Abstract
Alterations of DNA methylation play an important role in gliomas. In a genome-wide screen, we identified a CpG-rich fragment within the 5′ region of the tumor necrosis factor receptor superfamily, member 11A gene (TNFRSF11A) that showed de novo methylation in gliomas. TNFRSF11A, also known as receptor activator of NF-κB (RANK), activates several signaling pathways, such as NF-κB, JNK, ERK, p38α, and Akt/PKB. Using pyrosequencing, we detected RANK/TNFRSF11A promoter methylation in 8 (57.1%) of 14 diffuse astrocytomas, 17 (77.3%) of 22 anaplastic astrocytomas, 101 (84.2%) of 120 glioblastomas, 6 (100%) of 6 glioma cell lines, and 7 (100%) of 7 glioma stem cell-enriched glioblastoma primary cultures but not in four normal white matter tissue samples. Treatment of glioma cell lines with the demethylating agent 5-aza-2′-deoxycytidine significantly reduced the methylation level and resulted in increased RANK/TNFRSF11A mRNA expression. Overexpression of RANK/TNFRSF11A in glioblastoma cell lines leads to a significant reduction in focus formation and elevated apoptotic activity after flow cytometric analysis. Reporter assay studies of transfected glioma cells supported these results by showing the activation of signaling pathways associated with regulation of apoptosis. We conclude that RANK/TNFRSF11A is a novel and frequent target for de novo methylation in gliomas, which affects apoptotic activity and focus formation thereby contributing to the molecular pathogenesis of gliomas.
Introduction
Gliomas are the most common primary brain tumors in adults, with glioblastoma of World Health Organization (WHO) grade 4 being the most frequent and most malignant type of glioma. The incidence rate of primary glioblastoma is 3.18 per 100,000 person years, age adjusted to the 2000 US standard population. Gliomas can develop at any age with a peak incidence between 65 and 84 years. Glioblastomas (GBMs) are highly invasive and respond poorly to radiation therapy as well as most forms of chemotherapy. It, therefore, has a dismal prognosis with a median survival time of less than 12 months as shown in a population-based study [1,2]. Primary glioblastomas are characterized by multiple genetic alterations, commonly including loss of heterozygosity (LOH) on chromosome 10, EGFR amplification and overexpression, CDKN2A deletion and PTEN mutation [1,2]. Only 5% of glioblastomas develop through progression from preexisting astrocytomas of WHO grade 2 or 3 [3]. These so-called secondary glioblastomas typically lack EGFR amplification but frequently carry TP53 mutations. Recently, somatic mutations of the IDH1/IDH2 genes have been reported in most diffuse astrocytic and oligodendroglial tumors as well as secondary glioblastomas. These mutations, however, are rare in primary glioblastomas [4]. Interestingly, IDH1 mutation seems to be associated with increased DNA methylation at 5′-CpG islands of multiple genes, the so-called CpG island methylator phenotype [5]. To identify novel glioma-associated candidate genes subjected to epigenetic regulation, we applied a genome-wide methylation analysis using the differential methylation hybridization (DMH) technology. From this approach, we identified several novel candidate tumor suppressor genes showing altered promoter methylation patterns in gliomas [6]. Here, we report on the epigenetic inactivation of the RANK/TNFRSF11A gene in gliomas and derived cell lines. This gene encodes a type 1 membrane protein that, after binding its ligand RANKL, activates signaling pathways such as NF-κB, JNK, ERK, p38, and Akt/PKB, through TRAF protein phosphorylation. RANK/TNFRSF11A signaling is largely considered to be growth promoting and apoptosis reducing such as the effects observed in osteoclasts [7–9]. However, there is increasing evidence that in other cell systems the activity of RANK/TNFRSF11A may in fact trigger apoptosis and suppress proliferation [10,11]. Our data indicate that epigenetic silencing of RANK/TNFRSF11A was confirmed by treatment of glioma cells with demethylating agent 5-aza-2′-deoxycytidine, which resulted in increased expression of the corresponding transcript. Transient as well as stable transfection of RANK/TNFRSF11A into established glioblastoma cell lines resulted in a significant reduction of focus formation and increased apoptosis. Reporter assays of transfected glioblastoma cells reveal an up-regulation of NF-κB signaling but also unmask an activation of distinct signaling pathways associated with the regulation of apoptosis.
Materials and Methods
Tumor Samples and Cell Lines
We investigated glioma tumor specimens from 156 patients including 97 with primary glioblastomas WHO grade IV (pGBMIV), 23 with secondary glioblastomas WHO grade IV (sGBMIV), 22 with anaplastic astrocytomas WHO grade III (AAIII), and 14 with diffuse astrocytomas WHO grade II (AII). Primary as well as recurrent tumor samples of three patients were investigated. The patients included 63 women and 86 men. The mean age at surgery was 55 years (range, 24–80 years). All tumors were classified according to the WHO classification of tumors of the central nervous system [12]. Histologic assessment confirmed that all specimens used for extraction of nucleic acids consisted of at least 80% tumor cells. DNA and RNA were extracted from unfixed frozen tumor tissue as reported [13]. In brief, tumor samples were homogenized in 6 ml of 4 M guanidine isothiocyanate solution. The homogenate was then layered over 4 ml of CsCl and ultra-centrifuged at 170,000g for 16 hours. The RNA was recovered as a pellet and dissolved in diethylpyrocarbonate-treated water containing the RNase inhibitor RNasin (Promega, Mannheim, Germany). The DNA was purified from the CsCl phase using proteinase K digestion followed by phenol/chloroform extraction. Four normal white matter tissue samples were used to extract DNA as reference tissue for the DMH and focused methylation analyses. Glioma cell lines A172, U373MG, T98G, A178, U87MG, and LN229 were obtained from the LICR (San Diego, CA), and identities were confirmed by STR DNA profiling of 15 loci plus sex-determining marker amelogenin (Genetica DNA Laboratories, Inc, Cincinnati, OH). Seven stem cell- enriched primary glioblastoma cultures (15z, 21z, 25z, 35z, 46z, 78z, and 106z) were also included in our study [14]. All tissue samples were used in an anonymous manner as approved by the local ethics committee at Heinrich Heine University Düsseldorf and University Bonn.
DMH Analysis
The DMH procedure was performed as described [6,15]. CpG-rich DNA fragments were isolated from the human CGI (CpG island) library and screened for the presence of BstUI (NEB, Beverly, MA) and HpaII (NEB) restriction sites. A total of 16,000 suitable fragments were polymerase chain reaction (PCR) amplified using plasmid primer and spotted onto UltraGAPS microarrays (Corning, Acton, MA). For amplicon generation, 2 µg of genomic DNA of tumor and normal brain samples (white matter) was digested with MseI (NEB) according to the manufacturer's protocol. After ligation of linker H12/H24, fragments were digested with methylation-sensitive restriction enzymes BstUI and HpaII and amplified for 20 cycles with H24 as primer. PCR fragments were labeled by direct incorporation of Cy3-dCTP (normal brain) and Cy5-dCTP (tumor) using fluorescent dyes (Amersham, Buckinghamshire, United Kingdom) and the Klenow fragment (Invitrogen, Carlsbad, CA). Labeled amplicons were purified with Microcon YM-30 columns (Millipore, Bedford, MA), and equal amounts of Cy3 and Cy5 label and 20 µg of human CotI DNA were combined for hybridization on DMH microarrays. Hybridization and analysis procedure were carried out as described [6,15]. Data from single-copy sequences were normalized and loci with a Cy5/Cy3 ratio greater than 2 were scored as hypermethylated.
Analysis of the RANK/TNFRSF11A 5′-CpG Island Methylation Status by Pyrosequencing
Genomic DNA of gliomas, glioblastoma cell lines, stem cell-enriched primary glioblastoma cultures, and nonneoplastic white matter tissue was treated with sodium bisulfite using the EpiTect Bisulfite kit (Qiagen, Hilden, Germany). PCR products of RANK/TNFRSF11A containing 14 putative CpG methylation sites were amplified from sodium bisulfite-converted DNA using the primer RANK1 fw-5′-ggtaaggtaggagttagtgt-3′ and the biotinylated reverse primer RANK-Bio rev-5′-aatacccaaactcccctaattta-3′. The pyrosequencing primer was RANK-Pyro-5′-ggtaatttagggttggtttt-3′. Pyrosequencing was performed using the PyroMarkGold Reagents (Qiagen) on the Pyromark Q24 instrument (Qiagen) according to the manufacturer's instructions. Pyrogram outputs were analyzed using the PyroMark Q24 software.
Tissue Culture of Glioblastoma Cell Lines and Treatment with 5-Aza-2′-deoxycytidine
Glioblastoma cell lines A172 and U373MG were cultured as described [16]. To induce demethylation, cell lines were grown in the presence or absence of 1 to 5 µM 5-aza-2′-deoxycytidine (Sigma, St. Louis, MO). Medium was renewed daily. After 3 days, DNA and total RNA were extracted for RANK/TNFRSF11A methylation and expression analysis. This experiment was performed in triplicates.
RANK/TNFRSF11A mRNA Expression Analysis by Real-time Reverse Transcription-PCR
For complementary DNA (cDNA) synthesis, 1 µg of total RNA, random hexamers, and SuperScript II Reverse Transcriptase (Invitrogen) was used according to the instructions of the manufacturer.
RANK/TNFRSF11A expression was investigated by real-time reverse transcription-PCR (RT-PCR) using the ABI PRISM 5700 sequence detection system (Applied Biosystems). The transcript level of RANK/TNFRSF11A was normalized to the transcript level of ADP-ribosylation factor 1 (ARF1; GenBank accession no. M36340) [17]. To ensure that high-quality RNA was used for expression analysis, only tumors with Ct values of at most 30 cycles for the control locus ARF1 were used (range, 18.2–27.5). Primer sequences were RANK/TNFRSF11A-RT-F, 5′-tcaacaaggacacagtgtgc-3′, and RANK/TNFRSF11A-RT-R, 5′-tttggtggttttctagctgg-3′, resulting in a 181-bp fragment and ARF1-F, 5′-gaccacgatcctctacaagc-3′, and ARF1-R, 5′-tcccacacagtgaagctgatg-3′, resulting in a 111-bp fragment. RNA of normal white matter tissue and commercially available adult humanbrain RNA of corpus callosum, temporal lobe, parietal lobe, frontal lobe, and occipital lobe (BioChain Institute, Inc, Hayward, CA) was used as controls. Expression levels of individual tumors and cell lines are given as ratio = 2-ΔΔCt. Because of the limited availability of RNA expression analysis were performed in duplicates.
RANK/TNFRSF11A Information from TCGA and REMBRANDT Platform Analyses
The TCGA dat aportal does not show information on methylation or expression of RANK/TNFRSF11A in glioblastomas when assessing the TCGA data on the CGWB (The Cancer Genome Workbench) at the National Institutes of Health server https://cgwb.nci.nih.gov/ and https://cgwb.nci.nih.gov/cgi-bin/srstuff?prjid=41&db=hg18&op=methyl in March 2012 [1]. The REMBRANDT database (NCI2005; REMBRANDT version 1.5; http://rembrandt.nci.nih.gov/) contained expression data of RANK/TNFRSF11A. The Kaplan-Meier Survival plot of this data (343 glioma), however, does not show any significant correlation with up-or down-regulation of RANK/TNFRSF11A.
RANK/TNFRSF11A Cloning and Transfection
RANK/TNFRSF11A plasmid was purchased from OriGene (Rockville, MD) and subcloned into pcDNA4/myc-His expression vector (Invitrogen). U373MG and A172 cells were transfected by lipofection (FuGENE 6; Roche, Mannheim, Germany) and selected by adding 240 to 320 µg/ml zeocin (Invitrogen) to the growth medium after 24 hours. Approximately 2 weeks later, zeocin-resistant colonies were trypsinized, and total RNA were extracted for RANK/TNFRSF11A expression analysis. Assays were carried out at least in triplicates.
Colony Formation Assay
For colony formation assays, cells (2 x 105/well) were seeded in 12-well tissue culture plates and transfected with either RANK-pcDNA4/myc-His plasmid or pcDNA4/myc-His control vector (0.8 µg each) using FuGENE 6 transfection reagent (FuGENE 6; Roche). At 24 hours after transfection, cells were collected, seeded in 10-cm dishes, and exposed to 240 µg/ml (A172) or 320 µg/ml (U373MG) zeocin (Invitrogen) for a total of 14 days. Cells were fixed and stained in 20% methanol and crystal violet, and colonies with more than 50 cells were counted. Assays were carried out at least in triplicates.
Measurement of Cellular Proliferation and Viability by Flow Cytometry
U373MG cells stably or transiently expressing RANK-pcDNA4/myc-His plasmid or pcDNA4/myc-His control vector (in case of transient transfection cells were cotransfected with pMAX GFP) were serum starved for 24 hours. Flow cytometric immunophenotyping was performed as described [18,19] with an Alexa Fluor 647-labeled phospho-(Ser10)-histone H3 antibody (1:20; Cell Signaling Technology, Frankfurt am Main, Germany) staining proliferating cells and a phycoerythrin-labeled cleaved poly(ADP-ribose) polymerase (PARP) (Asp214) antibody (1:5; BD Biosciences, Heidelberg, Germany) to detect apoptotic cells [19–22].
At least 100,000 events were recorded per experiment. For transient transfection, only GFP-positive single cells were included in the analysis. Each experiment was carried out at least in triplicates.
Multipathway Reporter Array
U373MG cells overexpressing RANK-pcDNA4/myc-His plasmid or pcDNA4/myc-His control vector were reverse transfected with each of the reporter assays from the 45-Pathway reporter Array plate (SA Biosciences, Frederick, MD) using X-tremeGENE HP DNA transfection reagent (Roche) [23]. The reporter assay consists of a mixture of an inducible transcription factor responsive firefly luciferase reporter and constitutively expressing renilla construct. A mixture of noninducible firefly luciferase reporter and constitutively expressing renilla construct and a mixture of a constitutively expressing GFP construct constitutively expressing firefly luciferase and renilla luciferase construct was included as negative and positive controls. Reverse transfection was performed following the manufacturer's recommendation (SA Biosciences). After 48 hours, luciferase assay was performed using the Dual-Luciferase Reporter Assay following the manufacturer's protocol (Promega, Madison, WI), and results are expressed as fold change. The fold change was calculated by dividing the normalized luciferase activities of each pathway-focused reporter reverse transfected with U373MG cells overexpressing RANK-pcDNA4/myc-His plasmid by the normalized luciferase activity of each pathway-focused reporter reverse transfected with the negative control (U373MG cells expressing pcDNA4/myc-His plasmid). Reporters showing two-fold or higher changes up or down in relative luciferase units were considered as upregulated or downregulated, respectively. Experiments were performed in duplicates and analyzed using GloMax 96 Microplate Luminometer (Promega).
Statistical Analysis
All statistical analyses, including t test, Fisher exact test, and Mann-Whitney U test were carried out with the GraphPad Prism software (version 5; GraphPad Software, La Jolla, CA).
Results
Aberrant Methylation within the 5′-CpG Island of RANK/TNFRSF11A
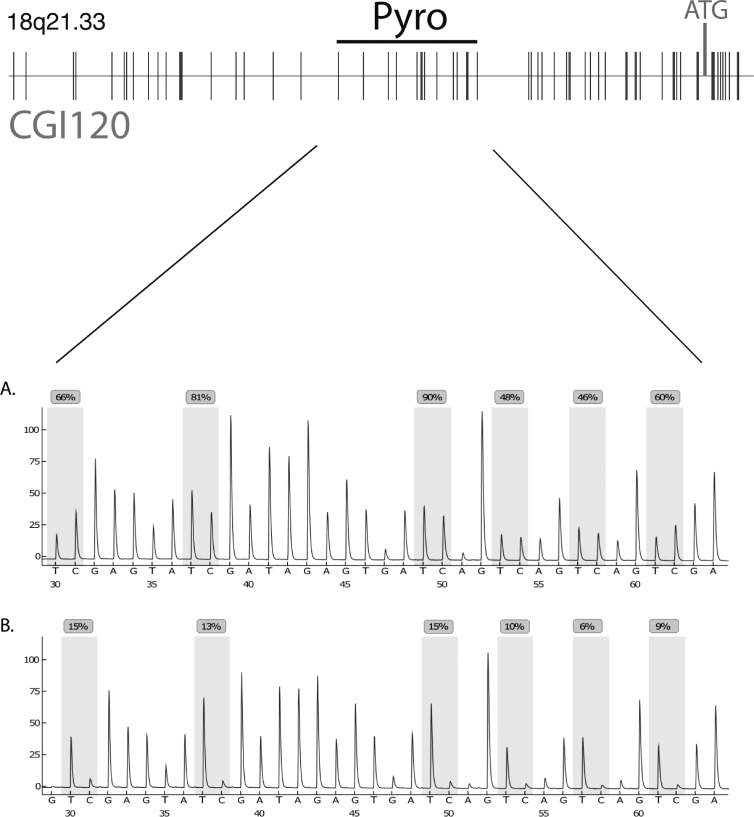
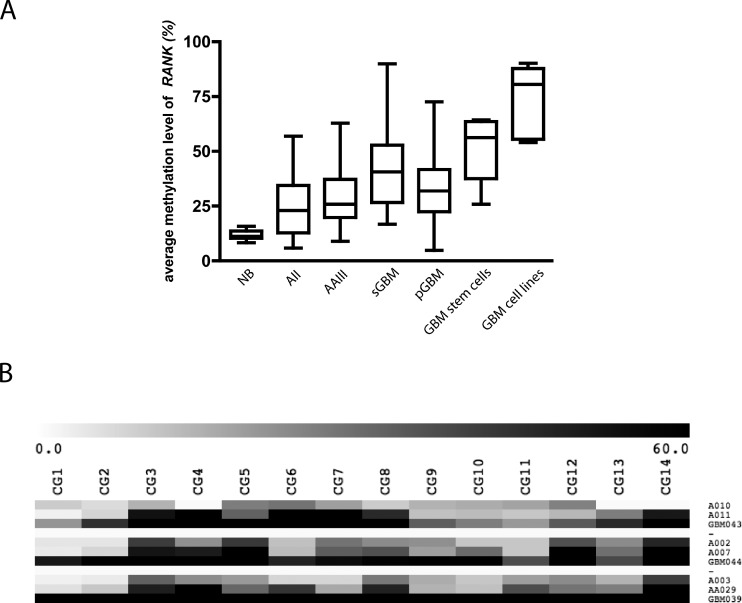
Using DMH analysis, we identified a CpG-rich fragment that was frequently hypermethylated in gliomas. This fragment is located within the 5′-CpG island 120 (CpG:120 chr18:59992070-59993556) starting at 477 bp upstream of the first exon of the receptor activator of NF-κB/tumor necrosis factor receptor superfamily 11A gene (RANK/TNFRSF11A; Figure 1). To determine the frequency and extent of de novo methylation within this fragment, pyrosequencing analyses were performed on 156 gliomas, 4 cerebral white matter samples, 6 glioma cell lines, and 7 stem cell-enriched glioma primary cultures. Figure 1 shows a section of representative pyrograms of CpG sites 1 to 14 within the investigated region in a primary glioblastoma (GBM042) and a normal brain white matter sample (NB2). The tumor sample carried a high content of methylated alleles with most CpG sites being strongly methylated, whereas white matter showed low levels of methylation in this region. Samples were scored methylated if they exceeded the two-fold SD of white matter control samples (20.3%). De novo methylation of RANK/TNFRSF11A was found in 8 (57.1%) of 14 AII, 17 (77.3%) of 22 AAIII, 20 (87%) of 23 sGBMIV, 81 (83.5%) of 97 pGBMIV, 6 (100%) of 6 glioma cell lines, and 7 (100%) of 7 stem cell-enriched glioma primary cultures but not in four normal white matter samples (Table W1). The overall methylation level as determined by taking the mean of the arithmetic means of pyrosequencing methylation grades of CpG sites was 25.4% in AII, 28.9% in AAIII, 40.2% in sGBMIV, 33.3% in pGBMIV, 50.3% in stem cell-enriched glioma primary cultures, 74.2% in glioblastoma cell lines, and 11.9% in normal white matter (Figures 2A, W1, and Table W1). These data demonstrate that methylation of RANK/TNFRSF11A is significantly more frequent in high-grade gliomas (Fisher exact test, P = .0246) compared with low-grade astrocytomas. Furthermore, the mean methylation levels of glioblastoma WHO grade IV are significantly stronger than those of low-grade astrocytomas WHO grade II and anaplastic astrocytomas WHO grade III (Table W1, t test: AII + AAII vs pGBMIV, P = .026; AII + AAIII vs sGBMIV, P = .0035), whereas AII and AAIII do not differ significantly (t test, P = .3767). This was confirmed in primary and recurrent tumors from three individual patients whereby the precursor lesions were of lower grade than the subsequent secondary glioblastomas. The observed methylation levels were three-to five-fold higher in the secondary glioblastomas than in the corresponding primary astrocytomas WHO grade 2 (t test, P =.0264; Figure 2B). The investigation of seven stem cell-enriched primary glioblastoma cultures by pyrosequencing showed even higher methylation levels (50.3%) especially surrounding the p53 binding site (Figure W1). This indicates that this epigenetic modification targets those cells that are considered to be the drivers of tumor growth and capable of forming gliomas in xenografted animals.
Figure 1.
Localization of the investigated DNA fragment within the CpG island at chromosome 18q21.33 (59991900-59992679). The sequence investigated by pyrosequencing, and the ATG start codon is indicated. Below, details of two representative pyrograms within the 5′ upstream region of RANK/TNFRSF11A are shown. The glioblastoma sample GBM042 (A) shows strong methylation, whereas low methylation levels were found in normal white matter tissue (B).
Figure 2.
Average methylation level of 156 primary glioma tissues, glioblastoma cell lines, stem cell-enriched primary glioblastoma cell cultures, and normal white matter tissues within the investigated region of RANK/TNFRSF11A (A). Heat map of quantitative methylation data of the 14 investigated CpG sites within the RANK/TNFRSF11A CpG island in three patients, for which tissue from the initial low-grade glioma and the high-grade relapse tumor were available. The color code shows the methylation range (0%–60%) (B).
Expression Analyses of RANK/TNFRSF11A in Glioma Tissues
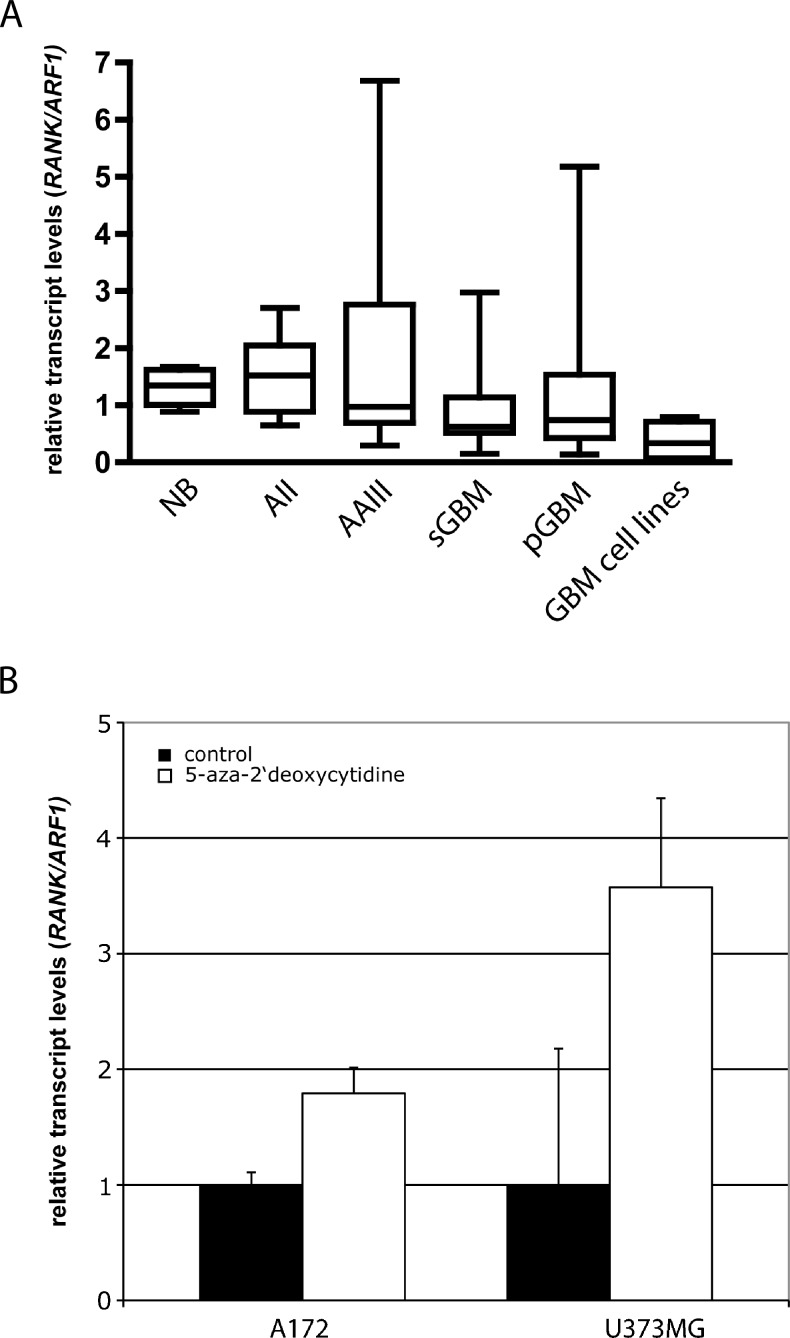
The mRNA expression of RANK/TNFRSF11A and the housekeeping gene ARF1 was investigated in 77 primary glioma tissues, 6 glioblastoma cell lines, and 9 normal white matter tissues (Figures 3A and W4). Compared with the normal brain tissues, there is a trend toward reduced transcript levels from diffuse astrocytomas, anaplastic astrocytomas to glioblastomas. In each group, however, there were few samples showing comparably high transcript levels. All investigated glioblastoma cell lines showed reduced RANK/TNFRSF11A transcript levels. Dividing the 77 samples into a methylated and an unmethylated group (samples were scored methylated if they exceeded the two-fold SD of white matter control samples [20.3%]) revealed a significant difference in the expression levels (Mann-Whitney U test, P = .0159; t test, P =.0017).
Figure 3.
Relative transcript levels of RANK/TNFRSF11A (RANK/TNFRSF11A/ARF1) analyzed by real-time RT-PCR in gliomas, glioblastoma cell lines, and normal white matter tissue (A). Relative transcript levels of RANK/TNFRSF11A in glioblatoma cell lines with (□) or without (■) 1 and 5 µM 5-aza-2′-deoxycytidine treatment (B).
Demethylation and Expression Analyses of RANK/TNFRSF11A in Glioblastoma Cell Lines
TNFRSF11A/RANK mRNA expression was downregulated in the glioblastoma cell lines A172 and U373MG. After treating the cells with 5-aza-2′-deoxycytidine, 1.7- and 3.5-fold increases in transcript levels were detected (Figure 3B). Bisulfite sequencing confirmed that 5-aza-2′-deoxycytidine treatment resulted in demethylation of the investigated region (Figure W2). These data indicate that aberrant 5′-CpG island methylation contributes to the down-regulation of RANK/TNFRSF11A transcription in glioblastoma cell lines.
Functional Analysis of RANK/TNFRSF11A in Glioma Cells
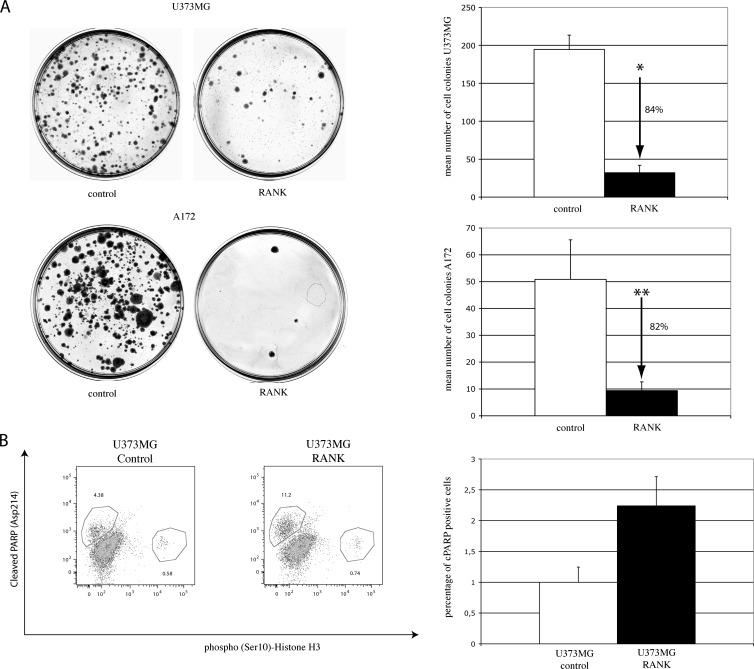
To study the influence of RANK/TNFRSF11A expression on growth and apoptotic activity of glioblastoma cells in vitro, we overexpressed RANK/TNFRSF11A in the glioblastoma cell lines U373MG and A172. RANK/TNFRSF11A expression was monitored by real-time RT-PCR (Figure W3). In colony formation assays, an average of 32 and 9 colonies formed in U373MG and A172 cells transfected with RANK/TNFRSF11A, in comparison to an average of 195 and 51 colonies in the control cells (Figure 4A). Thus, expression of RANK/TNFRSF11A led to an 84% and 82% decrease in colony number (t test, P < .0001 in U373MG cells and P = .0193 in A172 cells, respectively).
Figure 4.
Focus formation assay and flow cytometric analysis of U373MG glioblastoma cells transfected with RANK/TNFRSF11A or control vector pcDNA4/myc-His. Representative image of a focus assay of U373MG and A172 cells. Number of foci formed after transfection with RANK/TNFRSF11A (■) or pcDNA4/myc-His control (□); t test, * U373MG, P <.0001; **A172, P = .0193 (A). Representative image and summarized results of all flow cytometric analysis showing significant induction of apoptosis (t test, P <.0001) as measured by detection of cleaved PARP and analysis of phospho(Ser10)-histone H3 in U373MG cells transiently cotransfected with RANK-pcDNA4/myc-His plasmid and pMAX GFP or pcDNA4/myc-His control vector and pMAX GFP (B).
To further investigate the functional role of RANK/TNFRSF11A on cellular proliferation and apoptosis, we performed flow cytometric analyses using antibodies against Ser10-phosphorylated Histone H3 and PARP (Asp214). The apoptotic cellular population of U373MG cells transiently transfected with RANK/TNFRSF11A was significantly increased by 124% in U373MG cells (t test, P < .0001), when compared with the respective control cells, whereas the proliferation was slightly increased by 26% (Figure 4B). This experiment was repeated with U373MG cells stably expressing RANK/TNFRSF11A with similar results. The functional data indicate a role of RANK/TNFRSF11A in the regulation of apoptosis in glioblastoma cells. Because no significant reduction of proliferation was observed in flow cytometric analysis, it is likely that the reduced focus formation is not due to a reduced proliferative potential but rather to an increased apoptotic activity in the RANK/TNFRSF11A-transfected glioblastoma cells.
Activity of Downstream Signaling Pathways
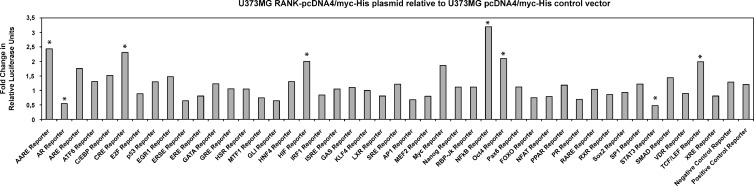
To identify molecular pathways responsible for transmitting signals downstream of RANK/TNFRSF11A, we have used a signal transduction reporter assay that allows the comprehensive analysis of 45 signaling pathways. U373MG cells overexpressing RANK-pcDNA4/ myc-His plasmid or pcDNA4/myc-His control vector were reverse transfected with each of the 45 reporter assays from the array plate. Our results indicate that overexpression of RANK/TNFRSF11A up-regulates amino acid response, cAMP/PKA (CRE), hypoxia (HIF), NF-κB, Oct4, and Wnt (TCF/LEF) signaling while downregulating androgen and STAT3 signaling in U373MG cells (Figure 5).
Figure 5.
Reporter array analysis of 45 signaling pathways in U373MG glioblastoma cells transfected with RANK-pcDNA4/myc-His plasmid and pcDNA4/myc-His control vector. The diagram depicts the fold change of reporter activity indicative for specific cancer-associated signaling pathways. *Signaling pathways showing significant up-regulation or down-regulation.
Relationship between Molecular Findings and Clinical Data
The comparison of RANK/TNFRSF11A methylation and clinical showed no significant correlation with survival data of pGBMIV patients (P =.355).
Mutation of the TP53 gene was assessed in 48 glioma samples (9 AAIII, 12 sGBMIV, and 27 pGBMIV) and was shown in 18 (40.9%) of the tumors studied, including 4 AAIII, 8 sGBM, and 6pGBM. TP53 mutation was not associated with RANK/TNFRSF11A methylation (Fisher exact test, P = .28). IDH1 mutations identified in 5 AII, 15 AAIII, 17 sGBMIV, and 9 pGBMIV do not correlated with RANK/TNFRSF11A methylation (Fisher exact test, P =1.0).
Discussion
Our global approach to identify novel genes silenced by promoter methylation in human gliomas uncovered the RANK/TNFRSF11A gene located on chromosome 18q21.33. The gene encodes a 616-amino acid transmembrane receptor that belongs to the TNFR family [24].
Binding of the ligand RANKL results in intracellular signal transduction through several TRAFs (TNF receptor-associated factor) and may lead to the activation of various signaling pathways, including the NF-κB, MAPK (JNK, p38, ERK) and PI3K/AKT cascades [7,9]. RANK/TNFRSF11A and RANKL are known to play a key role in the differentiation, activation, and survival of osteoclasts. The balance between RANK/TNFRSF11A, RANKL, and its decoy receptor OPG is pivotal for bone biology, but they are also involved in other biologic systems, for example, the immune and vascular systems [8,9]. Furthermore, there is also evidence that this receptor might have growth-suppressive and proapoptotic functions in other cell types [11,24]. Therefore, we decided to investigate the epigenetic regulation of this gene in gliomas and to elucidate its functional implications in glioma cells.
To our knowledge, this is the first study demonstrating epigenetic regulation of RANK/TNFRSF11A in cancer. We investigated the methylation status of RANK/TNFRSF11A in 156 astrocytic gliomas of different malignancy grades, established glioma cell lines and primary glioblastoma stem cell-enriched cultures. Our data revealed frequent hypermethylation of the promoter CpG island in both primary glioma tissues and cultured glioma cells. A significant increase in the overall methylation level and in the number of affected samples with WHO grade suggests a progression-associated advantage in growth or clonal expansion of glioma cells containing methylated RANK/TNFRSF11A alleles. Although the quantitative methylation analysis of the primary tumor tissues focused on a region adjacent to a putative p53 binding site in the RANK/TNFRSF11A promoter, there was a spread of methylation extending beyond this region in stem cell-enriched primary cultures and in established glioma cell lines. The fact that most of the investigated stem cell-enriched glioblastoma cultures contained strongly methylated RANK/TNFRSF11A alleles argues for an important role of this epigenetic modification in the molecular pathogenesis of glioblastomas. The strong methylation adjacent to the putative p53 binding site suggests that RANK/TNFRSF11A transcription might be altered in gliomas by perturbing p53 binding to the respective consensus sequence. Alternatively, the inactivation of p53 by somatic mutations, which are often seen in AII, AAIII, and sGBMIV gliomas, might prevent the binding of the protein to this site, thereby allowing CpG methylation to occur. A recent study indicated that a large fraction of diffuse and anaplastic gliomas as well as secondary glioblastoma display aberrant methylation in multiple gene promoters (CpG island methylator phenotype) and much less in primary glioblastomas [5]. However, RANK/TNFRSF11A promoter methylation cannot be attributed to the CpG island methylator phenotype alone because no significant correlation between IDH1 mutation and RANK/TNFRSF11A promoter methylation was observed (Fisher exact test, P = 1.0).
A functional role of the observed CpG island methylation in the transcriptional regulation of RANK/TNFRSF11A in gliomas is supported by the demonstration of reduced RANK/TNFRSF11A promoter methylation and increased expression in glioma cells treated with 5-aza-2′-deoxycytidine. RANK/TNFRSF11A mRNA is ubiquitously expressed in human tissues, and the protein was mainly detected in osteoclasts, fibroblasts, B and T lymphocytes, dendritic cells, and endothelial cells [2,9,25]. RANK/TNFRSF11A and RANKL mRNA and protein are also expressed in normal brain tissue of mice and rat primarily in astrocytes and microglia [26–28]. It is interesting to note that several members of the downstream cascade of RANK/TNFRSF11A are well-established target genes of malignant gliomas. In particular, AKT and PTEN alterations are common in glioblastomas [2] and may alter the RANK response of the affected tumor cells [8,9]. These observations further support that RANK/TNFRSF11A possesses tumor suppressor properties, which may be inhibited by epigenetic silencing of the gene. The lack of correlation between hypermethylation and survival despite the obvious impact of RANK expression on apoptosis and focus formation of GBM cells may be due to additional molecular alterations that may be present in the tumors of the investigated patients and affect the clinical course. In addition to the regulation of RANK/TNFRSF11A expression, its activity may also be altered by the expression of the extracellular modulator TNFRSF11B/OPG that serves as decoy receptor for RANKL and inhibits RANK/TNFRSF11A signaling. In concordance with a tumor suppressive function of RANK/TNFRSF11A, a strong expression of the RANKL decoy receptor TNFRSF11B/OPG was found to be associated with tumor invasion, nodal metastases, advanced tumor stage, and poor prognosis in gastric carcinoma patients [29]. Interestingly, it has been shown that a small subset of glioblastoma tissues as well as established glioblastoma cell lines express TNFRSF11B/OPG [30]. We analyzed OPG expression by real-time RT-PCR in our patient cohort and did not observe any correlation between OPG expression levels and RANK methylation or expression (data not shown). Furthermore, there are several observations indicating cell-type specific apoptotic and growth inhibitory effects of RANKL/RANK and/or their signaling targets NF-κB and JNK [10]. Müller et al. [11] observed that doxorubicin-induced apoptosis in the T-lymphoblastic cell line CEM was prevented by a neutralizing αRANKL antibody as well as by RANK-Fc fusion protein. The apoptotic effect in this context was mediated by NF-κB. Bharti et al. [10] showed that the treatment of RAW 264.7 monocyte cells with RANKL leads to induction of apoptosis and suppression of cellular proliferation. The antiproliferative effect in these cells was independent of NF-κB activation but required TRAF6 recruitment and was mediated through the MAP-kinase JNK.
The PI3K/Akt signaling pathway, which is frequently altered in high-grade gliomas, is influenced by RANKL/RANK, thereby affecting cellular apoptosis and survival. Gingery et al. [31] showed that blocking of Akt, PI3K, or NF-κB in murine osteoclasts maintained with RANKL resulted in cellular apoptosis. In addition, RANKL was shown to activate Akt and its antagonist PTEN and overexpression of PTEN can result in inhibition of Akt-induced survival [24,32].
The cytoplasmic domain of RANK/TNFRSF11A has been shown to interact with TRAF1, 2, 3, 5, and 6 [33–35]. Our observation that transfected variants of RANK/TNFRSF11A, lacking most of the TRAF binding sites, were equally capable of suppressing colony formation raises the question whether RANK/TNFRSF11A interacts with other ligands or transmembrane proteins to promote apoptotic responses of the cells observed in the focus formation assay and flow cytometric analysis (data not shown).
RANK/TNFRSF11A-transfected glioma cell lines showed a significant reduction in focus formation as well as an elevated apoptotic activity. The multipathway reporter array analysis revealed an activation of the NF-κB signaling pathway confirming accurate biologic activity of the construct. Beside this established signaling pathway, we observed an impact of RANK/TNFRSF11A expression on the activity of several other signaling cascades known to be deregulated in glioma molecular pathogenesis.
Signaling pathways initiated by amino acid deprivation (indicated by the AARE reporter) have already been associated with the induction of apoptosis and autophagy in vertebrate cells [36].
In addition, it is striking to note that the pathways showing the most prominent deregulation in RANK/TNFRSF11A-transfected cells are involved in regulation of the developmental potential or the proliferation of stem-like cells. This is especially interesting because the methylation profiling of primary stem cell-enriched glioblastoma cells that are considered to be the drivers of tumor growth exhibits particularly high levels of methylated RANK/TNFRSF11A alleles. These comprise the WNT, Oct4, and HIF signaling as indicated by a significant induction of respective specific target genes (TCF/LEF, Oct4, HIF). We also unmasked a significant down-regulation of the reporter activity of STAT3, which has been discussed as potential target for therapy for glioma patients [37,38]. Inhibition of STAT3 signaling pathway has also been associated with the growth regulation and apoptosis of glioma stem cells and medulloblastomas [39–41].
In conclusion, we identified RANK/TNFRSF11A as a novel and frequent target for de novo methylation in gliomas. The increased frequency and density of methylation in the 5′-CpG island of RANK/TNFRSF11A in glioblastoma tissues and stem cell-enriched glioblastoma cultures compared with lower-grade astrocytomas suggest a selection advantage for cells that inactivate RANK/TNFRSF11A through DNA methylation. Our functional in vitro results further suggest that this advantage might be due to a significantly reduced apoptotic activity of RANK/TNFRSF11A-silenced glioma cells. This assumption is further supported by the multi pathway reporter array that uncovered an involvement of RANK/TNFRSF11A in the regulation of proapoptotic signaling cascades as well as in the regulation of stem like cells within primary glioma tissues.
Supplementary Methods
Acknowledgments
The authors thank Peter Wurst, Dagmar Metzger, and Anne Woitecki for excellent technical assistance.
Footnotes
This study was supported by National Genome Research Network “NGFNplus,” Brain Tumor Network plus (grant 01GS08187, SP8) and the BONFOR program of the Medical Faculty of the University of Bonn. B.S. is supported by the Lichtenberg program of the VW Foundation. No potential conflicts of interest are disclosed.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.TCGA, author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waha A, Rodrigues F, Waha A, Meyer-Puttlitz B, Cavenee W, Huang T-M, Yan P. Methylation profiling identifies epigenetic markers for high-grade gliomas. Cancer Genomics Proteomics. 2004;1:209–214. [PubMed] [Google Scholar]
- 7.Darnay BG, Besse A, Poblenz AT, Lamothe B, Jacoby JJ. TRAFs in RANK signaling. Adv Exp Med Biol. 2007;597:152–159. doi: 10.1007/978-0-387-70630-6_12. [DOI] [PubMed] [Google Scholar]
- 8.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 9.Theoleyre S, Wittrant Y, Tat SK, Fortun Y, Redini F, Heymann D. The molecular triad OPG/RANK/RANKL: involvement in the orchestration of pathophysiological bone remodeling. Cytokine Growth Factor Rev. 2004;15:457–475. doi: 10.1016/j.cytogfr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Bharti AC, Takada Y, Shishodia S, Aggarwal BB. Evidence that receptor activator of nuclear factor (NF)-κB ligand can suppress cell proliferation and induce apoptosis through activation of a NF-κB-independent and TRAF6-dependent mechanism. J Biol Chem. 2004;279:6065–6076. doi: 10.1074/jbc.M308062200. [DOI] [PubMed] [Google Scholar]
- 11.Müller I, Pfister SM, Grohs U, Zweigner J, Handgretinger R, Niethammer D, Bruchelt G. Receptor activator of nuclear factor κB ligand plays a non-redundant role in doxorubicin-induced apoptosis. Cancer Res. 2003;63:1772–1775. [PubMed] [Google Scholar]
- 12.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichimura K, Schmidt EE, Goike HM, Collins VP. Human glioblastomas with no alterations of the CDKN2A (p16INK4A, MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996;13:1065–1072. [PubMed] [Google Scholar]
- 14.Glas M, Rath BH, Simon M, Reinartz R, Schramme A, Trageser D, Eisenreich R, Leinhaas A, Keller M, Schildhaus HU, et al. Residual tumor cells are unique cellular targets in glioblastoma. Ann Neurol. 2010;68:264–269. doi: 10.1002/ana.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 16.Waha A, Guntner S, Huang TH, Yan PS, Arslan B, Pietsch T, Wiestler OD, Waha A. Epigenetic silencing of the protocadherin family member PCDH-γ-A11 in astrocytomas. Neoplasia. 2005;7:193–199. doi: 10.1593/neo.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann W, Kuchler J, Koch A, Friedrichs N, Waha A, Endl E, Czerwitzki J, Metzger D, Steiner S, Wurst P, et al. Activation of phosphatidylinositol-3′-kinase/AKT signaling is essential in hepatoblastoma survival. Clin Cancer Res. 2009;15:4538–4545. doi: 10.1158/1078-0432.CCR-08-2878. [DOI] [PubMed] [Google Scholar]
- 19.Waha A, Felsberg J, Hartmann W, von dem Knesebeck A, Mikeska T, Joos S, Wolter M, Koch A, Yan PS, Endl E, et al. Epigenetic downregulation of mitogen-activated protein kinase phosphatase MKP-2 relieves its growth suppressive activity in glioma cells. Cancer Res. 2010;70:1689–1699. doi: 10.1158/0008-5472.CAN-09-3218. [DOI] [PubMed] [Google Scholar]
- 20.Friedrichs N, Kuchler J, Endl E, Koch A, Czerwitzki J, Wurst P, Metzger D, Schulte JH, Holst MI, Heukamp LC, et al. Insulin-like growth factor-1 receptor acts as a growth regulator in synovial sarcoma. J Pathol. 2008;216:428–439. doi: 10.1002/path.2438. [DOI] [PubMed] [Google Scholar]
- 21.Friedrichs N, Trautmann M, Endl E, Sievers E, Kindler D, Wurst P, Czerwitzki J, Steiner S, Renner M, Penzel R, et al. Phosphatidylinositol-3′-kinase/AKT signaling is essential in synovial sarcoma. Int J Cancer. 2011;129:1564–1575. doi: 10.1002/ijc.25829. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 23.Sherf BA, Navarro SL, Hannah RR, Wood KV. Dual-luciferase reporter assay: an advanced co-reporter technology integrating firefly and Renilla luciferase assays. Promega Notes. 1996;57:2–8. [Google Scholar]
- 24.Bharti AC, Aggarwal BB. Ranking the role of RANK ligand in apoptosis. Apoptosis. 2004;9:677–690. doi: 10.1023/B:APPT.0000045780.10463.c6. [DOI] [PubMed] [Google Scholar]
- 25.Min JK, Kim YM, Kim EC, Gho YS, Kang IJ, Lee SY, Kong YY, Kwon YG. Vascular endothelial growth factor up-regulates expression of receptor activator of NF-κ B (RANK) in endothelial cells. Concomitant increase of angiogenic responses to RANK ligand. J Biol Chem. 2003;278:39548–39557. doi: 10.1074/jbc.M300539200. [DOI] [PubMed] [Google Scholar]
- 26.Hanada R, Leibbrandt A, Hanada T, Kitaoka S, Furuyashiki T, Fujihara H, Trichereau J, Paolino M, Qadri F, Plehm R, et al. Central control of fever and female body temperature by RANKL/RANK. Nature. 2009;462:505–509. doi: 10.1038/nature08596. [DOI] [PubMed] [Google Scholar]
- 27.Kartsogiannis V, Zhou H, Horwood NJ, Thomas RJ, Hards DK, Quinn JM, Niforas P, Ng KW, Martin TJ, Gillespie MT. Localization of RANKL (receptor activator of NF κB ligand) mRNA and protein in skeletal and extraskeletal tissues. Bone. 1999;25:525–534. doi: 10.1016/s8756-3282(99)00214-8. [DOI] [PubMed] [Google Scholar]
- 28.Serrano EM, Ricofort RD, Zuo J, Ochotny N, Manolson MF, Holliday LS. Regulation of vacuolar H(+)-ATPase in microglia by RANKL. Biochem Biophys Res Commun. 2009;389:193–197. doi: 10.1016/j.bbrc.2009.08.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito R, Nakayama H, Yoshida K, Kuraoka K, Motoshita J, Oda N, Oue N, Yasui W. Expression of osteoprotegerin correlates with aggressiveness and poor prognosis of gastric carcinoma. Virchows Arch. 2003;443:146–151. doi: 10.1007/s00428-003-0845-8. [DOI] [PubMed] [Google Scholar]
- 30.Naumann U, Wick W, Beschorner R, Meyermann R, Weller M. Expression and functional activity of osteoprotegerin in human malignant gliomas. Acta Neuropathol. 2004;107:17–22. doi: 10.1007/s00401-003-0772-4. [DOI] [PubMed] [Google Scholar]
- 31.Gingery A, Bradley E, Shaw A, Oursler MJ. Phosphatidylinositol 3-kinase coordinately activates the MEK/ERK and AKT/NFκB pathways to maintain osteoclast survival. J Cell Biochem. 2003;89:165–179. doi: 10.1002/jcb.10503. [DOI] [PubMed] [Google Scholar]
- 32.Sugatani T, Alvarez U, Hruska KA. PTEN regulates RANKL- and osteopontin-stimulated signal transduction during osteoclast differentiation and cell motility. J Biol Chem. 2003;278:5001–5008. doi: 10.1074/jbc.M209299200. [DOI] [PubMed] [Google Scholar]
- 33.Darnay BG, Haridas V, Ni J, Moore PA, Aggarwal BB. Characterization of the intracellular domain of receptor activator of NF-κB (RANK). Interaction with tumor necrosis factor receptor-associated factors and activation of NF-κB and c-Jun N-terminal kinase. J Biol Chem. 1998;273:20551–20555. doi: 10.1074/jbc.273.32.20551. [DOI] [PubMed] [Google Scholar]
- 34.Galibert L, Tometsko ME, Anderson DM, Cosman D, Dougall WC. The involvement of multiple tumor necrosis factor receptor (TNFR)-associated factors in the signaling mechanisms of receptor activator of NF-κB, a member of the TNFR superfamily. J Biol Chem. 1998;273:34120–34127. doi: 10.1074/jbc.273.51.34120. [DOI] [PubMed] [Google Scholar]
- 35.Kim HH, Lee DE, Shin JN, Lee YS, Jeon YM, Chung CH, Ni J, Kwon BS, Lee ZH. Receptor activator of NF-κB recruits multiple TRAF family adaptors and activates c-Jun N-terminal kinase. FEBS Lett. 1999;443:297–302. doi: 10.1016/s0014-5793(98)01731-1. [DOI] [PubMed] [Google Scholar]
- 36.Martinet W, De Meyer GR, Herman AG, Kockx MM. Amino acid deprivation induces both apoptosis and autophagy in murine C2C12 muscle cells. Biotechnol Lett. 2005;27:1157–1163. doi: 10.1007/s10529-005-0007-y. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Li C, Lin J. STAT3 as a therapeutic target for glioblastoma. Anticancer Agents Med Chem. 2010;10:512–519. doi: 10.2174/187152010793498636. [DOI] [PubMed] [Google Scholar]
- 38.Natsume A, Kinjo S, Yuki K, Kato T, Ohno M, Motomura K, Iwami K, Wakabayashi T. Glioma-initiating cells and molecular pathology: implications for therapy. Brain Tumor Pathol. 2011;28:1–12. doi: 10.1007/s10014-010-0011-3. [DOI] [PubMed] [Google Scholar]
- 39.Li GH, Wei H, Lv SQ, Ji H, Wang DL. Knockdown of STAT3 expression by RNAi suppresses growth and induces apoptosis and differentiation in glioblastoma stem cells. Int J Oncol. 2010;37:103–110. [PubMed] [Google Scholar]
- 40.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F, Jove V, Xin H, Hedvat M, Van Meter TE, Yu H. Sunitinib induces apoptosis and growth arrest of medulloblastoma tumor cells by inhibiting STAT3 and AKT signaling pathways. Mol Cancer Res. 2010;8:35–45. doi: 10.1158/1541-7786.MCR-09-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.