding maximal affinity (0.05 μM)27 is comparable to that (0.1 μM) obtained in our runs 1–1 and 1–9. To further assess the effect of minor changes in force field parameters, we simulated MDM2 using the CHARMM general force field39 parameters for isopropanol (runs 1–7 and 1–8). Minor differences in atom types and partial charge distribution were observed (Table S1). However, the predicted maximal affinities remained practically unchanged, providing support to the robustness of our quantitative results with respect to variations in force field parameters or partial charge distributions within plausible limits.
The MDM2 solution structure, resolved by NMR spectroscopy,40 features 25 and 10 residue N- and C-terminal tails, respectively. By visual inspection of 24 models in this structure, we chose the second model in which the p53 binding site is the most occluded by the N-terminal tail. We performed two druggability simulations and one probe-free simulation. In the druggability simulations (runs 2–1 and 2–2 in Table S2), the p53 site was identified as a druggable site with maximal affinity ranging from 0.4 to 1.0 nM (Figure 3A), comparable to those from the simulations of a truncated structure. Locations of p53 hotspot residues W23 and F19 were also identified by the probe molecules (Figure 3B). We also compared the dynamics of the solution structure from druggability simulations to that from a probe-free simulation (run 2–3). In the absence of probe molecules, the N-terminal tail remained stretched over the p53 binding domain occluding the binding pocket (Figure 3D). On the other hand, the binding pocket was accessible to probe molecules in the druggability simulations (Figure 3C), demonstrating the adaptation in the protein structure to the presence of ligands.
Figure 3.
Druggability assessment of the p53 binding site when occluded by the MDM2 N-terminal tail. The MDM2 NMR model40 (1Z1M model 2), in ribbon representation, and interaction spots from probe mixture simulation 2–1 (Table S2), as spheres, are shown in panel A. The coloring scheme is the same as in Figure 2. The p53 pocket identified as druggable is indicated by the large spheres. (B) A closeup view of the p53 binding pocket from panel A is shown, with similar coloring as in Figure 2B. We note the overlap of p53 hotspot residues F19 and W23 with probe interaction spots. Panels C and D show 25 evenly spaced snapshots from druggability simulation 2–1 and probe-free simulation 2–3, respectively. In the presence of probe molecules, the p53 pocket was more accessible, whereas in their absence, the pocket remained occluded by the N-terminal tail. Coordinate files for interaction spots and simulation snapshots are provided in the Supporting Information.
PTP1B is a challenging target due to the highly basic character of its catalytic active site. Known inhibitors of this target are negatively charged. Consequently, it is not possible to identify a probe interaction spot at, or in the vicinity of, the catalytic cavity when only isopropanols are used as a probe (PTP1B run 3–1; Table S2).27 The probe-mixture/water simulations indeed indicate many acetate interaction spots at the catalytic cavity (Figure 4A) in addition to other probes, mostly isopropanols, in the vicinity. This analysis clearly highlights the need to adopt probes other than isopropanol for assessing the druggability of targets that contain polar/charged sites. Additionally, three more clusters that suggest additional druggable sites were found (enclosed in ellipses), one of which coincides with the allosteric site of PTP1B.
Figure 4.

Four druggable sites identified for PTP1B. (A) PTP1B structure80 (PDB ID: 1PH0) ribbon diagram and four druggable sites (indicated by color-coded, large interaction spots) found in PTP1B probe-mixture simulations. In addition to the catalytic site (rectangular box, magnified in panel B) and the allosteric site, two other potentially druggable sites (labeled as IRK interface and 4th site) were identified. (B) Closeup view of the catalytic site. An active site inhibitor (compound 6 in previous work80) with two negative charges is shown in stick representation. The interaction spots predicted to make the largest contribution to binding affinity overlap with the charged groups of the inhibitor. Acetate interaction spots are indicated by a negative sign.
The interaction spots that exhibit the highest binding affinity at the PTP1B catalytic site are coordinated by positively charged amino acids (K120, R221), along with polar side chains (Y46, S216) and backbone amides (Figure 4B). We predicted maximal affinity to range from 0.3 to 0.9 nM (runs 3–2 and 3–3), by allowing selected interaction spots to have up to 2 absolute total charge. When no more than one integral charge was allowed, the affinity dropped to 10.5 nM. These predictions are comparable to the experimental data reported for the best known catalytic site inhibitor, which is 2.2 nM.41 Our analysis shows that a major contribution to affinity is made by the acetate probes. Notably, acetate interaction spots were observed in the second aryl-phosphate binding cavity (coordinated by R24 and R254, Figure 4B), known to also bind catalytic site inhibitors.42 The computed binding affinity (−1.7 kcal/mol) at this site was weaker than that of acetates at the catalytic cavity (from −3.1 to −2.9 kcal/mol).
As to the PTP1B allosteric site (Figure 4A), the maximal affinity deduced from isopropanol-only simulation (2.8 μM; run 3–1) was comparable to earlier prediction27 (0.5 μM), whereas the probe-mixture/water simulations yielded an achievable affinity of 9.5 to 17.6 μM (runs 3–2 and 3–3). The best known inhibitor for this site has an IC50 of 8 μM.43 The interaction spots at this site are mostly contributed by isopropanols, with the highest affinity spots making contacts with L192, F196, and F280 side chains.
In addition to the catalytic and allosteric sites, we identified two more druggable sites on PTP1B. The first, at the insulin receptor kinase (IRK) interface,44 yields an achievable affinity of 43 nM. It mainly consists of favorable isopropanol and IPAM interactions with E4, F7, E8, D11, L272, and E276, resulting in positive total charge. Seco et al. reported a druggable site with an affinity of 180 nM in the vicinity of V184, P187, and R268 at the IRK interface,27 which is not reproduced in our isopropanol-only simulations. The other site (labeled fourth site in Figure 4A) was detected in both probe-mixture (4 nM) and isopropanol (200 nM) runs. Isopropanols favorably interacted with V113, M114, L119, and A122 and acetate molecules with R112, R156, and H175.
LFA-1 is a leukocyte cell surface glycoprotein that promotes intercellular adhesion and binds intercellular adhesion molecule 1.45 In this case, the binding site of interest is an allosteric pocket. We have used the ligand-free structure of LFA-146 in our simulations. In this structure, the allosteric pocket is occluded by K287 side-chain (Figure 5A), and the entry to the pocket is partly obstructed by a salt bridge between E284 and K305 (Figure 5B). Rearrangement of these side chains is essential to reaching the allosteric site by probe molecules. Hence, LFA-1 is one of the targets that substantiate the utility of MD-based druggability assessment.
Figure 5.
The only druggable site identified for LFA-1: a partially obstructed allosteric pocket. (A) LFA-1 structure46 (PDB ID: 1ZOP) in ribbon representation and probe interaction spots from the probe-mixture simulation as spheres are shown. The box indicates the location of the allosteric inhibitor binding pocket found to be the only druggable site. Larger spheres indicate interaction spots used to predict maximal affinity. (B) Close up view of the allosteric binding pocket, with the larger spots from panel A, superposed on the structure of a bound antagonist of LFA-1 (black sticks; compound 1 in Crump et al.47) in the bound LFA-1 structure (yellow; PDB ID: 1RD4).47 Bound (yellow) and unbound (green) conformations of E284, K287, E301, and K305 side chains are also compared. (C) Snapshots at 2 ns intervals from LFA-1 simulation 4–3 (rainbow, from blue to red) superposed onto the inhibitor-bound LFA-1 structure (gray), with the inhibitor shown in pink ball-and-stick. Probe molecules stabilize the C-terminal helix in bound form, break the salt bridge between K305 and E284, and induce the formation of the K287-E301 salt bridge. (D) Snapshots from probe free LFA-1 simulation 4–5 (coloring scheme same as panel C), showing the collapse of the C-terminal helix onto the allosteric binding pocket.
Our analysis found the allosteric site of LFA-1 as the only druggable site with a maximal achievable affinity in the range 0.8 to 0.03 nM irrespective of the probe type (Tables 2 and S2; runs 4–1 to 4–4). Like MDM2, most of the interaction spots are populated by isopropanols, consistent with the hydrophobic character of the LFA-1 allosteric site. Residues interacting with high affinity spots are L132, Y166, V233, I235, I259, and Y257. Previous simulations predicted a maximal affinity of 27 nM,27 and indeed the best inhibitor with known Kd reported at the time that binds this site has an affinity of 18.3 nM (it inhibits LFA-1/ICAM interaction with an IC50 of 10.3 nM).47 Our simulations suggest, however, that a higher affinity binding is achievable at this site. We searched the binding databases48 for better inhibitors of LFA-1 based on reported IC50 values. We found that a compound with an IC50 of 0.35 nM has been identified as a validated hit in a study of a series of meta-aniline based compounds (compound 20).49 Key to the stabilization of the ligand was the formation of a salt bridge between E301 and K287, allowing for amino-aromatic interaction between the K287 side chain and the ligand (Figure 5B and C).
Kinesin Eg5
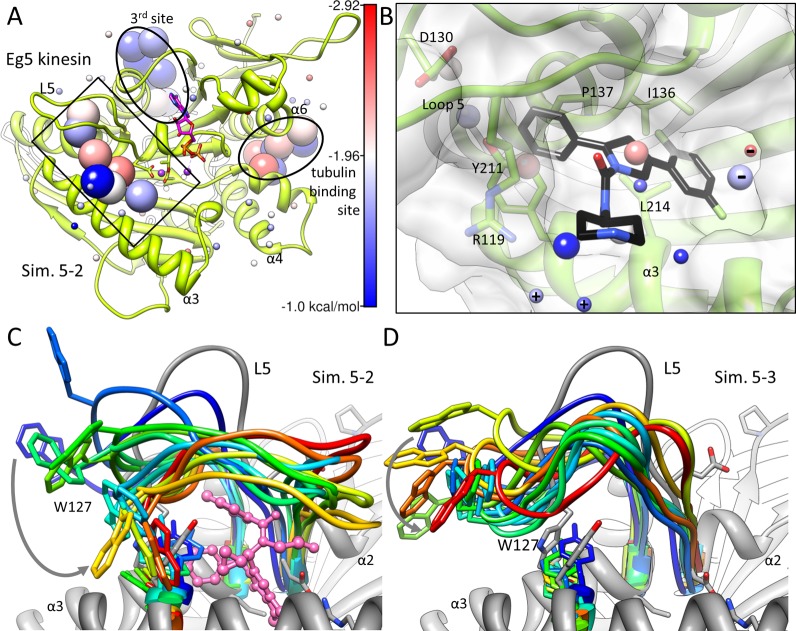
Allosteric inhibitors of Eg5 are known to bind a ligand-induced pocket 12 Å away from the catalytic cavity. The pocket is lined by helix α3 and the insertion loop 5 (L5) of helix α2 after its displacement by 7 Å toward helix α3 (rectangular box in Figure 6A).50 The absence of an accessible/open binding pocket in the unbound form constitutes a challenge for druggability assessment studies that use a static structure of Eg5. However, our simulations consistently located the allosteric pocket as a druggable site, irrespective of probe type or composition. The calculated achievable affinities were 0.3 nM in probe-mixture and 27 nM in isopropanol-only simulations. An IC50 of 0.2 nM has been reported for the best inhibitor of Eg5, in agreement with the probe-mixture prediction.51 High affinity probes were observed therein to be stabilized by interactions with E116, I136, and P137 on helix α2 and Y211 and L214 on helix α3. Other favorable interactions included those with W127, D130, and R119 on L5.
Figure 6.
Significance of conformational flexibility in identifying the allosteric pocket of Eg5 as a druggable site. (A) Interaction spots identified for Eg5 structure63 (PDB ID: 1II6) in probe-mixture simulation 5–2. Three druggable sites are observed: the allosteric site (0.3 nM; rectangular box) and two additional sites of maximal affinities 0.2 nM (tubulin binding site) and 47 nM (3rd site). (B) Closeup view of the inhibitor binding to the allosteric site. A pyrrolotriazine-4-one analog (compound 24 in Kim et al.;52 PDB ID: 2FKY) is shown in stick representation. Interaction spots include both negatively and positively charged probes as labeled. (C) Snapshots at 4 ns intervals for Eg5 probe-mixture simulation 5–2 (rainbow from blue to red) superposed onto the inhibitor-bound structure (gray; inhibitor in pink stick-and-ball). Inhibitor binding induces a displacement in the W127 side-chain by 10 Å (from blue to gray, see also in panel D) to approach its bound conformation. (D) Snapshots from probe-free Eg5 simulation 5–3. The loop 5 does not display rearrangements, in contrast to motions observed in the presence of probes in panel C.
Figure 6B shows the overlay of predicted interaction spots for Eg5 with a known inhibitor.52 The interaction spots identified in the allosteric site contain both positively and negatively charged probes, with the total charge not exceeding one electronic unit (absolute). However, in the isopropanol-only simulation, while the calculated achievable affinity was 27 nM, only four of the seven probes are actually located in the allosteric pocket; on the basis of these probes, the computed affinity at the allosteric site would be 45 μM. The additional probes that contributed to the calculated high affinity bound the exposed surface of helix α3. Thus, the probe-mixture is required here in order to accurately capture the binding affinity at the allosteric site. Another interesting observation is that two IPAM probes were stabilized at a position that closely mimics that of the nitrogen of the piperidine moiety in the ligand bound to Eg5 (Figure 6B). The closest interaction spot, 2.3 Å away from the nitrogen of piperidine, was contributed by the amine 23% of the time.
In addition, we identified two more nM sites (0.2–20 nM and 47–600 nM), enclosed by ellipses in Figure 6A. The first, labeled as the tubulin-binding site, is located between helices α4 and α6. It has a rather shallow surface geometry typical of protein–protein interfaces. Yet, a binding affinity of 0.2–20 nM site is predicted resulting from tight interactions with two groups of residues: (i) I272, L293, and S348 and (ii) G296, T300, Y352, and A356. A comparison of the Eg5 structure to tubulin-bound structures of other kinesins53,54 revealed that the first group is part of the tubulin-binding interface. The second group, on the other hand, is part of the binding site for the ATP competitive allosteric inhibitor GSK-1.55 The probe molecules, however, do not penetrate deep enough into the pocket to interact with the GSK-1-binding residues L295, I299, and I332. Finally, we detect a 47 nM site in proximity to Q20, M70, and V85, which has not yet been observed to bind a ligand.
p38 MAP kinase is involved in inflammatory diseases56 and is an extensively studied drug target. The best known p38 inhibitor has 50 pM affinity.57 The achievable affinity deduced from our isopropanol/water simulations is in the range 1–2 nM (Figure 7, Tables 2 and S2). Isopropanol molecules filled in only the adenine- and ribose-binding pockets in runs 6–1 and 6–2; they were not observed to bind the allosteric site, which is a hydrophobic cavity lined by L74, L75, M78, and F189.58 In probe-mixture/water simulations, on the other hand, probes were observed to bind the allosteric cavity (Figure 7B) with a maximal affinity of 10 to 120 pM (Tables 2 and S2), in agreement with experimental measurements for the best known inhibitor. Probes at high-affinity spots were observed to closely interact with A51, E71, L74, L75, L108, and M109. The adenine/ribose pocket was also occupied by all types of probes, but preferentially by acetamide, isopropanol, and IPAM molecules. The contribution of this pocket to affinity was −5.13 kcal/mol. The allosteric pocket, mainly populated with isopropanols, contributed an additional −4.4 kcal/mol.
Figure 7.

Identification of three potentially druggable sites on p38 kinase. (A) In addition to the ATP binding site (enclosed in rectangular box, magnified in panel B), two other druggable sites are identified at the MK2 binding site and at the MAPK insert. p38 structure81 (PDB ID: 1P38) was used in the simulations. (B) Close up view of the ATP binding site, with an inhibitor (compound 30 in Wrobleski et al.;82 PDB ID: 3BV2) in black stick representation. Adenine/ribose binding and allosteric pockets are indicated. Acetamide and IPAM interaction spots are labeled as A and + respectively.
These results again demonstrate that a mixture of polar probes better captures the druggability of the p38 allosteric site than isopropanol alone does. Our results diverge from previous work27 where isopropanol binding to the adenine/ribose pocket alone is estimated to contribute as much as −11.6 kcal/mol, leading to 2–3 orders of magnitude higher affinity than those found here with isopropanol-only simulations.27 This difference is attributed to the overestimation of atomic binding energies in their method, as will be discussed in the next subsection. In the present simulations, the binding free energy contributions are spread over a larger volume, and the positions of the interaction spot clusters show good overlap with the space experimentally observed to be occupied by inhibitors.
In addition to the ATP site, our simulations detected two more druggable sites on p38 (Figure 7A). The first is on the MAPK-activated protein kinase 2 (MK2 site in Figure 7A) activation loop and stabilizes the loop conformation assumed upon MK2 binding.59,60 The maximal affinity for this site is found to be 2 nM, mostly contributed by isopropanol interactions with T218, L222, L238, and V273 and acetate interactions with R220. The second site coincides with a lipid binding site formed by the MAP kinase insert (Figure 7A),61 which is also a binding site for some inhibitors.62 The maximal affinity for this site varied from 5 to 90 nM, contributed by isopropanol interactions with I229, L232, Y258, and I259. For these two sites, we did not find experimental affinity data.
Discussion of the Simulation Protocol and Length
The initial configurations of target systems contained very few probes interacting with the protein, and all known binding sites were free of probe molecules. Prior to the productive simulations, we performed 0.4 to 0.6 ns of annealing (protein non-hydrogen atoms were constrained to prevent unfolding) and 0.4 to 0.6 ns of equilibration (no constraints) simulations (details are given in Table S2). In the annealing step, the temperature of the system was raised to 600 K. This was particularly useful for targets with partially occluded binding sites such as the LFA-1 and MDM2 solution structure. Acceleration in solvent dynamics at high temperatures allowed probe molecules to locate drug/inhibitor binding sites before their collapse (LFA-1) or further occlusion (MDM2). The LFA-1 binding site (Figure 5D), for example, collapsed in a fraction of a nanosecond due to the hydrophobic nature of the binding pocket. The annealing step thus allowed for shorter equilibration times. As to the production runs, we performed 32 to 40 ns simulations. Multiple simulations have shown that the length was adequate for reproducing the binding site and maximal affinity predictions. It is also possible to combine independent simulations to get a consensus density map or probe interaction spot distribution. We have combined simulations performed using same probe compositions. The predicted affinities (Table S2) as well as the distribution of probe binding spots did not change in support of the adequacy of 32–40 ns simulations for sampling.
Intrinsic Dynamics of the Target Protein and the Effect of Probe Molecules
We presented results for six test cases exhibiting three different levels of flexibility at the binding sites: (i) local motions limited to side-chain fluctuations and isomerizations (MDM2 and PTP1B), (ii) local motions also including loop/helix rearrangements (LFA-1 and Eg5), and (iii) global motions involving lobe opening/closing (p38) or large scale terminal tail motions (MDM2 solution model).
At level i, comparable side-chain motions were observed in both druggability and probe-free simulations, as typified by MDM2 and PTP1B, although the recognition of druggable sites may require the use of a probe-mixture depending on the specificity of the detected surface (e.g., the catalytic site of PTP1B which is predominantly basic). At level ii, probe molecules successfully induced structural changes in the target to gradually approach the known inhibitor-bound forms. For example, the probes were able to locate and stabilize the allosteric pocket of LFA-1 in a conformation similar to its inhibitor-bound form (Figure 5C). Conversely, in probe-free simulations, the C-terminal helix of LFA-1 collapsed over the pocket within the first nanosecond of both runs and remained collapsed until the end of the simulations, thus obstructing the access to the binding site (Figure 5D). The presence of organic probe molecules thus appears to be a requirement for enabling the solvent-exposure of the allosteric site. For Eg5, we focused in particular on the tip residue Trp127 of loop 5 (L5) that is displaced by 10 Å between the inhibitor-bound and -unbound structures.52,63 Probe molecules induced a reconfiguration at this region, to sample conformations departing by 4 to 5 Å from the inhibitor-bound form (Figure 6C). In probe-free simulations, however, no L5 rearrangement was observed (Figure 6D). These observations point to the ability of Eg5 loop 5 to expose a druggable pocket that accommodates the bound inhibitor. The predisposition of the loop to undergo such conformational changes was higher with polar/charged probe molecules, as opposed to isopropanols exclusively.
The dynamics of p38 typifies group iii. p38 can undergo concerted opening/closing of N- and C-terminal lobes enabled by hinge-like flexibility near the ATP-binding site, complemented by movements at the glycine-rich loop, the C-helix and the activation loop, and side chain isomerizations.16,20 We compared the conformations sampled in druggability simulations to a set of 134 experimentally resolved structures (with a variety of inhibitors).64 We observed that residues at the binding pocket and glycine rich loop sampled conformations comparable to those assumed by the bound forms resolved by X-ray crystallography; e.g., probes were observed to bind to the allosteric pocket in the close neighborhood of the ATP-binding site. Although, the global closure of N-terminal and C-terminal lobes to form a compact/buried site upon inhibitor binding was not observed in the druggability simulations (6–1 to 6–4 in Table S2), the RMSD (averaged over all Cα-atoms) between the MD conformations and experimental bound structures came close to 1.0 Å. Two additional simulations in the absence of probe molecules also yielded RMSDs of 1.1 and 1.2 Å.64 The qualitative and quantitative agreement observed in these simulations with experimental data suggests that druggability runs of tens of nanoseconds do provide reasonable estimates of binding sites and affinities, despite their limitations with regard to sampling global motions.
Evaluation of Maximal Achievable Binding Affinity
This methodology utilizes a physics-based atomistic simulation and is therefore independent of any training set. In principle, the binding free energy of probe molecules should be comparable to the affinities typical of drug-like molecules. Ligand efficiency (LE)65 provides a metric for comparison of binding affinities. LE is defined as the binding free energy per atom, LE = −ΔG/Na, where Na is the number of non-hydrogen atoms in the molecule. For a drug-like molecule of 500 Da (∼30 heavy atoms) with an affinity of 10 pM to 10 μM (or a binding free energy of −15.10 to −6.86 kcal/mol, at T = 300K), the LE is 0.23 to 0.5 kcal/mol. The LEs for the interaction spots that we identified vary between 0.25 to 0.8 kcal/mol, consistent with the LE range typical of drug molecules. In previous work,27 the methyl carbons and hydroxyl oxygen of a given isopropanol molecule were taken as independent entities; i.e., their contributions to binding free energy were assumed to be additive. This resulted in an overestimation of binding affinity, leading to LE values higher than 1.5 kcal/mol, which has been estimated to be the limit for the contribution made per atom to binding energy.66 To avoid such overestimation in achievable affinities, the calculated free energies were rescaled, in addition to manual pruning of merged grid elements.27 In the present study, binding free energies are evaluated in a fully automated way, with no need for user intervention or additional rescaling. See the Methods section for a detailed description of the methodology.
Dealing with False Positive and False Negative Sites
Simulations of target proteins at high probe molecule concentration yield large numbers of binding spots. By considering energy and charge contributions of binding spots, the number of false positive sites, i.e., sites with no known binders, can be minimized. We observed that all binding sites with high affinity (submicromolar IC50 or Ki) inhibitors were bound by at least 6 probe interaction spots, chosen as roughly matching the volume of an average small molecule, with a relative binding free energy of −1.2 (LE 0.3) kcal/mol or better. Hence, we evaluated clusters of six or more probes with LEs of 0.3 kcal/mol or better. This threshold resulted in potential false positives for PTP1B (fourth site in Figure 4A) and eg5 kinesin (third site in Figure 6A). The selection of a subset of spots in a binding site for maximal affinity calculation is further described in the Methods section. A second criterion that helped eliminate potential false positives was limiting the number of charged probes (max. 3) and the absolute total charge (max. 2e–) in each cluster. For example, we eliminated a cluster of six high affinity acetate binding spots for eg5 kinesin located between side chains of four arginine residues (R192, R305, R312, and R317, available as Supporting Information).
The stringent criterion that we applied resulted in missing some sites with binding information but not with known high-affinity inhibitors. These sites include the IRK interface and allosteric site on PTP1B and the MK2 site and MAPK insert on p38. In line with the unavailability of high affinity inhibitors for these sites, the clusters that we identified at these sites contained four or less probes with a LE of 0.3 kcal/mol or better. We calculated the maximal affinity for these sites by linking clusters (MAPK insert and MK2 site) or augmented lower affinity spots (IRK interface and PTP1B allosteric site). Available binding spots in the PTP1B allosteric site with a LE of better than 0.25 kcal/mol covered only part of the site resulting in predicted maximal affinity of 75 μM, compared to the 8 μM IC50 of the best inhibitor. The incorporation of two lower affinity spots with energies of −0.5 and −0.33 kcal/mol (available as Supporting Information) improved the predicted maximal affinity to 17.6 μM.
Conclusion
Protein–ligand recognition is a complex phenomenon involving many balancing factors.67 It has been noted for some time now that one can identify hotspots in protein binding sites, i.e., regions where protein residue/ligand moiety interactions make the largest contributions to the overall binding free energy.68 However, a major obstacle to accurate estimation of binding affinities, and, in some cases, to identification of binding sites, has been the lack of adequate inclusion of the intrinsic dynamics of the target protein. The flexibility of the target usually allows for favorable enthalpic contributions due to the adaptability and optimal positioning of interacting groups, which, in many instances, more than offset the adverse entropic cost. Computationally inexpensive rigid docking methods perform poorly when benchmarking the computed affinities against experimental measurements.69 Nor do they provide an adequate description of the observed affinity of organic molecules on target proteins.70 Simulation-based methods and comprehensive sampling of the ligand and protein conformational space taking into account entropic effects are recognized to improve binding affinity predictions.71−73 The success of the current druggability method owes to the natural incorporation of target, solvent, and probe entropic effects into the estimation of binding affinities.
Notably, the use of a mixture of probes featuring diverse physicochemical properties is shown here to yield consistently better predictions of achievable affinities, in addition to a more complete identification of druggable sites on targets. The probe mixture simulations yielded more accurate ranges for the maximal achievable binding affinity for Eg5 and p38, and charged probes allowed us to accurately predict the achievable affinity for the PTP1B catalytic site. These observations suggest that the methodology suits a broad range of binding sites. This is an important improvement as marketed drug molecules more often than neutral are singly charged. For instance, a recent set of 2056 orally available drugs assembled for analysis included 35% neutral molecules with the remaining either charged or zwitterionic.74
The present study also highlights the utility of MD simulations with explicit probe molecules in the aqueous environment when dealing with flexible targets. In our test cases, high affinity interaction points are observed in pockets inaccessible in the unbound form, such as the LFA-1 induced site and p38 allosteric pocket in the ATP site. The approach rigorously accounts for the time-dependent molecular driving forces involved in ligand recognition, including entropic factors and desolvation. As it is physics-based and not trained on particular target classes, the method is virtually applicable to all types of protein targets within the limitations of current molecular mechanics force fields and solvent models. The simplified assumptions and refined algorithm makes intermediate results easily interpretable and applicable to guiding structure-based drug design. Using parallel platforms and software, calculations can be performed within a few days for most protein targets to make an assessment of druggability, potential for binding sites, and achievable affinities. This methodology appears to be especially useful for identifying allosteric binding sites with limited structural data on alternate protein conformations.
Methods
MD Simulations
Simulations were performed using NAMD75 software and the CHARMM76 force field. Productive simulation times ranged from 32 to 40 ns. See the Supporting Information and Table S2 for details.
Construction of Grids Based on Probe Locations
MD snapshots were superposed onto the reference PDB structure of the protein using Cα atoms. Probe molecules having a non-hydrogen atom within 4.0 Å of protein atoms were considered to interact with the protein. For each probe type, individual occupancy (number density) grids were calculated using their central carbon atoms with VMD77 Volmap. Grid calculations for combined trajectories (Table S2) were performed using Python packages ProDy78 and NumPy. In both cases, grid resolution was set to 0.5 Å. To reduce grid artifacts, the occupancy value in each voxel was averaged with its neighbors. When more than one type of probe was used, grids of individual probes were combined. In this case, each probe was assigned a fractional occupancy value (ranging from 0 to 1) for a given voxel. Fractional occupancy was obtained by dividing its occupancy by the total occupancy at the voxel.
Evaluation of Binding Free Energies of Interaction Spots
Occupancy grids are converted to binding free energy grids (Figure 1D) using eq 1 and the expected occupancy described in the Supporting Information. Interaction spots (Figure 1E) are defined as voxels satisfying three criteria: (i) An interaction spot does not overlap with other interaction spots. (ii) The binding free energy of the interaction spot is lower than a predefined upper limit for probe binding free energy. (iii) The binding free energy of an interaction spot is less than those of the surrounding voxels; that is, in a given volume matching the size of a probe molecule, the lowest energy voxel is selected as the interaction spot representing the probe. We set the value of the upper limit to −1 kcal/mol (LE = 0.25 kcal/mol). Starting from the voxel with lowest binding free energy value (central interaction spot), other voxels within the effective radius of the central interaction spot were eliminated. This was repeated for the next voxel with the lowest binding free energy until no pairs of overlapping voxels remained. When multiple probe types were used, the effective radius of a voxel was defined as the sum of effective radii (Table S3) of probes weighted by their fractional occupancies. In the case of charged probes, the effective charge of an interaction spot is calculated as the fractional occupancy weighted sum of probe charges. For example, an interaction spot occupied in half of the simulation time by isopropanol molecules, and the other half by acetate molecules, was assigned an effective charge of 0.5 electronic units.
Maximal Achievable Affinity Calculation
Druggable sites were identified by merging proximal interaction spots as follows: (i) The lowest energy interaction spot in a distinct binding site is selected as a seed. (ii) The next lowest energy interaction spot within 6.2 Å of the seed and satisfying the effective charge constraint is merged to the seed. (iii) The second step is repeated until a desired number of interaction spots are merged. The total effective charge in a druggable site was restricted to be less than or equal to 2e–. Maximal affinity of the druggable site is estimated from the sum of binding free energies of selected interaction spots.
Acknowledgments
The authors thank Drs. Eric Manas, Joy Yang, Guanglei Cui, Xuan Hong, and Alan Graves at GSK for their stimulating and fruitful discussions during the development of the methodology. Support from NIH grant # 2U19AI068021-06 and PA CURE grant via Project 0864104 is gratefully acknowledged by I.B. Simulations were performed using the Bahar Lab CPU cluster Liszt and Pittsburgh Supercomputing Center (PSC) CPU cluster Pople. The authors thank Dr. Levent S. Yilmaz for his help with using the PSC facilities.
Supporting Information Available
Details of probe molecules, force field parameters, simulations, computing times, and an extended discussion of the methodology are provided. In addition, the coordinates of proteins and probe molecules with predicted binding free energies are provided in PDB format files. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hopkins A. L.; Groom C. R. The druggable genome. Nat. Rev. Drug Discovery 2002, 1, 727–730. [DOI] [PubMed] [Google Scholar]
- Brown D.; Superti-Furga G. Rediscovering the sweet spot in drug discovery. Drug Discovery Today 2003, 8, 1067–1077. [DOI] [PubMed] [Google Scholar]
- Keller T. H.; Pichota A.; Yin Z. A practical view of ’druggability’. Curr. Opin. Chem. Biol. 2006, 10, 357–361. [DOI] [PubMed] [Google Scholar]
- Hajduk P. J.; Huth J. R.; Fesik S. W. Druggability indices for protein targets derived from NMR-based screening data. J. Med. Chem. 2005, 48, 2518–2525. [DOI] [PubMed] [Google Scholar]
- Allen K. N.; Bellamacina C. R.; Ding X.; Jeffery C. J.; Mattos C.; Petsko G. A.; Ringe D. An Experimental Approach to Mapping the Binding Surfaces of Crystalline Proteins. J. Phys. Chem. 1996, 100, 2605–2611. [Google Scholar]
- Cheng A. C.; Coleman R. G.; Smyth K. T.; Cao Q.; Soulard P.; Caffrey D. R.; Salzberg A. C.; Huang E. S. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007, 25, 71–75. [DOI] [PubMed] [Google Scholar]
- Halgren T. A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [DOI] [PubMed] [Google Scholar]
- Schmidtke P.; Barril X. Understanding and predicting druggability. A high-throughput method for detection of drug binding sites. J. Med. Chem. 2010, 53, 5858–5867. [DOI] [PubMed] [Google Scholar]
- Huang N.; Jacobson M. P. Binding-site assessment by virtual fragment screening. PLoS One 2010, 5, e10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenke R.; Kozakov D.; Chuang G. Y.; Beglov D.; Hall D.; Landon M. R.; Mattos C.; Vajda S. Fragment-based identification of druggable ’hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics 2009, 25, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan C.; Hall D. R.; Zerbe B.; Grove L. E.; Kozakov D.; Vajda S. FTSite: High accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics 2012, 28, 286–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. P.; Hajduk P. J. Effects of conformational dynamics on predicted protein druggability. ChemMedChem. 2006, 1, 70–72. [DOI] [PubMed] [Google Scholar]
- Ivetac A.; McCammon J. A. Mapping the druggable allosteric space of G-protein coupled receptors: a fragment-based molecular dynamics approach. Chem. Biol. Drug Des. 2010, 76, 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson H. A.; McCammon J. A. Accommodating protein flexibility in computational drug design. Mol. Pharmacol. 2000, 57, 213–218. [PubMed] [Google Scholar]
- Rueda M.; Bottegoni G.; Abagyan R. Consistent improvement of cross-docking results using binding site ensembles generated with elastic network normal modes. J Chem. Inf. Model. 2009, 49, 716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasotto C. N.; Abagyan R. A. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004, 337, 209–225. [DOI] [PubMed] [Google Scholar]
- Floquet N.; Marechal J. D.; Badet-Denisot M. A.; Robert C. H.; Dauchez M.; Perahia D. Normal mode analysis as a prerequisite for drug design: application to matrix metalloproteinases inhibitors. FEBS Lett. 2006, 580, 5130–5136. [DOI] [PubMed] [Google Scholar]
- May A.; Zacharias M. Protein-ligand docking accounting for receptor side chain and global flexibility in normal modes: evaluation on kinase inhibitor cross docking. J. Med. Chem. 2008, 51, 3499–3506. [DOI] [PubMed] [Google Scholar]
- Bahar I.; Lezon T. R.; Bakan A.; Shrivastava I. H. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem. Rev. 2010, 110, 1463–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan A.; Bahar I. The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles L. M.; Gur M.; Bakan A.; Bahar I.. Pre-existing Soft Modes of Motion Uniquely Defined by Native Contact Topology Facilitate Ligand Binding to Proteins. Protein Sci. 2011, not supplied. [DOI] [PMC free article] [PubMed]
- Lexa K. W.; Carlson H. A. Full Protein Flexibility Is Essential for Proper Hot-Spot Mapping. J. Am. Chem. Soc. 2010, 133, 200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H.; Grove L. E.; Yueh C.; Ngan C. H.; Kozakov D.; Vajda S. Robust identification of binding hot spots using continuum electrostatics: application to hen egg-white lysozyme. J. Am. Chem. Soc. 2011, 133, 20668–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp J. L. III; Kulp J. L. Jr.; Pompliano D. L.; Guarnieri F. Diverse fragment clustering and water exclusion identify protein hot spots. J. Am. Chem. Soc. 2011, 133, 10740–10743. [DOI] [PubMed] [Google Scholar]
- Guvench O.; Mackerell A. D. Jr. Computational fragment-based binding site identification by ligand competitive saturation. PLoS Comput. Biol. 2009, 5, e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman E. P.; Yu W.; Guvench O.; Mackerell A. D. Reproducing crystal binding modes of ligand functional groups using Site-Identification by Ligand Competitive Saturation (SILCS) simulations. J. Chem. Inf. Model. 2011, 51, 877–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seco J.; Luque F. J.; Barril X. Binding site detection and druggability index from first principles. J. Med. Chem. 2009, 52, 2363–2371. [DOI] [PubMed] [Google Scholar]
- Mattos C.; Ringe D. Locating and characterizing binding sites on proteins. Nat. Biotechnol. 1996, 14, 595–599. [DOI] [PubMed] [Google Scholar]
- Liepinsh E.; Otting G. Organic solvents identify specific ligand binding sites on protein surfaces. Nat. Biotechnol. 1997, 15, 264–268. [DOI] [PubMed] [Google Scholar]
- Knox C.; Law V.; Jewison T.; Liu P.; Ly S.; Frolkis A.; Pon A.; Banco K.; Mak C.; Neveu V.; Djoumbou Y.; Eisner R.; Guo A. C.; Wishart D. S. DrugBank 3.0: a comprehensive resource for ’omics’ research on drugs. Nucleic Acids Res. 2011, 39, D1035–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirtay C. K.; Mitchell J. B. O.; Lumley J. A.. Knowledge Based Potentials: the Reverse Boltzmann Methodology, Virtual Screening and Molecular Weight Dependence. QSAR Comb. Sci. 205, 24. [Google Scholar]
- Thomas P. D.; Dill K. A. Statistical potentials extracted from protein structures: how accurate are they?. J. Mol. Biol. 1996, 257, 457–469. [DOI] [PubMed] [Google Scholar]
- Haupt Y.; Maya R.; Kazaz A.; Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer 2003, 3, 102–109. [DOI] [PubMed] [Google Scholar]
- Kussie P. H.; Gorina S.; Marechal V.; Elenbaas B.; Moreau J.; Levine A. J.; Pavletich N. P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996, 274, 948–953. [DOI] [PubMed] [Google Scholar]
- Vassilev L. T.; Vu B. T.; Graves B.; Carvajal D.; Podlaski F.; Filipovic Z.; Kong N.; Kammlott U.; Lukacs C.; Klein C.; Fotouhi N.; Liu E. A. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [DOI] [PubMed] [Google Scholar]
- Zhong H.; Carlson H. A. Computational studies and peptidomimetic design for the human p53-MDM2 complex. Proteins 2005, 58, 222–234. [DOI] [PubMed] [Google Scholar]
- Yu S.; Qin D.; Shangary S.; Chen J.; Wang G.; Ding K.; McEachern D.; Qiu S.; Nikolovska-Coleska Z.; Miller R.; Kang S.; Yang D.; Wang S. Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem. 2009, 52, 7970–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Hatcher E.; Acharya C.; Kundu S.; Zhong S.; Shim J.; Darian E.; Guvench O.; Lopes P.; Vorobyov I.; Mackerell A. D. Jr. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrinova S.; Uhrin D.; Powers H.; Watt K.; Zheleva D.; Fischer P.; McInnes C.; Barlow P. N. Structure of free MDM2 N-terminal domain reveals conformational adjustments that accompany p53-binding. J. Mol. Biol. 2005, 350, 587–598. [DOI] [PubMed] [Google Scholar]
- Xin Z.; Liu G.; Abad-Zapatero C.; Pei Z.; Szczepankiewicz B. G.; Li X.; Zhang T.; Hutchins C. W.; Hajduk P. J.; Ballaron S. J.; Stashko M. A.; Lubben T. H.; Trevillyan J. M.; Jirousek M. R. Identification of a monoacid-based, cell permeable, selective inhibitor of protein tyrosine phosphatase 1B. Bioorg. Med. Chem. Lett. 2003, 13, 3947–3950. [DOI] [PubMed] [Google Scholar]
- Puius Y. A.; Zhao Y.; Sullivan M.; Lawrence D. S.; Almo S. C.; Zhang Z. Y. Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 13420–13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C.; Barr K. J.; Kung J.; Zhu J.; Erlanson D. A.; Shen W.; Fahr B. J.; Zhong M.; Taylor L.; Randal M.; McDowell R. S.; Hansen S. K. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Struct Mol. Biol. 2004, 11, 730–737. [DOI] [PubMed] [Google Scholar]
- Li S.; Depetris R. S.; Barford D.; Chernoff J.; Hubbard S. R. Crystal structure of a complex between protein tyrosine phosphatase 1B and the insulin receptor tyrosine kinase. Structure 2005, 13, 1643–1651. [DOI] [PubMed] [Google Scholar]
- Marlin S. D.; Springer T. A. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1). Cell 1987, 51, 813–819. [DOI] [PubMed] [Google Scholar]
- Qu A.; Leahy D. J. The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin. Structure 1996, 4, 931–942. [DOI] [PubMed] [Google Scholar]
- Crump M. P.; Ceska T. A.; Spyracopoulos L.; Henry A.; Archibald S. C.; Alexander R.; Taylor R. J.; Findlow S. C.; O’Connell J.; Robinson M. K.; Shock A. Structure of an allosteric inhibitor of LFA-1 bound to the I-domain studied by crystallography, NMR, and calorimetry. Biochemistry 2004, 43, 2394–2404. [DOI] [PubMed] [Google Scholar]
- Liu T.; Lin Y.; Wen X.; Jorissen R. N.; Gilson M. K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckian K. M.; Lin E. Y.; Silvian L.; Friedman J. E.; Chin D.; Scott D. M. Design and synthesis of a series of meta aniline-based LFA-1 ICAM inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 5249–5251. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Sardana V.; Xu B.; Homnick C.; Halczenko W.; Buser C. A.; Schaber M.; Hartman G. D.; Huber H. E.; Kuo L. C. Inhibition of a mitotic motor protein: where, how, and conformational consequences. J. Mol. Biol. 2004, 335, 547–554. [DOI] [PubMed] [Google Scholar]
- Roecker A. J.; Coleman P. J.; Mercer S. P.; Schreier J. D.; Buser C. A.; Walsh E. S.; Hamilton K.; Lobell R. B.; Tao W.; Diehl R. E.; South V. J.; Davide J. P.; Kohl N. E.; Yan Y.; Kuo L. C.; Li C.; Fernandez-Metzler C.; Mahan E. A.; Prueksaritanont T.; Hartman G. D. Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP. Bioorg. Med. Chem. Lett. 2007, 17, 5677–5682. [DOI] [PubMed] [Google Scholar]
- Kim K. S.; Lu S.; Cornelius L. A.; Lombardo L. J.; Borzilleri R. M.; Schroeder G. M.; Sheng C.; Rovnyak G.; Crews D.; Schmidt R. J.; Williams D. K.; Bhide R. S.; Traeger S. C.; McDonnell P. A.; Mueller L.; Sheriff S.; Newitt J. A.; Pudzianowski A. T.; Yang Z.; Wild R.; Lee F. Y.; Batorsky R.; Ryder J. S.; Ortega-Nanos M.; Shen H.; Gottardis M.; Roussell D. L. Synthesis and SAR of pyrrolotriazine-4-one based Eg5 inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 3937–3942. [DOI] [PubMed] [Google Scholar]
- Sindelar C. V.; Downing K. H. The beginning of kinesin’s force-generating cycle visualized at 9-A resolution. J. Cell Biol. 2007, 177, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodey A. J.; Kikkawa M.; Moores C. A. 9-Angstrom structure of a microtubule-bound mitotic motor. J. Mol. Biol. 2009, 388, 218–224. [DOI] [PubMed] [Google Scholar]
- Luo L.; Parrish C. A.; Nevins N.; McNulty D. E.; Chaudhari A. M.; Carson J. D.; Sudakin V.; Shaw A. N.; Lehr R.; Zhao H.; Sweitzer S.; Lad L.; Wood K. W.; Sakowicz R.; Annan R. S.; Huang P. S.; Jackson J. R.; Dhanak D.; Copeland R. A.; Auger K. R. ATP-competitive inhibitors of the mitotic kinesin KSP that function via an allosteric mechanism. Nat. Chem. Biol. 2007, 3, 722–726. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Boehm J.; Lee J. C. p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat. Rev. Drug Discovery 2003, 2, 717–726. [DOI] [PubMed] [Google Scholar]
- Pettus L. H.; Wurz R. P.; Xu S.; Herberich B.; Henkle B.; Liu Q.; McBride H. J.; Mu S.; Plant M. H.; Saris C. J.; Sherman L.; Wong L. M.; Chmait S.; Lee M. R.; Mohr C.; Hsieh F.; Tasker A. S. Discovery and evaluation of 7-alkyl-1,5-bis-aryl-pyrazolopyridinones as highly potent, selective, and orally efficacious inhibitors of p38alpha mitogen-activated protein kinase. J. Med. Chem. 2010, 53, 2973–2985. [DOI] [PubMed] [Google Scholar]
- Pargellis C.; Tong L.; Churchill L.; Cirillo P. F.; Gilmore T.; Graham A. G.; Grob P. M.; Hickey E. R.; Moss N.; Pav S.; Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 2002, 9, 268–272. [DOI] [PubMed] [Google Scholar]
- White A.; Pargellis C. A.; Studts J. M.; Werneburg B. G.; Farmer B. T. Molecular basis of MAPK-activated protein kinase 2:p38 assembly. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 6353–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E.; Prabhakar P.; Liu X.; Lepre C. Crystal structure of the p38 alpha-MAPKAP kinase 2 heterodimer. J. Biol. Chem. 2007, 282, 9733–9739. [DOI] [PubMed] [Google Scholar]
- Diskin R.; Engelberg D.; Livnah O. A novel lipid binding site formed by the MAP kinase insert in p38á. J. Mol. Biol. 2008, 375, 70–79. [DOI] [PubMed] [Google Scholar]
- Perry J. J.; Harris R. M.; Moiani D.; Olson A. J.; Tainer J. A. p38alpha MAP kinase C-terminal domain binding pocket characterized by crystallographic and computational analyses. J. Mol. Biol. 2009, 391, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.; Anderson R.; Guo J.; Beraud C.; Fletterick R.; Sakowicz R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J. Biol. Chem. 2001, 276, 25496–25502. [DOI] [PubMed] [Google Scholar]
- Bakan A.; Bahar I. Computational Generation of Inhibitor-Bound Conformers of p38 MAP Kinase and Comparison with Experiments. Pac. Symp. Biocomput. 2011 2011, 16, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A. L.; Groom C. R.; Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discovery Today 2004, 9, 430–431. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D.; Chen K.; Sharp K. A.; Kollman P. A. The maximal affinity of ligands. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 9997–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H. J.; Klebe G. What can we learn from molecular recognition in protein-ligand complexes for the design of new drugs?. Angew. Chem., Int. Ed. Engl. 1996, 35, 2588–2614. [Google Scholar]
- Clackson T.; Wells J. A. A hot spot of binding energy in a hormone-receptor interface. Science 1995, 267, 383–386. [DOI] [PubMed] [Google Scholar]
- Warren G. L.; Andrews C. W.; Capelli A. M.; Clarke B.; LaLonde J.; Lambert M. H.; Lindvall M.; Nevins N.; Semus S. F.; Senger S.; Tedesco G.; Wall I. D.; Woolven J. M.; Peishoff C. E.; Head M. S. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006, 49, 5912–5931. [DOI] [PubMed] [Google Scholar]
- English A. C.; Groom C. R.; Hubbard R. E. Experimental and computational mapping of the binding surface of a crystalline protein. Protein Eng. 2001, 14, 47–59. [DOI] [PubMed] [Google Scholar]
- Chang C. E.; Chen W.; Gilson M. K. Ligand configurational entropy and protein binding. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley D. L.; Graves A. P.; Chodera J. D.; McReynolds A. C.; Shoichet B. K.; Dill K. A. Predicting absolute ligand binding free energies to a simple model site. J. Mol. Biol. 2007, 371, 1118–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce S. E.; Mobley D. L.; Rocklin G. J.; Graves A. P.; Dill K. A.; Shoichet B. K. Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site. J. Mol. Biol. 2009, 394, 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeson P. D.; St-Gallay S. A.; Wenlock M. C. Impact of ion class and time on oral drug molecular properties. Med. Chem. Commun. 2011, 2, 91–105. [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Brooks C. L. III; Mackerell A. D. Jr.; Nilsson L.; Petrella R. J.; Roux B.; Won Y.; Archontis G.; Bartels C.; Boresch S.; Caflisch A.; Caves L.; Cui Q.; Dinner A. R.; Feig M.; Fischer S.; Gao J.; Hodoscek M.; Im W.; Kuczera K.; Lazaridis T.; Ma J.; Ovchinnikov V.; Paci E.; Pastor R. W.; Post C. B.; Pu J. Z.; Schaefer M.; Tidor B.; Venable R. M.; Woodcock H. L.; Wu X.; Yang W.; York D. M.; Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Bakan A.; Meireles L. M.; Bahar I.. ProDy: Protein Dynamics Inferred from Theory and Experiments. Bioinformatics. 2011, not supplied. [DOI] [PMC free article] [PubMed]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Liu G.; Xin Z.; Liang H.; bad-Zapatero C.; Hajduk P. J.; Janowick D. A.; Szczepankiewicz B. G.; Pei Z.; Hutchins C. W.; Ballaron S. J.; Stashko M. A.; Lubben T. H.; Berg C. E.; Rondinone C. M.; Trevillyan J. M.; Jirousek M. R. Selective protein tyrosine phosphatase 1B inhibitors: targeting the second phosphotyrosine binding site with non-carboxylic acid-containing ligands. J. Med. Chem. 2003, 46, 3437–3440. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Harkins P. C.; Ulevitch R. J.; Han J.; Cobb M. H.; Goldsmith E. J. The structure of mitogen-activated protein kinase p38 at 2.1-A resolution. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 2327–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobleski S. T.; Lin S.; Hynes J. Jr.; Wu H.; Pitt S.; Shen D. R.; Zhang R.; Gillooly K. M.; Shuster D. J.; McIntyre K. W.; Doweyko A. M.; Kish K. F.; Tredup J. A.; Duke G. J.; Sack J. S.; McKinnon M.; Dodd J.; Barrish J. C.; Schieven G. L.; Leftheris K. Synthesis and SAR of new pyrrolo[2,1-f][1,2,4]triazines as potent p38 alpha MAP kinase inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 2739–2744. [DOI] [PubMed] [Google Scholar]
- Yu S.; Qin D.; Shangary S.; Chen J.; Wang G.; Ding K.; McEachern D.; Qiu S.; Nikolovska-Coleska Z.; Miller R.; Kang S.; Yang D.; Wang S. Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem. 2009, 52, 7970–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.; Qin D.; Shangary S.; Chen J.; Wang G.; Ding K.; McEachern D.; Qiu S.; Nikolovska-Coleska Z.; Miller R.; Kang S.; Yang D.; Wang S. Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem. 2009, 52, 7970–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.