Abstract
Background
Analysis of plant cell dynamics over time, or four-dimensional imaging (4-DI), represents a major goal of plant science. The ability to resolve structures in the third dimension within the cell or tissue during developmental events or in response to environmental or experimental stresses (i.e. 4-DI) is critical to our understanding of gene expression, post-expression modulations of macromolecules and sub-cellular system interactions.
Scope
Microscopy-based technologies have been profoundly integral to this type of investigation, and new and refined microscopy technologies now allow for the visualization of cell dynamics with unprecedented resolution, contrast and experimental versatility. However, certain realities of light and electron microscopy, choice of specimen and specimen preparation techniques limit the scope of readily attaining 4-DI. Today, the plant microscopist must use a combinatorial strategy whereby multiple microscopy-based investigations are used. Modern fluorescence, confocal laser scanning, transmission electron and scanning electron microscopy provide effective conduits for synthesizing data detailing live cell dynamics and highly resolved snapshots of specific cell structures that will ultimately lead to 4-DI. This review provides a synopsis of such technologies available.
Keywords: Four-dimensional imaging, confocal laser scanning microscopy, fluorophores, transmission electron microscopy, plant cell biology
INTRODUCTION
Eukaryotic cells consist of multifunctional membrane-bound compartments and cytoskeletal elements residing in a complex cytoplasmic matrix surrounded by a plasma membrane and extracellular matrix (ECM). These components are precisely positioned in the cell's architectural design. They interact with each other via precisely co-ordinated signal cascades in response both to internal genetically programmed prompts and to external stresses. In most plants and other photosynthetic eukaryotes, this complexity is further heightened by the presence of plastids used for photosynthesis and an ECM that consists of a diverse assortment of polysaccharides, proteoglycans, proteins and polyphenolics, i.e. the cell wall. Since Robert Hooke, Anton van Leeuwenhoek and other early microscopy pioneers first examined cells with light microscopes four centuries ago (Chandler and Roberson, 2009), plant biologists have relied heavily on microscopy-based technology to resolve the structure, function and interactions of cells and sub-cellular components. Today, microscopy is even more valuable than ever. Over the past century, the introduction of laser-based, vibrational, electron and X-ray systems coupled with the rapid evolution of digital image capture and analysis technologies have revolutionized the capabilities and applications of ‘microscopy’. Coincidentally, new cell preparation techniques that encompass cryotechnology, immunological and genetic probes, especially when applied to carefully chosen model organisms, have moved modern microscopy closer to achieving perhaps its ultimate goal: resolving the 3-dimensional (3-D) structural and functional features of cellular life over time, i.e. 4-dimensional imaging or 4-DI.
4-DI represents the mechanism by which dynamic life processes are visualized throughout an organisms's developmental cycle or in response to external pressures (i.e. environmental and/or experimental). Ultimately, 4-DI will interface seamlessly with data derived from molecular or biochemical studies, thereby facilitating interpretation of an organism's gene expression programme and post-expression modulation events. Modern microscopy though is still limited by two major ‘reality checks’: (1) light microscopy (LM) and confocal laser scanning microscopy (CLSM) used to image dynamic events in live cells are inherently limited in resolution; and (2) electron microscopy (EM) cannot be used to view live cells. To address these limitations, combinatorial strategies employing multiple microscopy-based and/or other technologies (e.g. molecular and biochemical) are commonly used to gather, interpret and synthesize data that lead to 4-DI. Many outstanding reviews are currently available that describe the theory and practice of specific microscopy-based technologies and specimen preparation protocols (Inoue, 2006; Pawley, 2006; Chandler and Roberson, 2009; Frigault et al., 2009; Fang and Spector, 2010; Chen et al., 2012). This review will summarize how these diverse technologies may be integrated and they can be used to provide detailed information about the 4-DI of plant cells.
Current status and the ‘game plan’
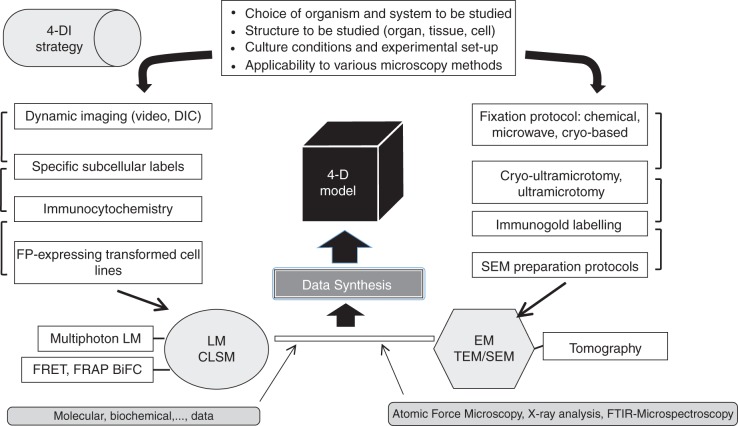
Plant biologists today are confronted with a very large assortment of microscopy-based instruments and strategies from which to choose when designing an experiment (Fig. 1). The phenotypic characteristics of the plant ‘specimen’ (e.g. cell, tissue, organ or whole plant) often play critical roles in the choice of a microscopy-based technology. Conversely, the physical limitations of any type of microscope and associated specimen preparation techniques often restrict how a plant may be manipulated experimentally and what may be ultimately visualized. Today, multiple microscopy technologies are most often combined in order to take advantage of particular optical benefits (Fig. 2). Subsequent interpretation of the resulting diverse data, often consisting of high-resolution snapshots and short video sequences, are then synthesized to yield 4-D models. While not ideal, these synthetic 4-D models often provide important insight into the specimen/process being studied and set the stage for further refinement when new microscopy technologies emerge. Today, it is essential for any cell scientist to develop a realistic ‘game plan’ that can satisfactorily test hypotheses and integrate all microscopy-generated data along with data derived from other methodologies. The main technologies that are available for microscopy-based analyses are now considered.
Fig. 1.
The strategy for 4-DI and subsequent development of models is complex, multifaceted and must consider the limitations of the different microscopy technologies. In initial planning, choice of specimen, its geometric characteristics and its applicability to various types of microscopy must be considered when planning experiments. Light microscopy (LM)-based visualization of dynamic processes including the use of fluorescent proteins (FPs) in live and transformed cells is typically balanced with specific labelling methodologies including immunocytochemistry. Images are then acquired with light or confocal laser scanning microscopy (CLSM) and many new technologies therein such as FRET, FRAP, BiFC and multiphoton microscopy. Electron microscopy (EM) is employed to acquire high-resolution snapshots not possible with light optics. Here, choices on the best methods for fixing specimens, sectioning and labelling for transmission electron microscopy (TEM) or specimen processing for scanning electron microscopy (SEM) must be determined. The snapshots are obtained via EM, and further analyses may be gained through technologies such as electron tomography. The data acquired by both LM and EM are then synthesized to develop models describing the 4-D features of the specimen. However, a larger combinatorial strategy that also includes molecular and biochemical data and information derived from atomic force microscopy or FTIR microspectroscopy is incorporated to yield a more complete model.
Fig. 2.
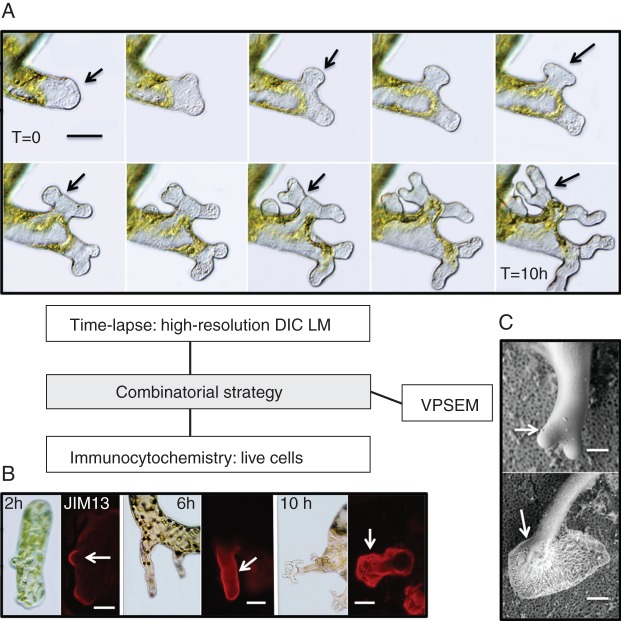
A combinatorial strategy for 4-DI. Multiple microscopy technologies and protocols are often necessary for studying 4-D features. In this example, a high-resolution time-lapse imaging of rhizoid formation in the green alga, Spirogyra, is undertaken. In A, each panel represents an image taken 1 h after filament wounding (total = 10 h). During this process, some specimens are removed and immunolabelled, in this case with the monoclonal antibody, JIM13, specific for arabinogalactan proteins (AGPs). AGP is first found in the wall at 2 and 6 h, and then in the sheath surrounding the wall at 10 h (B). In order to visualize the branching pattern and sheath production, variable pressure scanning electron microscopy (VPSEM) is employed. C demonstrates bifurcate branching (upper, arrow) and sheath production (lower, arrow). These multiple approaches together illustrate the production and release of AGP-like molecules to form a sheath that allows the rhizoid to attach to a substrate. Scale bars: (A, B) = 20 µm, (C) = 15 µm.
‘CONVENTIONAL’ TRANSMITTED LIGHT MICROSCOPY (LM)
Conventional transmitted LM including commonly used specialized optical variations such as differential interference contrast (DIC), polarization and phase contrast, sometimes partnered with staining with select chromatic dyes, represents a valuable tool in both overall qualitative screening and precise assessment of the quantitative characteristics of a specimen (Shaw, 2006; Wayne, 2009). Data derived from these more traditional approaches typically provide the foundation for a microscopy-based study and interface quite effectively with other more advanced microscopy technologies. Additionally, some conventional LM optics, when coupled with modern high-resolution video or time-lapse imaging, represent powerful tools for 4-DI of motile phenomena (e.g. cytoplasmic steaming or anisotropic growth) and developmental events of plants. The literature today abounds with many outstanding examples of plant cell structural and functional studies where more ‘traditional’ forms of microscopy are the main or important instruments of analysis, including, for example, those dealing with cuticle structure and function (Buda et al., 2009), anisotropic polar growth (Zonia and Minnik, 2008; McKenna et al., 2009; Suslov et al., 2009; Aouar et al., 2010; Fayant et al., 2010; Daher and Geitmann, 2011; Rounds et al., 2011) and cytoplasmic streaming (Goldstein et al., 2008; Verchot-Lubicz and Goldstein, 2010; Dodonova and Bulychev, 2011). Additionally, many outstanding reviews and practical guides for general LM analyses are currently available (e.g. Ruzin, 1999; Chandler and Roberson, 2009).
WIDE FIELD FLUORESCENCE MICROSCOPY (WFLM)
Fluorescence light microscopy and the development of fluorescent labels and probes (i.e. fluorophores) have profoundly enhanced the capabilities of LM, especially in the study of live plant cells. Co-labelling with multiple fluorophores has been especially important in defining the structural organization and interactions of sub-cellular components. Today, there is a huge library of fluorescent probes from which to choose. One need only to examine the Molecular Probes Handbook (http://www.invitrogen.com/site/us/en/home/References/Molecular-Probes-The-Handbook.html) to appreciate fully the astounding variety of fluorescent probes created for identifying specific sub-cellular components or accommodating the modern fluorescence microscope and particular filter sets chosen for any study. Today, fluorescence-based microscopy may be conveniently compartmentalized into two areas, WFLM (e.g. epifluorescence microscopy) and CLSM.
WFLM is a sensitive, convenient method using a relatively inexpensive instrument for acquisition of fluorophore-labelled data (Frigault et al., 2009). In fact, for many studies, it may use a more efficient instrument than CLSM especially in general screening of multiple specimens/fluorophores, microinjection studies and basic quantitative assessment of signal. Even for sensitive fluorescent protein (FP)-based labelling where high-resolution capture of the fluorescent signal is paramount, the introduction of new, high-efficiency neutral density filters that stabilize the fluorescent signal and, powerful deconvolution software that eliminates or reduces out-of-focus signal (Chen et al., 2012), make WFLM an important tool for plant cell studies. Several new variations of WFLM have recently been developed and are making an impact on biological microscopy. Total internal reflection fluorescence microscopy (TIRFM) or evanescent field microscopy captures the evanescent wave that occurs at the interface of two different media with different refractive indices. This yields superior signal-to-noise ratios and significantly reduces photobleaching (i.e. photo-induced alteration of a fluorophore that abolishes fluorescence) and phototoxicity. TIRFM has been used successfully for analyses of exocytosis, endocytosis and tubulin dynamics in plants (Knopka and Bednarek, 2008; Vizcay-Barrena et al., 2011). Variable angle epifluorescence microscopy (VAEM) uses illumination that penetrates into a sample and passes through it almost parallel to the cover slip. This technology yields superior visualization of surface phenomena and has been valuable in studies of secretory vesicle dynamics in pollen tubes (Wang et al., 2006), dynamin-related protein dynamics (Konopka and Bednarek, 2008) and single actin filament ‘behaviour’ (Blanchoin et al., 2011).
CONFOCAL LASER SCANNING MICROSCOPY (CLSM)
No other microscopy-based technology has contributed more recently than CLSM to biological imaging and 4-DI. CLSM uses laser-generated light along with high-efficiency filters, precisely tuned and automated z-plane control, precision apertures and advanced software technology to acquire serial optical sections of a specimen. CLSM allows for the optical dissection and high-resolution imaging of specific strata of a specimen, reduction or elimination of interfering out-of-focus light or glare from the focal plane and the post-imaging construction of 3-D profiles not possible with WFLM (see Pawley, 2006 for a comprehensive review of this technology). Most modern CLSM instruments are equipped with an acousto-optic-tunable filter (AOTF) that allows for superior attenuation of laser-generated light and the ability to turn off the laser during back-scanning to reduce damage to the specimen (Frigault et al., 2009). Additionally, specimen chambers and microscope stages that allow for carefully controlled experimental manipulation of live specimens have significantly enhanced acquisition of dynamic data. With new CLSM instruments, up to five different fluorophores can be monitored in one sample, and new cameras and software platforms have allowed for ‘time-lapse’ capture of developmental events that are essential for 4-DI. Today, CLSM is routinely used in plant cell biology to study the expression, targeting, co-localization, turnover and associations of diverse proteins via transformation-based FP technology. It has also been used to analyse other macromolecules and sub-cellular components through immunocytochemistry and a vast arsenal of organelle-specific fluorophores. Additionally, several newer CLSM-based technologies and modifications have recently evolved and contributed to enhanced imaging.
Spinning disc CLSM
In this CLSM ‘variation’, laser-generated light is sent through a disc containing multiple pinholes or slits and is coupled with a CCD camera that provides scanning rates as high as 1000 s−1. The displacement of the light is spread over larger areas of the specimen and phototoxicity is significantly reduced. These features make the spinning disc CLSM an excellent system for real-time observation of live and/or sensitive specimens as well as for the acquisition of highly resolved geometries that are not obtainable with a conventional CLSM. Spinning disc CLSM is commonly used in plant cell biology today, and examples of its utility may be found in recent studies estimating the trajectories of proteins, their velocity as well as their distribution and activity (Held et al., 2008) and elucidating the dynamic interactions between actin filaments and microtubules in arabidopsis (Sampathkumar et al., 2011).
Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP)
These two CLSM-based technologies are primarily used for studying intracellular components through precisely controlled photobleaching. In FRAP, a fluorescently labelled zone in the cell is bleached using a precisely focused high-intensity laser. Then, the displacement of surrounding unbleached fluorescent molecules into the bleached zone is monitored with low-intensity laser light. The resulting data that are acquired can then be used for determining a protein's diffusion coefficient especially in relation to its surrounding environment. In plant cell biology, FRAP has been used extensively for detailed studies of actin dynamics (Ketelaar et al., 2004; Smertenko et al., 2010), cellulose synthase-like proteins in the Golgi bodies (CSLD1 and CSLD4; Wang et al., 2011), vesicle dynamics at the polar tip of arabidopsis pollen tubes (Bove et al., 2008), functional analysis of constituitive and alternative pre-mRNA splicing mechanisms (Ali et al., 2008), the tracking of movement protein of the Tobacco mosaic virus to plasmodesmata (Wright et al., 2007) and endoplasmic reticulum (ER) dynamics (Martens et al., 2006; Forner and Binder, 2007). In FLIP, the photobleaching is repeated several times in alternating sequences with low-intensity laser imaging of the whole cell. FLIP has been important in elucidating cell compartment continuity and spatial features of immobile cell proteins (Martens et al., 2006; Held et al., 2008; De Blasio et al., 2010; Sparkes et al., 2011a, b).
Fluorescence or Förster resonance energy transfer (FRET), fluorescence lifetime imaging microscopy (FLIM) and bimolecular fluorescence complementation (BiFC)
4-DI entails not only high-resolution imaging of select individual sub-cellular components or macromolecules, but also the structural and temporal interactions between these entities. A powerful CLSM-based technology that is often used for this is Förster or fluorescence resonance energy transfer (FRET; Martens et al., 2006). FRET analyses the interaction of two fluorescently tagged molecules, a donor and an acceptor molecule, that are positioned in close proximity to each other (i.e. specifically within 10 nm). The donor and acceptor molecules are specifically chosen with the absorption spectrum of the acceptor overlapping the same wavelength region as the emission spectrum of the donor. Energy derived from the donor molecule upon excitation by a specific wavelength of light is transferred to the adjacent acceptor molecule that, in turn, leads to an increase in intensity of fluorescence (i.e. sensitized emission) that can be quantitatively monitored (Held et al., 2008; DeBlasio et al., 2010). In FRET, the choice of the pair of fluorophores is critical and, recently in plant studies, cyan fluorescent protein (CFP)/(yellow fluorescent protein (YFP) and TSapphire/mOrange have been successfully employed (Bayle et al., 2008). To avoid interference caused by signal overlap of the two molecules (or cross-talk), the acceptor may be photobleached and the donor fluorescence subsequently measured (Sparkes et al., 2011a). The resolution of the generated signal in FRET is typically on the order of angstroms (10−10 m) and therefore superior to conventional CLSM. This makes FRET a type of molecular ruler to measure distances between macromolecules or sub-cellular components. The FRET signal may also be quantitatively measured as the fluorescence decrease in the lifetime of the donor using fluorescence lifetime imaging (FLIM). In plant studies, FRET and FLIM have been valuable as tools providing information about the shape and dimension of proteins, analysing protein–protein interactions and tertiary protein complexes (Kwaaitaal et al., 2010), elucidating cortical ER (Sparkes et al., 2010, 2011b), tracing endocytosis (Griffing, 2008) and detecting/refining signals when autofluorescence in the sample is a problem (e.g. chlorophyll and phenolic compounds).
Another important innovation in the study of protein–protein interactions is bimolecular fluorescence complementation or BiFC (Walter et al., 2004; DeBlasio et al., 2010). Here, two potentially interacting proteins are each labelled with non-functional halves of a fluorescent protein such as YFP. Upon protein–protein interaction, the fluorescent protein reconstitutes and produces a fluorescent signal. The value of this technology is that it employs smaller fusion tags and therefore provides better resolution along with more stability over time than with conventional CLSM. BiFC has recently been used to analyse multiprotein complexes and membrane protein topology in plants (Sparkes et al., 2010).
SUB-CELLULAR LABELS FOR LM, WFLM AND CLSM
Dyes or stains have always been invaluable contrast-enhancing agents for microscopy-based imaging, and the number of labels available and their cellular or sub-cellular targets are indeed large and continue to grow (Krishnamurthy, 1999; Ruzin, 1999; Johnson, 2006; Tsien et al., 2006; Chen et al., 2012). These agents may be applied to live cells and/or fixed, embedded and sectioned specimens. Some of these labels yield chromatic or fluorescent signals and are derived from traditional plant histology, while others represent combinations of more recently developed fluorophores with specific binding agents. These labels continue to represent effective agents for enhancing microscopy-based imaging of sub-cellular components/processes/macromolecules/ions. Some of the more commonly used labels are presented in Table 1.
Table 1.
A sample list of general labels used for identifying particular components of plant cells
| Cell structure | Labels | References |
|---|---|---|
| Endomembrane system and membrane trafficking (exo- and endocytosis) | FM1-43, FM4-64 | Bolte et al. (2004); Griffing (2008); Van Gisbergen et al. (2008) |
| Endoplasmic reticulum | DiOC | Martens et al. (2006) |
| Nucleic acids and nucleus | DAPI, propidium iodide, hexidium iodide, SYTO dyes | Johnston et al. (1999); Haynes et al. (2004) |
| Mitochondria | Mitotracker | Arimura et al. (2004) |
| Cell walls/β-glucans | Calcofluor, Aniline Blue | Bougourd et al. (2000) |
| Cell walls/arabinogalactan proteins | Yariv reagent | Tang et al. (2006) |
| Actin cytoskeleton | Fluorophore-conjugated phalloidin | Iwano et al. (2007) |
| Calcium (Ca2+) | Fura-, Indo and Fluo-labels | Blancafluor and Gilroy (2000); Lazzaro et al. (2005); Hepler and Winship (2010) |
While antibodies and fluorescent proteins in transformed cells have become the primary tools in many investigations, these labels still play prominent roles in plant cell biology.
Over the past several decades though, newly engineered fluorophores have been developed partly in co-ordination with the astounding advancements made in light, laser and filter technologies and partly as a result of the relentless demand for fluorophores with maximum specificity and those yielding the highest resolution (e.g. fluorescein and rhodamine derivatives, Alexa, BODIPY; Bhat et al., 2006; Chen et al., 2012). Many of these agents have been used as the fluorophores conjugated to antibodies (immunocytochemistry). Immunocytochemistry is a versatile and powerful labelling technology that employs vertebrate-generated antibodies to localize macromolecules (antigens) or their specific sub-domains (epitopes). Antibodies are produced in response to, and bind to, architectural features that are relatively unique to an antigen (Chandler and Roberson, 2009). Monoclonal antibodies (mAbs) are epitope specific and are derived from individual antibody-producing cell lines of the immunized animal. The monospecificity of mAbs makes them exceptionally powerful tools for precise identification of macromolecules, but this also often limits their utility in cellular studies (Lipman et al., 2005). That is, minor changes to epitope structure in a specimen, high sensitivity to specific labelling protocols (e.g. buffer type and pH) and/or fluorophore-linking preparation techniques may significantly affect mAb applications. Polyclonal antibodies (pAbs) are comprised of multiple mAbs with unique specificities to an epitope of a particular antigen. They are easier/faster to obtain than mAbs and are more stable during labelling protocols. While mAbs are most often chosen for immunocytochemistry because of their high specificity, pAbs are convenient and relatively inexpensive markers that may be quite satisfactory for general screening of an antigen. In most applications, indirect immunolabelling protocols are employed whereby the specimen is first incubated in the primary mAb or pAb with specificity for a particular epitope or antigen. Then, the specimen is incubated with a fluorophore-conjugated secondary antibody with binding specificity toward the animal-specific immunoglobulin of the primary antibody. This labelling protocol also includes blocking to remove unspecific labelling, extensive washings and, sometimes, the inclusion of detergent, various solvents and/or wall-degrading enzymes or cryo-based wall fracturing to enhance permeability of the antibody. Production of a fluorescent signal is by the fluorophore attached to the secondary antibody attached to the primary antibody that is attached to the epitope (i.e. multi-'piggybacking'). The indirect labelling protocol sometimes suffers from high background labelling and false positives due to cross-reactivity of the secondary antibody to irrelevant immunoglobulins (i.e. those not containing the epitope) found in the primary antibody (Brelje et al., 1993). Direct labelling of a sample with a primary antibody conjugated to a fluorophore may be employed, but this method is less sensitive than the indirect approach and may be interfered with by amines, ammonium ions and some common buffers typically used in labelling processes. While immunofluorescence labelling has been successfully used to identify intracellular proteins and other biochemicals in many plant studies, the use of fluorescent proteins attached to intracellular proteins has subsumed a great deal of labelling (see below). However, immunofluorescence remains a critical technology for studying cells that have yet to be transformed or for studying non-protein macromolecules. Perhaps the best example of this is in studies of plant cell walls (Hervé et al., 2011; Fig. 3). Today, a large arsenal of mAbs raised against specific wall epitopes has been generated and are commercially available (Plant Probes, www.plantprobes.net/; the Complex Carbohydrate Research Center at the University of Georgia, www.ccrc.uga.edu/services/index.php; BioSupplies, www.biosupplies.com.au/). These mAbs have been used for both live cell and fixed samples, and are often used in combination with other immunobinding assays such as carbohydrate microarrays (Moller et al., 2007) or traditional biochemical studies. Correlative studies are also possible because of the adaptability of antibody labelling to thin sections for electron microscopy (i.e. immunogold labelling – see later). Recently, carbohydrate-binding modules or CBMs have been added to the library of cell wall labels. CBMs are non-catalytic protein domains found on many carbohydrate-active enzymes that act on cell walls (Blake et al., 2006; Kljun et al., 2011). CBMs are specific for the macromolecule to which they bind and have been developed as recombinant proteins with polyhistidine tags. This, in turn, allows them to be used like an antibody or as a fusion protein with green fluorescent protein (GFP) that allows for direct imaging. CBMs have great potential for the labelling of cellulose and hemicellulosic components of the cell wall, and future discovery of CBMs with specificity for pectins and other polymers offers great potential for their use in microscopy. Also, lectins, carbohydrate-binding glycoproteins/proteins, have been employed for characterization of glycan components of plant cells and as important tools in plant glycomics (Hirabayashi, 2004; Chandler and Roberson, 2009; Rhiel and Brock, 2012).
Fig. 3.
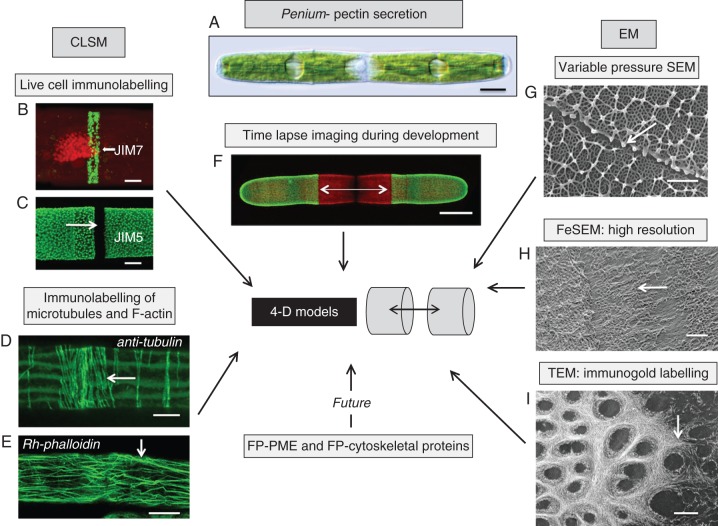
4-DI strategy of an emerging model organism, Penium margaritaceum (A). Penium is currently being used for 4-DI of pectin processing in plant cells. This alga can be live labelled with monoclonal antibodies specific for epitopes of homogalacturonans (HGs; B, C). JIM7 labels relatively high esterified HG while JIM5 labels low esterified HG. At the isthmus zone of the cell (C, arrow), the point of pre-cytokinetic wall expansion, high-esterified HG is released in a narrow band (B, arrow). As this HG is displaced outward toward both poles, it is de-esterified, most probably by the enzyme pectin methyl esterase (PME). Once de-esterified, Ca2+ binds with the HG to form a distinct lattice as noted by JIM5 labelling. The development of the wall closely corresponds with the cortical microtubule (D) and F-actin (E) networks at the isthmus. Tubulin was identified with an anti-tubulin antibody and F-actin was localized with rhodamine–phalloidin. Labelled cells are placed back in cultures and amounts of new growth can be monitored using CLSM (F, arrows). In order to elucidate further the formation of pectin, EM is also employed. To capture snapshots of rapidly developing events, cells were rapidly frozen and viewed with VPSEM (G). This allowed for structural analysis of the pectin lattice (arrow). FeSEM (H) and TEM (I) are also employed to obtain high-resolution images of the developing pectin fibrils in the isthmus (arrows). This story is only just beginning as transformed cells that are expressing FP–PME or FP–cytoskeletal proteins will yield vital dynamic information for the generation of 4-D models of pectin secretion. Scale bars: (A, F) = 17 µm, (B) = 3·5 µm, (C) = 4 µm, (D) = 3·1 µm, (E) = 14 µm, (G, H) = 250 nm, (I) = 200 nm.
FLUORESCENT PROTEINS AND WFLM/CLSM
No other labelling technology in LM has helped revolutionized 4-DI studies as much as the development and use of FPs. FPs attached to, and expressed with, a gene of choice (or targeting sequences therein) have revolutionized direct non-invasive in vivo visualization of live plant cells (Chan et al., 2011; Chen et al., 2012; Ckurshumova et al., 2011; Nair and DeBolt, 2011). Here, the genetic sequence for the FP is spliced onto the cDNA of a gene of choice or a partial sequence and inserted into a cell. Under the proper conditions, the FP–gene construct will be expressed, providing a fluorescence-based emission emanating from where the protein is localized. The first and most widely used FP is GFP derived from the jelly fish, Aequorea victoria (Rizzo et al., 2009). GFP is small (27 kDa), non-toxic and can be viewed in both live cells or those mildly fixed (DeBlasio et al., 2010). In plants, the successful expression of GFP was delayed until the removal of a cryptic intron (Haseloff et al., 1997). Today, a multitude of new FPs are being employed in the study of proteins expressed in transformed plant cells. These have been derived from other marine invertebrates (e.g. DsRedFP from Discosoma) and from mutagenesis of GFP (Shaner et al., 2005; Nelson et al., 2007; Geldner et al., 2009; Mathur et al., 2010). The use of FPs in microscopy is predicated on successful transformation of a cell type and subsequent expression of the FP, feats not especially common nor easy to accomplish. Large libraries of FP-expressing transformed lines are presently available for some model plants (e.g. http://arabidopsis.info/CollectionInfo?id=126) and major efforts to transform other organisms successfully make FP imaging technology the major tool in LM for the future. However, the microscopist must be aware of several problems that may result in FP-based experiments. These include low fluorescent signals, improper maturation of the FP, unusual or incorrect localization of the FP, and the FP directly affecting the dynamics of the cell under study. Ideally, the use of stable transformants and performing comparative studies of a protein containing different GFP variants or coral-derived proteins provide the best results. For more detailed information concerning the set-up and implementation of an FP study, numerous excellent reviews are available (Chalfie and Kain, 2005; Rizzo et al., 2009; Knapp et al., 2012)
OTHER LM TECHNOLOGIES
New technologies continue to emerge in LM-based optics and offer new ways to attain images that can be used for 4-DI.
Two- or multiphoton microscopy
In conventional CLSM, intense laser-generated light focused on a specimen may cause significant bleaching or damage, especially when using a UV laser to excite a fluorophore. In two- or multiphoton microscopy, wavelengths of light twice that of the typically used shorter wavelength are focused on the specimen for short periods of time that are less than the fluorescence decay time of the fluorophore being studied (Pawley, 2006). This results in fluorescence only at the focused zone (Inoue, 2006). The advantages of this type of instrument include avoidance of potentially damaging UV light and acquisition of the fluorescent signal from deeper tissues with greater light-gathering efficacy. Multiphoton microscopy has been successfully used in plant studies, for example in the visualization of DNA and the nucleus with 4′,6-diamidino-2-phenylindole (DAPI), Indo-1 labelling of calcium (Ca2+), imaging of ER dynamics and elucidation of pollen tube growth dynamics (Martens et al., 2006; Broess et al., 2009; Cheung et al., 2010; Wuyts et al., 2010).
Three-dimensional structured illumination microscopy (3D-SIM)
Axial resolution in both WFLM and CLSM is limited by diffraction of light (e.g. 500 nm in the z-plane for CLSM). This is above the resolution of many cellular components. 3D-SIM is used to image objects beyond the limit of diffraction. The specimen is illuminated with multiple interfering beams of light transmitted through a series of diffraction gratings that can produce resolution of 200 nm in the z-plane. Recently, this new technology has been used in plant cell studies including visualizing plasmodesmata and the viral movement protein in tobacco (Fitzgibbon et al., 2010) and Golgi bodies (Schoberer and Strasser, 2011).
Micromanipulation and microinjection
While both these technologies have been around for decades, they provide a direct means for assessing single cell systems. That is, one can directly assess the fate of an injected material in a single cell's developmental or experimental history. Microinjection has been very valuable in various areas of plant cell biology including elucidation of ion gradients, the cytoskeleton and developmental processes (Du and Ren, 2011).
Imaging technologies associated with LM
Several technologies that interface LM with spectroscopic or vibrational optics have made recent impacts in plant cell studies.
Fourier Transform Infrared microspectroscopy or FTIR microspectroscopy
FTIR microscopy employs LM to focus in on a specific locus of a sample and spectroscopy to measure the vibrations of molecular bonds therein. Recently, this technology has been valuable for analysis of cell wall polymers and specifically for high-throughput screening of cell wall mutants (Mouille et al., 2003, 2006; McCann et al., 2007).
Low-energy X-ray fluorescence microscopy or LEXRF
LEXRF is a recent technology developed for direct visualization of thick tissues that yields imaging data with both topographical and chemical information via X-ray acquisition (Kaulich et al., 2009). In plant studies, it has been successfully used by Regvar et al. (2011) in analysis of protein storage vacuoles in wheat aleurone.
Atomic force microscopy
The atomic force microscope (ATM) is a type of scanning probe microscope that generates an ‘image’ by measuring changes in the magnititude of the interaction between a vibrational probe and the specimen surface, in effect ‘feeling’ it. The ATM probe or microstylus is mounted on a cantilever, is run over a specimen and ultimately provides a direct measurement of the mechanical properties of that specimen (Yarbrough et al., 2010; Kirby, 2011; Milani et al., 2011). ATM requires little or no specimen preparation, but is somewhat limited in that it does not work well for specimens with significant contour (i.e. not flat).
ELECTRON MICROSCOPY (EM) AND TRANSMISSION ELECTRON MICROSCOPY (TEM) PREPARATION PROTOCOLS
The highest resolution that can be acquired in biological microscopy today is via EM. However, due to poor penetrative powers of electrons and exposure to both high vacuum and an irradiating electron beam, living things cannot be visualized with EM. Specimens must be fixed and, in TEM, sectioned before viewing. These limitations restrict the value of EM to static imaging. However, the high-resolution ‘snapshots’ obtained by EM (e.g. TEM is 4× better in z-plane resolution and 100× better in general than the best CLSM; McIntosh et al., 2004; Staehelin and Kang, 2008) continue to make EM a profoundly important instrument for 4-DI. As important, improved cryo-based specimen preparation technology for TEM and scanning electron microscopy (SEM), immunogold cytochemistry, the utilization of electron tomography in rendering high resolution 3-D images, the tremendous potential of new technologies including focused ion beam (FIB) dissection of cells and the development of EM-specific genetic markers make EM even more valuable for the future. However, presently, the microscopist still must deal with ‘fixing’ cells or tissues before viewing, and the choice of methods available are many.
For either TEM or SEM, cell/tissue preparation techniques (e.g. fixation) that enhance sub-cellular preservation and minimize specimen alteration are essential. In TEM, conventional chemical fixation employing aldehyde fixatives (e.g. glutaraldehyde and formaldehyde) and heavy metals [e.g. osmium tetroxide (OsO4)] followed by dehydration in organic solvents (e.g. ethanol or acetone) and embedding in various plastics are standard protocols prior to sectioning. These processes typically require days to complete and may result in artefact production, cytoplasmic shrinkage and cellular extractions, or may compromise antibody binding to its epitope. To combat these problems, special microwave instrumentation for specimen processing (Chebli et al., 2008; Zechmann and Zellnig, 2009) has been used and significantly reduces time afforded to fixation, dehydration and embedding. Likewise, for the elimination of OsO4 during fixation, the use of new plastics and embedding strategies and the introduction of energy-filtering lenses on many TEMs have significantly enhanced immunogold labelling and the visualization of low contrast structures (Zewail and Thomas, 2010). The basis of energy filtering is the selection and use of electrons of specific energies that, in turn, enhance contrast of the specimen. Additionally, these filters may be used in the acquisition of electron energy loss spectra (EELS) that may be exceptionally useful in determining the elemental composition of a specimen (Lutz-Meindl, 2007; Eder and Lutz-Meindl, 2008; Darehshouri and Lutz-Meindl, 2010).
However, cryo-fixation technology has made the greatest impact in TEM-based specimen preparation today. In many plant studies, specimens are rapidly frozen in a cryogen (e.g. liquid propane or ethane) cooled to –180°C or less. Common methods for ‘introducing’ the specimen to the cryogen including plunging, slamming and spraying are adequate for many single-celled organisms or for viewing the surface layers (i.e., the outer few micrometres) of a thicker specimen. However, these methods produce sufficiently slow freezing rates that result in damaging ice crystal formation occurring in an internal location of a specimen. Likewise, the loss of turgor occurring during specimen excision and trimming (i.e. to achieve an acceptable size for freezing) may also induce the formation of artefacts. High-pressure freezing, whereby the specimen is rapidly frozen under increased pressure at 2100 atmospheres within 100 ms (high hydrostatic pressure acts as a physical cryoprotectant), has greatly increased cryo-based fixation to the depths to as much as 500 µm in a specimen and is widely used for thick specimens (Mims et al., 2003; McDonald and Auer, 2006; Donohoe et al., 2007; Chandler and Roberson, 2009). Once frozen, several options exist for processing the specimen prior to sectioning and TEM viewing. Cryo-sectioning and visualization using a TEM equipped with a special cryo-specimen chamber is the most direct mode of imaging (Kuo, 2007), but the instrumentation required is extremely expensive. More often, a frozen specimen is freeze substituted (Staehelin and Kang, 2008; Takeuchi et al., 2010). Here, the cryo-fixed specimen is quickly transferred for various periods of time (24 h to 1 week) to an organic solvent cooled to –80 to –90°C and containing glutaraldehyde, formaldehyde, OsO4 and/or uranyl acetate. During this time, the fixatives slowly penetrate and fix the specimen. The specimen is then washed free of the fixative with fresh solvent cooled to –40 to –90°C. This is followed by infiltration and embedding of the specimen with plastic at low temperatures (–40 to –90°C) using UV light as the polymerizing catalyst. Alternatively, the freeze-substituted specimen can be warmed slowly to room temperature, washed with solvent and then infiltrated/embedded in plastic which is then polymerized by UV light or heat. Freeze substituion is commonly used today in plant cell biology and typically offers outstanding fixation quality.
Transmission electron microcopes of 120 kV are the most commonly used instruments in biology laboratories, and 50–120 nm sections are those that are typically viewed with these instruments. The thinness of these sections is somewhat limiting when considering the dimensions of a cell, but they do afford reasonable views of sub-cellular architecture and specific location identified via immunogold labelling. Higher kV transmission electron microcopes accommodate thicker sections (up to 200 nm at 200 kV or 400 nm sections with 300 kV), but these instruments are considerably more expensive than the 120 kV transmission electron microcope. Serial section imaging and 3-D reconstruction using various software programs are also used for analysing thicker volumes of cells, but presently resolution afforded here is restricted to 2× the thickness of the section (e.g. 120–200 nm z-axis resolution for 60–100 nm sections; Haas and Otegui, 2007). Recently though, electron tomography (ET) has proven to be an effective method for generating 3-D imaging or 3-DI. ET uses multiple 2-D projection images of a 3-D object over a wide range of viewing angles to create a tomogram. Typically, tomograms are derived from images taken at 1° intervals from 60° to –60°. To create more complete tomograms, dual-axis ET is used which stitches tomograms and provides information that is derived from 90° to –90°. ET has been of great value in plant cell biology in the elucidation of the cytokinetic mechanism of higher plants, cell wall development, Golgi vesicle structure and development, and chloroplast architecture (Shimoni et al., 2005; Donohoe et al., 2007; Haas and Otega, 2007; Staehelin and Kang, 2008; Austin and Staehlin, 2011; Otegui, 2011).
TEM-based studies often require the use of contrast-enhancing staining and/or labelling of a specific location. Many general staining protocols have been developed to enhance imaging of specific cellular structures (Kuo, 2007; Nakakoshi et al., 2011), and immunogold labelling technology (Takeuchi et al., 2010) is well established in this form of microscopy. Additionally, newly developing labelling methods offer great promise especially in correlative microscopy analyses. Here, a specimen may be viewed at both the LM and TEM levels, and especially promising are potential genetic tags to be used in TEM that are analogous to GFP in LM. These include fluoranogold, metallothionen and the small, genetically encodable protein module, mini-SOG (Haas and Otegui, 2007; Lee et al., 2011; Shu et al., 2011).
SCANNING SLECTRON MICROSCOPY (SEM)
Modern technological advancements in SEM have also helped refine the ultrastructural aspects of the surfaces of plants. SEM visualizes electrons derived from the surface or sub-surface layers of a specimen or captures X-rays generated from the interior that are subsequently used to identify elemental composition or create elemental maps of a specimen. For conventional SEM imaging, the specimen typically must be fixed, dehydrated/dried (e.g. by use of critical point drying or lyophilizer) and made conductive via sputter coating with a conductive material (e.g. gold, platinum or palladium) before viewing. This type of specimen preparation has been quite adequate for many plant specimens. However, as with TEM, the potential for specimen alteration, in this case extractions or morphological alteration of the surface to be studied, may be of concern. Recently, environmental SEM (ESEM) and variable pressure SEM (VPSEM) have emerged as vehicles for viewing non-fixed and non-conductive samples (Kuo, 2007; Chandler and Roberson, 2009). Here, the specimen is kept in a relatively high pressure chamber of the SEM column, the column vacuum is kept low and the working distance is kept short. The electron detectors of these instruments are also capable of working under the presence of water vapour. Positively charged ions created by interaction of the electron beam with gases in the column neutralize the negative charge on the specimen surface. For ESEM and VPSEM, specimens can be flash frozen, placed directly into the column using a cooled cryostub or Peltier stage, and viewed directly after sublimation of ice. Image quality is often comparable with conventional SEM preparations.
For high-resolution imaging of surfaces, field emission SEM or FeSEM is often used. FeSEM uses low voltage but high electron brightness that allows for high magnification, and high-resolution analyses (i.e. 1–2 nm or 3–6× better than conventional SEM). FeSEM has been an important imaging tool especially for cell wall studies dealing with cell wall porosity in pollen tubes (Derksen et al., 2011), cell wall extension (Marga et al., 2005), cellulose microfibril orientation in growth anisotropy (Baskin et al., 2004), cellulose orientation in relation to cortical microtubules and cellulose synthase (CESA) tracks (Chan et al., 2011; Crowell et al., 2011; Fujita et al., 2011) and development of wall ingrowths (Talbot et al., 2007).
SPECIMEN CHOICES, AND OLD AND NEW MODEL ORGANISMS
The choice of plant and/or part thereof, its phenotypic characteristics (e.g. size, thickness, location in the whole plant) and mode of maintenance (e.g. culture conditions) will define the limitations on the choice of microscope to be used and the associated experimental design. For example, a microscopy-based analysis of a specific cell/tissue type residing deeply within a large multicellular thallus or organ is often very difficult to accomplish and, for live cell studies, nearly impossible to do. While new technologies such as low energy X-ray fluorescence microscopy (LEXRF; Regvar et al., 2010) offer promise for deep tissue imaging of larger specimens, their large size still creates physical limitations for high-resolution imaging. Utilization of cell cultures derived from larger specimens via tissue culture methodology may address some of these limitations, but much care must be taken in interpreting data from cells/tissues maintained in such highly artificial conditions. However, unicellular plants or single-/thin-layered plant systems (i.e. organisms whose natural phenotype is unicellular or thin layered) may be conveniently used with great success in many areas of study of more generalized or ‘universal’ sub-cellular phenomena. In fact, many of these organisms are, or have the potential to be, model organisms in plant cell research. Model organisms represent well-studied and well-manipulated systems that provide the microscopist with a rich source of proven experimental and technical protocols as well as large pools of pre-existing data from which highly focused hypotheses may be formulated and for which results may be effectively compared. Additionally, the genomes and transcriptomes of many of these organisms have been or are being sequenced and many have been successfully transformed, allowing, in turn, for the incorporation of FP technology for live cell labelling. This allows for the critical integration of microscopy-based data with molecular data.
While several well-known model plants have been used extensively in microscopy-based studies (e.g. arabidopsis, tobacco cell culture lines), other taxa are quickly emerging as potential model organisms that are especially valuable for 4-DI. Though certainly not complete, the following represents a description of two model systems and one model group that are or will be especially valuable for 4-DI along with their unique features that may be used in the study of specific cellular phenomena:
Pollen tubes: growth anisotropy, endomembrane dynamics including exocytosis and endocytosis, cytoskeletal dynamics, cell wall development and ion gradients
When the male gametophyte or pollen of an angiosperm or gymnosperm germinates, a protuberance called the pollen tube emerges and grows toward the egg that is embedded in a female gametophyte and megasporangium (Boavida et al., 2005). The pollen tube is a spectacular structure ranging in size from 5 to 15 µm in diameter and achieving lengths of hundreds of micrometres or even centimetres. The tube serves as the ‘highway’ for directed transport of the sperm during fertilization. To attain such lengths, pollen tubes grow anisotropically at one rapidly expanding polar tip at rates of hundreds of micrometres to millimetres per hour. The basis of unipolar expansion is the precise control of wall polymer secretion and modulation at the tube tip. In the cytoplasm of the growing tip exists a confluence of dynamic sub-cellular activities including Golgi apparatus-derived production of wall precursor-carrying vesicles, cytoskeletal system-mediated membrane trafficking (i.e. exocytosis and endocytosis) and multiple signal transduction cascades (Samaj et al., 2006; Wilsen et al., 2006; Cheung and Wu, 2007; Bove et al., 2008; Lee et al., 2008; Zhang et al., 2010; Konrad et al., 2011; Kroeger and Geitmann, 2012). Pollen tubes are popular specimens for cell biology experiments as they are easily germinated and maintained in laboratory cultures and adapt well to microscopy and manipulation in microscopy-based devices (e.g. flow-through growth chambers, silicon isolators and microindentation devices; Cardenas et al., 2008; Zonia and Munnik, 2008). Pollen tubes may also be studied live with the application of various labels including antibodies and FPs (Cheung et al., 2008; Cai et al., 2011) and have become versatile systems for microinjection of Ca2+ dyes and ratiometric imaging (Feijo et al., 1999; Lovy-Wheeler et al., 2006, 2007; Cardenas et al., 2008) as well as mechanophysical studies including microidentation analyses (Parre and Geitmann, 2005). As a result of all of these attributes and associated research, detailed models of growth dynamics, including those incorporating 4-DI, have recently emerged (Aouar et al., 2010; Geitmann, 2010a, b; Kroeger et al., 2011) and have made pollen tubes arguably the most well-studied system in plant biology.
Physcomitrella patens: cell polarity, cytoskeleton and cell wall development
The moss, Physcomitrella patens, represents an emerging model organism for molecular and cell-based studies (Cove et al., 2006). This moss has a simple haploid phase-dominant life cycle, has an assembled and sequenced genome, and transformed cell lines have been established. It has become an excellent tool for gene targeting and RNA interference methods in order to study gene function (Cove et al., 2008). In microscopy-based studies, the protonemata life cycle phase of Physcomitrella has been exceptionally important for the study of fundamental cell and developmental mechanisms as it can be easily grown and experimentally manipulated. The protonemata have been used in studies of wall development (Lee et al., 2005), cytoskeletal dynamics during development (Vidali et al., 2007, 2009, 2010; Perroud and Quatrano, 2008; Augustine et al., 2011) and developmental responses to hormones and to environmental inputs (Marella et al., 2006; Sakata et al., 2010). The rapid accumulation of microscopy-based and genetic data from this organism make Physomitrella an important and convenient model in interpretation of basic sub-cellular phenomena in plants.
Charophycean green algae: cell wall development, cell morphogenesis, pattern development and cytoplasmic streaming
The Charophyceaen green algae or CGA represent the extant group of green algae that are most closely related and ancestral to land plants (Becker and Marin, 2009). Unique phenotypic features of these algae, such as a unicellular growth habit, large cell size or extraordinary developmental mechanisms, have launched several taxa as potentially important models for various types of cell-based research including 4-DI. For example, the unicellular desmid, Micrasterias, is an exceptional tool for elucidating cellular morphogenesis, the role of the cytoskeleton and the effects of environmental stressors (Affenzeller et al., 2009; Darehshouri and Lutz-Meindl, 2010). The large intermodal cells and rhizoids of the Charales, including Chara and Nitella, have served as outstanding models for studying the cellular phenomena of cytoplasmic streaming and cell wall growth mechanics (Shimmen and Yakota, 2004; Proseus and Boyer, 2005, 2006a, b, c; Shimmen, 2007; Wei and Lintilac, 2007; Goldstein et al., 2008). The plate-like thallus of Coleochaete has become a highly desirable system for elucidating pattern development is plant cell development (Dupuy et al., 2010).
A recent surge of research has shown that taxa of the late divergent CGA possess cell wall polymers remarkably similar to those of land plants (Popper and Fry, 2003; Domozych et al., 2007; Eder and Lutz-Meindl, 2008; Eder et al., 2008; Sørensen et al., 2010, 2011; Popper and Tuohy, 2010; Popper et al., 2011). Additionally, a unicellular desmid, Penium margaritaceum, has been shown to be a simple and convenient system for elucidating cell wall dynamics especially those dealing with pectins (Domozych et al., 2009, 2011). This alga is easily grown and manipulated in culture, exhibits a well-defined secretory mechanism during wall development and, most importantly, may be live-labelled with mAbs that bind to specific wall polymer epitopes. Labelled cells may then be returned to culture where subsequent wall development may be monitored and specific wall events studied via the addition of specific inhibitors or wall-altering enzymes. For example, high esterified homogalacturonans are secreted in a simple band in the cell centre. As these pectins are displaced outward toward both poles, they are de-esterified, bind to Ca2+ and form a distinct lattice (Domozych et al., 2009). These events, monitored in live cells by CLSM, may then be further studied using high-resolution EM. Figure 3 illustrates the versatility of this alga in cell wall research and the strategy in place for 4-DI studies. The next important steps in the study of Penium and other CGA will be gene sequencing and mapping, as well as successful stable transformation so that FP-based imaging may be employed.
CONCLUDING REMARKS
4-DI is of profound importance to our understanding of plant cell dynamics. When coupled with current efforts in functional genomics, 4-DI will yield critical insight into the foundations of plant cell structure, mechanics, developmental modulations and reactions to stress. These, in turn, may then be used in interpreting the manifestation of macroscopic phenomena, help in understanding how plants survive in our changing biosphere and also contribute to the design of plants and derived products in agriculture, biofuel production and other applied areas. Today, new and refined microscopy-based technologies and methods offer unprecedented imaging possibilities, but careful planning is required to maximize these benefits and minimize inherent limitations of certain approaches of microscopic investigation. With careful strategy and implementation of new technologies though, the outlook for 4-DI of plant cells is very promising indeed.
ACKNOWLEDGEMENTS
I thank Amanda Andreas, Carly Sacks, Hannah Brechka, Pia Ruisi-Besares, Korena Burgio (Skidmore College) and Zoe Popper (National University of Ireland, Galway) for their help and discussions. This work was supported by grants NSF-MCB-0919924 and NSF-MRI-0922805 from the National Science Foundation of the USA.
LITERATURE CITED
- Affenzeller MJ, Darehshouri A, Andosch A, Lutz C, Lutz-Meindl U. Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. Journal of Experimental Botany. 2009;60:939–954. doi: 10.1093/jxb/ern348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali GS, Prasad KVSK, Hanumappa M, Reddy ASN. Analysis of in vivo interaction and mobility of two spliceosomal proteins using FRAP and BiFC. PLoS ONE. 2008;3:e1953. doi: 10.1371/journal.pone.0001953. http://dx.doi.org/10.1371/journal.pone.0001953 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouar L, Chebli Y, Geitmann A. Morphogenesis of complex plant cell shapes – the mechanical role of crystalline cellulose in growing pollen tubes. Sexual Plant Reproduction. 2010;23:15–27. doi: 10.1007/s00497-009-0110-7. [DOI] [PubMed] [Google Scholar]
- Arimura S-i, Yamamoto J, Aida GP, Nkazonon M, Tsutsumi N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proceedings of the National Academy of Sciences, USA. 2004;101:7805–7808. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine RC, Pattavina KA, Tuzel E, Vidali L, Bezanilla M. Actin interacting protein1 and actin depolymerizing factor drive rapid actin dynamics in Physcomitrella patens. The Plant Cell. 2011;23:3696–3710. doi: 10.1105/tpc.111.090753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin JR, Staehelin LA. Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiology. 2011;155:1601–1611. doi: 10.1104/pp.110.170647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Beemster GTS, Judy-March JE, Marga F. Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiology. 2004;135:1–12. doi: 10.1104/pp.104.040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V, Nussaume L, Bhat RA. Combination of novel green fluorescent protein mutant TSapphire and DsRed variant mOrange to set up a versatile in planta FRET-FLIM assay. Plant Physiology. 2008;148:51–60. doi: 10.1104/pp.108.117358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Marin B. Streptophyte algae and the origin of the embryophytes. Annals of Botany. 2009;103:999–1004. doi: 10.1093/aob/mcp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RA, Lahaye T, Panstruga R. The visible touch: in planta visualization of protein–protein interactions by fluorophore-based methods. Plant Methods. 2006;2:12. doi: 10.1186/1746-4811-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchaflor EB, Gilroy S. Plant cell biology in the new millennium: new tools and new insights. American Journal of Botany. 2000;87:1547–1560. [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Henty JL, Khurana P, Staiger CJ. Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Current Opinion in Plant Biology. 2011;13:714–723. doi: 10.1016/j.pbi.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, et al. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. Journal of Biological Chemistry. 2006;281:29321–29329. doi: 10.1074/jbc.M605903200. [DOI] [PubMed] [Google Scholar]
- Boavida LC, Vieira AM, Becker JD, Feijo JA. Gametophyte interaction and sexual reproduction: how plants make a zygote. International Journal of Developmental Biology. 2005;49:615–632. doi: 10.1387/ijdb.052023lb. [DOI] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. Journal of Microscopy. 2004;214:159–173. doi: 10.1111/j.0022-2720.2004.01348.x. [DOI] [PubMed] [Google Scholar]
- Bougourd S, Marrison J, Haseloff J. An aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. The Plant Journal. 2000;24:543–550. doi: 10.1046/j.1365-313x.2000.00892.x. [DOI] [PubMed] [Google Scholar]
- Bove J, Vaillancourt B, Kroeger J, Hepler PK, Wiseman PW, Geitmann A. Magnitude and direction of vesicle dynamics in growing pollen tubes using spatiotemporal image correlation spectroscopy and fluorescence recovery after photobleaching. Plant Physiology. 2008;147:1646–1658. doi: 10.1104/pp.108.120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelje TC, Wessendorf MW, Sorenson RL. Multicolor laser scanning confocal immunofluorescence microscopy: practical application and limitations. Methods in Cell Biology. 1993;38:97–181. doi: 10.1016/s0091-679x(08)61001-8. [DOI] [PubMed] [Google Scholar]
- Broess K, Borst JW, van Amerogen H. Applying two-photon excitation fluorescence lifetime imaging microscopy to study photosynthesis in plant leaves. Photosynthetic Research. 2009;100:89–96. doi: 10.1007/s11120-009-9431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Faleri C, Del Casino C, Emons AMC, Cresti M. Distribution of callose synthase, cellulose synthase, and sucrose synthase in tobacco pollen tube is controlled in dissimilar ways by actin filaments and microtubules. Plant Physiology. 2011;155:1169–1190. doi: 10.1104/pp.110.171371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas L, Lovy-Wheeler A, Kunkel JG, Hepler PK. Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiology. 2008;146:1611–1621. doi: 10.1104/pp.107.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Kain SR. Green fluorescent protein: properties, applications and protocols. Methods of biochemical analysis. 2nd edn. Vol. 47. New York: Wiley; 2005. [Google Scholar]
- Chan J, Eder M, Crowell EF, Hampson J, Calder G, Lloyd C. Microtubules and CEAS tracks at the inner epidermal wall align independently of those on the outer wall of light-grown Arabidopsis hypocotyls. Journal of Cell Science. 2011;124:1086–1094. doi: 10.1242/jcs.086702. [DOI] [PubMed] [Google Scholar]
- Chandler DE, Roberson RW. Bioimaging. current concepts in light and electron microscopy. Sudbury, MA: Jones and Bartlett Publishers; 2009. [Google Scholar]
- Chebli Y, Daher FB, Sanyal M, Aouar L, Geitmann A. Microwave assisted processing of plant cells for optical and electron microscopy. Bulletin of the Microscopical Society of Canada. 2008;36:15–19. [Google Scholar]
- Chen T, Wang X, von Wangenheim D, et al. Probing and tracking organelles in living plant cells. Protoplasma. 2012 doi: 10.1007/s00709-011-0364-4. in press. http://dx.doi.org/10.1007/s00709-011-0364-4 . [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu H-M. Structural and functional compartmentalization in pollen tubes. Journal of Experimental Botany. 2007;58:75–82. doi: 10.1093/jxb/erl122. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Boavida LC, Aggarwal M, Wu H-M, Feijo JA. The pollen tube journey in the pistil and imaging the in vivo process by two-photon microscopy. Journal of Experimental Botany. 2010;61:1907–1915. doi: 10.1093/jxb/erq062. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Duan Q-h, Costa SS, et al. The dynamic pollen tube cytoskeleton: live cell studies using actin-binding and microtubule-binding reporter proteins. Molecular Plant. 2008;4:686–702. doi: 10.1093/mp/ssn026. [DOI] [PubMed] [Google Scholar]
- Churshumova W, Caragea AE, Goldstein RS, Berleth T. Glow in the dark: fluorescent proteins as cell and tissue-specific markers in plants. Molecular Plant. 2011;4:794–804. doi: 10.1093/mp/ssr059. [DOI] [PubMed] [Google Scholar]
- Cove DJ, Bezanilla M, Harries P, Quatrano RS. Mosses as model systems for the study of metabolism and development. Annual Review of Plant Biology. 2006;57:497–520. doi: 10.1146/annurev.arplant.57.032905.105338. [DOI] [PubMed] [Google Scholar]
- Cove DJ, Perroud P-F, Charron AJ, McDaniel SF, Khandelwal A, Quatrano RS. The moss Physcomitrella patens. A novel model system for plant development and genomic studies. In: Crotty DA, Gann A, editors. Emerging model organisms, a laboratory manual. Vol. I. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. pp. 69–104. [DOI] [PubMed] [Google Scholar]
- Crowell EF, Timpano H, Desprez T, et al. Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. The Plant Cell. 2011;25:2592–2605. doi: 10.1105/tpc.111.087338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher FB, Geitmann A. Actin is involved in pollen tube tropism through redfining the spatial targeting of secretory vesicles. Traffic. 2011;12:1537–1551. doi: 10.1111/j.1600-0854.2011.01256.x. [DOI] [PubMed] [Google Scholar]
- Darehshouri A, Lutz-Meindl U. H2O2 localization in the green alga Micrasterias after salt and osmotic stress by TEM-coupled electron energy loss spectroscopy. Protoplasma. 2010;239:49–56. doi: 10.1007/s00709-009-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlasio SL, Sylvester AW, Jackson D. Illuminating plant biology: using fluorescent proteins for high throughput analysis of protein localization and function in plants. Briefings in Functional Genomics. 2010;9:129–138. doi: 10.1093/bfgp/elp060. [DOI] [PubMed] [Google Scholar]
- Derksen J, Janssen G-J, Wolters-Arts M, et al. Wall architecture with high porosity is established at the tip and maintained in growing pollen tubes of Nicotiana tabacum. The Plant Journal. 2011;68:495–506. doi: 10.1111/j.1365-313X.2011.04703.x. [DOI] [PubMed] [Google Scholar]
- Dodonova SO, Bulychev FA. Cyclosis-related asymmetry of chloroplast–plasma membrane interactions at the margins of illuminated area in Chara corallina cells. Protoplasma. 2011;248:737–749. doi: 10.1007/s00709-010-0241-6. [DOI] [PubMed] [Google Scholar]
- Domozych DS, Serfis A, Keimle S, Gretz MR. The structure and biochemistry of the homogalacturonans of the cell wall of the desmid. Penium margaritaceum. Protoplasma. 2007;230:99–115. doi: 10.1007/s00709-006-0197-8. [DOI] [PubMed] [Google Scholar]
- Domozych DS, Lambiasse L, Kiemle SN, Gretz MR. Cell wall development and bipolar growth in the desmid Penium margaritaceum. Asymmetry in a symmetric world. Journal of Phycology. 2009;45:879–893. doi: 10.1111/j.1529-8817.2009.00713.x. [DOI] [PubMed] [Google Scholar]
- Domozych DS, Brechka H, Britton A, Toso M. Cell wall growth and modulation dynamics in a model unicellular green alga. Penium margaritaceum: live cell labeling with monoclonal antibodies. Journal of Botany. 2011;2011:632165. http://dx.doi.org/10.1155/2011/632165 . [Google Scholar]
- Donohoe BS, Kang B-H, Staehelin LA. Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proceedings of the National Academy of Sciences, USA. 2007;104:163–168. doi: 10.1073/pnas.0609818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Ren H. Development and application of probes for labeling the actin cytoskeleton in living plant cells. Protoplasma. 2011;248:239–250. doi: 10.1007/s00709-010-0202-0. [DOI] [PubMed] [Google Scholar]
- Dupuy L, Mackenzie J, Haseloff J. Coordination of plant cell division and expansion in a simple morphogenetic system. Proceedings of the National Academy of Sciences, USA. 2010;107:2711–16. doi: 10.1073/pnas.0906322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder M, Lütz-Meindl U. Pectin-like carbohydrates in the green alga Micrasterias characterized by cytochemical analysis and energy filtering TEM. Journal of Microscopy. 2008;231:201–214. doi: 10.1111/j.1365-2818.2008.02036.x. [DOI] [PubMed] [Google Scholar]
- Eder M, Tenhaken R, Driouich A, Lütz-Meindl U. Occurrence and characterisation of arabinogalactan-like proteins and hemicelluloses in Micrasterias (Streptophyta) Journal of Phycology. 2008;44:1221–1234. doi: 10.1111/j.1529-8817.2008.00576.x. [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL. Live cell imaging of plants. Cold Spring Harbor Protocols. 2010;2010(2) doi: 10.1101/pdb.top68. http://dx.doi.org/10:1101/pdb.top68 . [DOI] [PubMed] [Google Scholar]
- Fayant P, Girlanda O, Chebli Y, Aubin C-E, Villemure I, Geitmann A. Finite element model of polar growth in pollen tubes. The Plant Cell. 2010;22:2579–2593. doi: 10.1105/tpc.110.075754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijo JA, Sainhas J, Hackett GR, Kunkel JG, Hepler PK. Growing pollen tubes possess a constituitive alkaline band in the clear zone and a growth-dependent acidic tip. Journal of Cell Biology. 1999;144:483–496. doi: 10.1083/jcb.144.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K. Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiology. 2010;153:1453–1463. doi: 10.1104/pp.110.157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J, Binder S. The red fluorescent protein eqFP611: application in subcellular localization studies in plants. BMC Plant Biology. 2007;7:28. doi: 10.1186/1471-2229-7-28. http://dx.doi.org/10.1186/1471-2229-7-28 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigault MM, Lacoste J, Swift JL, Brown CM. Live-cell microscopy – tips and tools. Journal of Cell Science. 2009;122:753–767. doi: 10.1242/jcs.033837. [DOI] [PubMed] [Google Scholar]
- Fujita M, Himmelspach R, Hocart CH, Williamson RE, Mansfield SD, Wastenys GO. Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. The Plant Journal. 2011;66:915–928. doi: 10.1111/j.1365-313X.2011.04552.x. [DOI] [PubMed] [Google Scholar]
- Geitmann A. How to shape a cylinder: pollen tube as a model system for the generation of complex cellular geometry. Sexual Plant Reproduction. 2010a;23:63–71. doi: 10.1007/s00497-009-0121-4. [DOI] [PubMed] [Google Scholar]
- Geitmann A. Mechanical modeling and structural analysis of the primary plant cell wall. Current Opinion in Plant Biology. 2010b;13:696–699. doi: 10.1016/j.pbi.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Geldner N, De'nervaud-Tendon V, Hyman DL, Mayer U, Stierhof Y-D, Chory J. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. The Plant Journal. 2009;59:169–178. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RE, Tuval I. Microfluidics of cytoplasmic streaming and its implication for intracellular transport. Proceedings of the National Academy of Sciences, USA. 2008;105:3663–3667. doi: 10.1073/pnas.0707223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing LR. FRET analysis of transmembrane flipping of FM4-64 in plant cells: is FM4-64 a robust marker for endocytosis? Journal of Microscopy. 2008;231:291–298. doi: 10.1111/j.1365-2818.2008.02042.x. [DOI] [PubMed] [Google Scholar]
- Haas TJ, Otegui MS. Electron tomography in plant cell biology. Journal of Integrative Plant Biology. 2007;49:1091–1099. [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes JG, Czymmek KJ, Carlson CA, Veereshingam H, Dickstein R, Sherrier DJ. Rapid analysis of legume root nodule development using confocal microscopy. New Phytologist. 2004;163:661–668. doi: 10.1111/j.1469-8137.2004.01138.x. [DOI] [PubMed] [Google Scholar]
- Held MA, Boulaflous A, Bradizzi F. Advances in fluorescence protein-based imaging for the analysis of plant endomembranes. Plant Physiology. 2008;147:1469–1481. doi: 10.1104/pp.108.120147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Winship LJ. Calcium at the cell wall–cytoplast interface. Journal of Integrative Plant Biology. 2010;52:147–160. doi: 10.1111/j.1744-7909.2010.00923.x. [DOI] [PubMed] [Google Scholar]
- Herve C, Marcus SE, Knox JP. Monoclonal antibodies, carbohydrate-binding modules and the detection of polysaccharides in plant cell walls. Methods in Molecular Biology. 2011;715:103–113. doi: 10.1007/978-1-61779-008-9_7. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconjugate Journal. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- Inoue S. Foundations of confocal scanned imaging in light microscopy. In: Pawley JB, editor. Handbook of biological confocal microscopy. 3rd edn. New York: Springer; 2006. pp. 1–19. [Google Scholar]
- Iwano M, Shiba H, Matoba K, et al. Actin dynamics in papilla cells of Brassica rapa during self- and cross pollination. Plant Physiology. 2007;144:72–81. doi: 10.1104/pp.106.095273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ID. Practical considerations in the selection and application of fluorescent probes. In: Pawley JB, editor. Handbook of biological confocal microscopy. 3rd edn. New York: Springer; 2006. pp. 353–367. [Google Scholar]
- Johnston JS, Bennett MD, Rayburen AL, Galbraith DW, Price HJ. Reference standards for determination of DNA content of plant nuclei. American Journal of Botany. 1999;86:609–613. [PubMed] [Google Scholar]
- Kaulich B, Gianoncelli A, Beran A, et al. Low-energy X-ray fluorescence microscopy opening new opportunities for bio-related research. Journal of the Royal Society Interface. 2009;6:S641–S647. doi: 10.1098/rsif.2009.0157.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Anthony RG, Hussey PJ. Green fluorescent protein–mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin deplymerizing factor's actin depolymerizing activity in vitro. Plant Physiology. 2004;136:3990–3998. doi: 10.1104/pp.104.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby AR. Atomic force microscopy of plant cell walls. Methods in Molecular Biology. 2011;715:169–178. doi: 10.1007/978-1-61779-008-9_12. [DOI] [PubMed] [Google Scholar]
- Kljun A, Beninans TAS, Goubet F, Meulwaeter F, Knox JP, Blackburn RS. Comparative analysis of crystallinity changes in cellulose I polymers using ATR-FTIR diffraction, and carbohydrate-binding module probes. BioMacromolecules. 2011;12:4121–4126. doi: 10.1021/bm201176m. [DOI] [PubMed] [Google Scholar]
- Knapp E, Flores R, Scheibilin D, Modla S, Czymmek K, Yusibov V. A cryohistological protocol for preparation of large plant tissue sections for screening intracellular fluorescent protein expression. BioTechniques. 2012;52:31–37. doi: 10.2144/000113778. [DOI] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY. Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. The Plant Journal. 2008;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- Konrad KR, Wudick MM, Feijo JA. Calcium regulation of tip growth: new genes for old mechanisms. Current Opinion in Plant Biology. 2011;14:721–730. doi: 10.1016/j.pbi.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy KV. Methods in cell wall cytochemistry. Boca Raton, FL: CRC Press; 1999. [Google Scholar]
- Kroeger JH, Geitmann A. Pollen tube growth: getting a grip on cell biology through modeling. Mechanics Research Communications. 2012;42:32–39. [Google Scholar]
- Kroeger JH, Zerzour R, Geitmann A. Regulator or driving force? The role of turgor pressure in oscillatory plant cell growth. PLoS ONE. 2011;6:e18549. doi: 10.1371/journal.pone.0018549. http://dx.doi.org/10.1371/journal.pone.0018549 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J. Electron microscopy: methods and protocols. New York: Humana Press; 2007. [Google Scholar]
- Kwaaitaal M, Keinath NF, Pajonk S, Biskup C, Panstruga R. Combined bimolecular fluorescence complementation and förster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiology. 2010;152:1135–1147. doi: 10.1104/pp.109.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro MD, Cardenas L, Bhatt AP, et al. Calcium gradients in conifer pollen tubes; dynamic properties differ from those seen in angiosperms. Journal of Experimental Botany. 2005;56:2629–2628. doi: 10.1093/jxb/eri256. [DOI] [PubMed] [Google Scholar]
- Lee J-Y, Wang X, Weier C, et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. The Plant Cell. 2011;23:3353–3373. doi: 10.1105/tpc.111.087742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJD, Sakata Y, Mau S-L, et al. Arabinogalactan-proteins (AGPs) are required for apical cell extension in the moss Physcomitrella patens. The Plant Cell. 2005;17:3051–3065. doi: 10.1105/tpc.105.034413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Szumlanski A, Nielsen E, Yang Z. Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. Journal of Cell Biology. 2008;181:1155–1168. doi: 10.1083/jcb.200801086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman NS, Jackson LR, Trudel LJ, Weis-Garcia F. Monoclonal versus polyclonal antibodies distinguishing characteristics, applications and information resources. Institute of Laboratory Animal Resources. 2005;46:258–268. doi: 10.1093/ilar.46.3.258. [DOI] [PubMed] [Google Scholar]
- Lovy-Wheeler A, Kunkel JG, Allwood EG, Hussey PJ, Hepler PK. Oscillatory increases in alkalinity anticipate growth and may regulate actin dynamics in pollen tubes of lily. The Plant Cell. 2006;18:2182–2193. doi: 10.1105/tpc.106.044867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A, Cardenas L, Kunkel JG, Hepler PK. Differential organelle movement on the actin cytoskeleton in lily pollen tubes. Cell Motility and the Cytoskeleton. 2007;64:217–232. doi: 10.1002/cm.20181. [DOI] [PubMed] [Google Scholar]
- Lutz-Meindl U. Use of energy filtering transmission electron microscopy for image generation and element analysis in plant organisms. Micron. 2007;38:181–196. doi: 10.1016/j.micron.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Marella H, Sakata Y, Quatrano RS. Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. The Plant Journal. 2006;46:1032–1044. doi: 10.1111/j.1365-313X.2006.02764.x. [DOI] [PubMed] [Google Scholar]
- Marga F, Grandbois M, Cosgrove DJ, Baskin TI. Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. The Plant Journal. 2005;43:181–190. doi: 10.1111/j.1365-313X.2005.02447.x. [DOI] [PubMed] [Google Scholar]
- Martens HJ, Roberts AG, Oparka KJ, Schulz A. Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiology. 2006;142:471–480. doi: 10.1104/pp.106.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Radhamony R, Sinclair AM, et al. mEosFP-based green-to-red photoconvertible subcellular probes for plants. Plant Physiology. 2010;154:1573–1587. doi: 10.1104/pp.110.165431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Defernez M, Urbanowicz BR, et al. Neural network analysis of infrared spectra for classifying cell wall architecture. Plant Physiology. 2007;143:1314–1326. doi: 10.1104/pp.106.093054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, Auer M. High-pressure freezing, cellular tomography, and structural cell biology. BioTechniques. 2006;41:137–143. doi: 10.2144/000112226. [DOI] [PubMed] [Google Scholar]
- McIntosh R, Nicastro D, Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends in Cell Biology. 2004;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- McKenna ST, Kunkel JG, Bosch M, et al. Exocytosis precedes and predicts the increase in growth in oscillating pollen tubes. The Plant Cell. 2009;21:3026–3040. doi: 10.1105/tpc.109.069260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani P, Gholamirad M, Traas J, et al. In vivo analysis of local wall stiffness at the shoot apical meristem in Arabidopsis using atomic force microscopy. The Plant Journal. 2011;67:1116–1123. doi: 10.1111/j.1365-313X.2011.04649.x. [DOI] [PubMed] [Google Scholar]
- Mims CW, Celio GJ, Richardson EA. The use of high pressure freezing and freeze substitution to study host–pathogen interactions in fungal diseases of plants. Microscopy and Microanalysis. 2003;9:522–531. doi: 10.1017/S1431927603030587. [DOI] [PubMed] [Google Scholar]
- Moller I, Sorensen I, Bernal AJ, et al. High-throughput mapping of cell wall polymers within and between plants using novel microarrays. The Plant Journal. 2007;50:1118–1128. doi: 10.1111/j.1365-313X.2007.03114.x. [DOI] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Hofte H. Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. The Plant Journal. 2003;35:393–404. doi: 10.1046/j.1365-313x.2003.01807.x. [DOI] [PubMed] [Google Scholar]
- Mouille G, Witucka-Wall H, Bruyant M-P, et al. Quantitative trait loci analysis of primary cell wall composition in Arabidopsis. Plant Physiology. 2006;141:1035–1044. doi: 10.1104/pp.106.079384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair M, DeBolt S. Analysing cellulose biosynthesis with confocal microscopy. Methods in Molecular Biology. 2011;715:141–152. doi: 10.1007/978-1-61779-008-9_10. [DOI] [PubMed] [Google Scholar]
- Nakakoshi M, Nishioka H, Katayama E. New versatile staining reagents for biological transmission electron microscopy that substitute for uranyl acetate. Journal of Electron Microscopy. 2011;60:401–407. doi: 10.1093/jmicro/dfr084. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Otegui MS. Electron tomography and immunogold labelling as tools to analyse de novo assembly of plant cell walls. Methods in Molecular Biology. 2011;715:123–140. doi: 10.1007/978-1-61779-008-9_9. [DOI] [PubMed] [Google Scholar]
- Parre E, Geitmann A. More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiology. 2005;137:274–286. doi: 10.1104/pp.104.050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawley JB. Handbook of biological confocal microscopy. New York: Springer; 2006. [Google Scholar]
- Perroud P-F, Quatrano RS. BRICK1 is required for apical cell growth in filaments of the moss Physcomitrella patens but not for gametophore morphology. The Plant Cell. 2008;20:411–422. doi: 10.1105/tpc.107.053256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. Primary cell wall composition of bryophytes and charophytes. Annals of Botany. 2003;91:1–12. doi: 10.1093/aob/mcg013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Tuohy MG. Beyond the green: understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiogy. 2010;153:373–383. doi: 10.1104/pp.110.158055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, et al. Evolution and diversity of plant cell walls: from algae to flowering plants. Annual Review of Plant Biology. 2011;62:567–590. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Turgor pressure moves polysaccharides into growing cell walls of Chara corallina. Annals of Botany. 2005;98:967–979. doi: 10.1093/aob/mci113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Identifying cytoplasmic input to the cell wall of growing Chara corallina. Journal of Experimental Botany. 2006a;57:3231–3242. doi: 10.1093/jxb/erl087. [DOI] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Periplasm turgor pressure controls wall deposition and assembly of growing Chara corallina. Annals of Botany. 2006b;98:93–105. doi: 10.1093/aob/mcl098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Calcium pectate chemistry controls growth rate of Chara corallina. Journal of Experimental Botany. 2006c;57:3989–4002. doi: 10.1093/jxb/erl166. [DOI] [PubMed] [Google Scholar]
- Regvar M, Eichert D, Kaulich B, et al. New insights into globoids of protein storage vacuoles in wheat aleurone using synchroton soft X-ray microscopy. Journal of Experimental Botany. 2011;62:3929–3939. doi: 10.1093/jxb/err090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhiel E, Brock J. Lectin binding in Cryptomonas and Chroomonas (Cryptophyceae) Protoplasma. 2012 doi: 10.1007/s00709-011-0319-9. in press http://dx.doi.org/10.1007/s00709-011-0319-9 . [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Davidson MW, Piston DW. Fluorescent protein tracking and detection: applications using fluorescent proteins in living cells. Cold Spring Harbor Protocols. 2009 doi: 10.1101/pdb.top64. 2009(12). http://dx.doi.org/10.1101/pdb.top64 . [DOI] [PubMed] [Google Scholar]
- Rounds CM, Winship LJ, Hepler PK. Pollen tube energetics: respiration, fermentation and the race to the ovule. AoB PLANTS. 2011;2011:plr019. doi: 10.1093/aobpla/plr019. http://dx.doi.org/10.1093/aobpla/plr019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin SE. Plant microtechnique and microscopy. Oxford: Oxford University Press; 1999. [Google Scholar]
- Sakata Y, Nakamura I, Taji T, Tanaka S, Quatrano RS. Regulation of the ABA-responsive Em promoter by ABI3 in the moss Physcomitrella patens: role of the ABA response element and the RY element. Plant Signaling and Behavior. 2010;5:1–6. doi: 10.4161/psb.5.9.11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J, Muller J, Beck M, Bohm N, Menzel D. Vesicular trafficking, cytoskeleton and signaling in roots hairs and pollen tubes. Trends in Plant Science. 2006;11:594–600. doi: 10.1016/j.tplants.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Sampathkumar A, Lindeboom JJ, DeBolt S, et al. Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. The Plant Cell. 2011;23:2302–2313. doi: 10.1105/tpc.111.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Strasser R. Sub-compartmental organization of Golgi-resident N-glycan processing enzymes in plants. Molecular Plant. 2011;4:220–228. doi: 10.1093/mp/ssq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shaw SL. Imaging the live plant cell. The Plant Journal. 2006;45:573–598. doi: 10.1111/j.1365-313X.2006.02653.x. [DOI] [PubMed] [Google Scholar]
- Shimmen T. The sliding theory of cytoplasmic streaming: fifty years of progress. Journal of Plant Research. 2007;120:31–43. doi: 10.1007/s10265-006-0061-0. [DOI] [PubMed] [Google Scholar]
- Shimmen T, Yakota E. Cytoplasmic streaming in plants. Current Opinion in Cell Biology. 2004;16:68–72. doi: 10.1016/j.ceb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Shimoni E, Rav-Hon O, Ohad I, Brumfeld V, Reich Z. Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. The Plant Cell. 2005;17:2580–2586. doi: 10.1105/tpc.105.035030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biology. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. http://dx.doi.org/10.1371/journal.pbio.1001041 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Deeks MJ, Hussey PJ. Strategies of actin reorganization in plant cells. Journal of Cell Science. 2010;123:3019–3028. doi: 10.1242/jcs.071126. [DOI] [PubMed] [Google Scholar]
- Sørensen I, Domozych D, Willats WGT. How have plant cell walls evolved? Plant Physiology. 2010;153:366–372. doi: 10.1104/pp.110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen I, Pettolino F, Bacic A, et al. The Charophycean green algae provide insights into the early origins of plant cell walls. The Plant Journal. 2011;68:201–211. doi: 10.1111/j.1365-313X.2011.04686.x. [DOI] [PubMed] [Google Scholar]
- Sparkes I, Tolley N, Aller I, et al. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. The Plant Cell. 2010;22:1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Graumann K, Martinie're A, Scoberger J, Wang P, Osterrieder A. Bleach it, switch it, bounce it, pull it: using lasers to reveal plant cell dynamics. Journal of Experimental Botany. 2011a;62:1–7. doi: 10.1093/jxb/erq351. [DOI] [PubMed] [Google Scholar]
- Sparkes I, Hawes C, Frigerio L. FrontiERs: movers and shapers of the higher plant cortical endoplasmic reticulum. Current Opinion in Plant Biology. 2011b;14:658–665. doi: 10.1016/j.pbi.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Kang B-H. Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiology. 2008;147:1454–1468. doi: 10.1104/pp.108.120618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslov D, Verbelen J-P, Vissenberg K. Onion epidermis as a new model to study the control of growth anisotropy in higher plants. Journal of Experimental Botany. 2009;60:4175–4187. doi: 10.1093/jxb/erp251. [DOI] [PubMed] [Google Scholar]
- Tang X-C, He Y-Q, Sun M-X. The role of arabinogalactan proteins binding to Yariv reagents in the initiation, cell developmental fate, and maintenance of microspore embryogenesis in Brassica napus L. cv. Topas. Journal of Experimental Botany. 2006;57:2639–2650. doi: 10.1093/jxb/erl027. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Takab K, Mineyuki Y. Immunoelectron microscopy of cryofixed and freeze-substituted plant tissues. Methods in Molecular Biology. 2010;657:155–165. doi: 10.1007/978-1-60761-783-9_12. [DOI] [PubMed] [Google Scholar]
- Talbot MJ, Wastenys GO, Offler CE, McCurdy DW. Cellulose synthesis is required for deposition of reticulate wall ingrowths in transfer cells. Plant and Cell Physiology. 2007;48:147–158. doi: 10.1093/pcp/pcl046. [DOI] [PubMed] [Google Scholar]
- Tsien RY, Ernst L, Waggoner A. Fluorophores for general microscopy: photophysics and photochemistry. In: Pawley JB, editor. Handbook of biological confocal microscopy. 3rd edn. New York: Springer; 2006. pp. 338–352. In ed. [Google Scholar]
- Van Gisbergen PAC, Esseling-Ozdoba A, Vos JW. Microinjecting FM4-64 validates it as a marker of ther endocytic pathway in plants. Journal of Microscopy. 2008;231:284–290. doi: 10.1111/j.1365-2818.2008.02041.x. [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J, Goldstein RE. Cytoplasmic streaming enables the distribution of molecules and vesicles in large plant cells. Protoplasma. 2010;240:99–107. doi: 10.1007/s00709-009-0088-x. [DOI] [PubMed] [Google Scholar]
- Vidali L, Augustine RC, Kleinman KP, Bezanilla M. Profilin is essential for polarized growth in the moss Physcomitrella patens. The Plant Cell. 2007;19:3705–3722. doi: 10.1105/tpc.107.053413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Rounds C, Hepler PK, Bezanilla M. LifeAct–mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS One. 2009;4:e5744. doi: 10.1371/journal.pone.0005744. http://dx.doi.org/10.1371/journal.pone.0005744 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Burkart GM, Augusine RC, Kerdavid E, Tuzel E, Bezanilla M. Myosin XI is essential for tip growth in Physcomitrella patens. The Plant Cell. 2010;22:1868–1882. doi: 10.1105/tpc.109.073288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcay-Barrena G, Webb SED, Martin-Fernandez ML, Wilson ZA. Subcellular and single-molecule imaging of plant fluorescent proteins using total internal reflection fluorescence microscopy (TIRFM) Journal of Experimental Botany. 2011;62:5419–5428. doi: 10.1093/jxb/err212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, l Walter M, et al. Visualization of protein interactions in living plant cells using biomolecular fluorescence complementation. The Plant Journal. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Wang L, Chen C, et al. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. Journal of Experimental Botany. 2011;62:5161–5177. doi: 10.1093/jxb/err221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Teng Y, Wang Q, et al. Imaging of dynamic secretory vesicles in living pollen tubes of Picea meyeri using evanescent wave microscopy. Plant Physiology. 2006;141:1591–1603. doi: 10.1104/pp.106.080168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne R. Light and video microscopy. Amsterdam: Elsevier Academic Press; 2009. [Google Scholar]
- Wei C, Lintilhac PM. Loss of stability: a new look at the physics of cell wall behavior during plant cell growth. Plant Physiology. 2007;145:763–772. doi: 10.1104/pp.107.101964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsen KJ, Lovy-Wheeler A, Voight B, Menzel, Kunkel JG, Hepler PK. Imaging the actin cytoskeleton in growing pollen tubes. Sexual Plant Reproduction. 2006;19:51–62. [Google Scholar]
- Wright KM, Wood NT, Roberts AG, et al. Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic. 2007;8:21–31. doi: 10.1111/j.1600-0854.2006.00510.x. [DOI] [PubMed] [Google Scholar]
- Wuyts N, Palauqui J-C, Conejero G, Verdeil J-L, Granier C, Massonet C. High-contrast three-dimensional imaging of the Arabidopsis leaf enables the analysis of cell dimensions in the epidermis and mesophyll. Plant Methods. 2010;6:17. doi: 10.1186/1746-4811-6-17. http://dx.doi.org/10.1186/1746-4811-6-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough JM, Himmel ME, Ding S-Y. Plant cell wall characterization using scanning probe microscopy techniques. Biotechnology for Biofuels. 2010;2:17. doi: 10.1186/1754-6834-2-17. http://dx.doi.org/10.1186/1754-6834-2-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechmann B, Zellnig G. Microwave-assisted rapid plant sample preparation for transmission electron microscopy. Journal of Microscopy. 2009;233:258–268. doi: 10.1111/j.1365-2818.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- Zewail AH, Thomas JM. 4D electron microscopy. Imaging in space and time. London: Imperial College Press; 2010. [Google Scholar]
- Zhang Y, He J, Lee D, McCormick S. Interdependence of endomembrane trafficking and actin dynamics during polarized growth of Arabidopsis pollen tubes. Plant Physiology. 2010;152:2200–2210. doi: 10.1104/pp.109.142349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T. Vesicle trafficking dynamics and visualization of zones of exocytosis and endocytosis in tobacco pollen tubes. Journal of Experimental Botany. 2008;59:861–873. doi: 10.1093/jxb/ern007. [DOI] [PubMed] [Google Scholar]