Abstract
The Epstein-Barr nuclear antigen 1 (EBNA1) protein of Epstein-Barr virus (EBV) is expressed in both latent and lytic modes of EBV infection and contributes to EBV-associated cancers. Using a proteomics approach, we profiled EBNA1-host protein interactions in nasopharyngeal and gastric carcinoma cells in the context of latent and lytic EBV infection. We identified several interactions that occur in both modes of infection, including a previously unreported interaction with nucleophosmin and RNA-mediated interactions with several heterogeneous ribonucleoproteins (hnRNPs) and La protein.
TEXT
Epstein-Barr virus (EBV) is normally found in a latent mode of infection, which can contribute to the development of several cancers, including nasopharyngeal carcinoma (NPC) and gastric carcinoma (8, 17). Latent EBV can also be reactivated to enter the lytic cycle in which virions are produced. Epstein-Barr nuclear antigen 1 (EBNA1) is critical for latent infection, where it mediates the persistence of the EBV genomes and alters the cellular environment to promote cell proliferation and survival (7). All of these roles involve EBNA1 interactions with specific cellular proteins, and proteomics methods, including tandem affinity purification (TAP) tagging and affinity column profiling, have been particularly informative in identifying functional connections between EBNA1 cellular pathways (10). However, to date in vivo proteomics methods have been performed only for EBNA1 in 293T cells (due to their ease of transfection), which are not particularly relevant for EBV infection. In addition, interactions between EBNA1 and host proteins have not been globally examined in the context of latent or lytic viral infection. EBNA1 interactions in lytic infection are of interest because EBNA1 is also expressed in and contributes to lytic infection (25). In this study, we adapted proteomics methods to compare EBNA1-protein interactions in NPC and gastric carcinoma cell lines in the presence and absence of EBV latent and lytic infection.
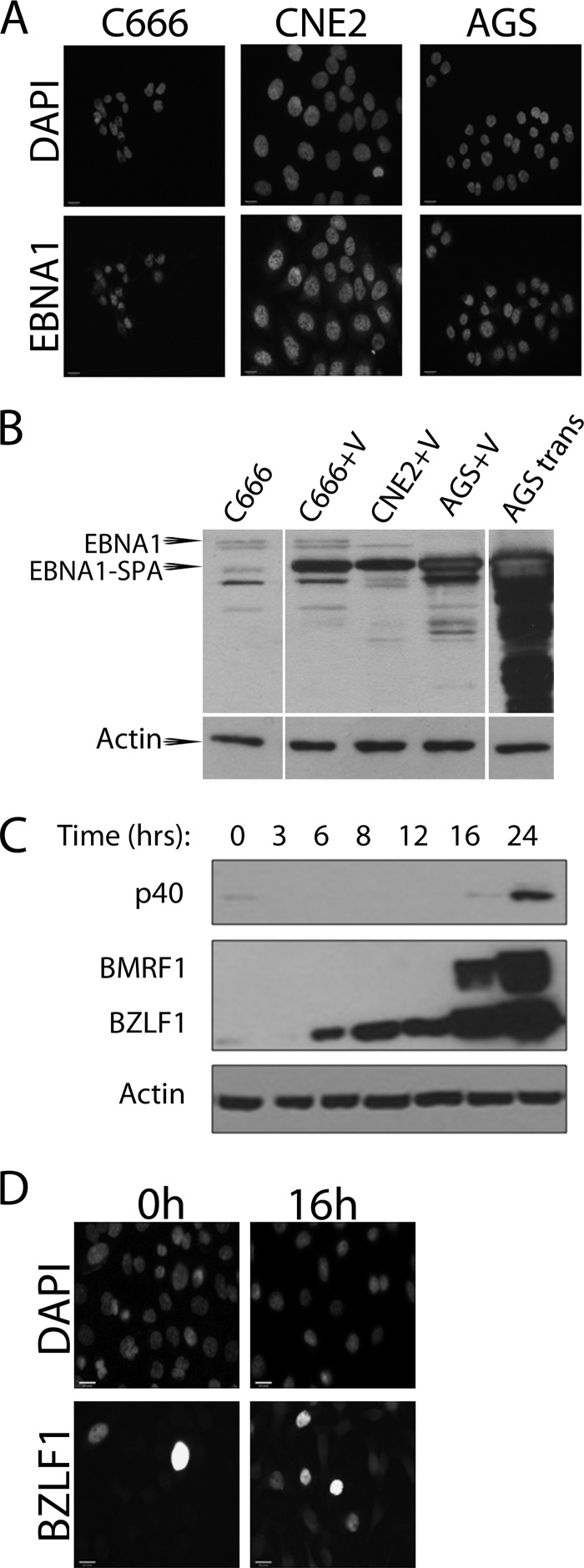
Our studies used both EBV-positive and EBV-negative NPC cell lines (C666 [3] and CNE2 [27], respectively), as well as AGS gastric carcinoma cells with (AGS-EBV) and without recombinant EBV episomes that can be reactivated to lytic infection by treatment with sodium butyrate (NaB) and tetradecanoyl phorbol acetate (TPA) (23, 25). The ViraPower adenoviral expression system (Invitrogen) was used to efficiently deliver low levels of EBNA1 cDNA or beta-galactosidase (β-Gal) cDNA (negative control) with a C-terminal sequential peptide affinity (SPA) tag, consisting of a calmodulin binding peptide and a triple-FLAG tag (31). This version of EBNA1 lacks most of the variable Gly-Ala repeat region and is identical to that used previously in TAP-tagging experiments (10). With this system, EBNA1 was expressed in almost all of the cells (Fig. 1A), at levels considerably lower than that typically seen with transfection of expression plasmids but somewhat higher than the native EBNA1 levels in C666 cells (Fig. 1B). Twenty-five 150-mm plates of cells were harvested 48 h postinfection, and lysates were generated as previously described (10). Lysates were mixed with 50 μl anti-FLAG M2 affinity gel (Sigma-Aldrich) for 4 h at 4°C, followed by washing, elution, and trypsinization of the bound proteins as described in Chen and Gingras (1). The peptides were analyzed by affinity chromatography coupled to liquid chromatography mass spectrometry (AP/MS) on a ThermoElectron LCQ Deca XP mass spectrometer coupled with an Agilent capillary HPLC 1100 series at the Advanced Protein Technology Centre (Hospital for Sick Children, Toronto, Canada).
Fig 1.
Expression of SPA-tagged EBNA1 and lytic cycle induction. (A) C666, CNE2, and AGS cells were grown on coverslips and infected with adenovirus to deliver SPA-tagged EBNA1. Forty-eight hours after infection, cells were fixed and stained as previously described (24) using FLAG antibody (Bethyl; 1:800) and goat anti-rabbit Alexa Fluor 555 secondary antibody (Invitrogen; 1:100). Coverslips were mounted onto slides using ProLong Gold antifade medium containing DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen). Images were obtained using the 40× oil objective on a Leica DM IRE2 inverted fluorescence microscope and processed using OpenLAB (version X.0) software. (B) C666, CNE2, and AGS cells were infected with the adenovirus expressing EBNA1-SPA (+V) and, 48 h later, lysed in 9 M urea and 5 mM Tris-HCl (pH 6.8) followed by brief sonication. Similar lysates were also generated of AGS cells 48 h posttransfection with pMZS3F.EBNA1 (22) expressing SPA-tagged EBNA1 (AGS tran) and of C666 cells (first lane). A total of 30 μg of each lysate was analyzed by Western blotting using K67 rabbit serum against EBNA1 (kindly supplied by Jaap Middeldorp). The positions of the native EBNA1 in C666 cells (EBNA1) and EBNA1-SPA are indicated. The latter is smaller than native EBNA1 because it lacks most of the nonessential Gly-Ala repeat region. (C) AGS-EBV cells were treated with 3 mM NaB and 20 ng/ml TPA and harvested 3 to 24 h later as indicated. Cells were lysed in 9 M urea 5 mM Tris-HCl (pH 6.8) and briefly sonicated. A total of 50 μg of total protein was analyzed by Western blotting using antibodies against BZLF1 (Santa Cruz; 1:1,000 dilution), BMRF1 (Chemicon; 1:5,000), VCA-p40 (1:500; kindly supplied by Jaap Middledorp), and actin (Santa Cruz; 1:5,000). (D) AGS-EBV cells grown on coverslips were fixed 16 h after NaB/TPA treatment, stained with antibody against BZLF1 (Santa Cruz; 1:50) followed by anti-mouse Alexa Fluor 488 (Invitrogen; 1:100), and counterstained with DAPI. Images were obtained as described for panel A.
The interactions shown in Table 1 are those identified with 100% probability (determined by Scaffold Proteomics Software) in two or more experiments for any cell line, where the peptide recovery (spectral counts) was above that seen with β-Gal (background). In addition, proteins that have been identified as common contaminants in FLAG tag pulldowns from 293 cells coupled to AP/MS (1) were eliminated, with three exceptions as discussed below. EBNA1 was found to have very similar interaction profiles in the different cell backgrounds, and many of these interactions corresponded to those previously identified in 293T cells (10), namely, interactions with ubiquitin-specific protease 7 (USP7); casein kinase 2 (CK2) subunits α, α′, and β; PRMT5; and P32/TAP. The interaction of EBNA1 with USP7 has been previously shown to lead to p53 destabilization and loss of promyelocytic leukemia (PML) nuclear bodies (that control apoptosis and DNA repair), and the EBNA1-CK2 interaction has also been shown to be important for inducing loss of PML nuclear bodies (18, 22–24). Therefore, our present observations suggest that EBNA1 can affect these processes both in the presence and absence of latent infection. The EBNA1 interaction with GMP synthetase that we detected was likely indirect and mediated by USP7, since we have previously shown that GMP synthetase binds to USP7 (19). PRMT5 and P32/TAP are common contaminants in proteomic studies (1); however, we included them in Table 1, as both have been previously confirmed as interactors of EBNA1 (2, 10, 21, 29). Note that the previously reported interaction of EBNA1 with EBP2 (12, 13, 20) could not be assessed by this method since EBP2 is largely insoluble and hence not present in the clarified lysate used for the FLAG pulldowns.
Table 1.
Summary of cellular proteins found to copurify with EBNA1 in NPC and gastric carcinoma cells
| Identified protein | Accession no. | Molecular mass (kDa) | No. of assigned spectraa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C666 |
CNE2 |
AGS |
AGS-EBV 0 h |
AGS-EBV 16 h |
||||||||
| β-Gal | EBNA1 | β-Gal | EBNA1 | β-Gal | EBNA1 | β-Gal | EBNA1 | β-Gal | EBNA1 | |||
| EBNA1 | GI:56566195 | 56 | 0 · 0 · 0 | 46 · 16 · 81 | 0 · 0 | 20 · 34 | 0 · 0 | 19 · 7 | 0 · 0 | 26 · 8 | 0 · 0 | 8 · 7 |
| CK2 subunit alpha | GI:4503095 | 45 | 0 · 0 · ·0 | 107 · 26 · 126 | 0 · 0 | 9 · 20 | 0 · 0 | 26 · 38 | 0 · 0 | 133 · 12 | 0 · 0 | 16 · 30 |
| CK2 subunit alpha′ | GI:4503097 | 41 | 0 · 0 · ·0 | 51 · 9 · 46 | 0 · 0 | 4 · 9 | 0 · 0 | 17 · 18 | 0 · 0 | 65 · 6 | 0 · 0 | 12 · 19 |
| CK2 subunit beta | GI:23503295 | 25 | 0 · 0 · 0 | 40 · 4 · 36 | 0 · 0 | 13 · 13 | 0 · 0 | 4 · 6 | 0 · 0 | 72 · 3 | 0 · 0 | 10 · 13 |
| GMP synthase | GI:4504035 | 77 | 0 · 0 · 0 | 0 · 4 · 7 | 0 · 0 | 0 · 0 | 0 · 0 | 17 · 0 | 0 · 0 | 5 · 2 | 0 · 0 | 0 · 1 |
| NAP1 | GI:37359287 | 45 | 2 · 3 · ·0 | 12 · 9 · 22 | 0 · 3 | 0 · 0 | 0 · 0 | 8 · 8 | 0 · 0 | 4 · 3 | 0 · 0 | 6 · 23 |
| Nucleophosminb | GI:10835063 | 33 | 1 · 0 · 0 | 32 · 9 · 38 | 1 · 3 | 1 · 11 | 0 · 0 | 16 · 14 | 2 · 1 | 30 · 9 | 0 · 3 | 3 · 22 |
| TAF-Iβ | GI:145843637 | 33 | 0 · 0 · 0 | 2 · 0 · 2 | 0 · 0 | 1 · 12 | 0 · 0 | 14 · 36 | 0 · 0 | 17 · 2 | 0 · 1 | 1 · 42 |
| P32b | GI:1096067 | 31 | 1 · 0 · 0 | 77 · 19 · 104 | 6 · 0 | 12 · 22 | 0 · 0 | 41 · 33 | 0 · 0 | 58 · 155 | 1 · 1 | 11 · 55 |
| PRMT5b | GI:20070220 | 73 | 7 · 2 · 10 | 62 · 3 · 44 | 19 · 11 | 17 · 29 | 2 · 12 | 38 · 9 | 11 · 0 | 25 · 12 | 0 · 7 | 4 · 19 |
| USP7 | GI:150378533 | 128 | 0 · 0 · 0 | 87 · 17 · 92 | 0 · 0 | 20 · 18 | 0 · 0 | 87 · 37 | 0 · 0 | 98 · 22 | 0 · 0 | 14 · 50 |
Total number of tryptic peptides identified in individual experiments. Data from individual experiments are separated by dots.
Identified as common contaminants in 293 cells (1).
We also detected interactions of EBNA1 with the related nucleosome assembly proteins TAF-Iβ (also called SET) and NAP1, previously isolated from HeLa lysates on EBNA1 affinity columns and subsequently shown to contribute to EBNA1 functions in transcriptional activation and DNA replication (10, 28). Interactions of EBNA1 with NAP1 and TAF-Iβ appeared to vary in different cell backgrounds, as the EBNA1-NAP1 interaction was not detected in CNE2 cells but was consistently detected in C666 and AGS cells, with highest recovery in C666 cells. However, EBNA1 was previously shown to immunoprecipitate with NAP1 in CNE2 cells (28), and therefore the lack of detection of this interaction in the current CNE2 experiments is likely due to the recovery of NAP1 being too small of a fraction of the total proteins to be detected during the mass spectrometry sampling period.
In addition, a higher number of TAF-Iβ peptides was recovered with EBNA1 in the AGS cells (than with the C666 and CNE2 cells), even though fewer EBNA1 peptides were recovered. The degree of recovery of NAP1 and TAF-Iβ does not appear to be due to their cellular expression levels since they are expressed in all of these cell lines at similar levels (data not shown).
We also identified an EBNA1 interaction with nucleophosmin (also called B23), a histone chaperone with multiple roles in regulating transcription, p53 function, and chromosome stability (4). While nucleophosmin is commonly isolated in FLAG pulldowns from 293 cells (1), we consistently found that it was recovered with EBNA1 at levels considerably higher than with β-Gal or typical background interactions. This interaction was not previously seen in TAP-tagging experiments in 293T cells where EBNA1-SPA was purified using both affinity tags, suggesting that it may not be stable enough to withstand the double purification but can be captured with the single affinity purification as conducted here. The EBNA1-nucleophosmin interaction was verified by coimmunoprecipitation in C666 cells and was not sensitive to RNase treatment (Fig. 2; B23).
Fig 2.
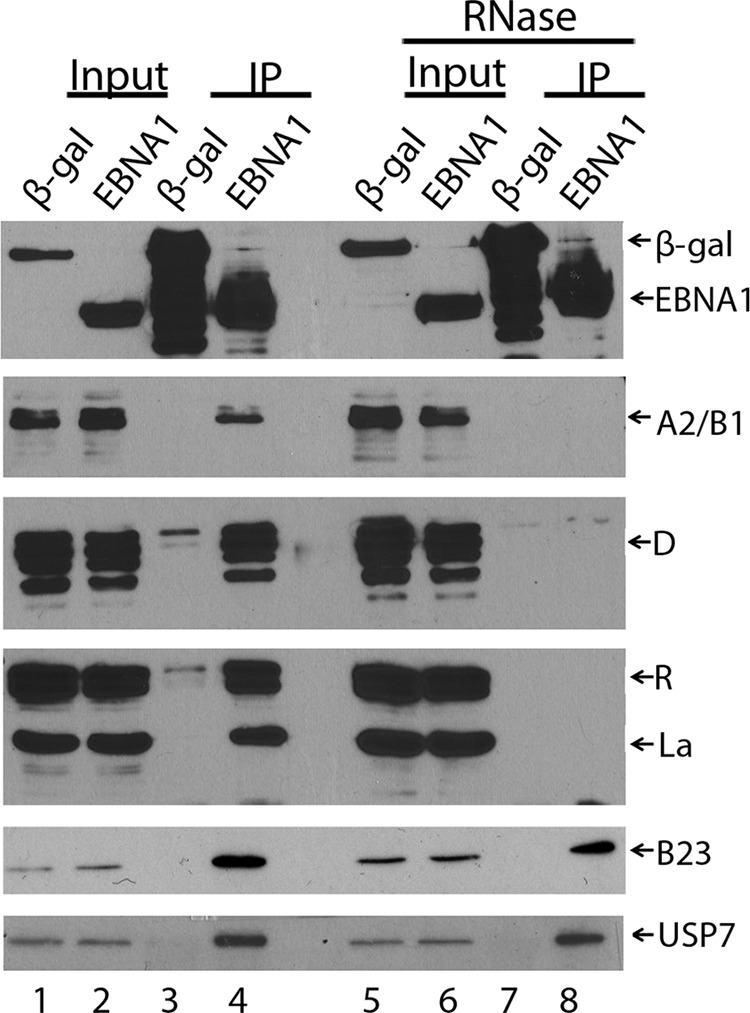
Coimmunoprecipitations of cellular proteins with EBNA1. EBNA1-SPA or β-Gal-SPA were immunoprecipitated from 1 mg of C666 cell lysates using M2 anti-FLAG resin, before (lanes 3 and 4) and after (lanes 7 and 8) treatment of the lysate with 50 μl/ml RNase at 25°C for 30 min. Immunoprecipitated proteins (IP) and samples of the input (1% of lysate) were analyzed by Western blotting using antibodies against FLAG (top); hnRNPs A2/B1 (Santa Cruz; 1:1,000), D (Upstate; 1:1,000), or R (Abcam; 1:1,000); La (Novus; 1:1,000); nucleophosmin (B23; Santa Cruz; 1:3,000); or USP7 (Bethyl; 1:5,000) as indicated.
In addition to studying EBNA1 interactions during latency, we also induced the lytic cycle in the AGS-EBV cells by NaB/TPA treatment 24 h after infection with the SPA-tagged EBNA1 or β-Gal adenoviruses. A time course after NaB/TPA treatment was first performed following expression of the BZLF1 immediate early, BMRF1 early, and VCA-p40 late viral proteins, to establish when the cells were in the early stage of infection. The Western blot in Fig. 1C indicates that cells were in the early stage of infection 16 h postinduction. In addition, immunofluorescence (IF) microscopy for BZLF1 showed that ∼75% of the cells had entered the lytic cycle at this time point (Fig. 1D). Therefore, FLAG pulldowns of EBNA1 and β-Gal were performed 16 h postinduction followed by mass spectrometry analysis as described above. As shown in Table 1 (last column), EBNA1 was found to mediate the same protein interactions in the lytic cycle as it did in latency (compare AGS-EBV 0 h to AGS-EBV 16 h).
Finally, we found that many RNA binding proteins copurified with EBNA1 (Table 2). In particular, a subset of heterogeneous ribonucleoproteins (hnRNPs) was isolated with EBNA1 but not with the β-Gal-negative control, as was La protein, a protein associated with the 3′ end of newly synthesized small RNA molecules (30). While hnRNPs are common interactors in AP/MS experiments (1), the peptide recovery for each of the proteins in Table 2 was considerably above that for the β-Gal control in at least one of the cell lines. Since EBNA1 can also associate with some RNA molecules (5, 15, 16, 26), we examined whether the RNA-binding proteins were tethered to EBNA1 by RNA. To this end, SPA-tagged EBNA1 or β-Gal were immunoprecipitated from C666 lysates using M2 anti-FLAG resin before and after RNase treatment, and coprecipitating proteins were analyzed by Western blotting using antibodies specific for hnRNPs A2/B1, D, or R and La (Fig. 2). While all of these protein interactions were confirmed in the absence of RNase, they were all abrogated after RNase treatment (compare lanes 4 and 8). In contrast, the EBNA1 interactions with USP7 (which is known to be direct) and nucleophosmin were not affected by RNase treatment (Fig. 2, bottom two panels). Therefore, the results indicate that EBNA1 interacts indirectly with these RNA binding proteins through RNA.
Table 2.
RNA binding proteins that copurify with EBNA1
| Identified protein | Accession no. | Molecular mass (kDa) | No. of assigned spectraa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C666 |
CNE2 |
AGS |
AGS-EBV 0 h |
AGS-EBV 16 h |
||||||||
| β-Gal | EBNA1 | β-Gal | EBNA1 | β-Gal | EBNA1 | β-Gal | EBNA1 | β-Gal | EBNA1 | |||
| EBNA1 | GI:56566195 | 56 | 0 · 0 · 0 | 46 · 16 · 81 | 0 · 0 | 20 · 34 | 0 · 0 | 19 · 7 | 0 · 0 | 26 · 8 | 0 · 0 | 8 · 7 |
| hnRNP A1 | GI:111306451 | 29 | 0 · 3 · 0 | 6 · 48 · 31 | 4 · 2 | 4 · 2 | 0 · 0 | 26 · 10 | 0 · 2 | 12 · 16 | 0 · 0 | 3 · 9 |
| hnRNP A2/B1 | GI:14043072 | 37 | 0 · 3 · 0 | 6 · 28 · 31 | 6 · 5 | 4 · 9 | 0 · 0 | 31 · 6 | 0 · 0 | 2 · 3 | 0 · 0 | 1 · 7 |
| hnRNP A/B | GI:33874222 | 31 | 0 · 0 · 0 | 2 · 1 · 3 | 1 · 0 | 0 · 0 | 0 · 0 | 5 · 0 | 0 · 0 | 13 · 20 | 0 · 0 | 2 · 2 |
| hnRNP A3 | GI:34740329 | 40 | 0 · 0 · 0 | 4 · 0 · 1 | 0 · 0 | 0 · 0 | 0 · 0 | 16 · 3 | 0 · 0 | 5 · 2 | 0 · 0 | 1 · 2 |
| hnRNP D | GI:14110420 | 38 | 0 · 2 · 0 | 6 · 7 · 3 | 3 · 0 | 1 · 0 | 0 · 0 | 13 · 3 | 0 · 0 | 5 · 6 | 0 · 0 | 6 · 4 |
| hnRNP Q | GI:228008291 | 70 | 0 · 0 · 0 | 26 · 3 · 11 | 2 · 2 | 1 · 1 | 0 · 0 | 54 · 9 | 0 · 0 | 50 · 12 | 0 · 0 | 17 · 22 |
| hnRNP R | GI:2697103 | 71 | 0 · 0 · 0 | 19 · 0 · 4 | 0 · 1 | 0 · 2 | 0 · 0 | 34 · 6 | 0 · 0 | 9 · 3 | 0 · 0 | 5 · 14 |
| La protein | GI:10835067 | 47 | 0 · 0 · 0 | 7 · 3 · 17 | 0 · 0 | 0 · 0 | 0 · 0 | 0 · 0 | 0 · 0 | 2 · 4 | 0 · 0 | 4 · 28 |
Total number of tryptic peptides identified in individual experiments. Data from individual experiments are separated by dots.
Interestingly, La has been shown to bind EBERs (stable RNA molecules produced by EBV) (11), and EBER1 has been shown to coimmunoprecipitate with EBNA1 (14), raising the possibility that EBERs may mediated an interaction between EBNA1 and La. In keeping with this hypothesis, we recovered only La with EBNA1 in cells containing EBV (C666 and AGS-EBV), whereas none of the EBNA1-hnRNP interactions were dependent on the presence of EBV. hnRNP proteins also have specificities for particular RNA sequences that are important for mediating their varied functions in splicing, nuclear export, transcription, and telomere maintenance (6, 9). Therefore, the RNA-mediated association of EBNA1 with specific hnRNPs may also reflect a tendency of EBNA1 to interact with the same RNA molecules or sequences as these hnRNPs. Interestingly, hnRNPs A2/B1 and A3 have all been shown to bind G-quadruplex RNA, and EBNA1 has also been found to bind this RNA sequence (15).
In summary, we have shown that several of the EBNA1-host protein interactions previously identified through proteomic studies in 293T and HeLa cells occur in the context of NPC and gastric carcinoma cells and that these interactions are maintained in both latent and lytic modes of EBV infection. We also identified EBNA1 interactions with nucleophosmin and several RNA-mediated interactions with specific RNA binding proteins that also occur in both latent and lytic infection.
ACKNOWLEDGMENTS
We thank Alan Cochrane for antibodies against the hnRNP proteins, Jaap Middeldorp for antibodies against VCA-p40 and EBNA1, and Anne-Claude Gingras for helpful discussions.
This work was funded by an operating grant to L.F. from the Canadian Cancer Society.
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Chen GI, Gingras AC. 2007. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42:298–305 [DOI] [PubMed] [Google Scholar]
- 2. Chen MR, Yang JF, Wu CW, Middeldorp JM, Chen JY. 1998. Physical association between the EBV protein EBNA-1 and P32/TAP/hyaluronectin. J. Biomed. Sci. 5:173–179 [DOI] [PubMed] [Google Scholar]
- 3. Cheung ST, et al. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 83:121–126 [DOI] [PubMed] [Google Scholar]
- 4. Colombo E, Alcalay M, Pelicci PG. 2011. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene 30:2595–2609 [DOI] [PubMed] [Google Scholar]
- 5. Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. 2009. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35:403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dreyfuss G, Kim VN, Kataoka N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195–205 [DOI] [PubMed] [Google Scholar]
- 7. Frappier L. 2012. Role of EBNA1 in NPC tumourigenesis. Semin. Cancer Biol. 22:154–161 [DOI] [PubMed] [Google Scholar]
- 8. Fukayama M. 2010. Epstein-Barr virus and gastric carcinoma. Pathol. Int. 60:337–350 [DOI] [PubMed] [Google Scholar]
- 9. He Y, Smith R. 2009. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell. Mol. Life Sci. 66:1239–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holowaty MN, et al. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987–29994 [DOI] [PubMed] [Google Scholar]
- 11. Howe JG, Shu MD. 1988. Isolation and characterization of the genes for two small RNAs of herpesvirus papio and their comparison with Epstein-Barr virus-encoded EBER RNAs. J. Virol. 62:2790–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kapoor P, Lavoie BD, Frappier L. 2005. EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol. Cell. Biol. 25:4934–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kapoor P, Shire K, Frappier L. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu CC, et al. 2004. Epstein-Barr virus nuclear antigen 1 is a DNA-binding protein with strong RNA-binding activity. J. Gen. Virol. 85:2755–2765 [DOI] [PubMed] [Google Scholar]
- 15. Norseen J, Johnson FB, Lieberman PM. 2009. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 83:10336–10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Norseen J, et al. 2008. RNA-dependent recruitment of the origin recognition complex. EMBO J. 27:3024–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431–441 [DOI] [PubMed] [Google Scholar]
- 18. Saridakis V, et al. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 18:25–36 [DOI] [PubMed] [Google Scholar]
- 19. Sarkari F, et al. 2009. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog. 5:e1000624 doi:10.1371/journal.ppat.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shire K, Ceccarelli DFJ, Avolio-Hunter TM, Frappier L. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shire K, et al. 2006. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J. Virol. 80:5261–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sivachandran N, Cao JY, Frappier L. 2010. Epstein-Barr virus nuclear antigen 1 hijacks the host kinase CK2 to disrupt PML nuclear bodies. J. Virol. 84:11113–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sivachandran N, et al. 2012. Contributions of the Epstein-Barr virus EBNA1 protein to gastric carcinoma. J. Virol. 86:60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sivachandran N, Sarkari F, Frappier L. 2008. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 4:e1000170 doi:10.1371/journal.ppat.1000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sivachandran N, Wang X, Frappier L. 2012. Functions of the Epstein-Barr virus latency protein, EBNA1, in the viral reactivation and lytic infection. J. Virol. 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Snudden DK, Hearing J, Smith PR, Grasser FA, Griffin BE. 1994. EBNA-1, the major nuclear antigen of Epstein-Barr virus, resembles ‘RGG’ RNA binding proteins. EMBO J. 13:4840–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Y, et al. 1992. An infrequent point mutation of the p53 gene in human nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. U. S. A. 89:6516–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang S, Frappier L. 2009. Nucleosome assembly proteins bind to Epstein-Barr virus nuclear antigen 1 and affect its functions in DNA replication and transcriptional activation. J. Virol. 83:11704–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y, Finan JE, Middeldorp JM, Hayward SD. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18–29 [DOI] [PubMed] [Google Scholar]
- 30. Wolin SL, Cedervall T. 2002. The La protein. Annu. Rev. Biochem. 71:375–403 [DOI] [PubMed] [Google Scholar]
- 31. Zeghouf M, et al. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463–468 [DOI] [PubMed] [Google Scholar]