Abstract
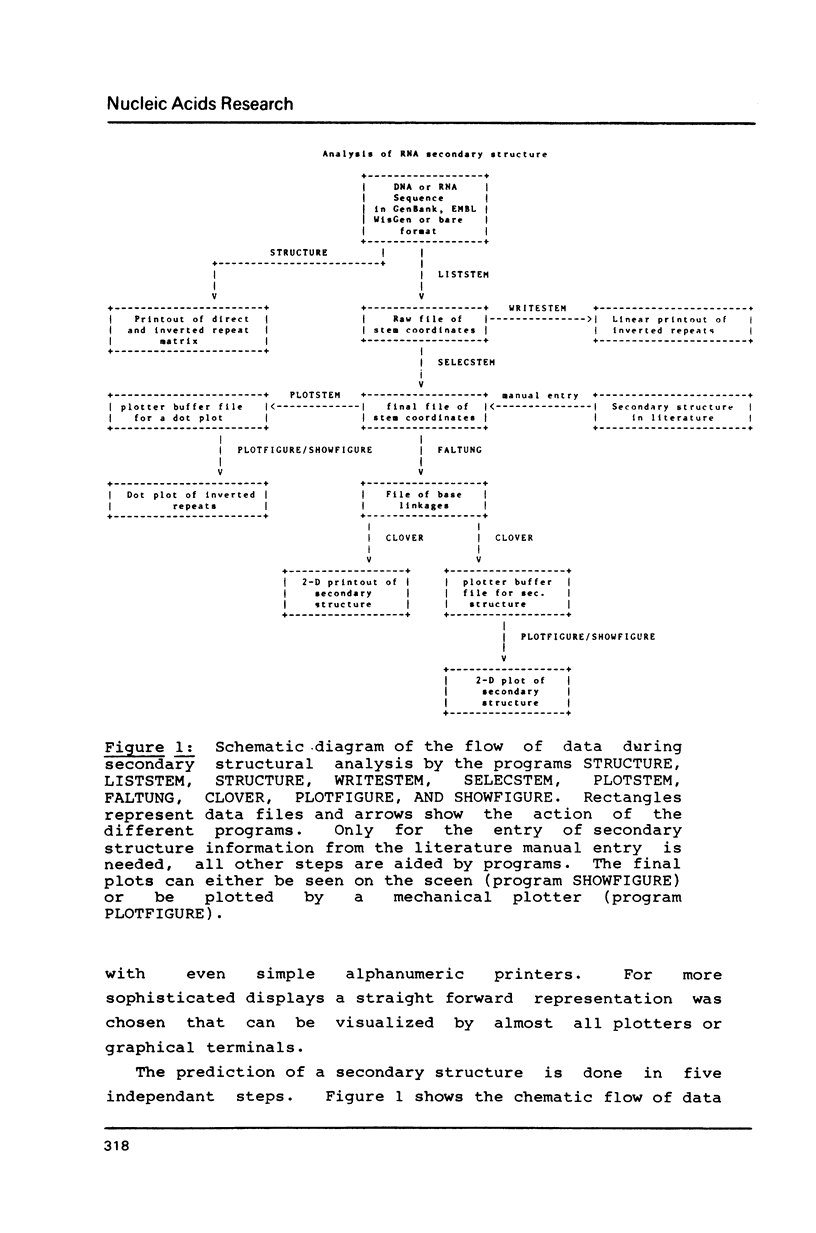
A set of programs has been developed for the prediction and display of nucleic acid secondary structures. Information from experimental data can be used to restrict or enforce secondary structural elements. The predictions can be displayed either on normal line printers or on graphic devices like plotters or graphic terminals.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez H. M. An RNA folding rule. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):323–334. doi: 10.1093/nar/12.1part1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Rogers J., Clarke P., Salser W. Sequence analysis of cloned cDNA encoding part of an immunoglobulin heavy chain. Nucleic Acids Res. 1979 Jul 25;6(10):3305–3321. doi: 10.1093/nar/6.10.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber K. Visualization of nucleic acid sequence structural information. Comput Appl Biosci. 1985;1(1):35–42. doi: 10.1093/bioinformatics/1.1.35. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Kitamura Y., Yoshikura H. Computation of statistical secondary structure of nucleic acids. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):335–346. doi: 10.1093/nar/12.1part1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


