Abstract
Transmissible plasmids are responsible for the spread of genetic determinants, such as antibiotic resistance or virulence traits, causing a large ecological and epidemiological impact. Transmissible plasmids, either conjugative or mobilizable, have in common the presence of a relaxase gene. Relaxases were previously classified in six protein families according to their phylogeny. Degenerate primers hybridizing to coding sequences of conserved amino acid motifs were designed to amplify related relaxase genes from γ-Proteobacterial plasmids. Specificity and sensitivity of a selected set of 19 primer pairs were first tested using a collection of 33 reference relaxases, representing the diversity of γ-Proteobacterial plasmids. The validated set was then applied to the analysis of two plasmid collections obtained from clinical isolates. The relaxase screening method, which we call “Degenerate Primer MOB Typing” or DPMT, detected not only most known Inc/Rep groups, but also a plethora of plasmids not previously assigned to any Inc group or Rep-type.
Introduction
Plasmids exert a great evolutionary impact in their bacterial hosts, allowing them to colonize new niches, obtain advantages against either natural competitors, or overcome artificial selective pressures. These beneficial characteristics easily spread between bacterial populations because of horizontal gene transfer. Among the clinically important disseminated traits are determinants for antibiotic resistance (AbR) and virulence [1], [2].
Basic physiological functions of plasmids are autonomous replication, stability and propagation (conjugation and establishment in new hosts) [3]. Differences in replication and stability constituted the basis for classifying plasmids, first by incompatibility (Inc) and later by replicon typing. Incompatibility (the inability of two plasmids to coexist within the same cell) is a phenotypic expression of the interactions in plasmid replication [4] or partition [5]. By Inc testing [6], enterobacterial plasmids were divided in 27 groups, with some further subdivisions [7]. Inc groups include historical R-plasmids, which largely contributed to AbR dissemination, together with xenobiotic biodegradation and virulence plasmids. The Inc classification did not always reflect true evolutionary divergence: highly similar plasmids can be compatible [8], [9], [10], [11], [12], [13], [14], while largely non homologous plasmids can be incompatible (e.g. IncX1 and IncX2 plasmids [15], [16], [17], some IncQ1 and IncQ2 plasmids [13]). As a consequence of the technical drawbacks of Inc testing, plasmid classification turned to molecular comparison of replication regions, leading to the development of two replicon typing methods. The first was based on DNA hybridization with specific plasmid probes (Inc/Rep-HYB) that contained either copy number control or partition DNA sequences of 19 Inc groups [18]. The second and presently most widely used method is called PCR-based replicon typing (PBRT). It was first used to identify five Inc groups of broad-host-range plasmids in environmental samples (IncW, IncP1, IncQ1, IncN [19], [20], [21] and IncP9 [22], [23]) and later on to detect replicons predominant in Enterobacteriaceae [24], [25], [26], [27], [28] as well as 19 groups of resistance plasmids of Acinetobacter baumanii [29]. Plasmid multilocus/double sequence type methods [27], [30], [31], [32], [33] and PCRs detecting plasmid genes other than replication/partition modules [19], [34] were also developed to detect some plasmid backbones. PBRT and these other methods allowed plasmid identification and circumvented the technical problems associated to Inc testing. As a drawback, they narrowed plasmid classification within the boundaries of Inc groups or small clusters of highly similar backbones. Thus, PBRT kept a significant fraction of plasmid groups out of assortment.
Around 50% of γ-Proteobacteria plasmids are potentially transmissible [35]. Conjugative plasmids encode all functions needed for transfer (i.e. origin of transfer locus (oriT), relaxase, coupling protein (T4CP) and type IV secretion system (T4SS)). Mobilizable plasmids code only for oriT, relaxase and nicking-accessory protein(s) (and only rarely for T4CP), requiring the help of a conjugative plasmid to be transferred. Thus, the only common component to all transmissible (conjugative and mobilizable) plasmids is the relaxase. Relaxases are multidomain proteins, the relaxase activity residing in their N-terminal domain [36]. The 3D structures of four relaxase domains have been solved: the MOBF relaxases TrwC_R388 [37] and TraI_F [38], the MOBQ relaxase MobA_R1162/RSF1010 [39] and the MOBV relaxase MobM_pMV158 (M. Espinosa, personal communication). In these proteins, the architecture of the active centre is highly similar in spite of the fact that they belong to three different MOB families [35]. Homology at the sequence level resides on three conserved motifs: motif I that contains the catalytic Tyr residue(s) involved in DNA cleavage-joining reactions; motif II that contains an Asp or Glu residue involved in activation of the nucleophilic hydroxyl of the catalytic Tyr, and the most conspicuous motif III, which contains a His triad that coordinates a divalent cation directly involved in the catalytic reactions [37], [40]. The evolutionary relationships among relaxase sequences were traced and transmissible plasmids distributed in six relaxase MOB families [35], [36]. Here, we developed a set of oligonucleotide primers for relaxase identification based on the relaxase protein phylogenies. The method is called “Degenerate Primer MOB Typing” (DPMT). As an application, we used DPMT to identify new relaxases and to classify plasmids isolated from clinical isolates of γ-Proteobacteria.
Results
Design and Validation of the DPMT Oligonucleotide Set
Phylogenetic trees of the five plasmid relaxase families which contained suitably populated and well supported subfamilies in γ-Proteobacteria were traced as shown in Figures 1, 2, 3, 4, 5, 6, 7. They served as guides for designing oligonucleotide primer pairs able to amplify relaxases clustered in those subfamilies. Each primer was partially degenerated, up to 24 degeneracy at its 3′ sequence, to encompass a relaxed codon usage. Primers for which the design resulted in degeneracy larger than 24, were reduced to degeneracy-24 by considering only the sequences present in the respective DNA relaxase alignment. Each primer pair was tested on a reference collection of 33 relaxases encoded by transmissible plasmids originally isolated from γ-Proteobacteria (Table 1). Once their specificity was validated, the set of validated primers was used to identify relaxases in plasmid collections from clinical isolates, leading to the identification of both known and non-previously reported relaxase sequences. Details for the design and range of substrates of the primer pairs selected for each MOB family follow.
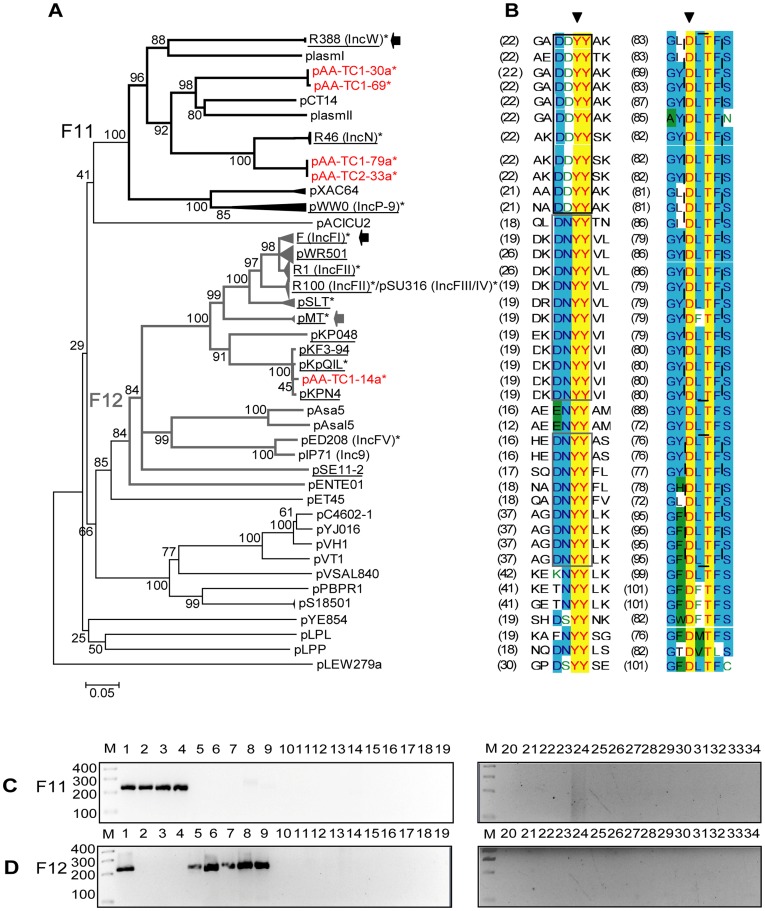
Figure 1. DPMT validation for MOBF relaxases.
A) Phylogenetic tree of MOBF relaxases. Triangles at the end of the branches represent a compressed group of very similar relaxases (>95%). A solid black arrow points to the prototype plasmid for each subfamily. Arrows point to plasmids that experimentally amplified, in spite of containing at least one mismatch in the 12 nucleotides of the CORE sequence. Relaxases contained in our reference collection (Table 1) are denoted by an asterisk. Plasmids detectable by PBRT amplification [19], [20], [21], [22], [24], [25], [27], [28], [29] are underlined. New relaxase sequences uncovered by DPMT are shown in red. B) Alignment of the relaxase motifs used to design the MOBF degenerate primers. Colour code: red on yellow = invariant amino acids; blue on blue = strongly conserved; black on green = similar; green on white = weakly similar; black on white = not conserved. Black arrowheads point to the key residues that define the relaxase motifs. Different rectangles embrace the conserved amino acids used to infer the 3′ degenerate core of each oligonucleotide (F11-f, continuous black; F12-f, continuous dark grey; and F1-r, dashed black). C) Amplicons obtained with primers for subfamily MOBF11 (F11-f and F1-r). Lane 1, pSU1588; 2, pSU4280; 3, pSU10013; 4, pSU10014; 5, pSU10017; 6, pSU10018; 7, pSU10021; 8, pSU316; 9, pSU10022; 10, pSU10010; 11, R751; 12, pSU10028; 13, pSU10029; 14, pSU10056; 15, pSU10055; 16, pSU10001; 17, pSU10012; 18, pSU10011; 19, pSU10009; 20, pSU4601; 21, pSU10006; 22, pSU10007; 23, pSU10064; 24, pSU10059; 25, pSU10008; 26, pSU10039; 27, pSU10040; 28, pSU10041; 29, pSU10004; 30, pSU10003; 31, pSU10043; 32, pSU4830; 33, pSU10002; 34, negative control. Lane M, molecular mass marker, HyperLadder IV (Bioline). D) Amplicons obtained with primers for subfamily MOBF12 (F12-f and F1-r). Lanes as in (C).
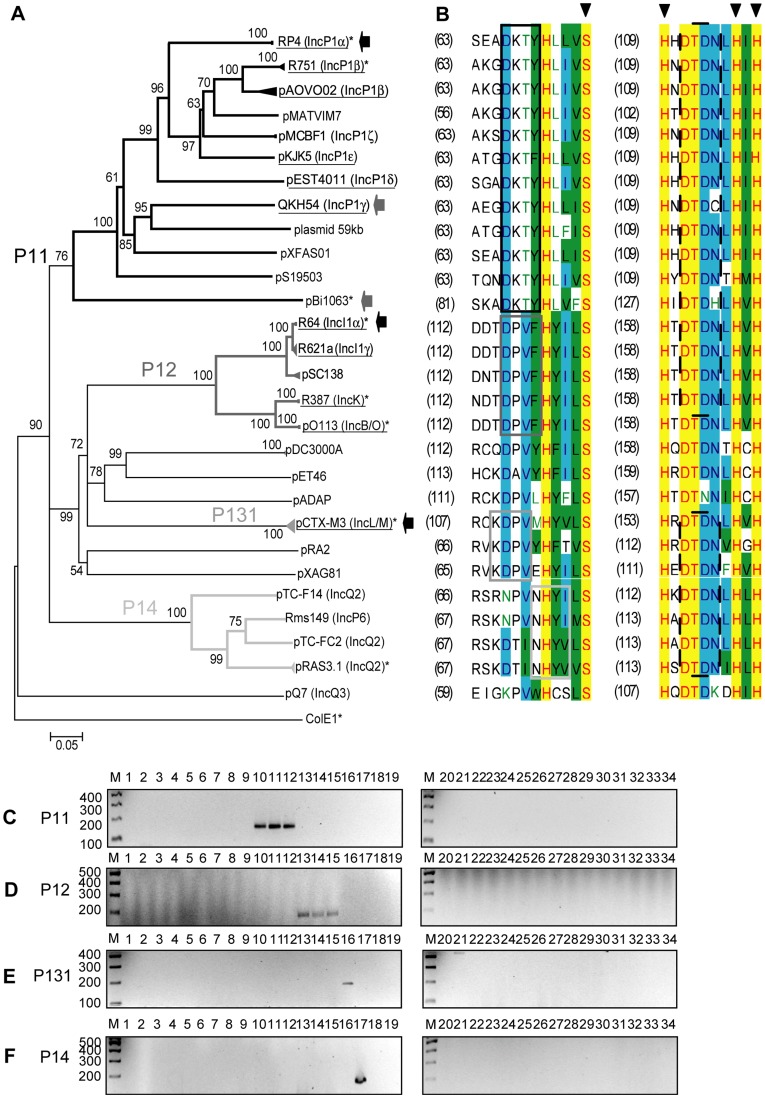
Figure 2. DPMT validation for MOBP1 relaxases.
A) Phylogenetic tree of MOBP1 relaxases. B) Alignment of the relaxase motifs used to design the MOBP1 degenerate primers (P11-f, continuous black; P12-f, continuous dark grey; P131-f, continuous grey; P14-f, continuous light grey; and P1-r, dashed black). C) Amplicons obtained with primers for subfamily MOBP11 (P11-f and P1-r). D) Amplicons obtained with primers for subfamily MOBP12 (P12-f and P1-r). E) Amplicons obtained with primers for subfamily MOBP13 (P131-f and P1-r). F) Amplicons obtained with primers for subfamily MOBP14 (P14-f and P1-r). Symbols, colour codes and lanes as in Figure 1.
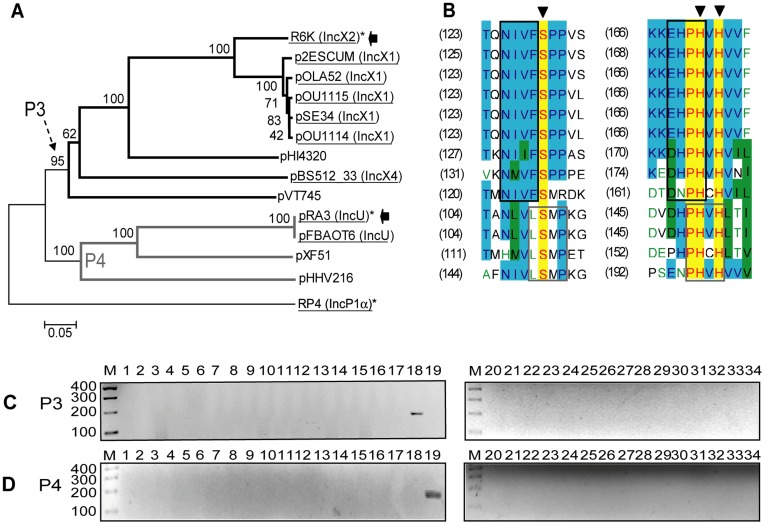
Figure 3. DPMT validation for MOBP3 and MOBP4 relaxases.
A) Phylogenetic tree of MOBP3 and MOBP4 relaxase families. B) Alignment of the relaxase motifs used to design the MOBP3 and MOBP4 degenerate primers (P3-f+P3-r, continuous black; and P4-f+P4-r, continuous dark grey). C) Amplicons obtained with primers for subfamily MOBP31 (P3-f and P3-r). D) Amplicons obtained with primers for subfamily MOBP42 (P4-f and P4-r). Symbols, colour codes and lanes as in Figure 1.
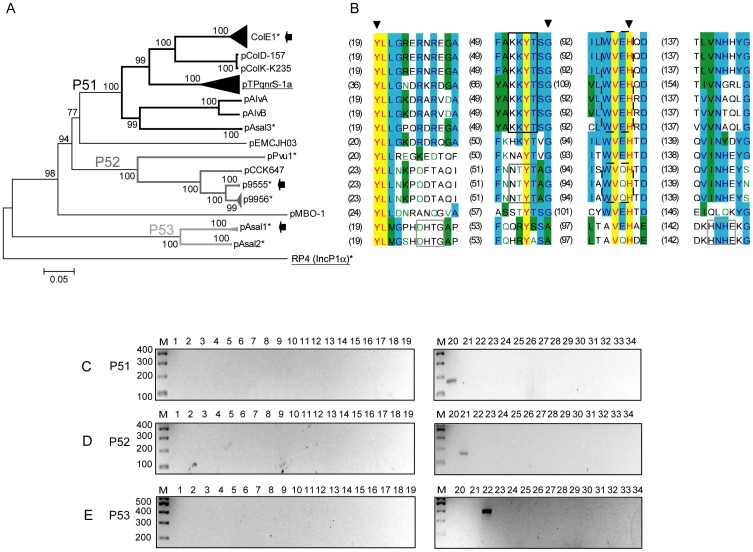
Figure 4. DPMT validation for MOBP5 relaxases.
A) Phylogenetic tree of MOBP5 relaxase family. B) Alignment of the relaxase motifs used to design the MOBP5 degenerate primers (P51-f, continuous black; P52-f, continuous dark grey; P5-r, dashed black; and P53-f+P53-r, continuous grey) C) Amplicons obtained with primers for subfamily MOBP51 (P51-f and P5-r). D) Amplicons obtained with primers for subfamily MOBP52 (P52-f and P5-r). E) Amplicons obtained with primers for subfamily MOBP53 (P53-f and P53-r). Symbols, colour codes and lanes as in Figure 1.
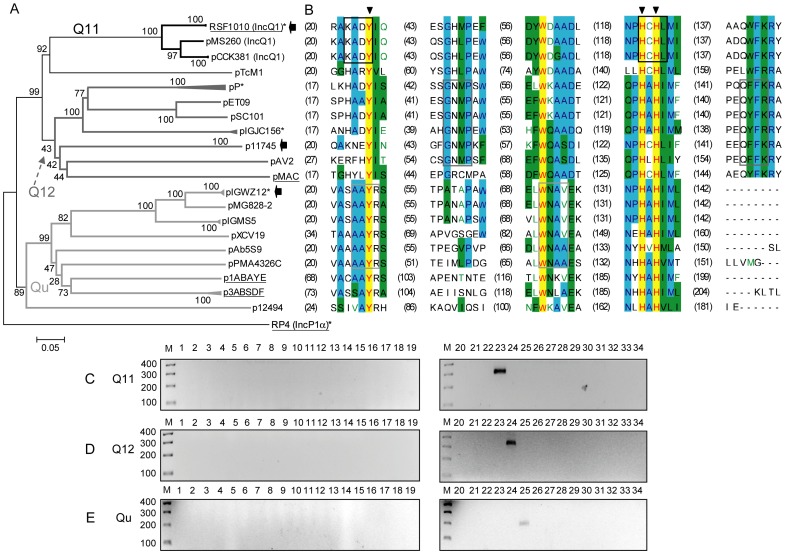
Figure 5. DPMT validation for MOBQ relaxases.
A) Phylogenetic tree of MOBQ relaxase family. B) Alignment of the relaxase motifs used to design the MOBQ degenerate primers (Q11-f+Q11-r, continuous black; Q12-f+Q12-r, continuous dark grey; and Qu-f+Qu-r, continuous grey). C) Amplicons obtained with primers for subfamily MOBQ11 (Q11-f and Q11-r). D) Amplicons obtained with primers for subfamily MOBQ12 (Q12-f and Q12-r). E) Amplicons obtained with primers for subfamily MOBQu (Qu-f and Qu-r). Symbols, colour codes and lanes as in Figure 1.
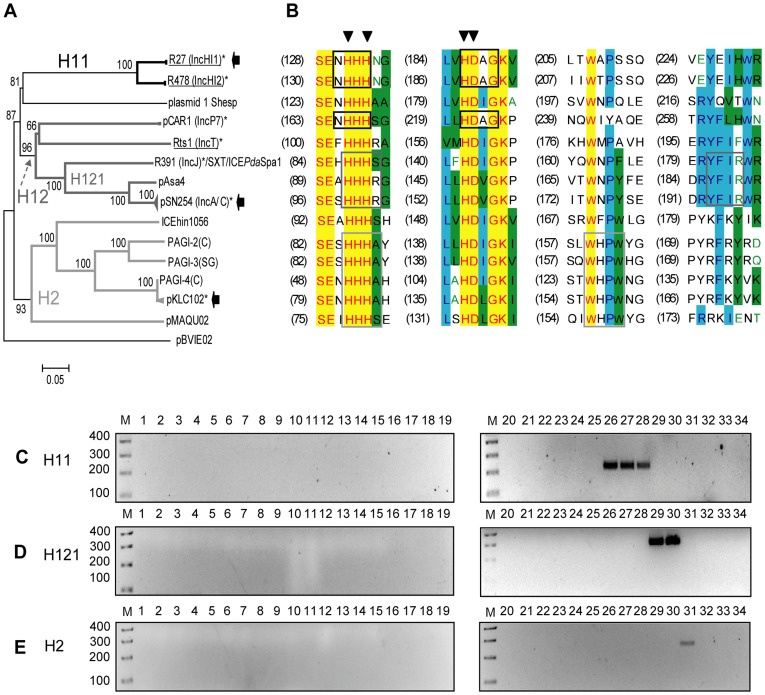
Figure 6. DPMT validation for MOBH relaxases.
A) Phylogenetic tree of MOBH relaxase family. B) Alignment of the relaxase motifs used to design the MOBH degenerate primers (H11-f+H11-r, continuous black; H121-f+H121-r, continuous dark grey; and H2-f+H2-r, continuous grey). C) Amplicons obtained with primers for subfamily MOBH11 (H11-f and H11-r). D) Amplicons obtained with primers for subfamily MOBH121 (H121-f and H121-r). E) Amplicons obtained with primers for subfamily MOBH2 (H2-f and H2-r). Symbols, colour codes and lanes as in Figure 1.
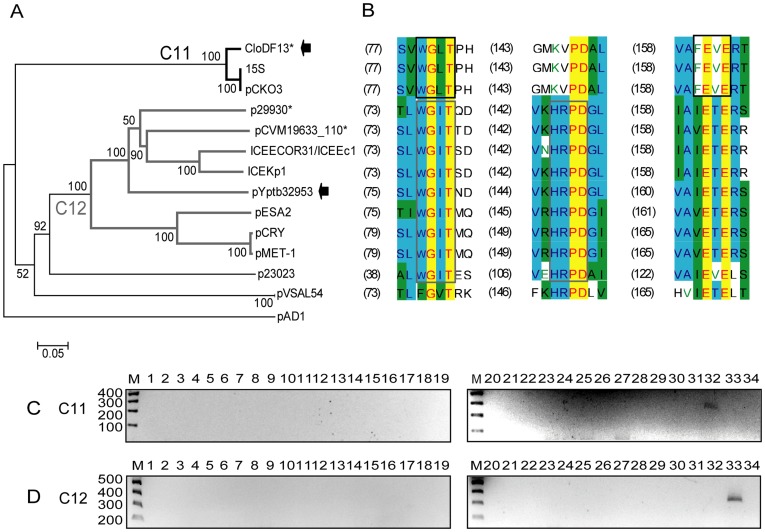
Figure 7. DPMT validation for MOBC relaxases.
A) Phylogenetic tree of MOBC relaxase family. B) Alignment of the relaxase motifs used to design the MOBC degenerate primers (C11-f+C11-r, continuous black; C12-f+C12-r, continuous dark grey). C) Amplicons obtained with primers for subfamily MOBC11 (C11-f and C11-r). D) Amplicons obtained with primers for subfamily MOBC12 (C12-f and C12-r). Symbols, colour codes and lanes as in Figure 1.
Table 1. Plasmids and relaxase genes used as controls in validation experiments.
| Plasmid | Cloned gene | Plasmid accession number a | Position b | Inc Group3 | MOB Subfamily4 | Reference |
| pSU1588 | trwC_R388 | BR000038 | 14128–15007* | IncW | F11 | [79] |
| pSU4280 | pKM101 complete MOB region | U09868 | 14810–20208* | IncN | F11 | [80] |
| pSU10013 | traC_pBi709 | AY299015 | 17902–18771* | – | F11 | This study |
| pSU10014 | traC_Pwwo | NC_003350 | 98516–99385 | IncP-9 | F11 | This study |
| pSU10017 | traI_F | NC_002483 | 92673–93590 | IncFI | F12 | This study |
| pSU10018 | traI_R100 | NC_002134 | 78466–79401 | IncFII | F12 | This study |
| pSU10021 | traI_pSLT | NC_003277 | 87282–88199 | IncFII | F12 | This study |
| pSU316 | – | M26937, X55894, M28097 | – | IncFIII-IncFIV | F12 | [81] |
| pSU10022 | traI_pED208 | AF411480 | 25650–26552 | IncFV | F12 | This study |
| pSU10010 | traI_RP4 | X54459 | 3389–4198* | IncP-1α | P11 | This study |
| R751 | – | NC_001735 | – | IncP-1β | P11 | |
| pSU10028 | traI_pBI1063 | AY299014 | 3848–4675* | – | P11 | This study |
| pSU10029 | nikB_R64 | NC_005014 | 67391–68350 | IncI1 | P12 | This study |
| pSU10056 | nikB_ R387 | M93063, X07848 | – | IncK | P12 | This study |
| pSU10055 | nikB_pO113 | NC_007365 | 62419–63393* | IncB/O | P12 | This study |
| pSU10001 | nikB_pCTX-M3 | NC_004464 | 32027–33049 | IncL/M | P131 | This study |
| pSU10012 | mobA_pRAS3.1 | NC_003123 | 10571–11395* | IncQ2 | P14 | This study |
| pSU10011 | taxC_R6K | Y10906, X95535 | – | IncX2 | P31 | This study |
| pSU10009 | nic_pRA3 | NC_010919 | 10360–11355 | IncU | P42 | This study |
| pSU4601 | ColE1::kan | NC_001371 | – | ColE1 | P51 | [82] |
| pSU10006 | mobA_p9555 | NC_010069 | 3368–4394 | – | P52 | This study |
| pSU10007 | mobA_pAsal1 | NC_004338 | 1052–2017 | – | P53 | This study |
| pSU10064 | mobA_RSF1010 | NC_001740 | 3250–3807 | IncQ1 | Q11 | This study |
| pSU10059 | ORF1_pP | NC_003455 | 9–1244 | – | Q12 | This study |
| pSU10008 | mobA/mobL_pIGWZ12 | NC_010885 | 1257–2240* | – | Qu | This study |
| pSU10039 | traI_R27 | NC_002305 | 106098–106934* | IncHI1 | H11 | This study |
| pSU10040 | traI_R478 | NC_005211 | 192385–193308 | IncHI2 | H11 | This study |
| pSU10041 | traI_pCAR1 | NC_004444 | 124079–125008 | IncP-7 | H11 | This study |
| pSU10004 | traI_pSN254 | NC_009140 | 46409–47593 | IncA/C | H121 | This study |
| pSU10003 | traI_R391 | AY090559 | 32341–33509 | IncJ | H121 | This study |
| pSU10043 | traI_2_pKLC102 | AY257538 | 99952–100788 | – | H2 | This study |
| pSU4830 | mobC_CloDF13 | NC_002119 | – | – | C11 | [83] |
| pSU10002 | triL_p29930 | AJ519722 | 31361–32107 | – | C12 | This study |
Accession number of the transmissible plasmid encoding the corresponding relaxase gene.
Nucleotide coordinates of the cloned relaxase fragment in the accession number of the original plasmid. An asterisk indicates that the relaxase gene is coded in the complementary strand of the original plasmid sequence.
Incompatibility group of the wt plasmid.
MOB subfamily of each relaxase gene.
MOBF family
Figure 1A shows the phylogenetic reconstruction of MOBF relaxases from γ-proteobacterial plasmids. Two subfamilies contain most MOBF relaxases found in clinically relevant plasmids. Subfamily MOBF11 includes, among others, relaxases of AbR plasmids from Inc groups W, N as well as metal-resistance and xenobiotic-biodegradation plasmids of Pseudomonas group IncP-9. Subfamily MOBF12 contains relaxases of AbR and virulence plasmids of the IncF complex (IncFI, IncFII, IncFIII and IncFV) and Inc9 (also known as com9), widely distributed among different genera of Enterobacteriaceae. Specific amplification of MOBF11 and MOBF12 plasmids was obtained with two forward primers (F11-f and F12-f) and one reverse primer (F1-r) (Table 2, Figure 1B–D). Since both forward primers differ only by a single nucleotide, cross-amplification was occasionally observed between MOBF11 and MOBF12 relaxases. Thus, the two amplification reactions identified the most relevant MOBF plasmids but did not discriminate among them.
Table 2. List of degenerate primers used for DPMT.
| Primer name | Primer sequence a | PCR conditions | Prototype b | Amplicon size (bp) c | Amplicon location d |
| F11-f | gca gcg tat tac ttc tct gct gcc gay gay tay ta | 25 cycles, 53°C | R388 | 234 | 13876–14047* |
| F1-r | act ttt ggg cgc gga raa btg sag rtc | ||||
| F12-f | agc gac ggc aat tat tac acc gac aag gay aay tay ta | 25 cycles, 55°C | F | 234 | 92744–92912 |
| F1-r | act ttt ggg cgc gga raa btg sag rtc | ||||
| P11-f | cgt gcg aag ggc gac aar acb tay ca | 25 cycles, 60°C | RP4 | 180 | 50361–50484* |
| P1-r | agc gat gtg gat gtg aag gtt rtc ngt rtc | ||||
| P12-f | gca cac tat gca aaa gat gat act gay ccy gtt tt | 30 cycles, 53.8°C, 1.5U Taq per reaction | R64 | 189 | 67744–67867 |
| P1-r | agc gat gtg gat gtg aag gtt rtc ngt rtc | ||||
| P131-f | aac cca cgc tgc aar gay ccv gt | 30 cycles, 59°C, 15 seconds of extension per cycle | pCTX-M3 | 180 | 32365–32491 |
| P1-r | agc gat gtg gat gtg aag gtt rtc ngt rtc | ||||
| P14-f | cgc agc aag gac acc atc aay cay tay rt | 25 cycles, 50°C | pRAS3.1 | 174 | 11053–11169* |
| P1-r | agc gat gtg gat gtg aag gtt rtc ngt rtc | ||||
| P3-f | cc gtg agc caa atc aca cag aat atk rtb tt | 25 cycles, 50°C | R6K | 177 | 38419–39573* |
| P3-r | cg aaa gcc aac atg aac atg hgg atk htc | ||||
| P4-f | gcg ttc agg atg gtc ytb tcs atg cc | 25 cycles, 64°C | pRA3 | 163 | 10695–10803 |
| P4-r | c ggt ttt gac cgt cag atg svm atg cgg | ||||
| P51-f | t acc acg ccc tat gcg aar aar tay ac | 30 cycles, 58°C, 20 seconds of extension per cycle | ColE1 | 167 | 572–688 |
| P5-r | cc ctt gtc ctg gtg yts nac cca | ||||
| P52-f | gat agc ctt gat ttt aat aac acc aay acy tay ac | 30 cycles, 58°C, 20 seconds of extension per cycle | p9555 | 175 | 3536–3652 |
| P5-r | cc ctt gtc ctg gtg yts nac cca | ||||
| P53-f | g ggc tcg cac gay cay acn gg | 30 cycles, 65°C | pAsal1 | 345 | 1136–1480 |
| P53-r | gc cca gcc ctt ttc rtg rtt rtg | ||||
| Q11-f | caa tcg tcc aag gcg aar gcn gay ta | 30 cycles, 50°C | RSF1010 | 331 | 3325–3606 |
| Q11-r | cg ctc gga gat cat cay ytg yca ytg | ||||
| Q12-f | ctg gaa tat act gaa cac ggn aay atg cc | 30 cycles, 52°C | pP | 341 | 975–1256 |
| Q12-r | atc ctt ggt gtt agc acg ttt raa rwa ytg | ||||
| Qu-f | agc gcc gtg ctg tcc gcb gcn tay cg | 30 cycles, 64°C | pIGWZ12 | 179 | 2034–2162* |
| Qu-r | ctc cgc agc ctc grc sgc rtt cca | ||||
| H11-f | ccg gcg tcg gag aay cay cay ca | Touchdown PCR: start at 65°C ΔTa = −1°C per cycle, 15 cycles at 55°C | R27 | 207 | 106380–106536 |
| H11-r | aag gtc gta tac ctt ycc kgc rtc rtg | ||||
| H121-f | g cca gct tcc gaa tca cay cay cay cg | 25 cycles, 59°C | pSN254 | 313 | 46714–46981 |
| H121-r | g tcg ctt gtc gcg cca ccg dat raa rta | ||||
| H2-f | ag ttc cca gcc tca gaa atc cay cay cay kc | 25 cycles, 68°C | pKLC102 | 264 | 100218–100428 |
| H2-r | g cgg acc gtg cca ngg rtg cca | ||||
| C11-f | gt cag gtc agc gtg tgg ggn ctn ac | Touchdown PCR: start at 65°C ΔTa = −1°C per cycle, 20 cycles at 55°C | CloDF13 | 283 | 2874–3106 |
| C11-r | ct ctt cac ggt gcc ctc nac ytc raa | ||||
| C12-f | gc acg act gga aaa ata tcg cta tgg ggn ath ac | 30 cycles, 59° | p29930 | 257 | 31594–31789 |
| C12-r | caa cgt gat aat ccc gtc rgg vcg rtg |
For each oligonucleotide, CORE nucleotides are in bold and CLAMP sequences in normal lettering. Underlined codons do not encompass all the possible variability to avoid excessive degeneracy. The sequences used are biased to accommodate the DNA sequences of existing elements.
Prototype plasmid for the given MOB subfamily.
Amplicon size obtained from the prototype plasmid relaxase gene.
Nucleotide coordinates of the prototype plasmid contained in the corresponding amplicon. An asterisk indicates that the relaxase gene is encoded in the complementary strand.
MOBP family
Within γ-Proteobacteria, MOBP contains relaxases of AbR plasmids belonging to the IncP1 complex (IncP1α, IncP1β, IncP1δ, IncP1γ, IncP1ε, and IncP1ζ), many of them recovered from soil and manure isolates [21], virulence and AbR plasmids of the IncI complex (IncI1α, IncI1γ, IncK, IncB/O), AbR plasmids IncL/M, IncQ2 (IncQ2α, IncQ2β, IncG/IncP-6, IncX1, IncX2, IncU and IncQ3 groups, plus several other branches that contained no Inc prototype. The ample diversity of this family was reflected in the MOBP phylogeny, which showed several well-resolved monophyletic groups, as well as additional, poorly-defined deep branches [35]. Thus, to construct the set of MOBP primers we had to manage each subfamily separately. Relaxases of IncP1α, IncP1β, IncP1δ, IncP1γ, IncP1ε, IncP1ζ, IncI1α, IncI1γ, IncK, IncB/O, IncL/M, IncQ2α, IncQ2β and IncG/IncP-6 plasmids -among others without Inc assignment- are grouped in the MOBP1 subgroup (Figure 2A); those of IncX1 and IncX2 plasmids are in group MOBP3 (Figure 3A); IncU plasmid relaxases are in group MOBP4 (Figure 3A), and relaxases of ColE1-related plasmids in MOBP5 (Figure 4A). Neither subfamily MOBP6, which contains a scarce number of γ-Proteobacteria relaxases (including those in IncI2 plasmids), nor other poorly resolved clades (as the one containing IncQ3 plasmids), were considered in this study.
MOBP1 subfamily
One reverse and four forward primers were needed for amplification of MOBP1 relaxases (Figure 2B, Table 2). The P11-f forward primer led to amplification of MOBP11 plasmids (including IncP1). Similarly, the P12-f forward primer identified MOBP12 plasmids (including IncI1, IncK, and IncB/O), P131-f forward primer identified MOBP13 plasmids (including IncL/M), and P14-f forward primer identified MOBP14 plasmids (including IncQ2 and IncG). Results are shown in Figure 2C–F. No cross-amplification was observed, except for P131-f + P1-r when using plasmid p9555 as template (Figure 2E). The non-specific amplicon was larger than that obtained from the reference MOBP131 relaxase gene nikB_pCTX-M3, so the interpretation of the data was unambiguous.
MOBP3 and MOBP4 subfamilies
MOBP3 relaxases correspond to IncX1 and IncX2 plasmids while MOBP4 contains relaxases of IncU plasmids (Figure 3A and Table S1). One primer pair was designed for each subfamily. No cross-amplification was observed (Figure 3C–D), except for the fortuitous amplification of some Salmonella chromosomes described in Methods, subsection “Validation and methodologies comparison”.
MOBP5 subfamily
Most MOBP5 (ColE1-like) relaxases lack the canonical 3H motif III, but contain a deviant HEN motif [41] (Figure 4B). Three primer pairs (P51, P52 and P53, Table 2) were designed to amplify this cluster (Figure 4), two pairs specific for plasmids with a HEN motif (P51 and P52) and one for plasmids with the 3H motif (P53).
MOBQ family
Phylogenetic reconstruction of γ-proteobacterial MOBQ relaxases showed two distinguishable MOBQ clades, MOBQ1 and MOBQu (Figure 5A). For amplifying the first broad clade, two primer pairs were designed, Q11 and Q12, and one primer pair, Qu, for the MOBQu cluster (Figure 5B, Table 2). Some phylogenetic overlapping between MOBQ and MOBP families has been reported [35]. Nevertheless, primers that hit each relaxase branch did not cross-amplify (Figure 5C–E).
MOBH family
MOBH relaxases are encoded by AbR IncHI1, IncHI2, IncA/C, IncT, and xenobiotic-biodegradation Pseudomonas P7 plasmids, as well as by some ICEs (e.g. R391/SXT-like, clc, PAPI-1, etc.) (Figure 6A, Table S1). R391-like elements, exhibiting incompatibility properties, were formerly considered as plasmids and classified as IncJ [42], [43]. MOBH relaxases have, besides the canonical conserved regions, additional motifs related to HD-hydrolases [44]. Three primer pairs were used to amplify MOBH relaxases (Figure 6C–D, Table 2): H11 (specific for IncHI1, IncHI2 and P-7 plasmids, represented by R27, R478 and pCAR1 respectively), H121 (amplifying IncA/C and R391-like elements, represented by pSN254 and R391 respectively) and H2 (amplifying a large set of relaxases from a family of ICEs, like pKLC102).
MOBC family
All MOBC relaxases encoded in γ-proteobacterial plasmids cluster in a single clade, MOBC1, when outgrouping with Firmicutes/Tenericutes MOBC relaxases (Figure 7A, Table S1). MOBC relaxases present in ICEs, such as ICEKp1 and ICEEc1 also cluster in clade C1. MOBC is a peculiar relaxase family that does not contain the three classical signature motifs present in all other MOB families. Two primer pairs were designed to amplify each MOBC1 subclade: C11 and C12 (Figure 7 B–D, Table 2).
Analysis of Clinical Plasmid Collections Using DPMT
Once validated by testing the reference collection of relaxases (Table 1), the set of 19 primer pairs was used to screen two plasmid collections from clinical samples as test cases (Table 3).
Table 3. Relaxases found in two test collections.
| Test collection a | MOBF | MOBP | MOBQ | MOBH | MOBC | Total | ||||||||||||||
| F11 | F12 | P11 | P12 | P13 | P14 | P3 | P4 | P51 | P52 | P53 | Q11 | Q12 | Qu | H11 | H121 | H2 | C11 | C12 | ||
| 1 | 14 | 60 | 4 | 39 | 6 | 0 | 3 | 0 | 71 | 0 | 0 | 0 | 5 | 11 | 13 | 10 | 0 | 0 | 1 | 237 |
| 2 | 0 | 30 | 2 | 6 | 0 | 0 | 7 | 0 | 18 | 0 | 0 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 6 | 77 |
| Total | 14 | 90 | 6 | 45 | 6 | 0 | 10 | 0 | 89 | 0 | 0 | 0 | 5 | 15 | 14 | 10 | 0 | 3 | 7 | 314 |
Isolate collections analyzed with DPMT.
Test collection 1 consisted of 135 isolates of Enterobacteriaceae, recovered in different countries (Canada, Portugal, Spain, France and Kuwait) from 1989 to 2008, and producing extended spectrum beta-lactamases (ESBL). 104 of them were E. coli transconjugants harbouring ESBL-coding plasmids from different Enterobacteriaceae donors while the remaining 31 were original donors unable to conjugate the ESBL determinant. The collection mainly included plasmid-encoded ESBLs from class A (SHV (4/135; [45], TEM (18/135; [46], [47], [48]) and CTX types (91/135; [45], [46], [47], [49], [50], [51], [52]). A total of 237 relaxases were identified in the 135 strains, distributed among the five MOB families targeted by the primer set. The resulting amplicons were sequenced. Out of 237 sequenced amplicons, only five corresponded to relaxase sequences not previously reported (we consider a relaxase new when it shows less than 95% amino acid sequence identity with the closest hit in the NCBI nr database). Two of them, corresponding to plasmids pAA-TC1-69 and pAA-TC1-30a (GenBank Accession numbers JN167247 and JN167248), respectively exhibited 62% and 64% amino acid identity to the MOBF11 relaxase of plasmid pCT14 (nearest hit). Two others, those of plasmids pAA-TC1-79a and pAA-TC2-33a, were 78% identical to R46 relaxase (details in Information S1), suggesting overall more diversity within the MOBF11 relaxase branch than anticipated from the analysis of present genome databases. Complete sequencing of the relaxase domain of these plasmid genes and the ensuing phylogenetic analysis classified them as well defined new branches in the MOBF11 phylogeny (incorporated to Figure 1 in red color). Similarly, a fifth relaxase, that of plasmid pAA-TC1-14a, was 87% identical to pKPN4 relaxase and was classified as MOBF12 (see Information S1). The finding of these five new relaxase sequences underscores the potency of DPMT to detect and classify plasmids unidentifiable by PBRT. The most represented MOB subfamilies in Test Collection 1 were MOBP5 (71 relaxases), MOBF12 (60), and MOBP12 (39), followed by MOBH (23), MOBQ (16) and MOBF11 (14). Finally, 7 out of 135 isolates, corresponding to transconjugants, did not render any relaxase amplicon. Since they probably code for relaxases of MOB subfamilies not considered in this work or new deviant relaxases, they were selected for complete sequencing and further investigation (work in progress).
Test collection 2 comprised E. coli isolates from urine cultures of Swedish women who suffered from uncomplicated, community-acquired urinary tract infections treated with pivmecillinam [53]. The isolates were assorted according to their PFGE profiles (Ellen Zechner, personal communication). We analyzed 49 representative isolates for the presence of relaxases using the same set of 19 MOB primer pairs. 30 out of the 49 primary strains gave positive amplification with at least one primer pair. The 19 isolates without positive DPMT results were used as donors in mating experiments. Transconjugants were obtained for 18 of them by using a battery of antibiotic resistances matching the donor AbR profiles. Selected transconjugants were tested again with the same set of primers. 13 out of 18 rendered amplicons with at least one primer pair, while five transconjugants remained unidentifiable. A total of 77 relaxase amplicons were obtained from the collection. 50 of them were sequenced, from which two corresponded to non-previously reported relaxase sequences; one MOBP12, pAA-A3201, was 80% identical to pO113 relaxase; and one MOBQu, pAA-A3488, was 72% identical to pSMS35_4 relaxase (see Information S1). Finally, a third relaxase, pAA-A3180 (Accession number JN167246), showed 97% amino acid identity to MOBF12 plasmid R1 relaxase. In summary, the analysis of this second collection identified two new relaxase sequences, representing in turn new branches in the MOB family trees. The most abundant MOB family was MOBP with 31 relaxases (18 belonging to subfamily MOBP5, 7 to MOBP3 and 6 to MOBP12), followed by MOBF, with 30 amplicons, all members of subfamily MOBF12. It is worth mentioning that the identification of 4 MOBQu and 9 MOBC plasmids of this collection would have not been possible by using the available PBRT or Inc/Rep-HYB probes.
Discussion
PBRT typing methods significantly improved the assignment of plasmids to Inc groups without the need to test for plasmid incompatibility despite some drawbacks like cross-hybridization between members of closely related Inc groups (such as IncI, IncK and IncB/O [18], [24]), false negative PCR results obtained when classifying more divergent plasmid groups (e.g. IncL/M [24]), and poor coverage of some groups (e.g. IncA/C [54], and ColE1-like [25]). PBRT identifies plasmids that belong to well-defined Inc groups. Nevertheless, a relevant part of the existing plasmid diversity, found in different ecological niches [55], [56], [57], [58], [59] that includes clinical settings [60], [61], remains elusive to PBRT classification (see Figure 8). In order to capture a broader range of plasmids, we considered groups of evolutionary related plasmid sequences instead of focussing on single sequences as PBRT usually does. Therefore, our set of primer pairs was not mainly designed to be used for screening purposes, but for the discovery of new relaxases and thus to expand and better delimit the known MOB subfamilies.
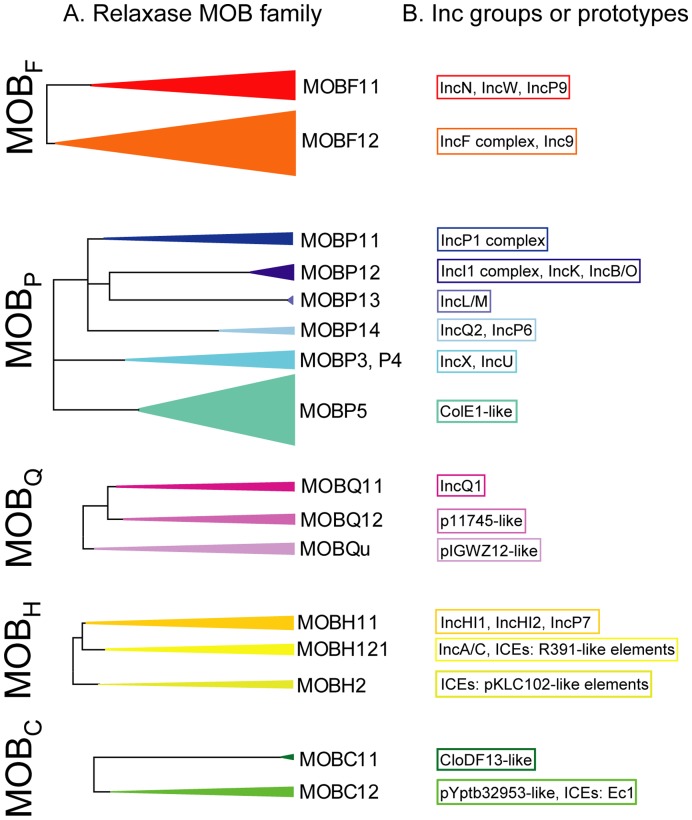
Figure 8. Correspondence between MOB and Rep types.
A) Simplified phylogenetic representation of the five relaxase MOB families considered in this study. Coloured triangles represent the MOB subfamilies amplified by DPTM. Their width and depth correspond, respectively, to the abundance and phylogenetic diversity of their relaxase sequences (Table S1). B) The Inc groups contained within each MOB subfamily are indicated at the right, boxed in the same colour. When no Inc group is contained, the name of a prototype plasmid is given.
For the purpose of discovering new relaxases, we took advantage of the phylogenetic studies carried out with relaxase sequences [35], [36], [62], [63]. According to their relaxase genes, plasmids belong to six MOB families: MOBF, MOBP, MOBQ, MOBH, MOBC, and MOBV [35], [62]. Within each MOB family, the different taxa form groups with high amino acid sequence identity that allow us to define robust phylogenetic branches. Due to the clinical and epidemiological importance of γ-Proteobacterial plasmids, the phylogenies of relaxase subfamilies of plasmids hosted in γ-Proteobacteria were updated for this work (Figures 1 to 7). All in all, 17 subfamilies contained most of the diversity found in γ-Proteobacterial plasmids (summarized in Figure 8): subfamilies F11 and F12 from MOBF (Figure 1); P11, P12, P13, P14, P3, P4 and P5 from MOBP (Figures 2, 3, 4); Q11, Q12 and Qu from MOBQ (Figure 5); H11, H12, and H2 from MOBH (Figure 6); and C11 and C12 from MOBC (Figure 7). Plasmids detected by PBRT are almost always included in these subfamilies (correspondences in Figure 8). The only exceptions are groups known not to contain relaxases (IncR, GR1/GR12, GR2/GR10, GR13, GR14, and GR17, whose prototype plasmids are pK245, pABSDF, pACICU1, p3ABAYE, p4ABAYE, and pAB1, respectively) or groups which do not contain any fully sequenced member or no relaxase in the known sequences (IncY (plasmid P1), IncFVI (pSU212), IncFVII (pSU221), GR3 (p736), GR4 (p844), GR5 (p537), GR8 (p11921), and GR16 (pAB49)). Families and subfamilies that contain only a few plasmid relaxases from γ-Proteobacteria, such as MOBV, MOBP6 (containing IncI2 plasmids), and some other poorly resolved clades (e.g. IncT and IncQ3 plasmids), were not considered in this study.
A computational protocol to search for conjugative and mobilizable genetic modules in a set of 1,730 completely sequenced plasmids recorded in the NCBI database, detected a relaxase in 260 out of the 503 plasmids hosted in γ-Proteobacteria [62]. We used that plasmid set to compare the detection capabilities of the available PBRT and DPMT probes (Table S1). Our set of 19 degenerate primer pairs was potentially able to detect 193 out of the 271 relaxases contained in the 260 transmissible γ-proteobacterial plasmids, that is, it would allow the classification of 186 out of these plasmids. Available PBRT probes (58 primer pairs) could potentially detect 153 plasmids in the total set, of which 98 were contained in the transmissible plasmid set. 87 out of 260 transmissible plasmids could be potentially detected by both PBRT and DPMT probes. This comparison suggests that DPMT is a powerful tool to detect and phylogenetically classify γ-proteobacterial transmissible plasmids.
A reference collection of 33 relaxases, containing representatives of the main MOB subfamilies, was used to test for specific amplification of the chosen primer pairs (Table 1). With few exceptions (see sections MOBF family and MOBP1 subfamily), no cross-amplification between MOB subfamilies was observed. Several DPMT primer pairs have already been successfully used conjointly with PBRT for identifying plasmids from clinical strains [47], [64], [65]. In this work we analyzed two enterobacterial plasmid collections by DPMT, capturing not only the known Inc plasmid groups but also a number of others undetected by PBRT, some of which contained new relaxase sequences. The DPMT method only failed to identify a MOB relaxase in 12 out of 122 transconjugants from these collections. Failure to find a relaxase in an experimentally verified transconjugant could be attributed to: i) the sequence bias introduced in some primers to avoid high degeneracy (see Table 2), ii) the presence of relaxases belonging to subfamilies not included as targets by our primer set, or iii) the existence of relaxases whose sequences could be largely deviant from the subfamily consensus. In any case, the results presented in this work suggest that the present implementation of the DPMT method identifies more than 90% of the transmissible R-plasmids in transconjugants of clinical isolates. Once less-populated or poorly-resolved relaxase phylogenetic clades become more robust by accretion of further data, our method could be expanded to allow the identification of a higher proportion of relaxases. Our ongoing work aims to do so, with the collaboration of a number of clinical research groups in Spain and Europe.
Detection of transmissible plasmids by PBRT and DPMT underscores their complementarities in focus and scope. While PBRT focuses in replication or partition regions shared by clusters of highly-related plasmids (>95% nucleotide identity), DPMT targets relaxase motifs conserved in large groups of plasmids with deep phylogenetic diversity. As shown in Results, we can detect relaxases with as little as 60% amino acid sequence identity to the nearest known hit in the databases. Thus, PBRT is useful at detecting blooms of redundant backbones that carry different cargo genes (“zoom in” strategy), while DPMT finds and classifies backbones that share a common relaxase ancestor (“zoom out” strategy). Most PBRT primers were designed for detecting plasmids from Enterobacteriaceae [24], [27], although there are a few available for detection of plasmids from other taxonomic families of γ-Proteobacteria, such as IncP-1 [19], [20], [21], IncP-9 [22], [23], or Acinetobacter baumannii replicons [29]. The vast diversity in the plasmid world makes the design of probes that target small groups of highly-related plasmids a strategy limited in practical terms for specific purposes, not suitable for studying global diversity neither for finding deviant plasmids from well-studied backbones. The DPMT strategy is more inclusive, allowing the detection of plasmids hosted by a larger number of taxonomic families. Nevertheless, it should be emphasized that it still recovers a higher proportion of plasmids from Enterobacteriaceae (85%) than from other γ-Proteobacterial families (51.4%). This is mostly due to the lack of a suitable number of related relaxase sequences to construct robust phylogenetic trees, as exemplified, for instance, by the Moraxellaceae, Vibrionaceae, Pseudomonadaceae and Aeromonadaceae plasmids [35], [62]. Perhaps investigators in public health surveillance, veterinary or environmental science should consider the interest of developing sets of oligonucleotide pairs more specifically adapted to their needs. Most clinically relevant transmissible plasmids detected by PBRT probes are also uncovered by DPMT, as shown in this work. On the contrary, no PBRT probes are available for many plasmids detected by DPMT such as the virulence plasmids IncFIII/IV (MOBF12), IncQ2 (MOBP14), IncP-7 (MOBH12), and a number of others out of Inc assignment. Of course, results obtained by DPMT can help PBRT to design primers for the assessment of the newly discovered plasmid groups. As an example, the classification of virulence plasmids in the IncF and IncI1 complexes, reviewed by [26], will obviously gain by a joint PBRT+DPMT analysis.
An added advantage of the DPMT method is its applicability in the identification of ICEs (see figures 6 and 7). ICEs are also vehicles that disseminate virulence and AbR genes [66], [67]. They are known to constitute an integral part of most bacterial genomes, outnumbering plasmids by 2 to 1 in sequence databases [63]. ICEs are beginning to be closely linked to some of the more powerful AbR mechanisms such as ESBL, metallo- and AmpC type β-lactamases. For instance, chromosomal MOBH121 (R391-like) elements putatively involved in bla CMY-2 mobilization were detected by DPMT in enterobacterial isolates [64]. The MOB families considered in our primer set are also abundant in ICEs of γ-Proteobacteria [63]. The expanded diversity that DPMT discovered in γ-proteobacterial plasmids (and ICEs) will help to populate poorly solved branches of the existent phylogenetic trees and, therefore, lead to better consensus sequences to improve the design of new primer sets and, eventually, to design a multiplex set of non-degenerate oligonucleotides for faster plasmid screening and identification procedures (work in progress). Additionally, and due to their broad amplification capabilities, the DPMT method could be used in the analysis of plasmids and ICEs in total community DNA. In this case, the DNA fragments obtained from amplification with the 19 DPMT primer pairs could be combined and subjected to deep sequencing methodology. As a result, all amplifying sequences could be identified and quantified, resulting in a quantitative description of the plasmid and ICE composition of the analyzed populations and given environmental conditions.
The analysis of relaxases and replicons of γ-proteobacterial plasmids carried out in this and previous works strongly suggests that there is a high correlation between the MOB and the Inc/Rep group. That is, in a single MOB subfamily, relaxases from different Inc plasmids can be grouped, but plasmids of such Inc groups do not contain relaxases dispersed in different MOB subfamilies. Some exceptions are observed, which can usually be explained by plasmid cointegration and secondary deletions. Thus, DPMT provides not only the relaxase identity but a quick inference of the phylogenetic relationships with other plasmids as well as an idea of the constitution of the plasmid backbone. In summary, the combination of both methods, DPMT and PBRT, could better serve in the identification and characterization of plasmid species which are relevant in human and animal medicine. We hope they will help to inspire more effective clinical and environmental policies to manage the dreadful increase of more virulent and multi-antibiotic resistant human pathogens.
Conclusions
The Degenerate Primer MOB Typing (DPMT) method allows rapid and accurate identification of transmissible plasmids based on their relaxase sequences. It detects a broader range of plasmids than the PCR-based replicon typing (PBRT) method and highlights a significant plasmid diversity that was underestimated. The DPMT method can be useful in the analysis of plasmids from both clinical and environmental isolates. The philosophy that guided the development of the γ-Proteobacteria MOB primer set can be easily extended to encompass relaxases of other taxonomical groups of bacteria.
Methods
Plasmids, Bacterial Strains, Growth Conditions and DNA Extraction
Relaxases representing five out of six MOB families described in Garcillán-Barcia, 2009 (MOBF, MOBP, MOBQ, MOBH, and MOBC) were used as standards for DPMT validation. MOBV relaxases were not included since they are barely represented in γ-Proteobacteria. The resulting reference collection included six conjugative or mobilizable plasmids and 27 recombinant plasmids containing cloned relaxase genes (Table 1). For their construction, relaxase domains were delimited by using PSIpred (http://bioinf.cs.ucl.ac.uk/psipred/) [68], [69] and GOR (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) [70]. Relaxase domains contained approximately the 300 N-terminal amino acids of these large multidomain proteins. Gene segments amplified by PCR were cloned either in the NdeI or NdeI/BamHI sites of vector pET3a (Novagen) or in the NcoI/BamHI sites of vector pET3d (Novagen), and introduced in E. coli DH5α by electroporation. Host strains were grown in Luria-Bertani broth (LB) in the presence of suitable antibiotics for plasmid selection. Total DNA was obtained using InstaGene Matrix (BioRad Laboratories), according to the manufactureŕs recommendations and starting from 100 µl saturated cultures.
Bacterial Matings
Donors (E. coli primary isolates) and recipients (either DH5α [71] or HMS174 [72]) were grown to saturation, mixed in ratio 1∶1 and mated o/n on LB-agar plates at either 30°C or 37°C. Cells were resuspended in LB and dilutions plated on appropriate antibiotics (recipient marker + plasmid marker) to select for transconjugants. Nalidixic acid (20 µg/ml) was used to select for DH5α and rifampicin (50 µg/ml) for HMS174.
Database Search
PSI-Blast [73] searches for relaxases were carried out using the N-terminal 300 amino acids of each MOB family prototype, following the method described in [36] and [35], but querying databases only for the subset of plasmids originally isolated from γ-Proteobacteria.
PCR Primer Design
For each MOB family, relaxase domains were aligned and their phylogenetic relationships traced as previously described [35]. For each well-populated and well-resolved subfamily, the corresponding protein alignment was used to find blocks of at least four contiguous, usually invariant amino acids located within or close to the conserved relaxase motifs. Among them, two blocks were finally chosen to design forward and reverse primers for each subfamily. Oligonucleotide pairs were selected that detected most subfamily members while minimizing codon degeneracy and resulting in amplicons smaller than 400 bp. When a single primer pair did not encompass all subfamily members, it was further subdivided (e.g., MOBC1 in C11 and C12). The primer pair for amplifying each MOB family was designed using CODEHOP [74] (Table 2). This strategy was already applied for the identification of DNA sequences of distantly related members of several gene families [75], [76], [77]. In CODEHOP, oligonucleotides derived from the selected blocks contain a 3′ partially-degenerate sequence, called CORE, comprising different codon variants of the highly conserved residues (11 nucleotides); and a 5′ non-degenerate sequence of variable size (around 14 nucleotides, to give a hybridization temperature of 55 to 60°C), called CLAMP, composed of the upstream contiguous nucleotides most conserved in the relaxase DNA alignment.
Validation and Methodologies Comparison
Each primer pair was tested for amplification of the collection of 33 reference plasmids in standard PCR reactions. Each reaction contained PCR buffer (50mM KCl, 10 mM Tris-HCl (pH 8.8), 0.1% Triton X-100), 1.5 mM MgCl2, 0.2 mM dNTP, 1 µM of the corresponding pair of degenerate oligonucleotides, 2–5 µl (0.4–1 µg) of total DNA, and 1 U of BioTaq polymerase (Bioline) in a final volume of 50 µl. Details of amplification conditions for each primer pair are described in Table 2. Generally, the standard PCR protocol involved a 4 min step at 94°C, 25–30 cycles of 30 sec at 94°C, 30 sec at the annealing temperature and 30 sec at 72°C (the extension time had to be varied to adapt to the expected size of some amplicons; see Table 2 for details), and a final extension step for 10 min at 72°C. A touchdown PCR protocol [78] was used for amplification of MOBH11 and MOBC11 groups, to avoid the appearance of aberrant amplification products. It should be noted that the P4 primer pair (Table 2) fortuitously amplified a segment of some Salmonella chromosomes (corresponding to gene fucO, for instance in S. typhimurium DT104), thus impeding relaxase identification in this genomic background. No additional fortuitous amplicons were obtained when using clinical samples from Escherichia, Salmonella or Klebsiella. Amplicons were visualized after 2% agarose gel electrophoresis, using a GelDoc (BioRad Laboratories) and, when appropriate, sequenced by Macrogen Laboratories (Seoul, South Korea).
Supporting Information
Plasmids from γ-Proteobacteria contained in the NCBI database.
(DOC)
Nucleotide sequences and their translated amino acid sequences of relevant relaxases obtained by DPMT from different test collections.
(DOC)
Acknowledgments
We thank Dr. Teresa M. Coque for critical reading of the manuscript, Val Fernández-Lanza for bioinformatics assistance, as well as all providers of plasmids listed in Table 1.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Spanish Ministry of Education (www.mec.es) (BFU2008-00995/BMC), RETICS research network, Instituto de Salud Carlos III, Spanish Ministry of Health (www.msps.es) (RD06/0008/1012) and grant n° 248919/FP7-ICT-2009-4 from the European VII Framework Program (http://cordis.europa.eu/fp7/home_en.html). AA was partially funded by the I Plan Regional de I+D+i de Cantabria (Ref. 20-1-2007). MPGB was funded by a JAE-Doc_2009 postdoctoral contract from Consejo Superior de Investigaciones Científicas (www.csic.es). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 2.Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2009;73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcillan-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev. 2011;35:936–956. doi: 10.1111/j.1574-6976.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- 4.Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin S, Nordstrom K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- 6.Datta N, Hedges RW. Compatibility groups among fi - R factors. Nature. 1971;234:222–223. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]
- 7.Taylor DG, Gibreel A, Lawley TD, Tracz DM. Funnell BE, Phillips GJ, editors. Antibiotic resistance plasmids. 2004. pp. 473–491. Plasmid Biology. Washington, DC: ASM Press.
- 8.Lopez J, Crespo P, Rodriguez JC, Andres I, Ortiz JM. Analysis of IncF plasmids evolution: nucleotide sequence of an IncFIII replication region. Gene. 1989;78:183–187. doi: 10.1016/0378-1119(89)90327-2. [DOI] [PubMed] [Google Scholar]
- 9.Nikoletti S, Bird P, Praszkier J, Pittard J. Analysis of the incompatibility determinants of I-complex plasmids. J Bacteriol. 1988;170:1311–1318. doi: 10.1128/jb.170.3.1311-1318.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sesma A, Sundin GW, Murillo J. Closely related plasmid replicons coexisting in the phytopathogen pseudomonas syringae show a mosaic organization of the replication region and altered incompatibility behavior. Appl Environ Microbiol. 1998;64:3948–3953. doi: 10.1128/aem.64.10.3948-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tietze E. Nucleotide sequence and genetic characterization of the novel IncQ-like plasmid pIE1107. Plasmid. 1998;39:165–181. doi: 10.1006/plas.1998.1343. [DOI] [PubMed] [Google Scholar]
- 12.Praszkier J, Wei T, Siemering K, Pittard J. Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J Bacteriol. 1991;173:2393–2397. doi: 10.1128/jb.173.7.2393-2397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner MN, Rawlings DE. Evolution of compatible replicons of the related IncQ-like plasmids, pTC-F14 and pTF-FC2. Microbiology. 2004;150:1797–1808. doi: 10.1099/mic.0.26951-0. [DOI] [PubMed] [Google Scholar]
- 14.Camps M. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat DNA Gene Seq. 2010;4:58–73. doi: 10.2174/187221510790410822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley DE. Further characterization of R485, and IncX plasmid that determines two kinds of pilus. Plasmid. 1982;7:95–100. doi: 10.1016/0147-619x(82)90031-2. [DOI] [PubMed] [Google Scholar]
- 16.Stalker DM, Helinski DR. DNA segments of the IncX plasmid R485 determining replication and incompatibility with plasmid R6K. Plasmid. 1985;14:245–254. doi: 10.1016/0147-619x(85)90008-3. [DOI] [PubMed] [Google Scholar]
- 17.Jones CS, Osborne DJ, Stanley J. Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J Gen Microbiol. 1993;139:735–741. doi: 10.1099/00221287-139-4-735. [DOI] [PubMed] [Google Scholar]
- 18.Couturier M, Bex F, Bergquist PL, Maas WK. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotz A, Pukall R, Smit E, Tietze E, Prager R, et al. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahl MI, Burmolle M, Meisner A, Hansen LH, Sorensen SJ. All IncP-1 plasmid subgroups, including the novel epsilon subgroup, are prevalent in the influent of a Danish wastewater treatment plant. Plasmid. 2009;62:134–139. doi: 10.1016/j.plasmid.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Heuer H, Binh CT, Jechalke S, Kopmann C, Zimmerling U, et al. IncP-1epsilon Plasmids are Important Vectors of Antibiotic Resistance Genes in Agricultural Systems: Diversification Driven by Class 1 Integron Gene Cassettes. Front Microbiol. 2012;3:2. doi: 10.3389/fmicb.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greated A, Thomas CM. A pair of PCR primers for Incp-9 plasmids. Microbiology 145 (Pt. 1999;11):3003–3004. doi: 10.1099/00221287-145-11-3003. [DOI] [PubMed] [Google Scholar]
- 23.Krasowiak R, Smalla K, Sokolov S, Kosheleva I, Sevastyanovich Y, et al. PCR primers for detection and characterisation of IncP-9 plasmids. FEMS Microbiol Ecol. 2002;42:217–225. doi: 10.1111/j.1574-6941.2002.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 24.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Fernandez A, Fortini D, Veldman K, Mevius D, Carattoli A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother. 2009;63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 26.Johnson TJ, Nolan LK. Plasmid replicon typing. Methods Mol Biol. 2009;551:27–35. doi: 10.1007/978-1-60327-999-4_3. [DOI] [PubMed] [Google Scholar]
- 27.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 28.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, et al. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Fernandez A, Chiaretto G, Bertini A, Villa L, Fortini D, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008;61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Fernandez A, Carattoli A. Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J Antimicrob Chemother. 2010;65:1155–1161. doi: 10.1093/jac/dkq101. [DOI] [PubMed] [Google Scholar]
- 32.Zong Z, Yu R, Wang X, Lu X. blaCTX-M-65 is carried by a Tn1722-like element on an IncN conjugative plasmid of ST131 Escherichia coli. J Med Microbiol. 2011;60:435–441. doi: 10.1099/jmm.0.026997-0. [DOI] [PubMed] [Google Scholar]
- 33.Phan MD, Kidgell C, Nair S, Holt KE, Turner AK, et al. Variation in Salmonella enterica serovar typhi IncHI1 plasmids during the global spread of resistant typhoid fever. Antimicrob Agents Chemother. 2009;53:716–727. doi: 10.1128/AAC.00645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heuer H, Kopmann C, Binh CT, Top EM, Smalla K. Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G+C content. Environ Microbiol. 2009;11:937–949. doi: 10.1111/j.1462-2920.2008.01819.x. [DOI] [PubMed] [Google Scholar]
- 35.Garcillan-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 36.Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, et al. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, et al. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 38.Datta S, Larkin C, Schildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure. 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD. The structure of the minimal relaxase domain of MobA at 2.1 A resolution. J Mol Biol. 2007;366:165–178. doi: 10.1016/j.jmb.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 41.Varsaki A, Lucas M, Afendra AS, Drainas C, de la Cruz F. Genetic and biochemical characterization of MbeA, the relaxase involved in plasmid ColE1 conjugative mobilization. Mol Microbiol. 2003;48:481–493. doi: 10.1046/j.1365-2958.2003.03441.x. [DOI] [PubMed] [Google Scholar]
- 42.Hedges RW, Jacob AE, Datta N, Coetzee JN. Properties of plasmids produced by recombination between R factors of groups J and FII. Mol Gen Genet. 1975;140:289–302. doi: 10.1007/BF00267320. [DOI] [PubMed] [Google Scholar]
- 43.Nugent ME. A conjugative ‘plasmid’ lacking autonomous replication. J Gen Microbiol. 1981;126:305–310. doi: 10.1099/00221287-126-2-305. [DOI] [PubMed] [Google Scholar]
- 44.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 45.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis. 2008;14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedrosa A, Novais A, Machado E, Canton R, Peixe L, et al. Recent Dissemination of blaTEM-52-producing Enterobacteriaceae in Portugal is caused by spread of IncI1 plasmids among Escherichia coli and Klebsiella pneumoniae clones. 18th European Congress of Clinical Microbiology and Infectious Diseases Poster presentation Barcelona, Spain. 2008.
- 47.Valverde A, Canton R, Garcillan-Barcia MP, Novais A, Galan JC, et al. Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother. 2009;53:5204–5212. doi: 10.1128/AAC.01706-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novais A, Baquero F, Machado E, Canton R, Peixe L, et al. International spread and persistence of TEM-24 is caused by the confluence of highly penetrating enterobacteriaceae clones and an IncA/C2 plasmid containing Tn1 and IS5075-Tn21. Antimicrob Agents Chemother. 2010;54:825–834. doi: 10.1128/AAC.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliver A, Coque TM, Alonso D, Valverde A, Baquero F, et al. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob Agents Chemother. 2005;49:1567–1571. doi: 10.1128/AAC.49.4.1567-1571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novais A, Canton R, Valverde A, Machado E, Galan JC, et al. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob Agents Chemother. 2006;50:2741–2750. doi: 10.1128/AAC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novais A, Canton R, Moreira R, Peixe L, Baquero F, et al. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob Agents Chemother. 2007;51:796–799. doi: 10.1128/AAC.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novais A, Viana D, Baquero F, Canton R, Coque TM. Contemporary spread of CTX-M-14-Escherichia coli in Spain is mainly associated with persistent IncK and IncI1 and emerging IncN plasmids. 20th European Congress of Clinical Microbiology and Infectious Diseases. Poster presentation. Vienna, Austria. 2010.
- 53.Ejrnaes K, Stegger M, Reisner A, Ferry S, Monsen T, et al. Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: phylogenetic groups, virulence factors and biofilm formation. Virulence. 2006;2:528–537. doi: 10.4161/viru.2.6.18189. [DOI] [PubMed] [Google Scholar]
- 54.Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, et al. Complete sequencing of the bla(NDM-1)-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6: e25334. 2011. Available: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0025334 Accessed 23 Septembre 2011. [DOI] [PMC free article] [PubMed]
- 55.Sobecky PA, Mincer TJ, Chang MC, Helinski DR. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol. 1997;63:888–895. doi: 10.1128/aem.63.3.888-895.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sobecky PA, Mincer TJ, Chang MC, Toukdarian A, Helinski DR. Isolation of broad-host-range replicons from marine sediment bacteria. Appl Environ Microbiol. 1998;64:2822–2830. doi: 10.1128/aem.64.8.2822-2830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Elsas JD, Gardener BB, Wolters AC, Smit E. Isolation, characterization, and transfer of cryptic gene-mobilizing plasmids in the wheat rhizosphere. Appl Environ Microbiol. 1998;64:880–889. doi: 10.1128/aem.64.3.880-889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gstalder ME, Faelen M, Mine N, Top EM, Mergeay M, et al. Replication functions of new broad host range plasmids isolated from polluted soils. Res Microbiol. 2003;154:499–509. doi: 10.1016/S0923-2508(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 59.Schluter A, Szczepanowski R, Kurz N, Schneiker S, Krahn I, et al. Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbors a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Appl Environ Microbiol. 2007;73:1952–1960. doi: 10.1128/AEM.02159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elhani D, Bakir L, Aouni M, Passet V, Arlet G, et al. Molecular epidemiology of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999–2005. Clin Microbiol Infect. 2010;16:157–164. doi: 10.1111/j.1469-0691.2009.03057.x. [DOI] [PubMed] [Google Scholar]
- 61.Sirichote P, Hasman H, Pulsrikarn C, Schonheyder HC, Samulioniene J, et al. Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar Choleraesuis isolates from patients in Thailand and Denmark. J Clin Microbiol. 2010;48:883–888. doi: 10.1128/JCM.01792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7: e1002222. 2011. Available: http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1002222. Accessed 18 August 2011. [DOI] [PMC free article] [PubMed]
- 64.Mata C, Miro E, Alvarado A, Garcillan-Barcia MP, Toleman M, et al. Plasmid typing and genetic context of AmpC beta-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: findings from a Spanish hospital 1999–2007. J Antimicrob Chemother. 2012;67:115–122. doi: 10.1093/jac/dkr412. [DOI] [PubMed] [Google Scholar]
- 65.Coelho A, Piedra-Carrasco N, Bartolome R, Quintero-Zarate JN, Larrosa N, et al. Role of IncHI2 plasmids harbouring bla(VIM-1), bla(CTX-M-9), aac(6′)-Ib and qnrA genes in the spread of multiresistant Enterobacter cloacae and Klebsiella pneumoniae strains in different units at Hospital Vall d’Hebron, Barcelona, Spain. Int J Antimicrob Agents. 2012;39:514–517. doi: 10.1016/j.ijantimicag.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 66.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 67.Toleman MA, Walsh TR. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev. 2011;35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 68.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 69.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 71.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campbell JL, Richardson CC, Studier FW. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc Natl Acad Sci U S A. 1978;75:2276–2280. doi: 10.1073/pnas.75.5.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, et al. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morant M, Hehn A, Werck-Reichhart D. Conservation and diversity of gene families explored using the CODEHOP strategy in higher plants. BMC Plant Biol. 2002;2:7. doi: 10.1186/1471-2229-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Provencher C, LaPointe G, Sirois S, Van Calsteren MR, Roy D. Consensus-degenerate hybrid oligonucleotide primers for amplification of priming glycosyltransferase genes of the exopolysaccharide locus in strains of the Lactobacillus casei group. Appl Environ Microbiol. 2003;69:3299–3307. doi: 10.1128/AEM.69.6.3299-3307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rose TM. CODEHOP-mediated PCR - a powerful technique for the identification and characterization of viral genomes. Virol J. 2005;2:20. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boer R, Russi S, Guasch A, Lucas M, Blanco AG, et al. Unveiling the molecular mechanism of a conjugative relaxase: The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol. 2006;358:857–869. doi: 10.1016/j.jmb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 80.Llosa M, Zunzunegui S, de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci U S A. 2003;100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andres I, Rodriguez JC, Ortiz JM. Physical and functional map of the hemolytic plasmid pSU316. Plasmid. 1984;11:96–98. doi: 10.1016/0147-619x(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 82.Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 83.Núñez B, de la Cruz F. Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol Microbiol. 2001;39:1088–1089. doi: 10.1046/j.1365-2958.2001.02308.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasmids from γ-Proteobacteria contained in the NCBI database.
(DOC)
Nucleotide sequences and their translated amino acid sequences of relevant relaxases obtained by DPMT from different test collections.
(DOC)