Abstract
It is widely accepted that a deranged immune system plays a key role in the onset and evolution of classic Kaposi sarcoma (CKS). Nevertheless, the usage of the T-cell receptor (TCR) β-variable (BV) chain repertoire expressed by peripheral blood lymphocytes in patients with CKS is still unknown. With the aim of providing some further insights into the complex role of the immune system in CKS pathogenesis, we performed an extensive analysis of the TCR BV repertoire in both CD4+ and CD8+ T cells in 30 human herpesvirus 8-positive Sardinian patients with CKS and an equal number of age-matched healthy controls. We used a panel of monoclonal antibodies covering approximately 70% of human BV subfamilies and third complementarity determining region (CDR3) spectratyping. Patients with CKS showed an increased frequency of BV expansions in both CD4+ and CD8+ lymphocytes, with no prevalent clones. On spectratyping analysis, most of the 720 BV CDR3 profiles obtained from both CD4+ and CD8+ T cells in patients with CKS were skewed. In particular, the surprising increase of BV skewing observed in CD4+ lymphocytes mimics the pattern of progressive TCR BV narrowing described in responses to persistent viral antigen stimulations. Our findings support the hypothesis that CKS evolution is associated with inadequate activation rather than impairment of the immune system.
Introduction
Kaposi sarcoma (KS) is an angioproliferative multifocal disease of the skin occurring in different clinical-epidemiological forms [1], all sharing the same histopathologic features [2] as well as the association with human herpesvirus 8 (HHV-8) infection [3]. As in other ethnic groups of Mediterranean descent [4], classic Kaposi sarcoma (CKS) is very common in the Sardinian population, in which the incidence of 4.06 per 100,000 persons per year among people older than 40 years represents one of the highest reported worldwide [5]. The onset of the disease in at-risk individuals is associated with CD8+ T-cell activation and increased T helper 1-type cytokine production. Such immunoactivation, mimicking a reactive inflammatory process, induces the extravasation of lymphomonocytes, spindle cell formation, and angiogenesis, the histologic hallmarks of KS lesions [6–8]. In this setting, the latent HHV-8 infection is then reactivated by the same inflammatory cytokines, which, instead of being effective against the virus, lead to HHV-8 spreading and progression of the disease [9]. Therefore, the immune response to HHV-8 paradoxically seems to exacerbate the reactive process, favoring its transition to true sarcoma lesions. If an acquired specific immunodeficiency occurring in both arms of the T-cell immune system modulates non-CKS initiation and progression [10–12], in the classic variant of KS, a peculiar impairment of the immune system has never been demonstrated. In addition, several studies focusing on the levels and functions of CD4+ and CD8+ T-lymphocyte subsets have shown conflicting results [13,14].
Because the antigen T-cell responses to infections and tumor antigens, as well as in the context of hypersensitivity and autoimmunity, are associated with a variety of biased profiles of T-cell receptors (TCRs) selected from a diverse, naive repertoire [15], we speculated that a comprehensive analysis of the TCR β-variable (BV) chain repertoire in isolated CD4+ and CD8+ peripheral blood T lymphocytes could provide further insights into the immunologic dysregulation characterizing CKS.
The overall expression of the TCR BV repertoire can be screened by flow cytometry using a panel of monoclonal antibodies directed against the variable domain of the majority of TCR BV families. Because robust reference values for the BV repertoire usage in a human healthy population are available [16], this rapid TCR BV analysis performed with an appropriate set of monoclonal antibodies is able to disclose most abnormal T-cell expansions. A further approach to investigate a possible bias in the TCR BV repertoire is provided by the so-called spectratyping analysis [17], which determines the profile of the third complementarity determining region (CDR3) length distribution in each BV subfamily.
The lack of studies addressing the TCR BV repertoire pattern in patients with CKS prompted us to investigate peripheral blood CD4+ and CD8+ subsets by combining flow cytometry and spectra-typing in a large series of patients with CKS. The presence in Sardinians of the highest TCR null allele BV20 polymorphism frequency [18], which could represent a functional bias in an otherwise normally preserved TCR BV repertoire [19], has been a further stimulus to investigate the TCR BV repertoire in an ethnically homogenous CKS group.
Materials and Methods
Patients and Healthy Controls
This study was approved by the local ethic committee. All patients and healthy controls gave informed consent to their participation to this study.
All the 30 patients with CKS (22 men and 8 women) were of Sardinian origin. Table 1 depicts the main clinical features of patients with CKS. Their median age was 70 years (range, 49–85 years). In each case, the diagnosis of CKS was confirmed by histologic examination. All patients were negative for HIV and none of them presented concomitant malignancies. Possible iatrogenic causes of KS were excluded. Because a universally accepted CKS staging system is still lacking, we classified our CKS series using the modified version [20] of the Mitsuyasu-Groopman classification [21]. Accordingly, 23 patients (77%) showing only cutaneous involvement were classified as having stage I to II disease, whereas the remaining 7 (23%) with systemic involvement were considered to have stage III to IV disease. The timing of the analysis from diagnosis varied from 1 to 240 months (mean, 59 months). Fifteen patients (50%) had no previous therapy, eight (27%) had undergone surgical excision or cryotherapy for their lesions, whereas seven (23%) had been treated with an anthracycline-based regimen. At the time of evaluation, almost all the patients had a stable disease and only one was in progression.
Table 1.
Summary of Patients and Healthy Controls' Characteristics.
| Characteristics | |
| Patients | |
| n | 30 |
| Age, median (range), y | 70 (49–85) |
| Sex, n (%) | |
| Male | 22 (73) |
| Female | 8 (27) |
| Disease stage, n (%) | |
| I/II | 23 (77) |
| III/IV | 7 (23) |
| Previous therapy, n (%) | |
| No therapy | 15 (50) |
| Surgery/cryotherapy | 8 (27) |
| Anthracycline-based regimen | 7 (23) |
| State of disease, n (%) | |
| Stable | 29 (97) |
| In progression | 1 (3) |
| Healthy controls | |
| n | 30 |
| Age, median (range), y | 72 (52–88) |
| Sex, n (%) | |
| Male | 15 (50) |
| Female | 15 (50) |
Thirty age-matched healthy Sardinian people (15 men and 15 women) were included in the study as normal controls. Their main characteristics are summarized in Table 1.
All patients and controls were tested for HHV-8 serostatus by using indirect immunofluorescence for the detection of antibodies to viral lytic antigens combined with enzyme-linked immunosorbent assay for the detection of specific antibodies to the K8.1 HHV-8 protein, as reported elsewhere [22]. Positivity was attributed to any sample that had positive results on both tests.
CD4+ and CD8+ T-cell Separation
Peripheral blood mononuclear cells were prepared by Ficoll-Hypaque (Sigma Diagnostic, St. Louis, MO) gradient centrifugation. Cells were separated into CD4+ and CD8+ subsets by positive selection using antibody-coated immunomagnetic beads (Dynabeads; Dynal, Oslo, Norway) for 30 minutes at 4°C on a rotating shaker followed by magnetic isolation. Guanidinium thiocyanate-phenol*chloroform (Invitrogen, Paisley, United Kingdom) was added to both cell fraction pellets, and they were stored at -80°C until RNA extraction.
Flow-Cytometry
We first determined the frequency of CD4+, CD8+ and natural killer T lymphocytes by using anti-CD3, CD4, CD8, and CD16/56 antibodies.
Flow cytometric analysis of the TCR BV repertoire was performed with the IOTest Beta Mark Kit (Beckman Coulter, San Diego, CA), according to the manufacturer's instructions. Briefly, 1 x 106 cells/sample were stained with a panel of 24 BV family-specific antibodies, combined in groups of three in eight tubes, one antibody being conjugated to fluorescein isothiocyanate, another to phycoerythrin, and the third to both fluorescein isothiocyanate and phycoerythrin. Costaining was performed with anti-CD4+ or anti-CD8+ peridinin chlorophyll protein complex (PerCp) (all antibodies from Becton Dickinson, San Jose, CA). Samples were acquired by a FACSort flow cytometer using CELLQuest software. A lymphocyte gate was established based on forward and side scatter characteristics. The relative representation of a given BV subfamily was expressed as the percentage of cells stained with the family-specific antibody among CD4+ or CD8+ T cells. A BV expansion was defined as any value of BV family expression higher than the mean plus 3 SD calculated in normal controls. Of note, the mean expression values of each BV subfamily in our healthy controls were very similar to the reported reference BV values [16]. Likely because of their older age, our controls showed a higher frequency of expanded BV subfamilies.
CDR3 Spectratyping
The RNA was isolated from each subset of cells as described elsewhere [23]. Complementary DNA was synthesized by using SuperScript III reverse transcriptase and random hexamer primers (Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions. Polymerase chain reaction (PCR) was performed in a volume of 25 µl comprising 1x PCR buffer, 2.5 mM magnesium chloride, 1 U of AmpliTaq Gold (Applied Biosystems, Foster City, CA), 200 µM deoxyribonucleoside triphosphate (dNTPs), and 500 nM of 1 of 24 TCR BV primers combined with 1 β-constant primer conjugated to the fluorescent dye 6-carboxyfluorescein amino hexy [24]. The sequences of the TCR BV and β-constant primers were described previously [17]. The PCR conditions were 95°C for 10 minutes followed by 36 cycles of 94°C for 20 seconds, 55°C for 40 seconds, 72°C for 40 seconds, and a final extension at 72°C for 5 minutes. The PCR fragments were then run on an ABI Prism 310 Genetic Analyzer (Applied Biosystems), and data were collected and analyzed by the ABI Prism GeneScan Analysis software version 3.7 (Applied Biosystems).
Spectratyping Analysis
Analysis of spectratyping data was performed by analyzing profiles by peak area and shape to establish the degree of skewing and oligoclonality. In particular, each spectratyping profile was assessed following previously described criteria [25]. A profile was defined as normal if showing a Gaussian, bell-shaped distribution, with discrete peaks spaced by three nucleotides. Evidence of oligoclonal expansion or skewing within each BV was assessed by calculating the relative fluorescence intensity (RI) of each peak (RI = peak area / total BV peak area). A profile was defined as skewed if: 1) a dominant peak with a RI greater than 50% of the total peak area was observed, 2) two dominant peaks were present and RI of each peak was greater than 25% of the total peak area, or 3) there were multiple peaks differing from a Gaussian pattern and the RI of dominant peaks was greater than 25%of the total peak area. The first of these three criteria (the presence of a dominant peak with an RI >50% of the total peak area) was used to specifically identify oligoclonal BVs. The percentages of skewed and oligoclonal BV subfamilies were calculated among the total number of BVs analyzed in each patient.
DNA Amplification
DNA was isolated from the peripheral blood mononuclear cells of 25 patients with CKS.
DNA samples were amplified by PCR using a DNA thermal cycler (Applied Biosystems). The PCR mix contained 2.5 µl of 10x PCR buffer (containing 25 mM magnesium chloride), 1 µl of deossi-nucleotide-triphosphates (final concentration, 10 mM each), 0.5 µl of Taq polymerase (10 U/µl), 50 pmol of each primer, and 500 ng of genomic DNA. The amplification profile was 40 cycles of 1 minute at 94°C, 1 minute at 60°C, and 2 minutes at 72°C, preceded by 1 minute at 94°C, and followed by 5 minutes at 72°C. The specific primers 5-ATTCATCAATGGCCAGCGAC-3 and 5-GGAGCTTCTTAGAACTCAG-3 were used as described elsewhere [26].
Restriction Enzyme Digestion Analysis
A 10-µl quantity of the amplified DNA was digested with 10 U of KpnI (Invitrogen, Carlsbad, CA) with the addition of the manufacturer's 10x digestion buffer. Restriction enzyme-digested PCR products were subjected to electrophoresis in a nondenaturant 3% agarose gel. The gel was stained with ethidium bromide and visualized by ultraviolet illumination. The analyzed single nucleotide polymorphism involves a C-to-T base transition at position 524 in the TCR BV20 gene segment. Hence, the resulting null allele (allele 2) carries in the BV20 gene segment the stop codon CGA>TGA responsible for the deletion of the restriction site for the KpnI enzyme. A single electrophoresis band at 235 bp corresponds to the homozygosity for allele 2. DNA from homozygotes for allele 1, that is, the BV20 gene segment nonmutated at position 524, is identified by the presence of two bands at 100 and 135 bp, whereas heterozygous subjects show three bands at 235, 135, and 100 bp. To exclude misinterpretation because of partial digestions, PCR products from subjects known to be homozygous for allele 1 and allele 2 were included in each set of digestions. Gene and genotype frequencies of TCR BV20 alleles were determined by counting.
Statistical Analysis
The comparison between the mean expression of each BV subfamily in patients and controls was performed using the nonparametric Mann-Whitney test. The Student's t test was used to assess the differences in the percentage of BV expansions determined by flow cytometry between patients and controls as well as in the percentage of skewed or oligoclonal BV subfamilies determined by spectratyping. All quoted P values are two-sided with P < .05 considered statistically significant.
Results
HHV-8 Serostatus of Patients with CKS and Healthy Controls
All the patients with CKS had antibodies against HHV-8, whereas among the 30 healthy controls, 13 (43%) were positive for HHV-8.
Patients with CKS Show an Increased Frequency of Lymphocyte Expansions in Both CD4+ and CD8+ T cells
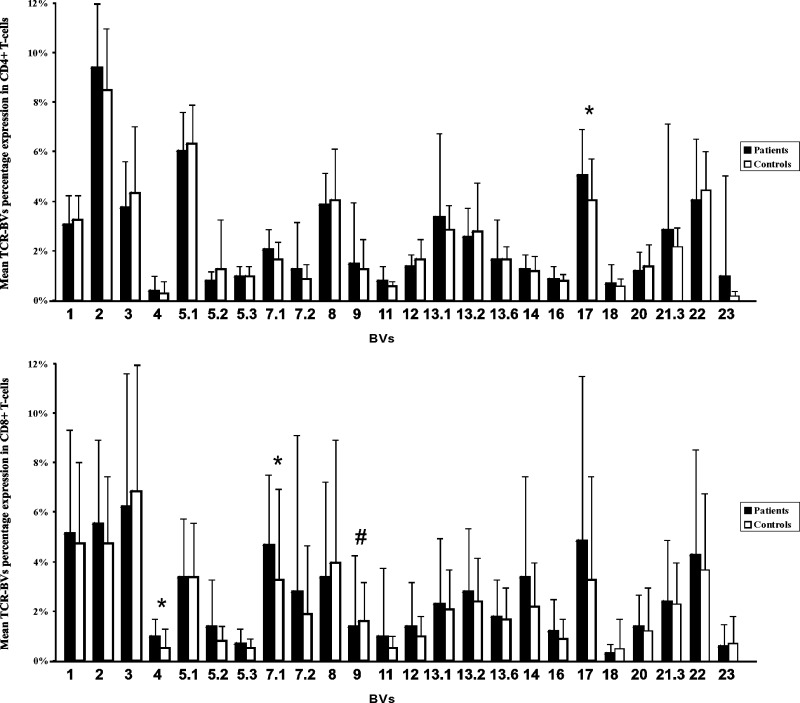
The mean frequencies of CD3+, CD4+, CD8+, and CD16/CD56+ T cells were superimposable in patients and controls (65% vs 68%, 40% vs 41%, 22% vs 23%, and 20% vs 21%, respectively). On the whole, the mean expression levels of almost all the BV subfamilies did not show a significant difference among patients and controls in both CD4+ and CD8+ T cells. Only the BV17 subfamily in the CD4+ fraction and BV4 and BV7-1 in CD8+ T cells in patients with CKS showed significantly increased expression levels in comparison with healthy controls, whereas a decreased expression of BV9 in CD8+ T cells of patients with CKS was also demonstrated (Figure 1). This variability could just reflect the expansions of several BV subfamilies described below.
Figure 1.
Mean BV subfamily percentage expression on flow cytometry in CD4+ and CD8+ T cells in patients and controls. Patients and controls showed similar expression in most BVs. Nonetheless, in patients, we found a significantly increased expression (BV marked with an asterisk [*]) only in the BV17 (mean 5.1% vs 4.1%, P < .05) of the CD4+ T cells and in the BV4 (mean 1.0% vs 0.5%, P < .001) and the BV7.1 (mean 4.3% vs 3.3%, P < .05) of the CD8+ subset. Conversely, we found a decreased expression (BV marked with a pound sign [#]) only in the BV9 (mean 1.4% vs 1.6%, P < .05) of CD8+ T cells.
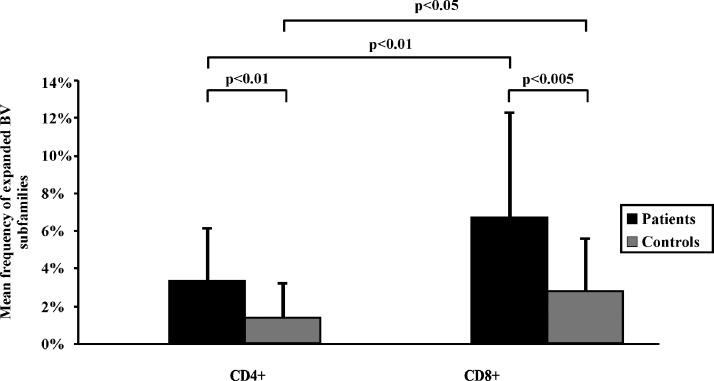
Because there were no significant differences in the mean frequencies of the expanded BV subfamilies between HHV-8+ and HHV-8- healthy controls (data not shown), we could assume that, in such a cohort, the expression of the T-cell repertoire is rather homogenous. In comparison with controls, patients with CKS were characterized by a significantly higher frequency of expanded BV subfamilies in both the CD4+ subset (3.6% vs 1.5%, P < .01) and the CD8+ one (7.3% vs 3.0%, P < .005; Figure 2). The relative size of the whole 79 T-cell expansions detected in patients with CKS ranged from 1.1% (involving BV11) to 25.7% (BV21.3) in the CD4+ subset and from 1.1% (BV4) to 36.6% (BV17) in the CD8+ one. As already reported [16], the frequency of expanded BV subfamilies in controls was significantly higher in CD8+ than in the CD4+ T cells (P < .05), and this was also observed in patients with CKS (P < .01; Figure 2). No relationship was found between the frequency of BV expansions and staging or previous treatment of patients with CKS. Finally, no preferential BV subfamily expansions were found among patients.
Figure 2.
Mean frequency of the expanded BV subfamilies detected by flow cytometry in CD4+ and CD8+ T cells in patients and controls. Patients were characterized by a higher frequency of expanded BV subfamilies than normal controls, both in the CD4+ subset (3.6% vs 1.5%, P < .01) and in the CD8+ one (7.3% vs 3.0%, P < .005). In both controls and patients, the frequency of expanded BV subfamilies was significantly higher in CD8+ than in CD4+ cells. A BV expansion was defined as any value of BV family expression higher than the mean +3 SD calculated in normal controls.
Patients with CKS Show a Contracted TCR Repertoire on CDR3 Spectratyping Analysis
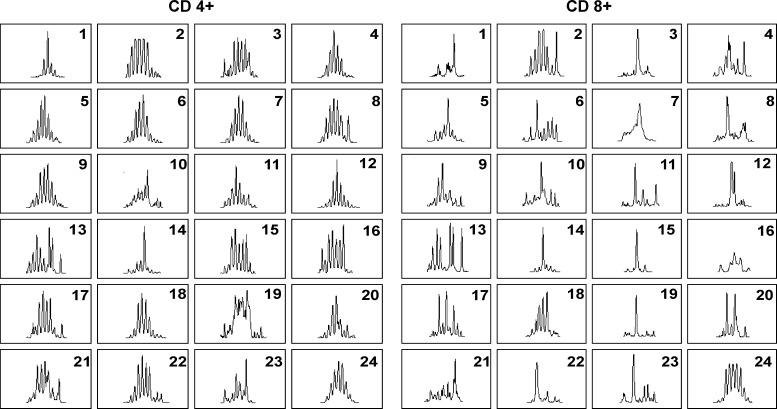
Of note, the spectratyping method we used could detect a mean of 22 (range, 15–24) and 23 (range 16–24) specific CDR3 BV profiles per sample in CD4+ and CD8+ T cells, respectively. A representative CDR3 TCR BV spectratyping profile obtained in the CD4+ and CD8+ T cells isolated from the peripheral blood of one of the patients is shown in Figure 3. On the whole, 632 and 595 BVs were available for statistical comparison in CD4+ T cells and 633 and 585 BVs in CD8+ T cells in controls and patients with CKS, respectively.
Figure 3.
Representative spectratyping profiles of the CDR3 in CD4+ and CD8+ T cells of one of the patients. According to the criteria described in Materials and Methods, BVs 8, 10, 13, 15, 16, 19, 20, 21, and 23 were skewed; BVs 1 and 14 were oligoclonal; whereas the remaining BVs were Gaussian in CD4+ T cells. In CD8+ T cells, BVs 1, 3, 14, 15, 19, and 22 were oligoclonal, whereas all the other BVs were skewed with the exception of BV24, which was Gaussian.
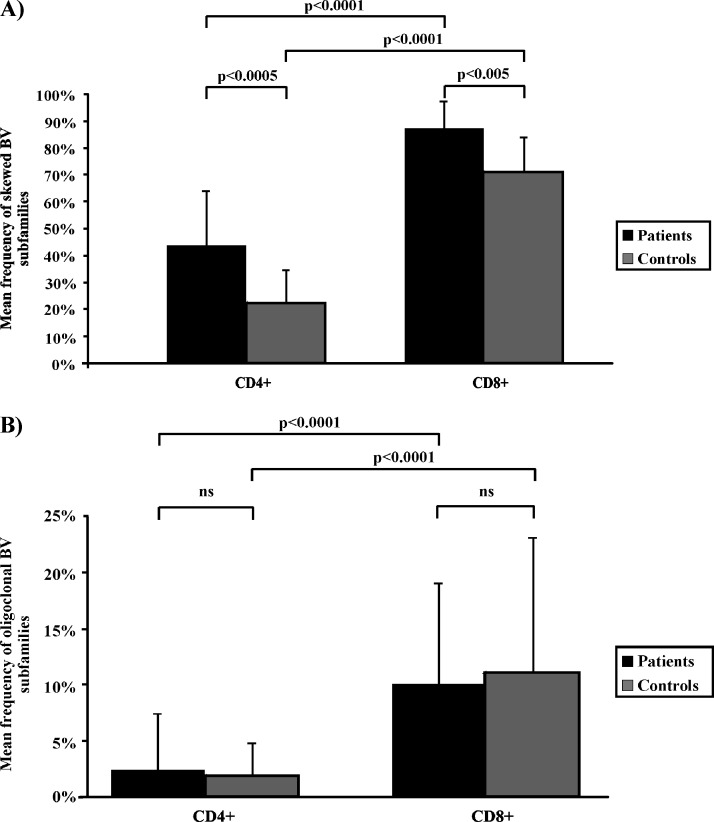
Taking into account that HHV-8+ and HHV-8- healthy controls did not show significant differences in the mean incidence of BVs showing a skewed or oligoclonal profile in both CD4+ and CD8+ T cells (data not shown), we compared the degree of BV skewing between patients with CKS and the whole group of controls. A significantly higher frequency of skewness in both the CD4+ subset (43% vs 25%, P < .0005) and CD8+ T cells (86% vs 75%, P < .005) was found in patients with CKS (Figure 4A). As expected, the occurrence of skewed BV subfamilies was significantly more frequent in CD8+ than in CD4+ T cells (P < .0001) in both groups (Figure 4A).
Figure 4.
Representation of the mean frequency of CDR3 spectratyping profiles in the BV subfamilies observed in patients with CKS and controls. (A) Patients were characterized by a significantly higher frequency of BV subfamilies showing a skewed profile in both CD4+ T cells (43% vs 25%, P < .0005) and in the CD8+ subset (86% vs 75%, P < .005). As expected, the occurrence of skewed BV subfamilies was significantly more frequent in CD8+ than in CD4+ cells in both controls and patients (P < .0001). (B) We found a similar incidence of BVs with an oligoclonal CDR3 spectratyping profile between patients and controls in both T-cell subpopulations (2.0% vs 1.8% in CD4+ T cells and 10.4% vs 11.3% in CD8+ ones). Once again, the frequency of oligoclonal BV subfamilies was significantly higher in CD8+ than in CD4+ cells in both controls and patients (P < .0001). NS indicates not significant.
Nevertheless, the incidence of BVs with a clear oligoclonal profile was similar between patients and controls (2.0% vs 1.8% in CD4+ T cells and 10.4% vs 11.3% in CD8+ T cells, respectively; Figure 4B). Once again, in both patients and controls, the mean frequency of oligoclonal BVs was significantly higher in the CD8+ than in the CD4+ subset (P < .0001; Figure 4B).
We then compared the findings obtained by spectratyping and flow cytometry in patients to dissect if expanded lymphocyte subpopulations were characterized by either a polyclonal or an oligoclonal pattern. According to the criteria we used to define T-cell expansions on flow cytometry and the degree of BV CDR3 profile skewness on spectratyping analysis, we found that, of the 27 expanded BVs in CD4+ T cells, 61% of them showed a skewed CDR3 profile, 8% were oligoclonal, and 31% were Gaussian. In CD8+ T cells, of the 52 expanded BVs, 66%, 23%, and 11% showed a skewed, oligoclonal, and Gaussian CDR3 profiles, respectively (data not shown).
Finally, the frequency of skewed and oligoclonal BVs did not correlate with the stage of the disease or with previous treatments (data not shown). Moreover, when we investigated the prevalence of skewing or oligoclonal patterns involving specific BV subfamilies in patients with CKS, we could not detect any prevalent usage of the TCR repertoire.
Genotypic Analysis in Patients with CKS Showed a Frequency of Homozygotes for the BV20S1 Null Allele Similar to That Reported in the Whole Sardinian Population
After DNA amplification and KpnI enzyme digestion, 4 (17%) of the 24 patients with CKS analyzed resulted homozygous for the so-called null allele (allele 2) and 12 of them (50%) were heterozygous. These incidences correspond to those previously reported in the whole Sardinian population [18].
Discussion
CKS is a reactive inflammatory-angiogenic process of the skin of polyclonal origin which can progress into a true sarcoma involving visceral organs in rare cases. The HHV-8 persistent infection characterizes the course of this rare disease, but it is well recognized that an altered cellular immune response also plays an important role in its pathogenesis and evolution [2,10,11,27]. Because the TCR BV repertoire generated within an immune response can undergo some well-defined alterations, such as restrictions and immunodominant expansions, we performed an extensive TCR repertoire analysis on CD4+ and CD8+ circulating cells of patients with CKS by combining flow cytometry and CDR3 spectratyping. We found that, in patients with CKS, peripheral blood lymphocytes are characterized by an increased frequency of expanded subpopulations occurring in both the CD4+ and the CD8+ T-cell subsets. Moreover, we could not find any preferential usage of specific BV subfamilies. These findings recall the lack of TCR immunodominance and the weak immunogenicity of HHV-8-specific epitopes emerging from preliminary in vitro and ex vivo studies on CD8+ T-cell responses against these epitopes [27–30]. Whether these lymphocyte expansions are triggered in the course of CKS by aberrantly expressed oncogenes and fusion genes in neoplastic cells or by HHV-8—alone or in association with other infectious agents—is yet to be solved.
Because the specificity of T lymphocytes is a function of the CDR3 BV structure, the characterization of CDR3 sequence variations should be considered a measure of T-cell diversity in an antigen driven T-cell repertoire. Being aware of the limits of the spectratyping, which is able to merely mirror the range of CDR3 length variations [31], we tried to further dissect the TCR BV repertoire pattern by comparing the CDR3 BV profiles obtained in each patient by flow cytometry and spectratyping. In comparison with the healthy population, patients with CKS show an increased degree of skewing within the TCR BV repertoire not only in CD8+ lymphocytes but also in CD4+ T cells, suggesting that this constrained TCR BV repertoire is either improperly or heterogeneously stimulated in the various phases of CKS. This hypothesis is supported by the observation that CD8+ T cells infiltrating the CKS cutaneous lesions are polyclonal [32] and mostly not epitope specific [27]. Noteworthy, a nonspecific activation of the T-cell immune system occurs in diseases, which, although quite different from CKS, are characterized by an evident immune dysregulation [33–35]. Although all these cases are dominated by an evident restriction of the TCR BV repertoire confined to the CD8+ T-cell compartment, our CDR3 spectratyping data show that the TCR BV repertoire is largely restricted also in circulating blood CD4+ lymphocytes. To note, the increased CD4+ TCR BV repertoire skewness occurs in both the expanded and the nonexpanded BV subfamilies, suggesting that the entire TCR repertoire is somehow limited in diversity in CKS. Moreover, it has been experimentally shown that the development of TCR diversity during thymic selection can be limited to only a few possible gene rearrangements [36]. These findings obtained from CD4+ lymphocyte analysis seem to mimic the pattern of progressive TCR BV narrowing described in response to persistent viral antigens [37,38].
In conclusion, our CDR3 spectratyping data, providing some further clues on the complex role of the immune system in CKS pathogenesis, could support the hypothesis that an inadequate activation rather than an overall impairment of the immune system could characterize the evolution of CKS.
Acknowledgments
The authors thank Francesco Dazzi (Department of Haematology, Division of Experimental Medicine, Imperial College, London, United Kingdom) for providing a critical review of the article.
Abbreviations
- BV
β-variable
- CDR3
third complementarity determining region
- CKS
classic Kaposi sarcoma
- HHV-8
human herpesvirus 8
- KS
Kaposi sarcoma
- RI
relative fluorescence intensity
- TCR
T-cell receptor
Footnotes
This work was partially supported by the “Fondazione Banco di Sardegna” (Sassari, Italy). The authors declare no competing financial interests.
References
- 1.Hengge UR, Ruzicka T, Tyring SK, Stuschke M, Roggendorf M, Schwartz RA, Seeber S. Update on Kaposi's sarcoma and other HHV8 associated diseases. Part 1: Epidemiology, environmental predispositions, clinical manifestations, and therapy. Lancet Infect Dis. 2002;2:281–292. doi: 10.1016/s1473-3099(02)00263-3. [DOI] [PubMed] [Google Scholar]
- 2.Ensoli B, Sgadari C, Barillari G, Sirianni MC, Sturzl M, Monini P. Biology of Kaposi's sarcoma. Eur J Cancer. 2001;37:1251–1269. doi: 10.1016/s0959-8049(01)00121-6. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 4.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans: human immunodeficiency viruses and human T-cell lymphotropic viruses. Lyon, France, 1–18 June 1996. IARC Monogr Eval Carcinog Risks Hum. 1996;67:1–424. [Google Scholar]
- 5.Cottoni F, Masala MV, Pattaro C, Pirodda C, Montesu MA, Satta R, Cerimele D, de Marco R. Classic Kaposi sarcoma in northern Sardinia: a prospective epidemiologic overview (1977–2003) correlated with malaria prevalence (1934) J Am Acad Dermatol. 2006;55:990–995. doi: 10.1016/j.jaad.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Sirianni MC, Vincenzi L, Fiorelli V, Topino S, Scala E, Uccini S, Angeloni A, Faggioni A, Cerimele D, Cottoni F, et al. gamma-Interferon production in peripheral blood mononuclear cells and tumor infiltrating lymphocytes from Kaposi's sarcoma patients: correlation with the presence of human herpesvirus-8 in peripheral blood mononuclear cells and lesional macrophages. Blood. 1998;91:968–976. [PubMed] [Google Scholar]
- 7.Fiorelli V, Gendelman R, Samaniego F, Markham PD, Ensoli B. Cytokines from activated T cells induce normal endothelial cells to acquire the phenotypic and functional features of AIDS-Kaposi's sarcoma spindle cells. J Clin Invest. 1995;95:1723–1734. doi: 10.1172/JCI117849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samaniego F, Markham PD, Gendelman R, Watanabe Y, Kao V, Kowalski K, Sonnabend JA, Pintus A, Gallo RC, Ensoli B. Vascular endothelial growth factor and basic fibroblast growth factor present in Kaposi's sarcoma (KS) are induced by inflammatory cytokines and synergize to promote vascular permeability and KS lesion development. Am J Pathol. 1998;152:1433–1443. [PMC free article] [PubMed] [Google Scholar]
- 9.Monini P, Colombini S, Sturzl M, Goletti D, Cafaro A, Sgadari C, Butto S, Franco M, Leone P, Fais S, et al. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi's sarcoma. Blood. 1999;93:4044–4058. [PubMed] [Google Scholar]
- 10.Cattelan AM, Calabro ML, Aversa SM, Zanchetta M, Meneghetti F, De Rossi A, Chieco-Bianchi L. Regression of AIDS-related Kaposi's sarcoma following antiretroviral therapy with protease inhibitors: biological correlates of clinical outcome. Eur J Cancer. 1999;35:1809–1815. doi: 10.1016/s0959-8049(99)00161-6. [DOI] [PubMed] [Google Scholar]
- 11.Nagy S, Gyulai R, Kemeny L, Szenohradszky P, Dobozy A. Iatrogenic Kaposi's sarcoma: HHV8 positivity persists but the tumors regress almost completely without immunosuppressive therapy. Transplantation. 2000;69:2230–2231. doi: 10.1097/00007890-200005270-00053. [DOI] [PubMed] [Google Scholar]
- 12.Barozzi P, Bonini C, Potenza L, Masetti M, Cappelli G, Gruarin P, Whitby D, Gerunda GE, Mondino A, Riva G, et al. Changes in the immune responses against human herpesvirus-8 in the disease course of posttransplant Kaposi sarcoma. Transplantation. 2008;86:738–744. doi: 10.1097/TP.0b013e318184112c. [DOI] [PubMed] [Google Scholar]
- 13.Brown EE, Whitby D, Vitale F, Marshall V, Mbisa G, Gamache C, Lauria C, Alberg AJ, Serraino D, Cordiali-Fei P, et al. Virologic, hematologic, and immunologic risk factors for classic Kaposi sarcoma. Cancer. 2006;107:2282–2290. doi: 10.1002/cncr.22236. [DOI] [PubMed] [Google Scholar]
- 14.Pellet C, Kerob D, Dupuy A, Carmagnat MV, Mourah S, Podgorniak MP, Toledano C, Morel P, Verola O, Dosquet C, et al. Kaposi's sarcoma-associated herpesvirus viremia is associated with the progression of classic and endemic Kaposi's sarcoma. J Invest Dermatol. 2006;126:621–627. doi: 10.1038/sj.jid.5700083. [DOI] [PubMed] [Google Scholar]
- 15.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 16.van den Beemd R, Boor PP, van Lochem EG, Hop WC, Langerak AW, Wolvers-Tettero IL, Hooijkaas H, van Dongen JJ. Flow cytometric analysis of the Vβ repertoire in healthy controls. Cytometry. 2000;40:336–345. doi: 10.1002/1097-0320(20000801)40:4<336::aid-cyto9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 17.Gorski J, Yassai M, Zhu X, Kissela B, Kissella B, Keever C, Flomenberg N. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping. Correlation with immune status. J Immunol. 1994;152:5109–5119. [PubMed] [Google Scholar]
- 18.Bonfigli S, Fozza C, Contini S, Buzzetti R, Cucca F, Longinotti M. High frequency of the TCRBV20S1 null allele in the Sardinian population. Hum Immunol. 2007;68:426–429. doi: 10.1016/j.humimm.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Bonfigli S, Doro MG, Fozza C, Derudas D, Dore F, Longinotti M. T-cell receptor repertoire in healthy Sardinian subjects. Hum Immunol. 2003;64:689–695. doi: 10.1016/s0198-8859(03)00086-7. [DOI] [PubMed] [Google Scholar]
- 20.Stratigos AJ, Malanos D, Touloumi G, Antoniou A, Potouridou I, Polydorou D, Katsambas AD, Whitby D, Mueller N, Stratigos JD, et al. Association of clinical progression in classic Kaposi's sarcoma with reduction of peripheral B lymphocytes and partial increase in serum immune activation markers. Arch Dermatol. 2005;141:1421–1426. doi: 10.1001/archderm.141.11.1421. [DOI] [PubMed] [Google Scholar]
- 21.Mitsuyasu RT, Groopman JE. Biology and therapy of Kaposi's sarcoma. Semin Oncol. 1984;11:53–59. [PubMed] [Google Scholar]
- 22.Santarelli R, De Marco R, Masala MV, Angeloni A, Uccini S, Pacchiarotti R, Montesu MA, Satta R, Cerimele D, Faggioni A, et al. Direct correlation between human herpesvirus-8 seroprevalence and classic Kaposi's sarcoma incidence in Northern Sardinia. J Med Virol. 2001;65:368–372. doi: 10.1002/jmv.2043. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Barfield R, Benaim E, Leung W, Knowles J, Lawrence D, Otto M, Shurtleff SA, Neale GA, Behm FG, et al. Prediction of T-cell reconstitution by assessment of T-cell receptor excision circle before allogeneic hematopoietic stem cell transplantation in pediatric patients. Blood. 2005;105:886–893. doi: 10.1182/blood-2004-04-1405. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Basu A, Melenhorst JJ, Young NS, Brown KE. Analysis of T-cell repertoire in hepatitis-associated aplastic anemia. Blood. 2004;103:4588–4593. doi: 10.1182/blood-2003-11-3959. [DOI] [PubMed] [Google Scholar]
- 26.Charmley P, Wang K, Hood L, Nickerson DA. Identification and physical mapping of a polymorphic human T cell receptor V β gene with a frequent null allele. J Exp Med. 1993;177:135–143. doi: 10.1084/jem.177.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert M, Gannage M, Karras A, Abel M, Legendre C, Kerob D, Agbalika F, Girard PM, Lebbe C, Caillat-Zucman S. Differences in the frequency and function of HHV8-specific CD8 T cells between asymptomatic HHV8 infection and Kaposi sarcoma. Blood. 2006;108:3871–3880. doi: 10.1182/blood-2006-03-014225. [DOI] [PubMed] [Google Scholar]
- 28.Guihot A, Dupin N, Marcelin AG, Gorin I, Bedin AS, Bossi P, Galicier L, Oksenhendler E, Autran B, Carcelain G. Low T cell responses to human herpesvirus 8 in patients with AIDS-related and classic Kaposi sarcoma. J Infect Dis. 2006;194:1078–1088. doi: 10.1086/507648. [DOI] [PubMed] [Google Scholar]
- 29.Wang QJ, Huang XL, Rappocciolo G, Jenkins FJ, Hildebrand WH, Fan Z, Thomas EK, Rinaldo CR., Jr Identification of an HLA A*0201-restricted CD8(+) T-cell epitope for the glycoprotein B homolog of human herpesvirus 8. Blood. 2002;99:3360–3366. doi: 10.1182/blood.v99.9.3360. [DOI] [PubMed] [Google Scholar]
- 30.Stebbing J, Bourboulia D, Johnson M, Henderson S, Williams I, Wilder N, Tyrer M, Youle M, Imami N, Kobu T, et al. Kaposi's sarcoma-associated herpesvirus cytotoxic T lymphocytes recognize and target Darwinian positively selected autologous K1 epitopes. J Virol. 2003;77:4306–4314. doi: 10.1128/JVI.77.7.4306-4314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miqueu P, Guillet M, Degauque N, Dore JC, Soulillou JP, Brouard S. Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol Immunol. 2007;44:1057–1064. doi: 10.1016/j.molimm.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Alaibac M, Congedo M, Barbarossa G, Bottiglieri A, Fillippi ED, Marzullo F, Quarta G, Schittulli F. Analysis of clonal antigen receptor gene rearrangements in T-cells involved with Kaposi's sarcoma. Anticancer Res. 1997;17:1205–1207. [PubMed] [Google Scholar]
- 33.Fozza C, Contini S, Galleu A, Simula MP, Virdis P, Bonfigli S, Longinotti M. Patients with myelodysplastic syndromes display several T-cell expansions, which are mostly polyclonal in the CD4(+) subset and oligoclonal in the CD8(+) subset. Exp Hematol. 2009;37:947–955. doi: 10.1016/j.exphem.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Risitano AM, Kook H, Zeng W, Chen G, Young NS, Maciejewski JP. Oligoclonal and polyclonal CD4 and CD8 lymphocytes in aplastic anemia and paroxysmal nocturnal hemoglobinuria measured by V β CDR3 spectratyping and flow cytometry. Blood. 2002;100:178–183. doi: 10.1182/blood-2002-01-0236. [DOI] [PubMed] [Google Scholar]
- 35.Arons E, Sorbara L, Raffeld M, Stetler-Stevenson M, Steinberg SM, Liewehr DJ, Pastan I, Kreitman RJ. Characterization of T-cell repertoire in hairy cell leukemia patients before and after recombinant immunotoxin BL22 therapy. Cancer Immunol Immunother. 2006;55:1100–1110. doi: 10.1007/s00262-005-0099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correia-Neves M, Waltzinger C, Mathis D, Benoist C. The shaping of the T cell repertoire. Immunity. 2001;14:21–32. doi: 10.1016/s1074-7613(01)00086-3. [DOI] [PubMed] [Google Scholar]
- 37.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, Bonneville M. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 38.Wynn KK, Crough T, Campbell S, McNeil K, Galbraith A, Moss DJ, Silins SL, Bell S, Khanna R. Narrowing of T-cell receptor β variable repertoire during symptomatic herpesvirus infection in transplant patients. Immunol Cell Biol. 2010;88:125–135. doi: 10.1038/icb.2009.74. [DOI] [PubMed] [Google Scholar]