Abstract
Tetherin (BST2) is the host cell factor that blocks the particle release of some enveloped viruses. Two putative feline tetherin proteins differing at the level of the N-terminal coding region have recently been described and tested for their antiviral activity. By cloning and comparing the two reported feline tetherins (called here cBST2504 and cBST2*) and generating specific derivative mutants, this study provides evidence that feline tetherin has a shorter intracytoplasmic domain than those of other known homologues. The minimal tetherin promoter was identified and assayed for its ability to drive tetherin expression in an alpha interferon-inducible manner. We also demonstrated that cBST2504 is able to dimerize, is localized at the cellular membrane, and impairs human immunodeficiency virus type 1 (HIV-1) particle release, regardless of the presence of the Vpu antagonist accessory protein. While cBST2504 failed to restrict wild-type feline immunodeficiency virus (FIV) egress, FIV mutants, bearing a frameshift at the level of the envelope-encoding region, were potently blocked. The transient expression of the FIV envelope glycoprotein was able to rescue mutant particle release from feline tetherin-positive cells but did not antagonize human BST2 activity. Moreover, cBST2504 was capable of specifically immunoprecipitating the FIV envelope glycoprotein. Finally, cBST2504 also exerted its function on HIV-2 ROD10 and on the simian immunodeficiency virus SIVmac239. Taken together, these results show that feline tetherin does indeed have a short N-terminal region and that the FIV envelope glycoprotein is the predominant factor counteracting tetherin restriction.
INTRODUCTION
All the viruses belonging to the family Lentiviridae are characterized by specific properties, such as the ability to infect macrophages and nondividing cells and a slow disease progression. In addition to these common features, feline immunodeficiency virus (FIV) shares additional relevant similarities (14) with human immunodeficiency virus (HIV), the most important human pathogen belonging to this viral genus. Indeed, while in nondomestic felids, FIV is minimally pathogenic (6, 8, 27, 62, 63), as is the simian immunodeficiency virus (SIV) SIVagm and other naturally occurring SIVs in their natural hosts (66), the relatively recent jump to a new host species, i.e., Felis catus (42, 64), has led to high immune virulence and to a severe immunodeficiency syndrome similar to the one caused by HIV-1 in humans.
HIV-1 and HIV-2 similarly resulted from cross-species transmissions from chimpanzees/gorillas and sooty mangabeys, respectively, to humans (9, 15, 18, 25, 66). Thus, FIV is the only nonprimate lentivirus that causes an AIDS-like disease in its natural host, the domestic cat (3, 43, 44, 72). In addition, FIV enters target cells via CD134 (57), a T cell-costimulatory protein, and CXCR4 (48, 69, 70), a coreceptor, and its genome encodes a factor, called Vif, as is the corresponding one in HIV-1, that is required for the production of fully infectious virions (61).
Given the close similarity of HIV and FIV in terms of genome structure, mechanism of transmission, course of infection, as well as pathogenicity, the domestic cat is considered the smallest available natural animal model for the study of AIDS in humans and for the development of potential therapeutic strategies (62, 69, 70).
In addition to conventional innate and acquired immune responses, humans and other mammals have evolved different antiviral factors to defend themselves from retroviral infection. Among these, the so-called “host restriction factors” are host cellular proteins constitutively expressed or induced by interferon (IFN) in response to viral infection. Host restriction factors represent a crucial aspect of innate immunity, defined as intrinsic immunity (4, 19). The properties that mainly characterize these proteins and have contributed to their discovery are their species and virus specificities (28, 52, 53). The species-specific expression and activity of restriction factors limit viral host tropism and constitute a barrier to cross-species transmission events (64). In order to efficiently replicate, retroviruses need to overcome restriction factors and have thus evolved countermeasures or strategies to antagonize them. In the case of HIV and SIV, different counteracting factors have been identified among accessory and structural proteins, including Vif, Vpu, Nef, and the envelope glycoprotein (32).
To date, three major types of restriction factors, acting at specific steps of the retroviral life cycle, have been discovered: APOBEC3G (52, 55, 56), which targets reverse transcription; TRIM5α (59), which interferes with the uncoating of incoming capsids; and, more recently, tetherin/BST2 (40, 65), which blocks the release of viral particles. Tetherin is constitutively expressed in human cell lines such as HeLa cells (22), several cancer cell lines (41), B cells, T cells, monocytes, macrophages, and plasmacytoid dendritic cells (5, 36, 67), and its expression can be induced by type I and type II interferon treatment (36, 38–40, 65). Tetherin also causes the retention of fully formed, mature virions on the surface of cells infected with Vpu-deficient HIV-1 (40). The HIV-1 Vpu protein antagonizes tetherin by causing its degradation and sequestration into a perinuclear compartment away from virus assembly sites (12, 13, 20, 26, 33). Moreover, the Nef and envelope proteins from some SIVs (24, 29, 51, 54, 73) and the HIV-2 envelope protein (26, 31) function as antagonists of tetherin in a species-specific manner.
It was recently reported in two independent studies (11, 17) that the feline genome carries a gene with significant identity to the known primate, rodent, and canine tetherin sequences, indicating that cats may harbor a homologue of BST2/tetherin. Interestingly, those two studies differed in one crucial aspect, i.e., the N-terminal region of the feline protein supposed to function as tetherin. In particular, Fukuma and coworkers (17) expressed a protein displaying an N-terminal portion that was shorter than the one cloned and characterized by Dietrich and collaborators (11), which, instead, closely resembled human tetherin with regard to the length of the intracytoplasmic domain. Even though both studies showed that feline tetherin has an evident effect on the release of viral particles, the viruses tested were different. Indeed, while the shorter version of tetherin was tested only against the feline endogenous retrovirus RD114, the one characterized by a longer N-terminal region was assayed against HIV-1 (both Vpu positive [Vpu+] and Vpu negative [Vpu−]) as well as against vesicular stomatitis virus G protein (VSV-G)-pseudotyped FIV. Interestingly, the latter version of feline tetherin had not effect on wild-type FIV replication, suggesting that the FIV genome expresses a factor capable of counteracting the action of feline tetherin. Dietrich and coworkers did not, however, identify such a factor (11).
In the light of those two reports and bearing in mind that a virus might evolve a specific countermeasure in response to a restriction factor, the goal of the present study was to characterize feline tetherin and to investigate the mechanism by which FIV counteracts this cellular defense. Here we provide strong evidence that the feline protein that displays tetherin activity and that is effective against different lentiviruses has an N-terminal region shorter than that of the human tetherin orthologue. The minimal promoter region sensitive to IFN-α treatment and responsible for feline tetherin expression was also characterized. Finally, we identified the main viral factor able to counteract the tetherin function in the FIV envelope glycoprotein.
MATERIALS AND METHODS
Bioinformatic analysis and prediction.
Different homologous tetherins and the promoter region sequences were aligned by using the CLUSTALW2 algorithm (http://www.ebi.ac.uk/Tools/clustalw2) and were adjusted manually. The topology of cat BST2 (cBST2) was predicted by using the following programs: HMMTOP (http://www.enzim.hu/hmmtop/), to analyze the transmembrane region; big-PI Predictor GPI Modification Site Prediction (http://mendel.imp.ac.at/sat/gpi/gpi_server.html), to identify the glycosylphosphatidylinositol (GPI) anchor site modification; and NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/), to recognize putative N-glycosylation sites. The transcription initiation start site and the different cis-acting elements present in the putative cBST2 promoter region were predicted by using the following programs: the TSSW prediction program (Softberry), TFsitescan (MIRAGE [Molecular Informatics Resource for the Analysis of Gene Expression] website), TFsearch (http://www.cbrc.jp/research/db/TFSEARCH.html), and Transcription Element Search System (TESS) (http://www.cbil.upenn.edu/cgi-bin/tess/).
Mammalian expression plasmids.
The human BST2 (hBST2) construct was generated by cloning the human tetherin-encoding region first into the pCR2.1-TOPO vector (Invitrogen, Milan, Italy) and then into the pcDNA3.1 mammalian expression vector (Invitrogen). The human tetherin-encoding region was obtained by reverse transcription-PCR (RT-PCR) performed on total RNA extracted from HeLa cells by means of forward primer 5′-GCCACCATGGCATCTACTTCGTATGAC-3′ and reverse primer 5′-TCACTGCAGCAGAGCGCTGAGG-3′. The construct was enriched with a 3′ Flag epitope, as described elsewhere previously (40).
The pCMV-eGFP construct was generated by BamHI and NotI digestion of the pEGFP1 plasmid (EuroClone, Milan, Italy) and the insertion of the enhanced green fluorescent protein (eGFP)-encoding sequence into the pcDNA3.1 plasmid (Invitrogen) digested with the same enzymes.
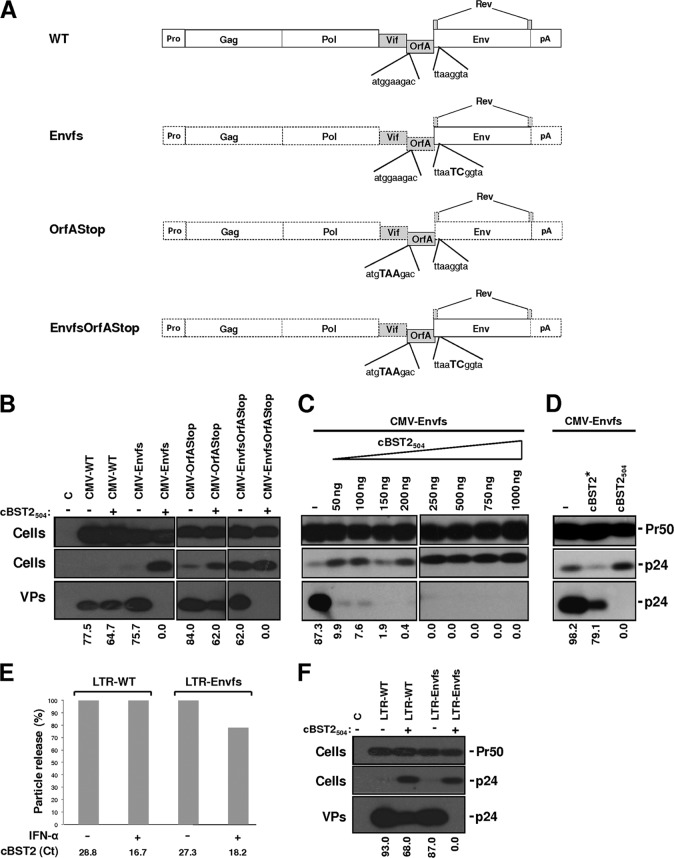
As described elsewhere previously (46), plasmid pΔ00 contains the complete FIV proviral genome encompassing the gag, pol, env, vif, orfA, and rev genes of the Petaluma strain. Plasmid pΔLTR is a derivative of plasmid pΔ00, characterized by 5′ and 3′ long terminal repeats (LTRs) replaced by the cytomegalovirus (CMV) early promoter (Pro) and bovine growth hormone poly(A), respectively (46). By using these constructs as a starting point, a series of mutants was generated at the OrfA- and envelope-encoding regions, as follows. The sequences encoding the viral envelope or OrfA were obtained from plasmid pΔLTR by enzymatic restrictions and inserted into pBluescript KS II (Agilent Technologies, Milan, Italy). The QuikChange II site-directed mutagenesis kit (Agilent Technologies) was then employed to introduce a frameshift immediately after nucleotide 268 of the viral envelope-encoding sequence, maintaining the overlapping rev frame and a stop codon after the ATG of the OrfA-encoding sequences. The fragments carrying the mutations were inserted back into pΔ00 or pΔLTR, replacing the wild-type (WT) counterparts, either alone or in combination, thus obtaining the relative CMV-Envfs, CMV-OrfAStop, and CMV-EnvfsOrfAStop mutants as well as the LTR-Envfs construct. The presence of mutations was confirmed by DNA sequencing. A schematic representation of the FIV-derived constructs employed in this study is outlined in Fig. 6A.
Fig 6.
Effect of cBST2 on particle release of FIV Env and/or OrfA mutants. (A) Schematic representation of the FIV derivative constructs. The complete FIV proviral genome encompassing the gag, pol, env, vif, orfA, and rev genes of the p34TF10 Petaluma strain, either the wild type (WT) or characterized by the mutations indicated (Envfs, OrfAStop, and EnvfsOrfAStop), is under the transcriptional control of either the FIV 5′ LTR or the CMV promoter (Pro). The polyadenylation signals (pA) are provided by either the 3′ end of the FIV 3′ LTR or that of the bovine growth hormone transcript. (B) 293T cells were transfected with constructs expressing either the wild-type FIV genome (CMV-WT) or its derivatives, characterized by a frameshift in the envelope-encoding sequence (CMV-Envfs), a stop codon at the amino acid 2 position of the OrfA sequence (CMV-OrfAStop), or a combination of both these changes (CMV-EnvfsOrfAStop), alone (−) or along with (+) 1,500 ng of the cBST2504 construct. (C and D) 293T cells were transfected with the CMV-Envfs construct alone (−), along with increasing amounts of plasmids expressing cBST2504 (50, 100, 150, 200, 250, 500, 750, and 1,000 ng) (C), or with 1,500 ng of cBST2* or cBST2504 (D). Cell lysates (Cells) and purified virus particles (VPs) were analyzed 24 h after transfection by SDS-PAGE, followed by Western blotting employing the anti-FIV Gag monoclonal antibody (Pr50 and p24), as indicated. C represents cells transfected with the pcDNA3.1 empty vector. Numbers below each lane indicate the percentages of particle release. (E) CrFK cells were transfected with constructs expressing either the wild-type FIV genome (LTR-WT) or its derivative, characterized by a frameshift in the envelope-encoding sequence (LTR-Envfs), and treated with IFN-α (+) or mock treated (−). Cell lysates and purified virus particles were analyzed 24 h after IFN-α treatment by SDS-PAGE, followed by Western blotting employing the anti-FIV Gag monoclonal antibody. The histograms display the percent particle release and are representative of one out of three independent experiments. Values obtained for LTR-WT and LTR-Envfs in the absence of IFN-α treatment were set as 100%. The induction of tetherin expression was confirmed by real-time PCR (cBST2 Ct). (F) CrFK cells were transfected with either LTR-WT or its LTR-Envfs derivative alone (−) or along with 1,500 ng of the cBST2504 construct (+). Cell lysates (Cells) and purified virus particles (VPs) were analyzed 24 h after transfection by SDS-PAGE, followed by Western blotting employing the anti-FIV Gag monoclonal antibody (Pr50 and p24), as indicated. C represents cells transfected with the pcDNA3.1 empty vector. Numbers below each lane indicate the percent particle release.
The EVA232 pROD10 reagent containing the proviral DNA of the HIV-2 ROD10 strain (10) was obtained from the Centre for AIDS Reagents. Plasmids p239SpSp5′ and p239SpE3′, encoding the SIVmac239 molecular clone 5′ and 3′ ends, respectively (30, 49), were obtained through the AIDS Research and Reference Reagent Program (ARRP), Division of AIDS, NIAID, NIH. The plasmid encoding the envelope of the in vivo-readapted FIV strain Petaluma (FIV-Petaluma) (pEE14-Env) was described elsewhere previously (47).
The pSVC21 construct expressing the HIV-1 HXBc2 molecular clone (16) was kindly provided by Joseph Sodroski (Harvard Medical School, Boston, MA). SVC Vpu+ is a vpu-positive variant of HXBc2 (22).
All plasmid DNA expression constructs were prepared by using a QIAFilter kit (Qiagen, Milan, Italy), quantified, and stored.
Cell cultures and transfection.
Crandell-Rees feline kidney (CrFK) (ATCC CCL-94TM) cells as well as human 293T (ATCC CRL-11268TM) and HeLa (ATCC CCL-2) cells were grown in Dulbecco's modified Eagle's medium (Gibco, Invitrogen) with the addition of 10% heat-inactivated fetal calf serum (Invitrogen) (complete medium). When necessary and according to the specific experimental procedures, cells were transfected either by the calcium-phosphate method (7) or with Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's instructions.
Identification and cloning of feline BST2.
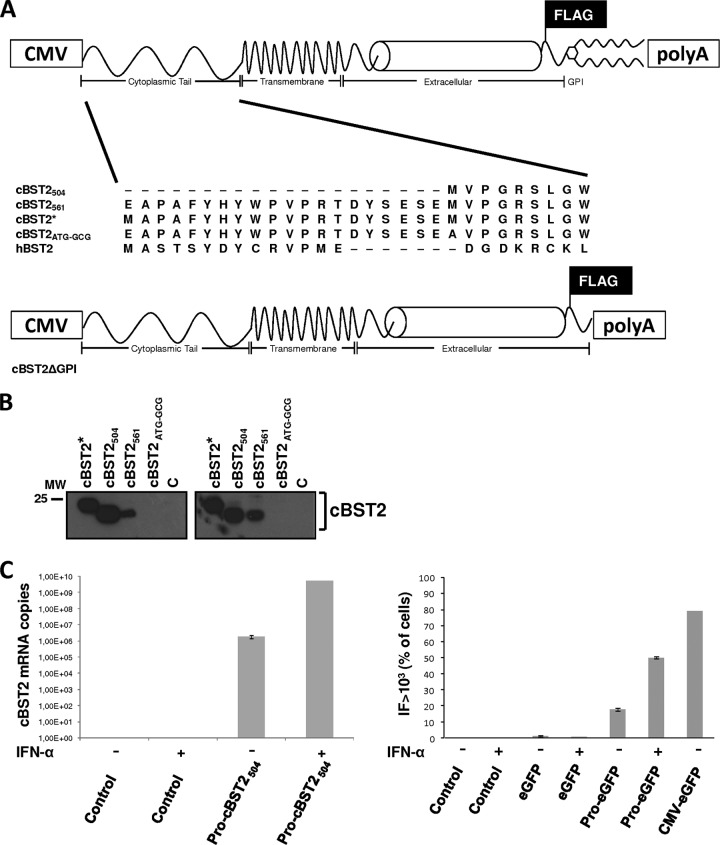
In order to identify a genomic sequence potentially encoding feline tetherin, a CLUSTALW2 algorithm-based analysis was performed by aligning the 2× domestic cat (Felis catus) genome sequence with that of the human tetherin (GenBank accession no. NM_004335). The candidate feline tetherin gene was identified on Felis catus c430601298.contig 1 (GenBank accession no. ACBE01053987). Primers were synthesized in the genomic region: a forward primer located 57 nucleotides upstream of the putative initiation start site (primer A [5′-GCCACCGAGGCACCTGCTTTTTACCACTATTGG-3′]), a second forward primer mapping in the region corresponding to the predicted start codon (primer B [5′-CTACTAGCTAGCGCCACCATGGTGCCAGGTCGGAGTCTTGGGT-3′]), and the reverse primer located in the region corresponding to the stop codon (rev-primer [5′-TCAGGCCAGCAGAGCAACGAAG-3′]) of feline tetherin. Total RNA was extracted from CrFK cells by employing the RNeasy minikit (Qiagen), according to the manufacturer's instructions, and subjected to RT-PCR. The amplified fragments were inserted into the pCR2.1-TOPO vector (Invitrogen), confirmed by DNA sequencing, and cloned into the pcDNA3.1 vector (Invitrogen) to obtain cat tetherin (cBST2) expression vectors. Specifically, two different constructs, cBST2504 and cBST2561, were generated by employing forward primers A and B, respectively. An additional construct, cBST2*, was obtained by introducing a start codon (ATG) in place of the first nucleotide triplet (GAG) of the feline sequence inserted into cBST2561, employing the QuikChange II site-directed mutagenesis kit (Agilent Technologies). Finally, the cBST2561 construct was also subjected to site-directed mutagenesis in order to replace the first ATG of the coding sequence with the GCG triplet, thus obtaining a plasmid called cBST2ATG-GCG. All the constructs were enriched by a PCR-based strategy with a Flag epitope inserted between the predicted extracellular coiled-coil domain of the protein and the predicted GPI anchor signal.
A version of the protein lacking the putative GPI anchor was also generated by RT-PCR using total RNA from CrFK cells and the following primers: forward primer 5′-CTACTAGCTAGCGCCACCATGGTGCCAGGTCGGAGTCTTGGGT-3′ and reverse primer 5′-CTTATCGTCGTCATCCTTGTAATCAGTCCCTTTTCTCAG-3′. A construct encoding a truncated form of cBST2 enriched with a 3′ Flag tag was thus obtained (cBST2ΔGPI).
Identification and analysis of the minimal cBST2 promoter.
By using a CLUSTALW2 algorithm (http://www.ebi.ac.uk/Tools/clustalw2)-based analysis, starting from the putative minimal promoter of hBST2 (41), a corresponding nucleotide sequence was identified on c430601298.contig 1 of the feline genome. The sequence identified spans nucleotides −2192 to +1, considering nucleotide +1 the A of the first cBST2 ATG present in the coding sequence. This region was amplified by PCR performed on genomic DNA extracted from CrFK cells by employing the DNeasy blood and tissue kit (Qiagen). The PCR product was cloned into the pEGFP1 reporter vector (EuroClone) upstream of the eGFP-encoding sequence, at the EcoRI/KpnI sites, generating the Pro-eGFP construct. An additional construct was obtained, Pro-cBST2504, by PCR joining the feline genomic region spanning nucleotides −2192 to +1 with the cBST2504-encoding sequence and by inserting the fragment obtained into the pEGFP1 vector (EuroClone) in place of the eGFP-encoding region.
In order to test the activity of the identified region, 293T cells (3.5 × 105 cells) were seeded into 6-well plates and transfected with either Pro-eGFP or Pro-cBST2504 by using Lipofectamine 2000 reagent (Invitrogen). Transfected cells were either incubated in the presence of 1,000 U/ml of human recombinant alpha interferon A/D (IFN-αA/D) (Sigma, Milan, Italy) or mock incubated. Twenty-four hours after IFN treatment, cells transfected with the Pro-eGFP construct and the appropriate controls (the pEGFP1 reporter vector and pCMV-eGFP) were subjected to flow cytometry analysis by employing a FACSCalibur cytometer (Becton Dickinson, Milan, Italy). The data were analyzed with WinMDI 2.8 software. Mean and peak fluorescence intensities were calculated.
Twelve hours after IFN treatment, cells transfected with Pro-cBST2504 and with the appropriate controls (pcDNA3.1 and cBST2504) were lysed, total RNA was extracted, and quantitative real-time PCR was performed. Briefly, to remove contaminating DNA, i.e., plasmid DNA, RNA was treated with 30 U of RNase-free DNase I (Roche Diagnostics, Milan, Italy) at 37°C for 30 min and heat inactivated at 95°C for 15 min, followed by quick cooling on ice. cDNA was made from 2 μg of total RNA in a 50-μl reaction mixture containing 1 mM Tris-HCl (pH 8.3), 5 mM KCl, 5.5 mM MgCl2, 2.5 mM primer annealing to the Flag epitope sequence, 0.5 mM (each) deoxynucleoside triphosphate (dNTP), 40 U of RNase inhibitor, and 125 U of Moloney murine leukemia virus (Mo-MuLV) reverse transcriptase (Applied Biosystems, Monza, Italy). RNA samples were incubated in the reaction mixture at 25°C for 10 min, at 42°C for 50 min, and then at 95°C for 5 min. The quantitative real-time method was based on the real-time detection of accumulated fluorescence (Taq-Man). The selected target sequence was mapped to cBST2 exon 1. Real-time PCR primers and probe were selected by using Primer-Express software (Applied Biosystems). The forward and reverse primer sequences were 5′-GGCGGTGCTGGGTCTGT-3′ and 5′-TTTGCTGTTGGCCTTGACAA-3′, respectively. The probe (5′-CCCTGGTCATCTTT-3′) contains a fluorescent reporter dye (6-carboxyfluorescein [FAM]) at the 5′ end and a fluorescent quencher dye (6-carboxytetramethylrhodamine [TAMRA]) at the 3′ end. A 58-bp fragment was amplified. The cDNA extracted was assayed with a sequence detector system (ABI Prism 7700; Applied Biosystems) in 50 μl of a PCR mixture containing 25 μl TaqMan Universal Master Mix, 15 pmol each primer, and 10 pmol probe. Thermal cycling conditions were 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s and 60°C for 1 min. A 58-bp PCR fragment, amplified from the cBST504 plasmid, was ligated into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions. The resulting plasmid, pTA-cBST2, was quantified by using a fluorimetric Picogreen method, and a serial dilution was used as a reference for cBST2 quantification. A six-point standard curve, which had a linear course within the set limits, was plotted by using a serial dilution (from 5 × 106 to 50 copies) of plasmid pTA-cBST2. The cBST2 mRNA copy number was calculated automatically with 7700 ABI Prism SDS software (Applied Biosystems).
Western blotting.
For immunoblot analysis, proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electroblotted onto a Protran membrane (Whatman, Milan, Italy). The membranes were incubated with the appropriate primary antibody, namely, a mouse monoclonal anti-FIV capsid antiserum (anti-FIV p24 Gag antibody; AbD Serotec, Milan, Italy); a rabbit polyclonal anti-HIV-1 capsid antiserum (anti-HIV-1 p24 Gag antiserum; ABi Advanced Biotechnologies, Monza, Italy); a mouse monoclonal anti-Flag antibody (Sigma); a mouse monoclonal anti-HIV-1 p24 antibody, which cross-reacts with SIV p27 and HIV-2 p24 (183-H12-5C; ARRP, NIAID, NIH) (68); or a monoclonal anti-FIV envelope antibody (anti-FIV SU alfaenv-71.2 monoclonal antibody; kindly provided by Brian Willett). Blots were visualized with a peroxidase-conjugated anti-mouse or anti-rabbit IgG secondary antibody (GE Healthcare, Milan, Italy) and developed with enhanced chemiluminescence reagents (GE Healthcare), as described elsewhere previously (58). When necessary, the pixel intensities of the visualized bands were determined by using Image J software (W. S. Rasband, NIH, Bethesda, MD [http://rsb.info.nih.gov/ij/]).
Analysis of viral particle release.
Viral particle release assays were performed by transfecting either 293T cells or CrFK cells with the appropriate proviral construct. In particular, 1.2 × 106 293T cells were transfected by the calcium-phosphate technique into 25-cm2 tissue culture flasks with 1.25 μg of the appropriate proviral construct (58). In cotransfection experiments, along with the viral construct, different amounts (from 50 to 1,500 ng) of feline or human tetherin expression vectors were transfected. In all cases, the total amount of transfected DNA was brought to a total of 3.25 μg (293T cells) by the addition of the pcDNA3.1 empty vector (Invitrogen). When required, pcDNA3.1 was also employed as a negative control. On the other hand, 1.5 × 106 CrFK cells in 25-cm2 tissue culture flasks were transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. In this case, 6.5 μg of the appropriate viral construct and, when required, 1,500 ng of feline tetherin expression vectors were transfected. The total amount of DNA was always brought to 8 μg with the pcDNA3.1 empty vector. In both cases, at 24 h posttransfection, the culture supernatants were collected, and the cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS). Supernatants were clarified by low-speed centrifugation and passaged through 0.45-μm-pore-size filters. Released virus-like particles (VLPs) were spun through 20% sucrose cushions for 2 h at 4°C at 125,000 × g in a Beckman SW41 Ti rotor. Pelleted virion particles were lysed in RIPA buffer, and viral proteins were analyzed by SDS-PAGE, followed by Western blotting. Cell lysates were subjected to the same procedure to examine the intracellular Gag and tetherin expression levels. The percentages of the particles released were determined as the ratio between the pixel intensity (evaluated as explained above) of the extracellular capsid Gag band and the relative total capsid Gag pixel intensity (calculated by summing the pixel intensity of the extracellular capsid Gag and that of the corresponding intracellular capsid Gag) and are outlined in the figures. The reduction in the viral particle release, reported as percentages in Results, was obtained from the values described above, taking into consideration the appropriate controls.
Immunofluorescence analysis.
293T cells (3.5 × 105) were grown on glass coverslips. The cells were transfected with the appropriate tetherin construct by employing Lipofectamine 2000 reagent (Invitrogen). Twenty-four hours later, the cells were fixed in paraformaldehyde (2% in phosphate-buffered saline [PBS]) for 20 min at room temperature. Samples were then incubated with the anti-Flag primary mouse monoclonal antibody (Sigma) and with a secondary fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G antibody (DBA, Milan, Italy) diluted with bovine serum albumin (1% [wt/vol] in PBS). The cells were observed with a confocal microscope (Leica).
Coimmunoprecipitation assay.
The cells were lysed in RIPA buffer containing protease inhibitors (0.1 mM N-p-tosyl-l-lysine chloromethyl ketone–0.1 mM tosylsulfonylphenylalanylchloromethyl ketone). Coimmunoprecipitations were carried out with the appropriate antibodies from lysates of transfected cells, as described elsewhere previously (58). The immunocomplexes were harvested with protein G-Sepharose (GE Healthcare).
RESULTS
Feline tetherin/BST2 is characterized by an N-terminal region that is shorter than those of other mammalian and nonmammalian homologues.
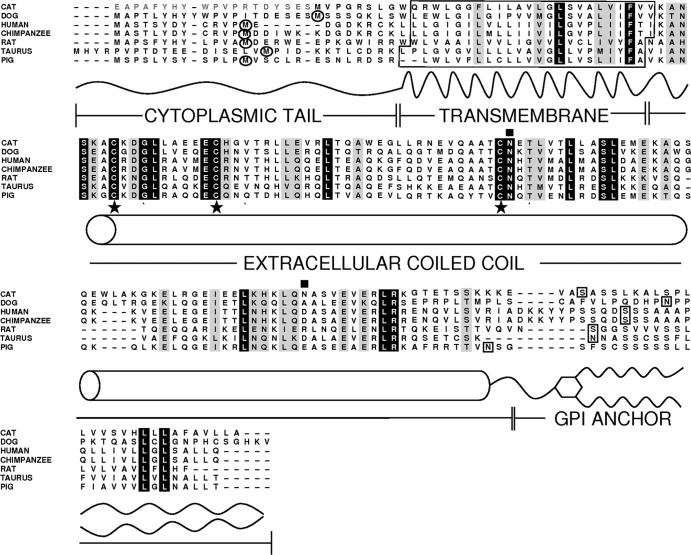
When the 2× Felis catus genome was compared to the sequence of human tetherin using megaBLAST, a region on Felis catus c430601298.contig 1 (GenBank accession no. ACBE01053987) with significant homology to human tetherin/BST2 was identified, potentially encoding a protein called here cat BST2 (cBST2). On the basis of bioinformatic predictions (CLUSTALW2, HMMTOP, big-PI Predictor GPI Modification Site Prediction, and NetNGlyc 1.0 Server), cBST2 appears to have a topology comparable to those of other mammalian and nonmammalian tetherins. In particular, a C-terminal glycosylphosphatidylinositol (GPI) anchor, an extracellular coiled-coil domain, and an N-terminal transmembrane domain have been identified (Fig. 1). When the cBST2 amino acid sequence was aligned, starting from the first available methionine, with those of tetherins from dog (GenBank accession no. XP_865603.1), human (GenBank accession no. NP_004326.1), chimpanzee (GenBank accession no. NP_001177409.1), rat (GenBank accession no. NP_937767), taurus (GenBank accession no. XP_876152.2), and pig (GenBank accession no. NP_001155227.1), an N-terminal cytoplasmic tail that is significantly shorter than those of all the other BST2s considered was found (Fig. 1). On the other end, upstream of the first ATG, the cBST2 sequence identified is characterized by 57 in-frame nucleotides, starting with a GAG codon, which can potentially encode 19 amino acids (highlighted in gray in Fig. 1), displaying significant homology with the N-terminal regions of all the other BST2s analyzed. Dietrich and coworkers recently characterized a putative feline homologue of human tetherin and assessed its effect on viral replication (11). That study was based on a protein containing the above-mentioned 19 amino acids upstream of the first methionine and, most likely, generated by the positioning of an ATG in the place of the GAG codon during the cloning process (11). Indeed, by means of genomic sequencing performed on DNA extracted from Crandell-Rees feline kidney (CrFK) cells, we were able to confirm that there is no canonical start codon at the −57 position (data not shown). In a previous attempt to characterize the antiviral activity of cat tetherin, Fukuma and colleagues identified the initiation site of its coding sequence by using the 5′ rapid amplification of cDNA ends (5′-RACE) technique on total RNA extracted from interferon (IFN)-treated CrFK cells and cloned a protein starting from the first methionine (17). While this putative cBST2 was found to be capable of blocking the production of cat infectious RD114 endogenous retrovirus, its effect on FIV and other lentiviruses was not analyzed (17). In order to investigate the potential relevance of the tetherin N-terminal region, we asked whether physiologically encoded tetherin starts from the first reported methionine or whether an unconventional triplet, located along the sequence upstream of the annotated methionine, functions as an initiation site. Three different constructs (cBST2504, cBST2561, and cBST2*) were thus generated by appropriate amplification from total RNA extracted from CrFK cells (Fig. 2A). Specifically, cBST2561 was obtained by adopting a 5′ primer that anneals to the c430601298.contig 1 sequence from nucleotide −57 to nucleotide −37, where nucleotide +1 is represented by the A of the first annotated ATG (forward primer A) (see Fig. S1 in the supplemental material). The PCR product obtained encompasses 561 nucleotides, as specified in the name of the generated construct; cBST2504 contains the sequence starting from the first ATG present in the cBST2-encoding region identified (forward primer B) (Fig. S1). In this case, the PCR product obtained encompasses 504 nucleotides, as specified in the name of the generated construct; cBST2* was obtained from cBST2561 by the introduction of an ATG in the place of the GAG (from positions −57 to −55) (Fig. S1). In addition, a construct characterized by the ATG replaced by a GCG (from positions +1 to +3) (Fig. S1) was designed as a control (cBST2ATG-GCG). In all cases, a Flag tag was introduced at the 3′ end before the GPI modification signal. Human embryonic 293T and feline kidney CrFK cells were transfected with the different constructs, and 24 h later, cell lysates were analyzed by Flag-specific Western blotting. Figure 2B clearly shows that while cBST2561 and cBST2504 encode a protein characterized by a similar molecular weight, cBST2* leads to the expression of a higher-running product. These results suggest that the first annotated ATG does function as the start codon. In addition, the GAG does not represent a noncanonical start codon in feline cells, as shown by the transfection of 293T and CrFK cells with the cBST2ATG-GCG construct (Fig. 2B). Thus, feline tetherin does indeed have a shorter N-terminal region than those of other homologues described.
Fig 1.
Amino acid sequence alignment of tetherins of cat, dog, human, chimpanzee, rat, taurus, and pig origins. Identical amino acids are shaded in black, and conserved or similar residues are shown in gray. Dashes represent gaps. The predicted topology of feline tetherin is reported and includes an amino-terminal cytoplasmic tail, a transmembrane region, an extracellular coiled-coil domain, and an addiction site for the GPI anchor, boxed in black. Putative N-glycosylation sites in the extracellular coiled-coil domain are indicated by black squares, while cysteines potentially involved in disulfide bounds are marked by stars. The black circles indicate a second methionine mapping at the level of the cytoplasmic tail.
Fig 2.
Characterization of the feline tetherin-encoding region. (A) Schematic representation of the BST2 constructs employed in this study. The predicted BST2 topology is shown. The alignment of the cytoplasmic tails of the different constructs developed is outlined. The cBST2ΔGPI construct represents cBST2 lacking the putative GPI anchor signal. The Flag tag position is indicated. (B) The −57 nucleotide region upstream of the first annotated ATG of feline tetherin does not contain noncanonical initiation start sites. 293T (left) and CrFK (right) cells were transfected with a construct either (i) containing the cBST2-encoding region starting from the first annotated ATG (cBST2504); (ii) encompassing the −57 nucleotides upstream of the first annotated ATG (cBST2561); (iii) characterized by the first nucleotide triplet (GAG) of the cBST2 sequence, cloned into cBST2561, mutagenized into an ATG (cBST2*); or (iv) in which the annotated ATG was replaced by a GCG (cBST2ATG-GCG). At 24 h posttransfection, cell lysates obtained from either 293T or CrFK cells were loaded onto an SDS-polyacrylamide gel, followed by Western blotting with an anti-Flag antibody (cBST2). C represents cells transfected with the pcDNA3.1 empty vector. MW, molecular weight marker (in thousands). (C) The feline genomic region starting 2,192 nucleotides upstream of the cBST2 ATG shows promoter activity and is sensitive to IFN-α treatment. (Left) 293T cells were transfected with the Pro-cBST2504 construct and were either incubated with IFN-α (+) or mock incubated (−). Twelve hours later, quantitative real-time RT-PCR analysis was performed on the total RNA extracted. The cBST2 mRNA copies were calculated by means of a standard curve. (Right) 293T cells were transfected with either Pro-eGFP, CMV-eGFP, or pEGFP1 (eGFP) constructs, as indicated. Cells were either treated with IFN-α (+) or mock incubated (−). Twenty-four hours later, cells were collected for fluorescence-activated cell sorter (FACS) analysis. Histograms show the percentages of cells displaying a mean intensity of GFP fluorescence (IF) above the threshold of 103. Control represents cells transfected with the pcDNA3.1 empty vector.
Characterization of the cBST2 minimal promoter region.
It was previously reported that like those of the other homologues, cBST2 expression can be responsive to alpha interferon (IFN-α) treatment (11, 17). The alignment of the 5′ nucleotide sequence upstream of the functional cBST2 ATG with the putative minimal promoter human sequence (41) made it possible to identify the corresponding region in the feline genome (see Fig. S1 in the supplemental material). As expected, this region contains, in addition to canonical cis elements (CCAAT box, GC box, TATA box, and SP-1), at least two interferon-responsive elements (IRF-1/2) as well as interferon-activated transcription factor binding sites (STAT3 and ISFG3) (Fig. S1). Interestingly, the TSSW prediction program (Softberry) identified the transcription start site 2 nucleotides downstream of the GAG triplet and 55 nucleotides upstream of the ATG. A construct (Pro-cBST2504) was thus generated in which the cBST2-encoding sequence is under the transcriptional control of the feline genomic region spanning from nucleotide −2192 to the annotated ATG. 293T cells were transfected with the construct obtained, either treated with IFN-α or left untreated, and, 12 h later, subjected to total RNA extraction. By using specific real-time RT-PCR analysis, it was possible to demonstrate that the nucleotide sequence selected did indeed show promoter activity and was inducible by IFN-α treatment (Fig. 2C, left). Western blot analysis consistently showed a protein of the size that was expected for cBST2504. The level of expression was, however, very low and not comparable with that achieved by the expression of cBST2504 under the transcriptional control of the CMV promoter and appreciable only after IFN-α treatment (data not shown). When an eGFP-encoding sequence was inserted under the transcriptional control of the putative cBST2 promoter (Pro-eGFP), the reporter protein was expressed in an IFN-α-inducible manner but at a significantly lower level than that achieved under the transcriptional control of the CMV promoter (CMV-eGFP) (Fig. 2C, right). Altogether, these results are consistent with the hypothesis that the feline genomic region selected is a minimal promoter whose activity can be induced by IFN-α.
The N-terminal cytoplasmic region of tetherin plays a role in BST2 antiviral activity and in its response to the viral counteracting factor.
Since these data clearly indicate that feline tetherin has a shorter N-terminal region than those of other homologues described (Fig. 1), it was important to investigate whether this structural feature could have an impact on the protein's biological and functional characteristics. With this aim, 293T cells were transfected with either cBST2*, cBST2504, or human BST2 (hBST2), as a control. Twenty-four hours later, the cells were lysed, and the proteins were run in SDS-polyacrylamide gels both in the presence and in the absence of β-mercaptoethanol (βME). It was found that both feline proteins were able to dimerize, a feature which has been demonstrated to play a role in the ability of human tetherin to restrict HIV-1 particle release (1, 45) (Fig. 3A), and were correctly expressed on the cell surface (Fig. 3B to M).
Fig 3.
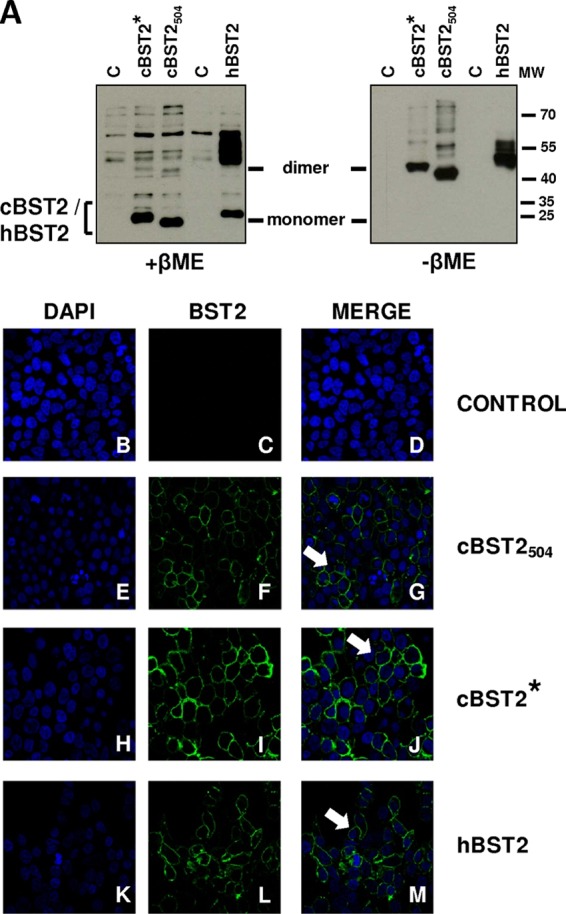
cBST2 dimerizes and is expressed on the cell surface, independently from the N-terminal cytoplasmic tail length. (A) 293T cells were transfected with either cBST2*, cBST2504, or hBST2 as a control. Twenty-four hours later, cell lysates were run in denaturing β-mercaptoethanol (+βME) (left) or nondenaturing β-mercaptoethanol (−βME) (right) gels, followed by Western blotting with an anti-Flag monoclonal antibody. C represents cells transfected with the pcDNA3.1 empty vector. MW, molecular weight marker (in thousands). (B to M) 293T cells were transfected with the tetherin constructs described above. Twenty-four hours later, the cells were stained with a monoclonal antibody to Flag and analyzed by confocal microscopy (magnification, ×40). The arrows point to membrane-localized tetherin. Control represents cells transfected with the pcDNA3.1 empty vector. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
A cBST2 protein resembling our cBST2* protein was previously reported to be active against HIV-1 in a Vpu-independent manner (11). Human 293T cells were cotransfected with 1.25 μg of either the HIV-1 vpu-minus (Vpu−) strain or the isogenic Vpu-encoding HIV strain (HIV-1 Vpu+) along with the Flag-tagged cBST2504- or cBST2*-encoding construct to investigate the restriction abilities of cBST2* and cBST2504. cBST2504 displayed activity against the HIV-1 Vpu− strain (Fig. 4A) as well as the HIV-1 Vpu+ strain (Fig. 4B) in a dose-responsive manner. Interestingly, while 250 ng of the cBST2504 construct was sufficient to cause roughly 50% and 40% reductions in HIV-1 Vpu+ and Vpu− strain budding, respectively (Fig. 4A and B), cBST2* was not so effective. In agreement with this finding, while 1,000 ng of the cBST2504 construct reduced HIV-1 Vpu− strain particle release by approximately 80% compared to the control (Fig. 4A), with the same amount of cBST2*, a reduction in particle release of roughly 30% was achieved (Fig. 4C). Even when 1,500 ng of the cBST2* construct was employed, a roughly 45% reduction in viral release with respect to that of the HIV-1 Vpu− strain (Fig. 4D) and an approximately 25% reduction with respect to that of the HIV-1 Vpu+ strain (Fig. 4E) were observed. As demonstrated previously in the case of human tetherin (40, 45), the deletion of the GPI anchor abrogated the ability of feline tetherin to block HIV-1 release regardless of the presence of Vpu (Fig. 4D and E, lane 6).
Fig 4.
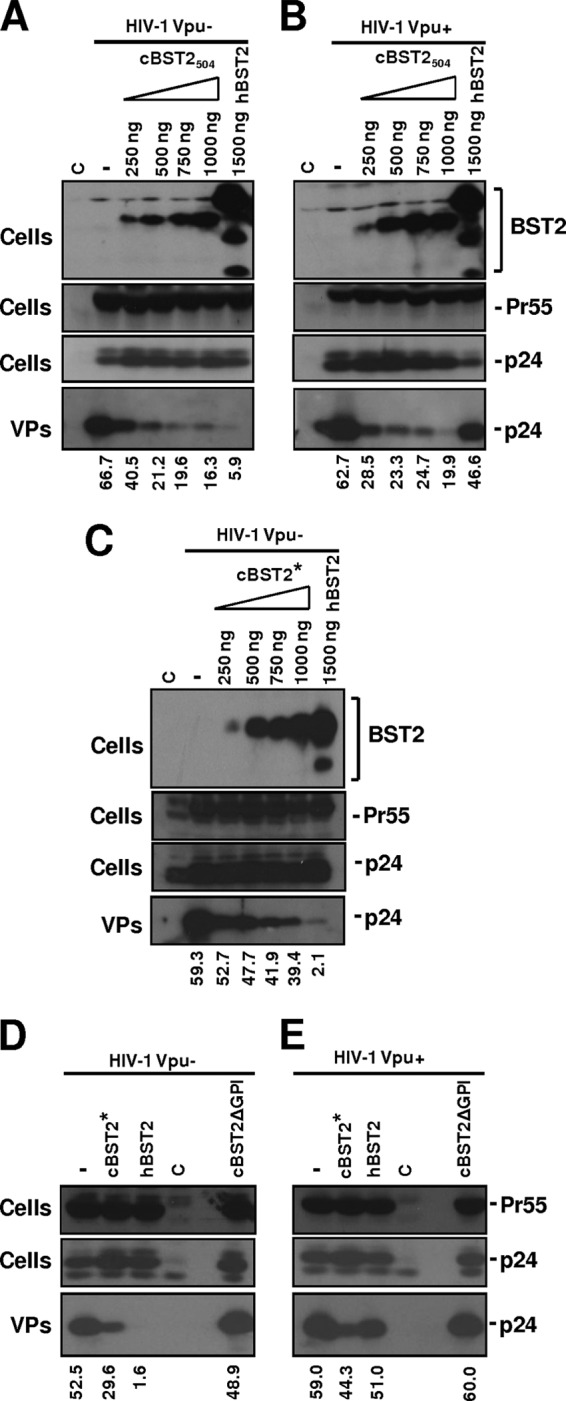
Effect of cBST2 on Vpu− and Vpu+ HIV-1 strain particle release. (A to C) The HIV-1 Vpu−- or HIV-1 Vpu+-expressing constructs (1.25 μg) were transfected into 293T cells alone (−), in the presence of 1,500 ng of hBST2, or along with increasing amounts (250, 500, 750, and 1,000 ng) of cBST2504 (A and B) or cBST2* (C), as indicated. (D and E) 293T cells were transfected with either HIV-1 Vpu−- or HIV-1 Vpu+-expressing plasmids alone (−) or along with 1,500 ng of either the cBST2*, the hBST2, or the cBST2ΔGPI plasmid. At 24 h posttransfection, the cells were lysed, and the viral particles (VPs) were spun down by ultracentrifugation. Proteins derived from the cell lysates (Cells) and from viral particles were analyzed by SDS-PAGE, followed by Western blotting with an anti-Flag monoclonal antibody (BST2) or an anti-HIV-1 Gag monoclonal antibody (Pr55 and p24), as indicated. The numbers below each panel represent the percentage values of particle release, as specified in Materials and Methods. C represents cells transfected with the pcDNA3.1 empty vector.
Overall, these data highlight the relevant role played by the N-terminal cytoplasmic region of feline tetherin in terms of antiviral activity and the response to the viral counteracting factor.
The envelope glycoprotein is the FIV factor that counteracts the effect of cBST2.
BST2 has been shown to be targeted and functionally inactivated by the evolution of antagonistic accessory proteins (23, 24, 29, 31, 35, 73). Primate lentiviruses have indeed evolved the ability to use three proteins to antagonize tetherin, i.e., HIV-1 Vpu, Nef, and envelope proteins from some simian immunodeficiency viruses as well as the HIV-2 envelope protein. Thus, it was important to determine if, like other lentiviruses, FIV encodes a factor capable of counteracting the effect of cBST2 on particle release. The first step was to analyze the activity of the cellular protein upon wild-type virus budding. 293T cells were cotransfected with constructs expressing either cBST2504, cBST2*, or hBST2, along with a plasmid expressing the full FIV genome, under the transcriptional control of the CMV promoter (Fig. 5). As expected, while hBST2 efficiently blocked FIV budding (Fig. 5B), cBST2504 did not efficiently inhibit viral particle release (Fig. 5A), even when 1,000 ng of the expression plasmid was transfected. Indeed, the percent budding reduction observed was comparable to that obtained when a similar amount of hBST2 was expressed in HIV Vpu+ strain-positive cells (data not shown). Moreover, no clear dose-response effect was observed. Finally, the inability of cBST2* to efficiently restrict FIV budding was also evident (Fig. 5B). These data indicate that the FIV genome encodes a viral counteracting factor.
Fig 5.
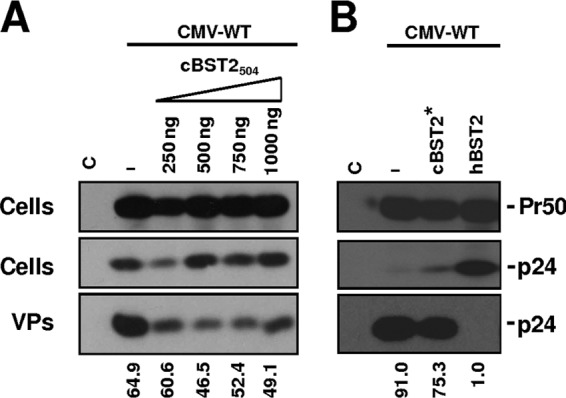
Effect of cBST2 on wild-type FIV particle release. 293T cells were transfected with the construct expressing the wild-type FIV genome (CMV-WT) alone (−) or along with either increasing amounts of the cBST2504 plasmid (250, 500, 750, and 1,000 ng) (A) or 1,500 ng of the cBST2* or hBST2 construct (B). At 24 h posttransfection, the cells were lysed, and the viral particles (VPs) were spun down by ultracentrifugation. Proteins derived from the cell lysates (Cells) and from viral particles were analyzed by SDS-PAGE, followed by Western blotting with an anti-FIV Gag monoclonal antibody (Pr50 and p24), as indicated. The numbers below each panel represent the percentage values of particle release. C represents cells transfected with the pcDNA3.1 empty vector.
Given the absence of an FIV open reading frame with significant homology to vpu, we focused our attention on the envelope and OrfA proteins. Series of viral mutants expressing the full FIV genome containing a frameshift introduced at the level of nucleotide 268 of the envelope-encoding region (Envfs) or a stop codon at the level of amino acid 2 of the OrfA sequence (OrfAStop) were generated, either alone or in combination (EnvfsOrfAStop) (Fig. 6A), and their particle release, both in 293T and in CrFK cells, in the presence of cBST2504 was assayed. As shown in Fig. 6, the expression of cBST2504 potently blocked the particle release of FIV mutants, characterized by a frameshift mutation in the envelope-encoding sequence in 293T cells (Fig. 6B). On the other hand, a significant contribution of FIV OrfA as a tetherin-counteracting factor was not observed. The dose-response effect of cBST2504 on CMV-Envfs particle release further supports these conclusions (Fig. 6C). In contrast, the effect was significantly less evident when cBST2* was adopted (Fig. 6D). To further confirm these results, CrFK cells were transfected with either the LTR-WT or LTR-Envfs constructs and incubated with IFN-α or mock incubated. Twenty-four hours later, VLPs were purified from the cell supernatants. As shown in Fig. 6E, IFN-α treatment slightly impaired LTR-Envfs particle release, while LTR-WT was not affected. Since IFN-α is expected to display pleiotropic antiviral activities suppressing viral growth at different levels (11), to further correlate the observed phenotype with feline tetherin activity against LTR-Envfs particle release, CrFK cells were cotransfected with either LTR-WT or LTR-Envfs constructs along with feline tetherin. Under these experimental conditions, a potent inhibition of the LTR-Envfs particle release was observed (Fig. 6F). Overall, these data strongly indicate that the envelope represents the predominant viral gene product by which FIV counteracts restriction by tetherin.
Moreover, as shown in Fig. 7A, the expression of the FIV envelope glycoprotein in trans was capable of rescuing the budding of CMV-Envfs particles when cells were cotransfected with 150 ng of the cBST2504-expressing construct. Interestingly, the FIV envelope glycoprotein derived from in vivo-readapted FIV-Petaluma (Env) was unable to rescue CMV-Envfs particle release, even when human tetherin was expressed at a very low level (50 and 30 ng) (Fig. 7B and C, respectively). This result indicates that the counteracting activity of the viral glycoprotein is specific to feline tetherin. To support these findings, we used an immunoprecipitation assay and showed that feline tetherin interacts with the FIV-Petaluma envelope glycoprotein in 293T cells transfected with a construct expressing the entire viral genome (CMV-WT), while human tetherin does not (Fig. 7D and E, respectively).
Fig 7.
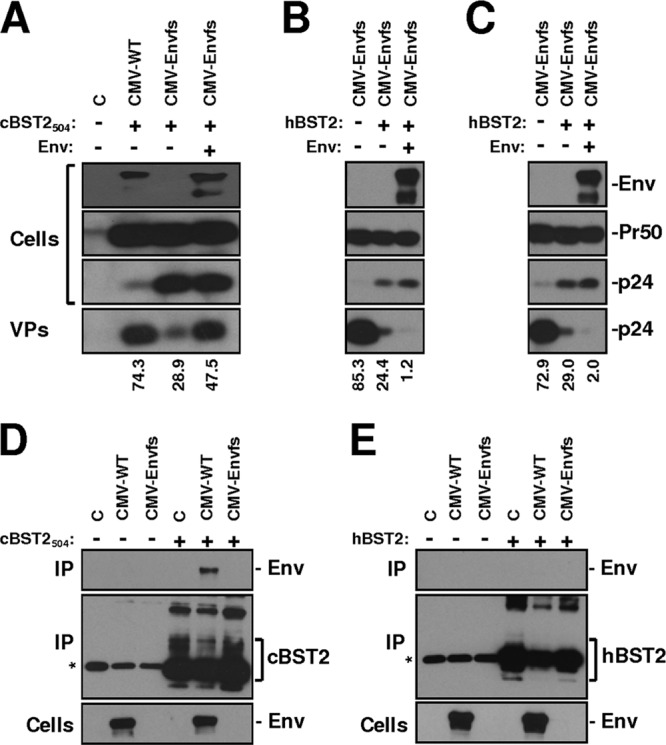
The FIV envelope glycoprotein represents the viral factor counteracting cBST2 activity. (A) 293T cells were transfected with constructs expressing either the wild-type FIV genome (CMV-WT) or its derivative, characterized by a frameshift in the envelope-encoding sequence (CMV-Envfs), along with 150 ng of the cBST2504 construct either alone or in the presence of 500 ng of the pEE14-Env construct (Env). (B and C) 293T cells were transfected with CMV-Envfs along with either 50 ng (B) or 30 ng (C) of the hBST2 construct alone or in the presence of 500 ng of the pEE14-Env construct (Env). Cells (Cells) and purified virus particles (VPs) were analyzed 24 h after transfection by SDS-PAGE, followed by Western blotting employing an anti-FIV Env (Env) or an anti-FIV Gag (Pr50 and p24) monoclonal antibody. Numbers below each lane indicate the percent particle release. (D and E) 293T cells were transfected with 4 μg of either the CMV-WT or CMV-Envfs constructs alone (−) or along with (+) either 4 μg of the cBST2504 (D) or 500 ng of the hBST2 plasmid (E). Twenty-four hours later, cells were lysed, and samples were immunoprecipitated with a monoclonal antibody to Flag and analyzed by Western blotting with a monoclonal antibody to FIV Env (Env) or to Flag (cBST2 and hBST2), as indicated (IP). Cell lysates were also loaded and assayed by Western blotting with a monoclonal antibody to FIV Env (Cells). C represents cells transfected with the pcDNA3.1 empty vector. The asterisks point to the chain of the antibody.
cBST2 potently inhibits wild-type HIV-2 ROD10 and SIVmac239.
It has been demonstrated that host restriction factors, such as tetherin, can impede viral cross-species transmission because the virus-encoded antagonists are usually species specific. As shown above, cBST2 efficiently blocks HIV-1 budding independently from the presence of the Vpu viral protein. To gain a better understanding of this aspect, the effect of cBST2504 expression was investigated by using 293T cells cotransfected with constructs expressing either HIV-2 ROD10, SIVmac293, or the HIV-1 Vpu+ strain, as a control. At 24 h posttransfection, cell supernatants were collected, and the viral particles released were analyzed. As shown in Fig. 8, cBST2504 blocked the budding of all the lentiviruses tested, thus demonstrating that, like the other homologues previously characterized, feline tetherin has a broad spectrum of antiviral activity.
Fig 8.
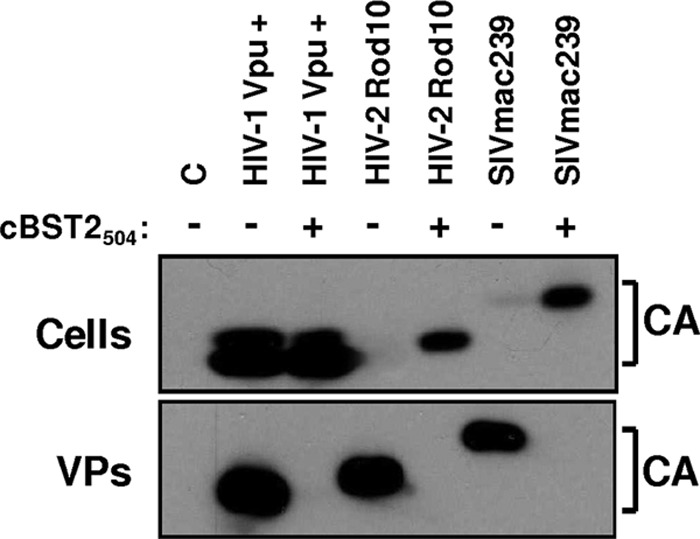
Effect of cBST2 on HIV-2 ROD10 and SIVmac239 particle release. 293T cells were transfected with constructs expressing either the HIV-1 Vpu+, HIV-2 ROD10, or SIVmac239 proviral DNA alone (−) or along with 1,500 ng of cBST2504 (+). Twenty-four hours later, cell lysates (Cells) and purified viral particles (VPs) were analyzed by Western blotting with an anti-capsid HIV-1/HIV-2/SIV polyclonal antibody (CA). C represents cells transfected with the pcDNA3.1 empty vector.
DISCUSSION
The expression of specific cellular proteins capable of interfering in various steps of the viral replication cycle is one of the most intriguing innate immunity mechanisms that have evolved in human and nonhuman primates to limit viral replication. These proteins are known collectively as “host restriction” factors and are often expressed under the transcriptional control of interferons. In particular, the role of three restriction proteins, APOBEC3G, TRIM5α, and, more recently, tetherin/BST2, has been extensively investigated in the context of retroviral life cycle control, with particular attention to the main human pathogen belonging to the Lentiviridae subfamily, human immunodeficiency virus (HIV). Although divergent at the amino acid level, feline immunodeficiency virus (FIV) shares several structural and pathophysiological similarities with HIV. Indeed, the domestic cat (Felis catus), the natural host for FIV, is considered a useful model for AIDS research to prevent and treat HIV-1 infection of humans (14, 62, 69–71). Feline restriction factors, key proteins that potently defend mammals against viral infection, are being studied with the prospect of developing innovative therapeutic approaches based on antiviral proteins in the domestic cat (11, 37).
Two independent studies recently reported that the feline genome carries a gene with significant identity to the known primate, rodent, and canine tetherin sequences, indicating that cats may harbor a homologue of tetherin/BST2 (11, 17), and described two different forms of feline tetherin. In one of those works, Fukuma and coworkers characterized a protein whose N-terminal region is shorter than those of most of the homologues described previously and demonstrated that it blocks the production of infectious feline endogenous retrovirus RD114. The effect of this feline tetherin on FIV and other lentiviruses, however, was not investigated (17). Dietrich and collaborators instead expressed a longer form of feline tetherin, closely resembling human tetherin, and analyzed its effect on HIV and FIV particle release as well as on FIV replication (11). Using these studies as a starting point, we investigated which of the two feline tetherins is the physiological one and further assessed their abilities to interfere with lentiviral particle release. The results of our study indicate the following.
(i) According to the Felis catus c430601298.contig 1 sequence reported in GenBank (GenBank accession no. ACBE01053987) and according to the 5′-RACE data outlined previously by Fukuma and coworkers (17), the first ATG present in the tetherin-encoding region is located 57 nucleotides downstream of the start codon proposed by Dietrich and coworkers (11). It was found that this ATG leads to the production of a protein, termed cBST2504, which is able to dimerize and localizes correctly to the plasma membrane.
(ii) The GAG triplet located 57 nucleotides upstream of the first ATG does not function as an unconventional start codon in feline or human cells, at least under our experimental conditions. Indeed, the transfection of CrFK and 293T cells with a construct containing the cBST2-encoding sequence starting from the GAG and mutated at the first ATG (cBST2ATG-GCG) did not lead to the production of detectable levels of feline tetherin. In contrast, the use of the same construct characterized by the presence of wild-type ATG (cBST2561) led to the expression of a protein having the expected size of cBST2504. Finally, the mutation of the GAG to an ATG within a Kozak-optimized context (cBST2*) led to the expression of a larger protein, which likewise dimerizes and localizes to the plasma membrane.
(iii) The feline genomic sequence from nucleotide −2192 to the first ATG displays promoter activity and is inducible by IFN treatment. As expected, in addition to canonical promoter cis elements (CCAAT box, GC box, TATA box, and SP-1), this region contains at least two interferon-responsive elements (IRF-1/2) as well as interferon-activated transcription factor binding sites (STAT3 and ISFG3) (see Fig. S1 in the supplemental material). Moreover, prediction analyses localized the putative transcription start site 2 nucleotides downstream of the GAG codon and 55 nucleotides upstream of the first ATG. Interestingly, when attempts were made to visualize the protein expressed by a construct in which the cBST2-encoding sequence was under the transcriptional control of the feline genomic region from nucleotide −2192 to the annotated ATG, a protein that was the same size as that expected for cBST2504 (data not shown) was consistently observed. However, the level of expression was very low compared to that achieved by the overexpression of the protein under the transcriptional control of the CMV promoter and was appreciable only after IFN-α treatment. Notwithstanding the responsiveness to IFN treatment, this result indicates that regions other than the ones analyzed contribute to cBST2 transcription. On the other hand, it was shown previously that endogenous transcripts in IFN-treated cells display significantly lower levels than those under the transcriptional control of strong promoters (i.e., CMV) (11).
Overall, our findings support the conclusion that the shorter form of feline tetherin is the physiological one. It is noteworthy that a shorter N-terminal region implies the lack of different putative sorting and ubiquitination motifs (21), which, as recently reported (60), may be important for the responsiveness to the viral counteracting factor. With respect to mammalian tetherins, cBST2 does not, however, represent the first case of a protein with a short N-terminal region lacking putative trafficking motifs. Indeed, Arnaud and coworkers demonstrated previously that the sheep genome contains two BST2 genes in tandem generated by gene duplication, encoding two tetherins (2). Interestingly, these forms are expressed in vivo in different tissues, and they both display antiviral activity. In addition, they both lack the two tyrosines in the cytoplasmic tail, which represents, in the case of human tetherin, a noncanonical internalization motif responsible for protein endocytosis from the cell surface (34, 50). Moreover, oBST2A, one of the two ovine tetherins, displays a cytoplasmic tail 11 amino acids shorter than the N-terminal region of human tetherin. This shorter isoform is, however, characterized by a stronger antiviral activity than that found for the longer one (2). Those findings indicate that a short cytoplasmic tail lacking putative functional motifs can be encoded and can display antiviral activity. Our observation that some tetherin homologues contain a second methionine within the cytoplasmic tail downstream of the double tyrosine motifs supports this conclusion, as it suggests that a shorter functional version of tetherin can potentially be encoded, even in mammals other than cats and sheep.
(iv) Neither cBST2504 nor cBST2* significantly blocks the budding of wild-type FIV particles. By contrast, both cBST2504 and cBST2* inhibited HIV particle release, with the only appreciable difference being that there was a stronger effect of the shorter version. cBST2504, moreover, potently inhibited HIV-2 ROD10 and SIVmac239 particle release. These data indicate that both cBST2504 and cBST2* display antiviral activities.
(v) cBST2504 and, to a lesser extent, cBST2* expression, both in human and in feline cells, potently inhibited the particle release of an FIV mutant containing a frameshift in the envelope-encoding sequence (FIV CMV/LTR-Envfs). Moreover, IFN-α treatment also reduced LTR-Envfs particle release from CrFK cells. This evidence indicates that the FIV envelope glycoprotein is the viral factor counteracting tetherin. The finding that the transient expression of the FIV envelope glycoprotein significantly rescues viral mutant budding in the presence of tetherin supports this hypothesis. To date, three viral proteins have been described as tetherin-counteracting factors: Vpu, Nef, and envelope. The FIV genome does not contain a Vpu- or a Nef-encoding sequence, and OrfA expression does not significantly affect tetherin antiviral activity (11; this report). Thus, the possibility that the viral envelope can display an antitetherin function is not a surprising one. While it is true that the FIV envelope glycoprotein did not exhibit this kind of activity in the study by Dietrich et al., the discrepancy between their and our conclusions might be explained by the different approaches employed. Indeed, while we utilized viral mutants that are unable to express the envelope glycoprotein and tested their abilities to produce particles in the context of tetherin-expressing cells, Dietrich et al. performed an assay in which VSV-G/FIV pseudotypes were prepared in the presence or absence of both feline tetherin and a replication-defective molecular clone of FIV (CMVG8MΔpol4) and then titrated on 293T cells. Moreover, while we tested the ability of transiently expressed FIV envelope glycoprotein to rescue CMV-Envfs particle release from cells expressing tetherin, Dietrich et al. tested the ability of the FIV surface glycoprotein to rescue HIV-1 particle release in the presence of tetherin. Finally, Dietrich and coworkers employed a form of tetherin that resembles our cBST2*. While we could still see the effect of this tetherin version on LTR/CMV-Envfs budding, it was not as strong as the one obtained by the expression of cBST2504. Finally, under our experimental conditions, cBST2ATG-GCG was unable to block the release of LTR/CMV-Envfs (data not shown), which provides the final evidence that the first annotated ATG located in the feline tetherin-encoding region is the functional translation start codon and that the upstream GAG does not function as an unconventional one.
It has to be mentioned that, also under the experimental conditions of Dietrich et al., feline tetherin showed a potent effect on particle release driven by a Gag/Pol-expressing construct lacking different viral accessory proteins as well as, and most significantly for our conclusions, the viral envelope glycoprotein. This finding along with the fact that in the present study no effect of feline tetherin on wild-type FIV replication could be observed strongly support the hypothesis that there is a gene within the feline genome that expresses a factor that counteracts the host defense.
(vi) The FIV envelope interacts with feline tetherin in coimmunoprecipitation assays. As the viral glycoprotein was unable to immunoprecipitate human tetherin and to counteract the inhibitory effect displayed by human tetherin on FIV particle egress, this strongly suggests that its effect is specific for the feline antiviral factor.
In conclusion, our findings provide strong evidence that feline tetherin has an intracytoplasmic N-terminal region that is shorter than those of most homologues characterized so far and that it is capable of efficiently inhibiting FIV particle release in the absence of the viral envelope glycoprotein. Further studies are required to identify the molecular mechanisms underlying the envelope protein's ability to counteract tetherin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Istituto Superiore di Sanità (grant number 40H98 to C.P.), the University of Padova (Ex 60 to A.C., G.P., and C.P.), and Regione Veneto.
We thank the Centre for AIDS Reagents, NIBSC HPA UK, supported by the EC FP6/7 Europrise Network of Excellence; the NGIN consortia; the Bill and Melinda Gates GHRC-CAVD project; and the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, for providing some of the reagents employed in this study. We are grateful to Samuele Asnicar for technical assistance with the real-time experiments.
Footnotes
Published ahead of print 18 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Andrew AJ, Miyagi E, Kao S, Strebel K. 2009. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology 6:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnaud F, et al. 2010. Interplay between ovine bone marrow stromal cell antigen 2/tetherin and endogenous retroviruses. J. Virol. 84:4415–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bendinelli M, et al. 1995. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin. Microbiol. Rev. 8:87–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115 [DOI] [PubMed] [Google Scholar]
- 5. Blasius AL, et al. 2006. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naïve mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. 177:3260–3265 [DOI] [PubMed] [Google Scholar]
- 6. Brown EW, Yuhki N, Packer C, O'Brien SJ. 1994. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J. Virol. 68:5953–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calistri A, et al. 2009. Role of the feline immunodeficiency virus L-domain in the presence or absence of Gag processing: involvement of ubiquitin and Nedd4-2s ligase in viral egress. J. Cell. Physiol. 218:175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carpenter MA, et al. 1996. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor). J. Virol. 70:6682–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Z, et al. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 71:3953–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clavel F, et al. 1986. Molecular cloning and polymorphism of the human immune deficiency virus type 2. Nature 324:691–695 [DOI] [PubMed] [Google Scholar]
- 11. Dietrich I, et al. 2011. Feline tetherin efficiently restricts release of feline immunodeficiency virus but not spreading of infection. J. Virol. 85:5840–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Douglas JL, et al. 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/tetherin via a betaTrCP-dependent mechanism. J. Virol. 83:7931–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dube M, et al. 2009. Suppression of tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J. Virol. 83:4574–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elder JH, Lin YC, Fink E, Grant CK. 2010. Feline immunodeficiency virus (FIV) as a model for study of lentivirus infections: parallels with HIV. Curr. HIV Res. 1:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fadel HJ, Poeschla EM. 2011. Retroviral restriction and dependency factors in primates and carnivores. Vet. Immunol. Immunopathol. 143:179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fisher AG, Collati E, Ratner L, Gallo RC, Wong-Staal F. 1985. A molecular clone of HTLV-III with biological activity. Nature 316:262–265 [DOI] [PubMed] [Google Scholar]
- 17. Fukuma A, Abe M, Morikawa Y, Miyazawa T, Yasuda J. 2011. Cloning and characterization of the antiviral activity of feline tetherin/BST-2. PLoS One 6:e18247 doi:10.1371/journal.pone.0018247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao F, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 19. Goff SP. 2004. Retrovirus restriction factors. Mol. Cell 16:849–859 [DOI] [PubMed] [Google Scholar]
- 20. Goffinet C, et al. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285–297 [DOI] [PubMed] [Google Scholar]
- 21. Goffinet C, et al. 2010. Antagonism of CD317 restriction of human immunodeficiency virus type 1 (HIV-1) particle release and depletion of CD317 are separable activities of HIV-1 Vpu. J. Virol. 84:4089–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1993. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. U. S. A. 90:7381–7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gupta RK, et al. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5:e1000443 doi:10.1371/journal.ppat.1000443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta RK, et al. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Natl. Acad. Sci. U. S. A. 106:20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614 [DOI] [PubMed] [Google Scholar]
- 26. Hauser H, et al. 2010. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hofmann-Lehmann R, et al. 1996. Prevalence of antibodies to feline parvovirus, calicivirus, herpesvirus, coronavirus, and immunodeficiency virus and of feline leukemia virus antigen and the interrelationship of these viral infections in free-ranging lions in east Africa. Clin. Diagn. Lab. Immunol. 3:554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huthoff H, Towers GJ. 2008. Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha. Trends Microbiol. 16:612–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jia B, et al. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429 doi:10.1371/journal.ppat.1000429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kestler H, et al. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109–1112 [DOI] [PubMed] [Google Scholar]
- 31. Le Tortorec A, Neil SJ. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 83:11966–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malim MH, Emerman M. 2008. HIV-1 accessory proteins ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 33. Mangeat B, et al. 2009. HIV-1 Vpu neutralizes the antiviral factor tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 5:e1000574 doi:10.1371/journal.ppat.1000574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Masuyama N, et al. 2009. HM1.24 is internalized from lipid rafts by clathrin-mediated endocytosis through interaction with alpha-adaptin. J. Biol. Chem. 284:15927–15941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McNatt MW, et al. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5:e1000300 doi:10.1371/journal.ppat.1000300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miyagi E, Andrew AJ, Kao S, Strebel K. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. U. S. A. 106:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neagu MR, et al. 2009. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J. Clin. Invest. 119:3035–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. 2006. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2:e39 doi:10.1371/journal.ppat.0020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 41. Ohtomo T, et al. 1999. Molecular cloning and characterization of a surface antigen preferentially overexpressed on multiple myeloma cells. Biochem. Biophys. Res. Commun. 258:583–591 [DOI] [PubMed] [Google Scholar]
- 42. Pecon-Slattery J, Troyer JL, Johnson WE, O'Brien SJ. 2008. Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology. Vet. Immunol. Immunopathol. 123:32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pedersen NC, Ho EW, Brown ML, Yamamoto JK. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790–793 [DOI] [PubMed] [Google Scholar]
- 44. Pedersen NC, Yamamoto JK, Ishida T, Hansen H. 1989. Feline immunodeficiency virus infection. Vet. Immunol. Immunopathol. 21:111–129 [DOI] [PubMed] [Google Scholar]
- 45. Perez-Caballero D, et al. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pistello M, et al. 2007. Streamlined design of a self-inactivating feline immunodeficiency virus vector for transducing ex vivo dendritic cells and T lymphocytes. Genet. Vaccines Ther. 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pistello M, et al. 2010. Env-expressing autologous T lymphocytes induce neutralizing antibody and afford marked protection against feline immunodeficiency virus. J. Virol. 84:3845–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poeschla EM, Looney DJ. 1998. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J. Virol. 72:6858–6866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Regier DA, Desrosiers RC. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 6:1221–1231 [DOI] [PubMed] [Google Scholar]
- 50. Rollason R, Korolchuk V, Hamilton C, Schu P, Banting G. 2007. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J. Cell Sci. 21:3850–3858 [DOI] [PubMed] [Google Scholar]
- 51. Sauter D, et al. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sawyer SL, Emerman M, Malik HS. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:e275 doi:10.1371/journal.pbio.0020275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Serra-Moreno R, Jia B, Breed M, Alvarez X, Evans DT. 2011. Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9:46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 56. Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 57. Shimojima M, et al. 2004. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science 303:1192–1195 [DOI] [PubMed] [Google Scholar]
- 58. Strack B, Calistri A, Accola MA, Palù G, Gottlinger HG. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. U. S. A. 97:13063–13068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stremlau M, et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 60. Tokarev AA, Munguia J, Guatelli JC. 2011. Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J. Virol. 85:51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tomonaga K, et al. 1992. Identification of a feline immunodeficiency virus gene which is essential for cell-free virus infectivity. J. Virol. 66:6181–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Troyer JL, et al. 2004. Patterns of feline immunodeficiency virus multiple infection and genome divergence in a free-ranging population of African lions. J. Virol. 78:3777–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Troyer JL, et al. 2005. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J. Virol. 79:8282–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Troyer JL, et al. 2008. FIV cross-species transmission: an evolutionary prospective. Vet. Immunol. Immunopathol. 123:159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Van Damme N, et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. VandeWoude S, Apetrei C. 2006. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 19:728–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vidal-Laliena M, et al. 2005. Characterization of antibodies submitted to the B cell section of the 8th Human Leukocyte Differentiation Antigens Workshop by flow cytometry and immunohistochemistry. Cell. Immunol. 236:6–16 [DOI] [PubMed] [Google Scholar]
- 68. Wehrly K, Chesebro B. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12:288–293 [DOI] [PubMed] [Google Scholar]
- 69. Willett BJ, Flynn JN, Hosie MJ. 1997. FIV infection of the domestic cat: an animal model for AIDS. Immunol. Today 18:182–189 [DOI] [PubMed] [Google Scholar]
- 70. Willett BJ, Hosie MJ, Neil JC, Turner JD, Hoxie JA. 1997. Common mechanism of infection by lentiviruses. Nature 385:587. [DOI] [PubMed] [Google Scholar]
- 71. Wongsrikeao P, Saenz D, Rinkoski T, Otoi T, Poeschla E. 2011. Antiviral restriction factor transgenesis in the domestic cat. Nat. Methods 8:853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yamamoto JK, et al. 1988. Pathogenesis of experimentally induced feline immunodeficiency virus infection in cats. Am. J. Vet. Res. 49:1246–1258 [PubMed] [Google Scholar]
- 73. Zhang F, et al. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.