Abstract
Naturally occurring G-quadruplex structural motifs, formed by guanine-rich nucleic acids, have been reported in telomeric, promoter and transcribed regions of mammalian genomes. G-quadruplex structures have received significant attention because of growing evidence for their role in important biological processes, human disease and as therapeutic targets. Lately, there has been much interest in the potential roles of RNA G-quadruplexes as cis-regulatory elements of post-transcriptional gene expression. Large-scale computational genomics studies on G-quadruplexes have difficulty validating their predictions without laborious testing in ‘wet’ labs. We have developed a bioinformatics tool, QGRS-H Predictor that can map and analyze conserved putative Quadruplex forming 'G'-Rich Sequences (QGRS) in mRNAs, ncRNAs and other nucleotide sequences, e.g. promoter, telomeric and gene flanking regions. Identifying conserved regulatory motifs helps validate computations and enhances accuracy of predictions. The QGRS-H Predictor is particularly useful for mapping homologous G-quadruplex forming sequences as cis-regulatory elements in the context of 5′- and 3′-untranslated regions, and CDS sections of aligned mRNA sequences. QGRS-H Predictor features highly interactive graphic representation of the data. It is a unique and user-friendly application that provides many options for defining and studying G-quadruplexes. The QGRS-H Predictor can be freely accessed at: http://quadruplex.ramapo.edu/qgrs/app/start.
INTRODUCTION
Nucleic acids containing runs of guanines can form highly stable quadruple helices stabilized by cyclic Hoogsteen hydrogen bonds involving non-Watson–Crick interactions (1). The G-quadruplex structure can result from intra- or intermolecular associations, and is composed of stacked G-tetrads that are square, co-planar arrays of four guanine bases each. (The current work focuses on intra-molecular G-quadruplexes). G-quadruplexes have been the focus of a large number of studies in recent years that have been extensively reviewed (2–11). Naturally occurring G-quadruplexes have been reported in telomeric, promoter and transcribed regions of mammalian genomes. Although telomeric G-quadruplexes could inhibit DNA replication, a very recent study reports downregulation of the relaxin gene expression by a G-quadruplex located as far as 3.8 kb upstream of the gene (12). G-quadruplex forming sequences in the promoter regions of several oncogenes have been associated with regulation of transcription, and hence have also been identified as targets for anti-cancer therapy. Several recent studies, reviewed in References (7–9), have focused on G-quadruplex involvement in human disease or as targets for therapies.
Although DNA G-quadruplexes have gained significant attention from researchers, the RNA G-quadruplex structure is less well understood. Nevertheless, RNA G-quadruplexes are likely to be formed in vivo (13) and are more stable than the DNA G-quadruplexes (14–16). Several recent studies, reviewed earlier (9–11,17,18), report cis-regulatory roles for RNA G-quadruplexes in post-transcriptional gene expression. For example, G-quadruplex structures located in the 5′-UTR (untranslated region) of human fibroblast growth factor 2 (FGF2) (19) and human VEGF (Vascular Endothelial Growth Factor) (20) mRNAs act as internal ribosomal entry sites (IRES) for cap independent translation initiation. On the other hand, formation of G-quadruplexes can also play inhibitory roles for translation of NRAS (Neuroblastoma Ras) oncogene (21), Ying Yang 1 (YY1) involved in tumorigenesis (22) and ADAM10 responsible for anti-amyloidogenic processing of the amyloid precursor protein (APP) (23). A 3′-UTR polymorphism that affects G-quadruplex structure has been shown to modulate gene expression of the KiSS1 mRNA (24). A G-quadruplex is directly involved in regulated alternative splicing of fragile X mental retardation 1 (FMR1) transcripts (25).
G-quadruplex-mediated regulation of post-transcriptional regulation is suggested to be the result of their interactions with specific binding proteins. Whereas human fragile X mental retardation protein (FMRP) apparently disturbs translation by binding to G-quadruplexes on its target mRNAs (26,27), nucleolin binding to G-Rich sequences in the coding, as well as UTRs positively influences protein translation (28). Proteins involved in (P)-bodies formation (29), and mRNA decay (30) also show preference for RNA G-quadruplexes, suggesting their roles in mRNA turnover.
G-quadruplex structures need to be dynamically formed and resolved for them to be involved in regulating gene expression. RNA helicases like RHAU (31) and DHX9 (32) have been shown to be preferentially associated with RNA G-quadruplexes. The RHAU helicase also physiologically targets a G-quadruplex in the 5′ region of a non-coding TERC (Telomerase RNA Component) RNA, which is part of the telomerase holoenzyme (33).
Clearly, RNA and genomic G-quadruplex structures seem to play a wide variety of roles in important biological processes. Therefore, there is a need to study their overall distribution in the genomes, as well as transcriptomes. Consequently, there have been several computational efforts for large scale predictions of G-quadruplexes (3,34–38). However, most computational methods have difficulty validating their predictions without laborious testing in ‘wet’ labs. We have developed a bioinformatics tool, QGRS-H Predictor that can map and analyze conserved putative Quadruplex forming ‘G’-Rich Sequences (QGRS) in mRNAs, ncRNAs and other nucleotide sequences, e.g. promoter, telomeric and gene flanking regions. Identifying conserved motifs helps validate computations and enhances accuracy of predictions. QGRS-H Predictor is particularly useful for mapping homologous G-quadruplex forming sequences in the 5′- and 3′-UTRs of aligned mRNA sequences. Prediction of evolutionarily conserved G-quadruplexes in otherwise inherently variable regions like UTRs emphasizes their functional significance.
To the best of our knowledge, QGRS-H Predictor is the first known freely available web server for predicting conserved G-quadruplexes in a variety of nucleotide sequences.
METHODS
G-quadruplex mapping
The QGRS-H Predictor identifies Quadruplex forming G-rich sequences (QGRS; predicted G-quadruplexes) in the nucleotide sequences based on established algorithms previously described in detail (39,40). Briefly, the putative G-quadruplexes are identified using the motif GxNy1GxNy2GxNy3Gx. The motif consists of four guanine tracts of equal size interspersed by three loops. The size of each G-tract corresponds to the number of stacked G-tetrads forming the quadruplex structure. The default maximum QGRS length for QGRS-H Predictor is 30 nt.
The stability, as well as regulatory characteristics of G-quadruplexes are dependent on the number of tetrads (41). Whereas quadruplexes with at least three G-tetrads have been accepted as highly stable structures, two G-tetrad quadruplexes with moderate stability are not uncommon (20,42,43). In fact, two-tetrad G-quadruplexes can influence gene expression by acting as thermoregulators (13). In addition, two-tetrad G-quadruplexes are expected to be far more prevalent in the genomes than the ones with three G-tetrads, reflecting their potentially broad impact on biological processes. QGRS-Homology Predictor is meant to be a flexible and comprehensive tool for investigating G-quadruplexes; hence it has been designed to consider QGRS motifs with two or higher numbers of tetrads.
The QGRS-H Predictor weeds out potential false-positives with two strategies. It ignores the QGRS predictions with a G-score (39) <13, representing the bottom 25% of all the G-quadruplexes in the entire human transcriptome predicted in our lab (data not presented). Second, our program only reports conserved QGRS motifs, which also validates our predictions.
G-score
QGRS-H Predictor uses a scoring system previously devised in our lab (40) that evaluates a QGRS for its likelihood to form a stable G-quadruplex. Higher scoring sequences will make better candidates for G-quadruplexes. The G-scoring method that has been described in detail previously (40), uses principles that are based on published studies performed on the stability and structure of G-quadruplexes. Briefly: the G-score is a function of G-tetrads and loop lengths. Greater number of G-tetrads, shorter loops and evenly sized loops lead to higher G-scores.
Homology score
Our homology scoring strategy evaluates the evolutionary conservation of two QGRS instances found in two aligned orthologous nucleotide sequences. The biological roles of G-quadruplexes are expected to involve structure and specific location within the gene rather than just their sequence. The G-quadruplex structure is primarily dependent on the G-tetrads and loop lengths. The determinants of G-quadruplex homology are expected to be similarities in their specific locations on the aligned transcripts, number of tetrads, loop lengths and overall lengths. Consequently, the QGRS Homology score calculation is based on these parameters.
QGRS-H Predictor calculates the percentage of position overlap between pairs of QGRS on the aligned nucleotide sequences and weighs it at 65% of the overall homology score. The G-tetrad similarity in the two QGRS being analyzed, makes up 20% of the homology score. The loop length similarity compares the length of each loop of the two G-quadruplexes, respectively and the average of these values is weighted at 10% of the homology score. Finally, the total lengths of the two G-quadruplexes are compared and make up the last 5% of the homology score. The result is a homology score between 0 and 1, where 1 indicates a very high level of evolutionary conservation. (See Supplementary File 1 for some examples of QGRS Homology score calculations).
OVERVIEW OF QGRS-H PREDICTOR FEATURES
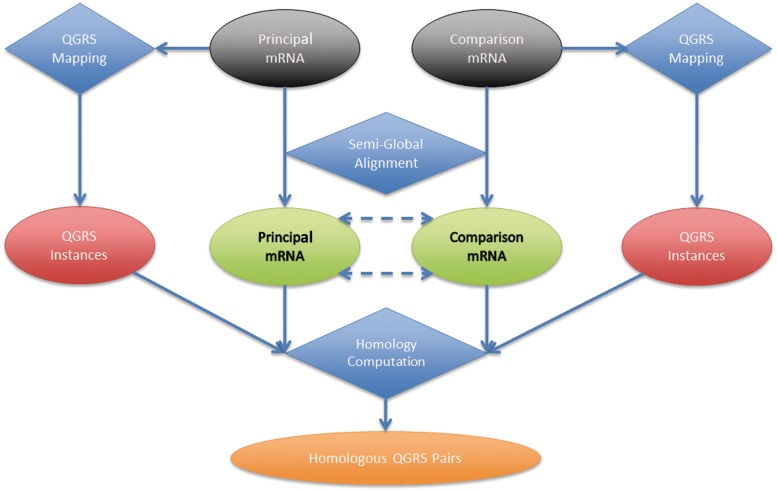
The computation is comprised of three main stages: QGRS identification within the individual sequences, a semi-global alignment calculation to associate the sequences and a final homology computation on each QGRS pairing. The computational workflow is outlined in Figure 1.
Figure 1.
Computational workflow of QGRS-H Predictor. The QGRS Predictor performs a three-stage computation to produce homology results given two sequences. The results of the QGRS identification stage (performed individually on each sequence) is combined with the results of the semi-global alignment stage to perform the last stage, homology computation. The results of the last stage are filtered according to settings specified in the homology map and presented to the user.
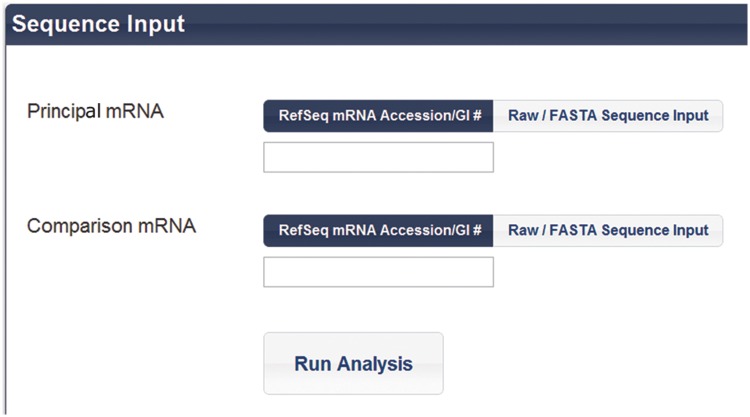
At the start page, the user enters two mRNA sequences, a principal and comparison, which typically would be known to be homologous. The program accepts four modes of data entry (Figure 2). An mRNA sequence can be identified by entering its NCBI accession or GI number, in which case the gene sequence data will be automatically downloaded and utilized. In these input modes, the sequence-associated meta-data, including the annotations and sequence features are also downloaded and displayed. Alternatively, the user may enter the sequence in FASTA format or by simply entering the raw sequence string. When entering FASTA format, the program will always use the sequence data entered, but it will also download meta-data using the provided accession number in the FASTA entry. All of the above input methods can also be used to analyze non-coding RNA entries in the NCBI-based nucleotide sequence databases. Raw sequence input can be used to predict conserved QGRS motifs in promoter regions, telomeric sequences and other genomic sequences up to 10 000 nt in length.
Figure 2.
Input fields for entering the pair of nucleotide sequences in a variety of formats. The program accepts four modes of data entry, NCBI accession, GI number, FASTA and the raw format. Using the first two methods, nucleotide sequence data, along with annotations and sequence features, will be automatically downloaded and utilized. When entering FASTA format, the program will always use the sequence data entered, but will still download the associated meta-data using the provided accession number.
Users may select any method of entry for each input—the same method need not be used for both sequences. The start page also provides several sample homologous pairs for users to run in order to become better acquainted with the tool.
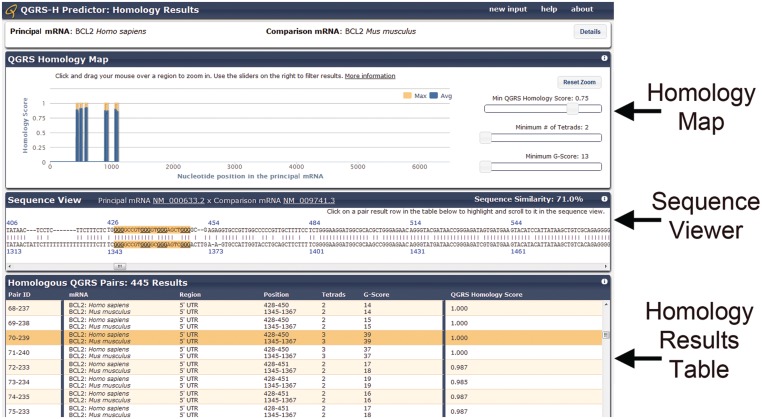
Once the homology analysis is complete, the user is presented with a results page (Figure 3). The results page contains detailed information about the given mRNA sequences, a homology map to show the location of highly homologous QGRS pairs within the aligned sequences and a complete view of the aligned sequences. Finally, a table is shown at the bottom containing the complete listing of homology results (predicted QGRS pairs) based on the user’s current filter settings.
Figure 3.
Results page. The top panel can be expanded to show details about the sequences entered, including species, common names and sequence features such as UTRs, CDS and poly(A) sites. The second panel contains a homology map that can be used to filter the homology pairing results (bottom of the page) according to their locations and characteristics. The third and fourth panels provide a sequence viewer and a tabular listing of the homology results.
Homology map
The highly interactive QGRS Homology map (Figure 4) shows a view of the principal nucleotide sequence. The map serves two roles:
It allows the user to quickly identify regions of interest within the aligned sequences, where the vertical bars indicate locations where G-quadruplexes have been found that have homologous G-quadruplexes in the comparison sequence.
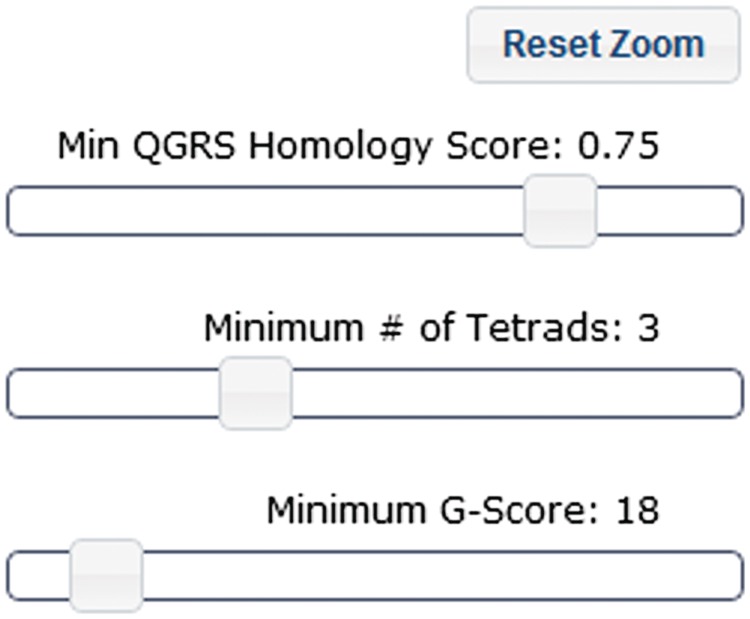
It allows the user to control which QGRS pairings are visible within the homology results table. The user can click and drag regions within the homology map to filter the results list to only include QGRS pairs within the selected region (see bottom of Figure 3). The map also provides filtering controls to further refine the types of QGRS pairs appearing in the results table (Figure 5).
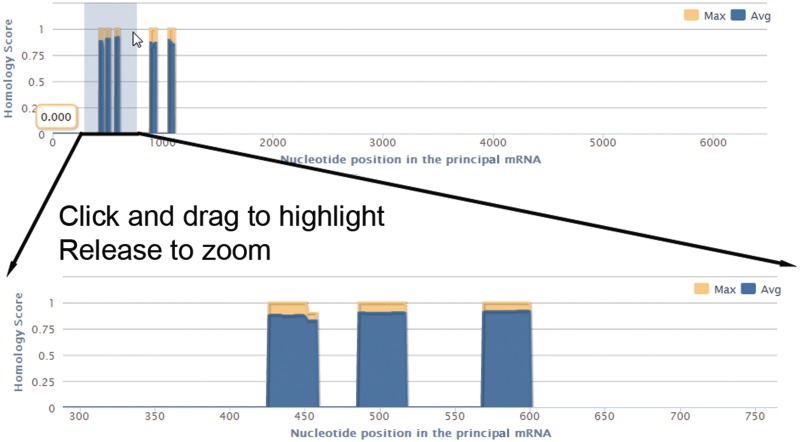
Figure 4.
The highly interactive homology map allows the user to click and drag to zoom into regions of interest within the aligned sequences. The homology map provides an overview of the regions of the principal sequence containing highly conserved QGRS instances. The vertical bars represent the maximum and average homology score of all QGRS pairs found at the given location within the sequence. The user can highlight individual or contiguous sets of regions by dragging the mouse on them, which will filter the results table accordingly.
Figure 5.
Filtering controls for defining characteristics of homologous QGRS motifs. Homology results can be filtered based on their characteristics. The top slider controls the minimum homology score to be shown (a homology score of 1 is the highest value possible). The other sliders control the minimum number of tetrads and minimum G-score of which each QGRS instance in a pair must exhibit in order to be included in the results. (A minimum default G-score of 13 is necessary to weed out potentially false positive instances of QGRS motifs. Please see ‘Methods’ section for details).
Sequence viewer
The Sequence viewer shows the actual sequence of both the principal and comparison mRNAs with the semi-global alignment transformation applied. QGRS-H Predictor alignment method is based on a well-established semi-global pair-wise alignment algorithm (44,45).
This view is scrollable, the user can view any location within the aligned sequences—however, it is best navigated by clicking on results in the table below it. When clicking on a homologous QGRS pair in the results table, the sequence viewer will automatically scroll to the location of the pair and highlight the corresponding QGRS instances in both sequences.
Homology results table
The results table is also interactive and contains a listing of all the QGRS pairings across the principal and comparison mRNA sequences that fall within the range (selected region) and filter settings (minimum QGRS Homology score, number of tetrads and G-score) set in the QGRS Homology map.
The results in the table provide a wealth of information about the QGRS pair, including its base position and region within the nucleotide sequence, number of tetrads, G-score and a composite homology score. In addition, clicking on a row will bring the corresponding QGRS instances into view within the sequence viewer, allowing the user to examine their exact sequence composition.
The Homology results table displays data for all regions of the aligned nucleotide sequences predicted to be capable of forming homologous G-quadruplexes. Each region typically consists of several overlapping QGRS which may differ in their number of tetrads, total length, loop sizes and loop nucleotide sequence. These sequence arrangements have the potential to form intra-molecular quadruplexes in the cell, many of which may be stable. The user may apply appropriate filter settings to select high scoring QGRS motifs and reduce the displayed overlaps.
Online help materials
The application includes an extensive help page that fully describes the various features of the tool, a tutorial for using it, and a glossary of terms that commonly appear on its pages. Context-specific help is also included in the form of clickable icons on each of the sections of the results page to further aid the user in understanding how to take advantage of the data presented.
IMPLEMENTATION
The QGRS-H Predictor is a J2EE web application. The user interface is written entirely in standards compliant HTML + CSS with a significant reliance on JavaScript and the jQuery library for interactivity and XSLT/XML for the presentation logic. Users may access the application using any modern browser, including Internet Explorer 8 and above or any version of Google Chrome, Apple Safari and Mozilla Firefox.
The backend of the application is written in Java 6 using the standard J2EE servlet API, hosted within an Apache Tomcat instance running on Linux. The tool makes use of the BioJava library (version 1.8) (46) for accessing the NCBI databases to retrieve sequence data and meta-data when the user enters accession or GI numbers as FASTA data inputs. Pair-wise semi-global sequence alignment, QGRS mapping and homology computations are performed using Java routines developed at Ramapo College.
Performance
QGRS-H Predictor supports multiple simultaneous users, however due to memory constraints during the sequence alignment phase of the computation, concurrent users are temporarily batched while their jobs pass through the alignment stage. All other computational stages (e.g. QGRS mapping, homology computation) are performed in parallel. Users need not consider these concurrency issues when accessing the system. Overall computation time for a given mRNA pair averages <30 s, whereas pairings that include extremely high numbers of QGRS instances within the mRNA sequences can take several minutes to complete.
Validation
The accuracy of the QGRS-H Predictor depends on its ability to locate QGRS instances within mRNA sequences along with an accurate semi-global alignment procedure. We have confirmed that the QGRS identification stage (with default settings) exhibits 100% precision, as well as sensitivity when compared with previously published results. The homology scores for pairing are based on the accurate computation of QGRS structure (loop lengths, number of tetrads and overall length) along with positional information. These computations have been validated through extensive automated testing of 162 orthologous nucleotide sequence pairs using JUnit. In addition to automated tests, we have manually examined 50 QGRS instances (that were not used in the training) spread over the 5′-UTR, CDS (Coding Sequence) and 3′-UTR regions of 22 orthologous mRNA pairs and an additional 27 orthologous mRNA pairs have been verified by individuals outside our research group.
CONCLUSIONS
QGRS-H Predictor is a bioinformatics web server that can map and analyze conserved putative QGRSs in the mRNAs, ncRNAs and genomic sequences like promoter, telomeric and gene flanking regions. This application is freely accessible on the web and does not require a login procedure. It has been extensively tested for performance, sensitivity, specificity and precision. It works well on multiple web browsers. QGRS-H Predictor is a user friendly application. The website provides a tutorial for a quick start, as well as context-specific help. To the best of our knowledge, this is the only freely accessible web-based server of its kind at present.
This program is capable of searching NCBI-based databases for direct retrieval of user-identified nucleotide sequence pairs for analysis. It can also accept FASTA or raw sequences as input. The QGRS-H Predictor has been designed for predicting homologous G-quadruplex forming sequences in the context of 5′- and 3′-UTRs and CDS sections of pair-wise aligned mRNA sequences. The result outputs provide data on composition, location, predicted stability and homology rating of G-quadruplex pairs, relative to important sites in the mRNA sequences with the help of highly interactive graphic representations. The server can also be used to identify and analyze conserved G-quadruplexes in non-coding RNAs and genomic sequences, thereby significantly extending its utility for analysis of a wide variety of nucleotide sequences.
The QGRS-H Predictor was applied in our lab to test previously reported regulatory G-quadruplexes in the 5′-UTRs of human Zic-1 (47) and NRAS (21) genes. Our program successfully identified 5′-UTR G-quadruplexes in both the genes to be conserved across multiple mRNA orthologs. Additionally, we were able to predict a new G-quadruplex conserved in the 5′-UTR of the human and mouse Bcl2 gene (Figure 3). We tested several nucleolin targets (28) located in the CDS and 3′-UTRs with the QGRS-H Predictor and found them to be capable of forming conserved G-quadruplexes (data not shown). Our program was also able to successfully identify a previously reported G-quadruplex motif, which is the target of a specific RNA helicase RHAU, in the human telomerase non-coding RNA template TERC (33) and determined it to be phylogenetically conserved (data not shown). We are currently using QGRS-H Predictor to develop a database of transcriptome-wide G-quadruplex motifs conserved across multiple mammalian mRNA orthologs. The database will be used to study G-quadruplex composition, as well as distribution in the untranslated and translated regions with a view to investigate their biological roles as cis-regulatory elements.
The researchers will find QGRS-H Predictor to be very useful for investigating the functional role of G-quadruplexes as cis-regulatory elements in the post-transcriptional, as well as transcriptional gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary File 1.
FUNDING
The Provost Office of Ramapo College of New Jersey. Funding for open access charge: Rampo College of New Jersey.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank RCNJ Information Technology Services and Michael J. Skafidas for technical assistance with the web servers. We acknowledge Matt Crum for help with extensive testing of the QGRS-H Predictor application and thank Vladislav Todorov for his technical assistance. We wish to thank Manuel Viotti of Cornell University, Viktor Vasilev of Boston University, and Gadareth Higgs of Yale University, for help with extensive testing of the QGRS-H Predictor.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis JT. G-quartets 40 years later: from 5'-GMP to molecular biology and supramolecular chemistry. Angew Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 3.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Huppert JL. Structure, location and interactions of G-quadruplexes. Febs J. 2010;277:3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 7.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Brosh RM., Jr G-quadruplex nucleic acids and human disease. Febs J. 2010;277:3470–3488. doi: 10.1111/j.1742-4658.2010.07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collie GW, Parkinson GN. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 10.Ji X, Sun H, Zhou H, Xiang J, Tang Y, Zhao C. Research progress of RNA quadruplex. Nucleic Acid Ther. 2011;21:185–200. doi: 10.1089/nat.2010.0272. [DOI] [PubMed] [Google Scholar]
- 11.Halder K, Hartig JS. RNA quadruplexes. Met. Ions Life Sci. 2011;9:125–139. doi: 10.1039/9781849732512-00125. [DOI] [PubMed] [Google Scholar]
- 12.Lin S, Gu H, Xu M, Cui X, Zhang Y, Gao W, Yuan G. The formation and stabilization of a novel g-quadruplex in the 5'-flanking region of the relaxin gene. PLoS One. 2012;7:e31201. doi: 10.1371/journal.pone.0031201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Mergny JL, De Cian A, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 16.Zhang AY, Bugaut A, Balasubramanian S. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry. 2011;50:7251–7258. doi: 10.1021/bi200805j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugaut A, Balasubramanian S. 5'-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 21.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K, Wan M, Giri B, Lei M, Nagamine Y, et al. Yin Yang 1 contains G-quadruplex structures in its promoter and 5'-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2011;40:1033–1049. doi: 10.1093/nar/gkr849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lammich S, Kamp F, Wagner J, Nuscher B, Zilow S, Ludwig AK, Willem M, Haass C. Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5'-untranslated region. J. Biol. Chem. 2011;286:45063–45072. doi: 10.1074/jbc.M111.296921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huijbregts L, Roze C, Bonafe G, Houang M, Bouc YL, Carel JC, Leger J, Alberti P, Roux ND. DNA polymorphisms of the KiSS1 3' untranslated region interfere with the folding of a G-rich sequence into G-quadruplex. Mol. Cell Endocrinol. 2011;351:239–248. doi: 10.1016/j.mce.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Didiot MC, Tian Z, Schaeffer C, Subramanian M, Mandel JL, Moine H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 27.Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- 28.Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, Kim MM, Selimyan R, Martindale JL, Yang X, Carrier F, et al. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011;39:8513–8530. doi: 10.1093/nar/gkr488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marnef A, Maldonado M, Bugaut A, Balasubramanian S, Kress M, Weil D, Standart N. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA. 2010;16:2094–2107. doi: 10.1261/rna.2295410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair. 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Lattmann S, Stadler MB, Vaughn JP, Akman SA, Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–9404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huppert JL. Hunting G-quadruplexes. Biochimie. 2008;90:1140–1148. doi: 10.1016/j.biochi.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 38.Kikin O, Zappala Z, D'Antonio L, Bagga PS. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:D141–148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Antonio L, Bagga PS. Proceedings of the IEEE Conference on Computational Systems Bioinformatics (CSB 2004) 2004. Computational methods for predicting intramolecular G-quadruplexes in nucleotide sequences; pp. 561–562. [Google Scholar]
- 40.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halder K, Wieland M, Hartig JS. Predictable suppression of gene expression by 5'-UTR-based RNA quadruplexes. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kankia BI, Barany G, Musier-Forsyth K. Unfolding of DNA quadruplexes induced by HIV-1 nucleocapsid protein. Nucleic Acids Res. 2005;33:4395–4403. doi: 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gotoh O. An improved algorithm for matching biological sequences. J. Mol. Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 45.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 46.Holland RC, Down TA, Pocock M, Prlic A, Huen D, James K, Foisy S, Drager A, Yates A, Heuer M, et al. BioJava: an open-source framework for bioinformatics. Bioinformatics. 2008;24:2096–2097. doi: 10.1093/bioinformatics/btn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]