Abstract
The SEQALIGN programs1 described in this report aid in the assembly of up to 100 individual overlapping DNA sequences generated by M-13 subcloning and sequencing methods. The program produces a printout of the aligned sequences presented in register. Use of the program will be facilitated because 1) it is written with the Microsoft BASIC interpreter, 2) sequence data may be entered and edited using WORDSTAR or similar word processing programs, and 3) hardware requirements for execution of the program on CP/M or MS-DOS (IBM-PC compatible) systems are minimal.
Full text
PDF






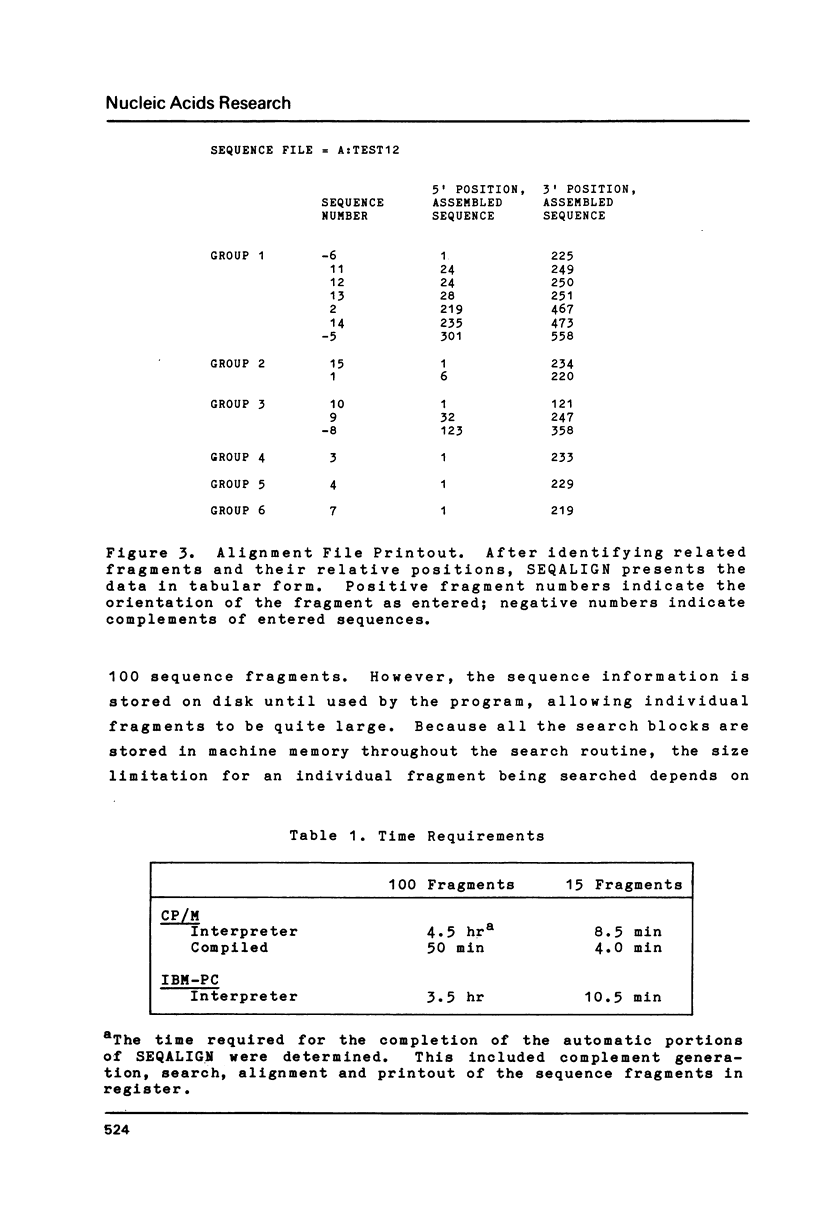



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clayton J., Kedes L. GEL, a DNA sequencing project management system. Nucleic Acids Res. 1982 Jan 11;10(1):305–321. doi: 10.1093/nar/10.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Milazzo J. P., Sciaky D., Roberts R. J. Computer programs for the assembly of DNA sequences. Nucleic Acids Res. 1979 Sep 25;7(2):529–545. doi: 10.1093/nar/7.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malthiery B., Bellon B., Giorgi D., Jacq B. Apple II PASCAL programs for molecular biologists. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):569–579. doi: 10.1093/nar/12.1part2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Parks T. D., Dougherty W. G., Levings C. S., Timothy D. H. Identification of two methionine transfer RNA genes in the maize mitochondrial genome. Plant Physiol. 1984 Dec;76(4):1079–1082. doi: 10.1104/pp.76.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola H., Söderlund H., Ukkonen E. SEQAID: a DNA sequence assembling program based on a mathematical model. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):307–321. doi: 10.1093/nar/12.1part1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]


