Abstract
A fast restriction sites search algorithm using a quadruplet look-ahead feature has been written in 6502 assembly language code. The search time, tested on the sequence of pBR322, is 4.1 s/kilobase using a restriction site library including 112 specificities corresponding to a total site length of over 700 bases. The search for a short sequence (less than 36 bases) within a longer one (up to 9999 bases) with a given number of mismatches or gaps allowed has also been written in assembly language. Typical run time for the search of a 12 base sequence with 1, 2 or 3 gaps allowed are 6.2, 9.4 or 13.6 s/kilobase, respectively. The dot matrix analysis needs 7.5 minutes per square kilobase when using a stringency of 15 matched bases out of 25. A 7/21 matrix of two 500 amino acid proteins is obtained in 3 minutes. These three routines are included in DPSA, a general package of programs allowing manipulation and analysis of DNA and protein sequences.
Full text
PDF


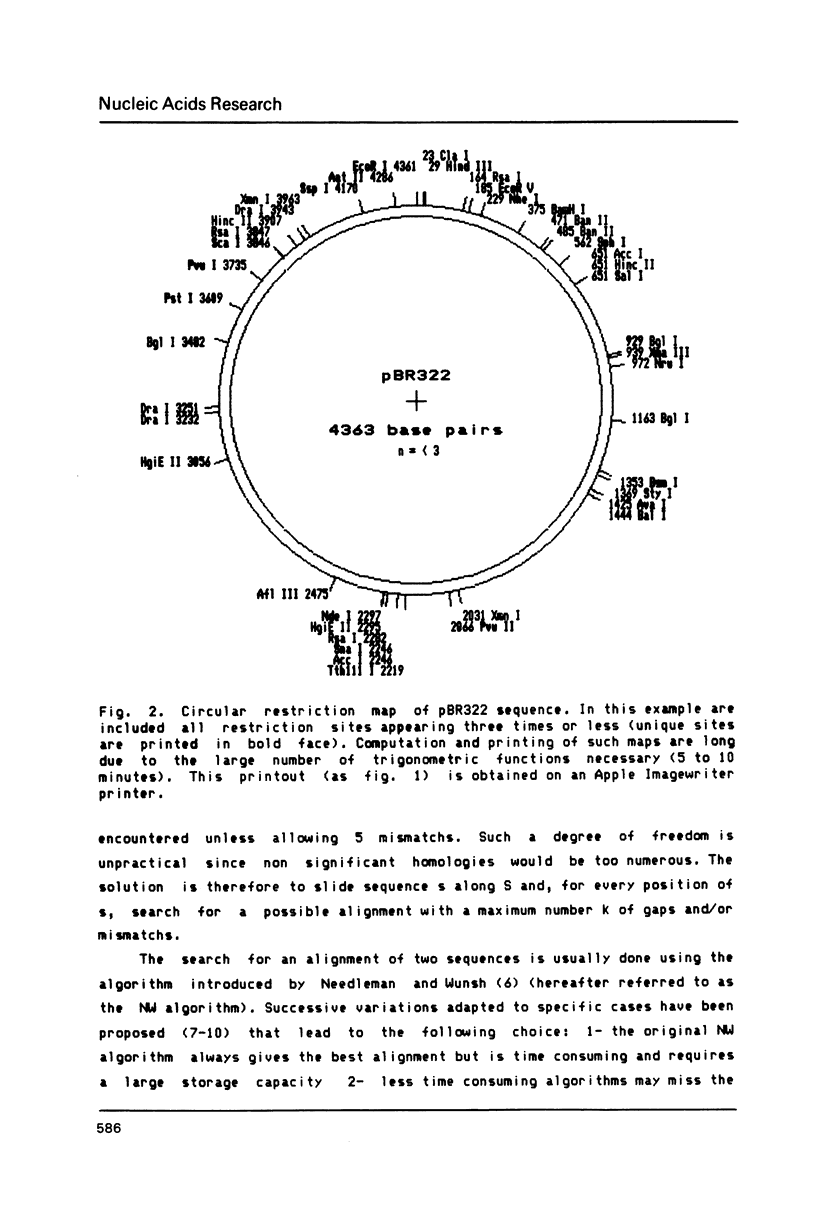




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cottrelle P., Thiele D., Price V. L., Memet S., Micouin J. Y., Marck C., Buhler J. M., Sentenac A., Fromageot P. Cloning, nucleotide sequence, and expression of one of two genes coding for yeast elongation factor 1 alpha. J Biol Chem. 1985 Mar 10;260(5):3090–3096. [PubMed] [Google Scholar]
- Fickett J. W. Fast optimal alignment. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):175–179. doi: 10.1093/nar/12.1part1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P. A., Max E. E., Seidman J. G., Maizel J. V., Jr, Leder P. Cloned human and mouse kappa immunoglobulin constant and J region genes conserve homology in functional segments. Cell. 1980 Nov;22(1 Pt 1):197–207. doi: 10.1016/0092-8674(80)90168-3. [DOI] [PubMed] [Google Scholar]
- Lagrimini L. M., Brentano S. T., Donelson J. E. A DNA sequence analysis package for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):605–614. doi: 10.1093/nar/12.1part2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1985;13 (Suppl):r165–r200. doi: 10.1093/nar/13.suppl.r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. T., Hardies S. C., Hutchison C. A., 3rd, Edgell M. H. The diagonal-traverse homology search algorithm for locating similarities between two sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):751–766. doi: 10.1093/nar/12.1part2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]


