Abstract
We described a new computer program for calculation of RNA secondary structure. Calculation of 20 viral RNAs with this program showed that genomes of the icosahedral capsid viruses had higher folding probabilities than those of the helical capsid viruses. As this explains virus assembly quite well, the information of capsid structure must be imprinted not only in the capsid protein structures but also in the base sequence of the whole genome. We compared folding probability of the original sequence with that of the random sequence in which base composition was the same as the original. All the actual genomes of RNA viruses were more folded than the corresponding random sequences, even though most transcripts of chromosomal genes tended to be less folded. The data can be related to encapsidation of viral genomes. It was thus suggested that there exists a relation between actual sequences and random sequences with the same base ratios, and that the base ratio itself has some evolutional meaning.
Full text
PDF

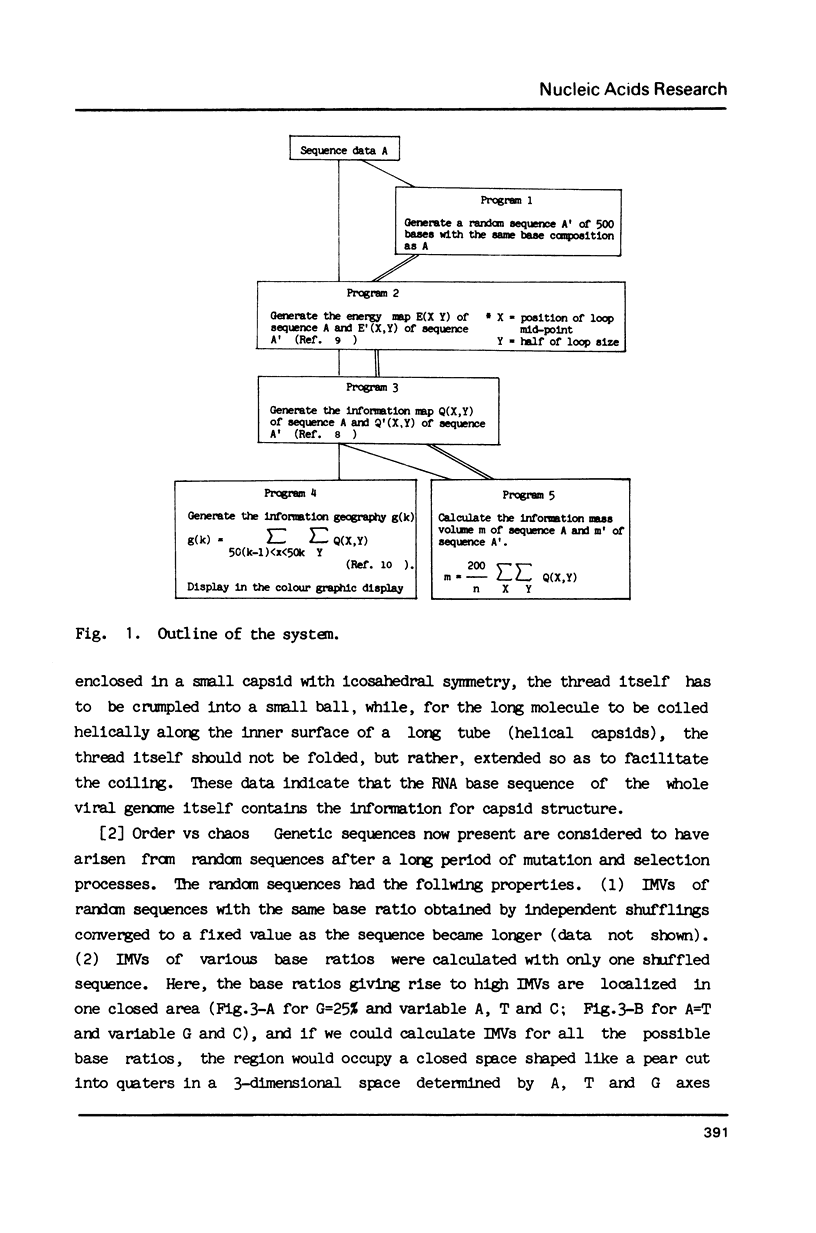
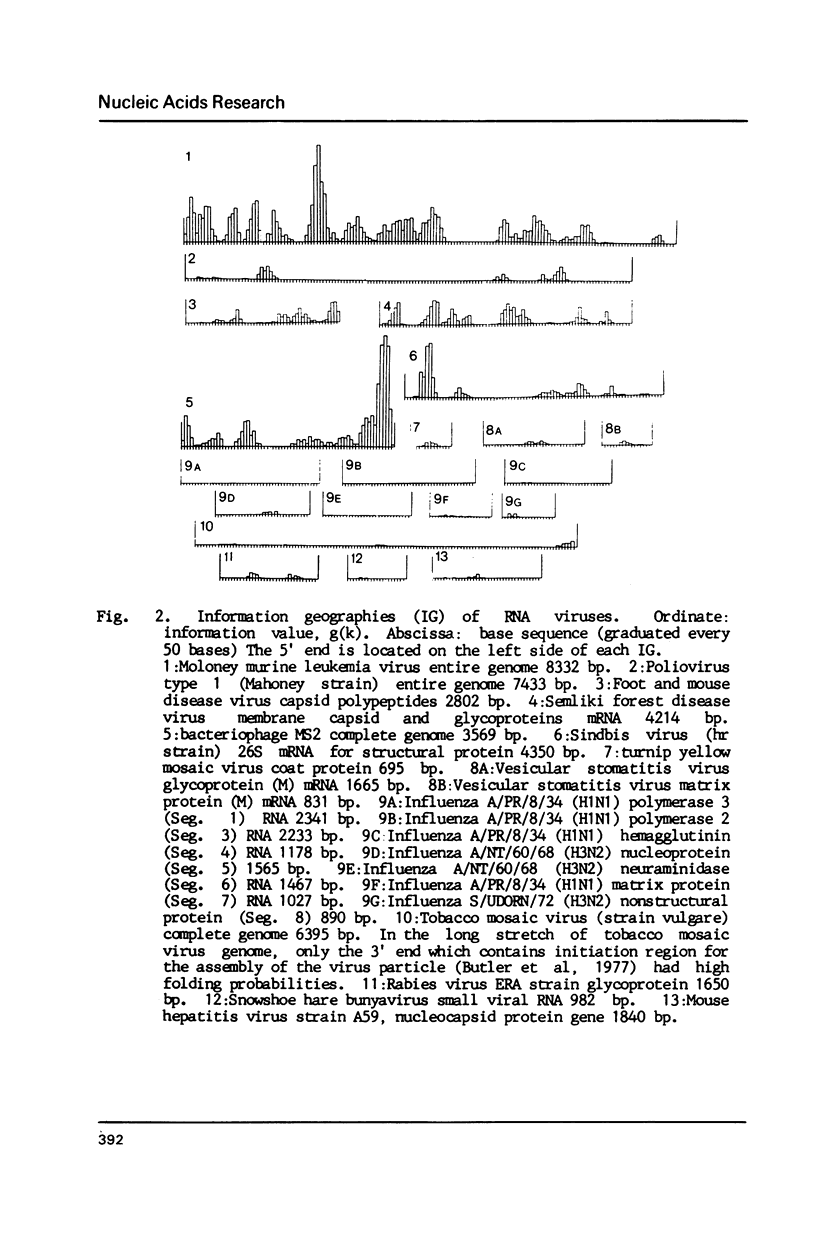
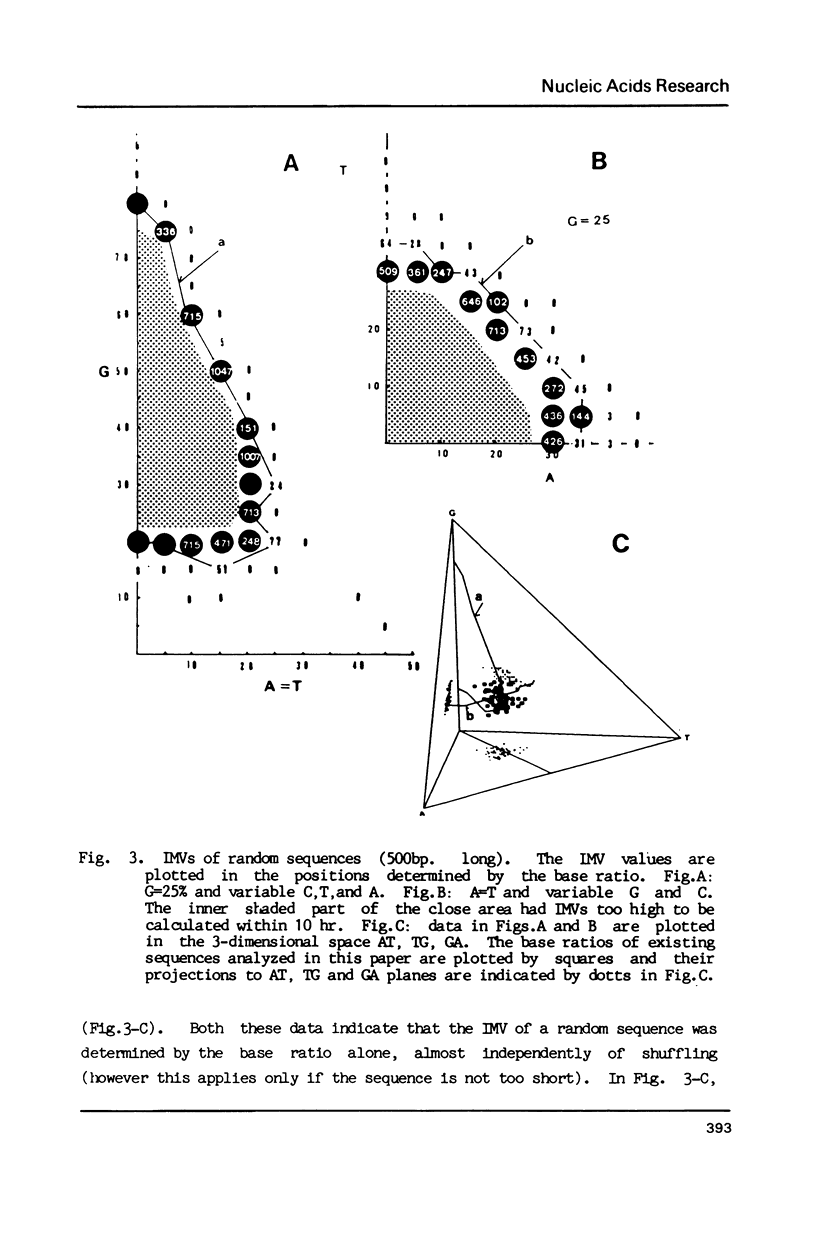



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Comay E., Nussinov R., Comay O. An accelerated algorithm for calculating the secondary structure of single stranded RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):53–66. doi: 10.1093/nar/12.1part1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeweg P., Hesper B. Energy directed folding of RNA sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):67–74. doi: 10.1093/nar/12.1part1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou C., Gouy M., Ninio J. An energy model that predicts the correct folding of both the tRNA and the 5S RNA molecules. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):31–44. doi: 10.1093/nar/12.1part1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Kitamura Y., Yoshikura H. Computation of statistical secondary structure of nucleic acids. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):335–346. doi: 10.1093/nar/12.1part1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Yoshikura H. Computer programme statistically predicting folding structure of nucleic acids of any length: and examples of calculation. Jpn J Exp Med. 1984 Dec;54(6):241–247. [PubMed] [Google Scholar]


