Abstract
Primary sensory nuclei of the thalamus process and relay parallel channels of sensory input into the cortex. The developmental processes by which these nuclei acquire distinct functional roles are not well understood. To identify novel groups of genes with a potential role in differentiating two adjacent sensory nuclei, we performed a microarray screen comparing perinatal gene expression in the principal auditory relay nucleus, the medial geniculate nucleus (MGN), and principal visual relay nucleus, the lateral geniculate nucleus (LGN). We discovered and confirmed groups of highly ranked, differentially expressed genes with qRT-PCR and in situ hybridization. A functional role for Zic4, a transcription factor highly enriched in the LGN, was investigated using Zic4-null mice, which were found to have changes in topographic patterning of retinogeniculate projections. Foxp2, a transcriptional repressor expressed strongly in the MGN, was found to be positively regulated by activity in the MGN. These findings identify roles for two differentially expressed genes, Zic4 and Foxp2, in visual and auditory pathway development. Finally, to test whether modality-specific patterns of gene expression are influenced by extrinsic patterns of input, we performed an additional microarray screen comparing the normal MGN to “rewired” MGN, in which normal auditory afferents are ablated and novel retinal inputs innervate the MGN. Data from this screen indicate that rewired MGN acquires some patterns of gene expression that are present in the developing LGN, including an upregulation of Zic4 expression, as well as novel patterns of expression which may represent unique processes of cross-modal plasticity.
Introduction
Sensory information is processed along pathways comprised of modality-specific brain regions (Kiecker and Lumsden, 2005; Sur and Rubenstein, 2005; Lim and Golden, 2007). These regions arise during embryogenesis, when signaling centers induce graded patterns of gene expression to instruct local differentiation programs (Figdor and Stern, 1993; Ragsdale and Grove, 2001; O'Leary and Nakagawa, 2002; Grove and Fukuchi-Shimogori, 2003; Shimogori et al., 2004). The mechanisms linking early patterning events and later processes of functional differentiation are not well understood in the dorsal thalamus, the principal relay and processing center for all incoming sensory information to the cortex except for olfaction (Jones, 2007).
The lateral geniculate nucleus (LGN) and medial geniculate nucleus (MGN) are primary sensory thalamic nuclei, receiving visual and auditory afferents from the retina and inferior colliculus (IC), respectively, which in turn send respective projections to the primary visual cortex (V1) and primary auditory cortex (A1) (Fig. 1A). They parcellate as neighbors in relative dorsocaudal and ventroanterior positions (Kiecker and Lumsden, 2004), which likely affect their exposure to molecular signals during development and subsequent patterns of gene expression (Kataoka and Shimogori, 2008; Vue et al., 2009).
Figure 1.
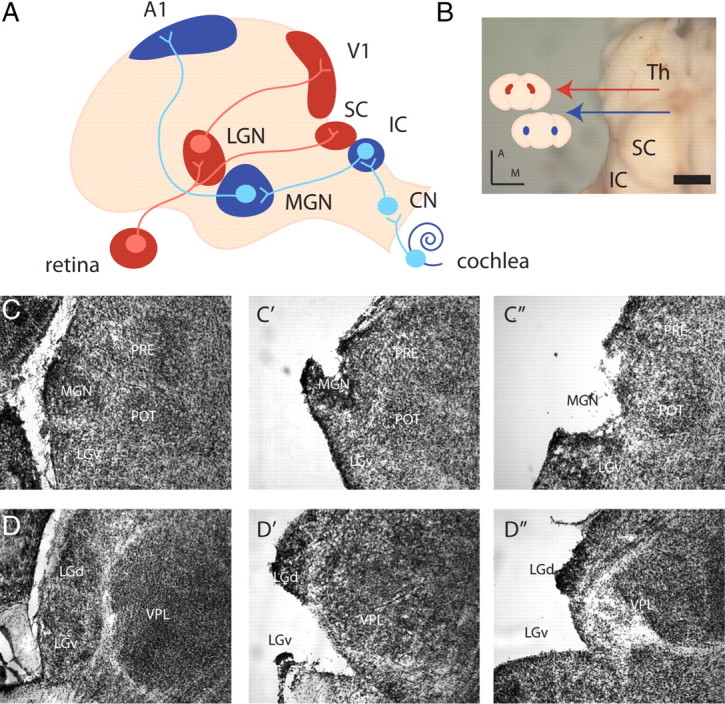
Primary visual and auditory sensory pathways to the cortex. A, Retinal ganglion cells project to the LGN, which in turn projects to V1. Auditory afferents from the cochlea terminate in the cochlear nucleus (CN) of the brainstem, which then innervates the IC. Cells from the IC project to the MGN, which in turn projects to A1. B, Dorsal view of the neonatal thalamus (the cortex has been peeled off), red arrow, LGN; blue arrow, MGN; Th, thalamus. Scale bar, 1 mm. Colored lines show coronal planes of section for Nissl stains and point to schematics of coronal sections featuring the MGN (blue) and LGN (red). C, D, Coronal sections of cresyl violet stains reveal the nuclear structure of the MGN (C) and LGN (D) at P0. Two examples of MGN (C′, C″) and LGN (D′, D″) dissected brain are representative of the tissue samples taken for microarray analysis. PRE, Pretectal nucleus; POT, posterior thalamic nucleus; VPL, ventral posterior thalamic nucleus, lateral part; LGv, ventral subdivision of the LGN; D, dorsal; M, medial. Scale bar, 150 μm.
To identify genetic programs that contribute to the functional specification of the LGN and MGN, we performed a gene microarray screen comparing LGN and MGN at postnatal day 0 (P0) and P5, a time after structural parcellation has occurred but during afferent ingrowth and continued cellular differentiation and synapse formation (Jones, 2007). To test the sensitivity of our screen in isolating genes with a sensory-specific role in thalamic differentiation, we characterized functional roles of two novel candidates, the zinc-finger transcription factor, Zic4, and the transcriptional repressor, Foxp2, in visual and auditory pathway development, respectively.
A persistent question in developmental neurobiology is the extent to which intrinsic lineage-derived programs and extrinsic factors from inputs instruct the differentiation of functional regions (Dehay et al., 1993, 2001; Miyashita-Lin et al., 1999). We tested whether specific sensory inputs contribute to the genetic differences between the LGN and MGN by comparing gene expression between normal MGN and “rewired MGN” (rwMGN), which has surgically been induced to receive visual input. In mice, hamsters and ferrets, ablating the IC early in development is sufficient to induce retinal afferents to innervate the MGN and drive the auditory pathway to process visual information (Schneider, 1973; Sur et al., 1988; Lyckman et al., 2001; Newton et al., 2004). Although structural features of the MGN and A1 persist, functional features of visual processing occur (Sharma et al., 2000; von Melchner et al., 2000). We compared normal MGN and rwMGN at P5, the earliest age at which retinal afferents are detected in MGN after IC ablation. Finally, we tested whether loss of a downregulated gene, Slc6a4, was sufficient for rewiring in the absence of IC ablation.
Materials and Methods
Animals.
Wild-type mice of background strain 129/SvEv were used for the microarray analysis. Zic4-null mice had been generated previously and obtained as a gift from the Millen laboratory (Grinberg and Millen, 2004). These mice were maintained on a 129/SvIMJ background and were viable and fertile. Slc6a4-null mice were generated previously and obtained from Taconic (B6.129-Slc6a4tm1Kpl) (Bengel et al., 1998). This line was maintained on a C57BL/6 background. All experiments were approved by Massachusetts Institute of Technology's Institutional Animal Care and Use Committee and performed in compliance with National Institutes of Health guidelines.
Dissection of thalamic nuclei and RNA preparation.
Fifteen to twenty neonatal (P0) and perinatal (P5) 129/Svev mice were used for each of three (P5) to four (P0) biological replicates of LGN and MGN (14 groups total, n = 45–60 mice per group). Brains were dissected and submerged under RNase-free conditions in RNeasy (Qiagen). Under light microscopy, the cortex was peeled off exposing the thalamus. The LGN and MGN were removed bilaterally with micro-dissection spring scissors and stored in 1 ml of Trizol (Invitrogen). Dissection of appropriate nuclei was confirmed with coronal sections of dissected thalami. Approximately 0.5 mg of nucleus-specific tissue was acquired from each animal. Anatomical localization of the LGN relative to the MGN was inferred from thalamus specimens of representative animals subjected to intraocular WGA-HRP injections and tetramethyl benzadine reaction. Tissue was homogenized by sequential pulverization through 18, 22, and 26 gauge needles and a 1 cc syringe. RNA was tissue extracted using a Qiagen RNAeasy kit, and a cDNA copy of the RNA was made using an Invitrogen Superscript T7 in vitro reverse transcription kit and T7 oligo-dT primers. Labeled cRNA was synthesized using biotinylated nucleotides with an Enzo Biolabeling Kit.
Gene microarray hybridization and analysis.
Labeled cRNA (15–20 μg) from each of three (P5) to four (P0) replicates for P0 and P5 LGN and MGN was applied to Affymetrix murine U74v2 or 430 2.0 microarray chips by the Massachusetts Institute of Technology Biopolymers lab.
Microarray cel files were normalized and subjected to robust multichip average (RMA) preprocessing using RMAexpress software (Bolstad et al., 2003; Irizarry et al., 2003). Significance analysis of microarrays (SAM) was used to identify enriched genes by setting a minimum fold change at 1.5 and adjusting the delta value (a threshold value for the difference between the observed relative difference and the expected relative difference, as an average over n permutations) to present <0.01% false discovery rate (FDR) (Tusher et al., 2001). Many genes are represented on the Affymetrix chips by multiple probes. All probes mapping onto the same gene identified through this screen showed consistent patterns of differential expression between LGN and MGN.
Quantitative real-time PCR.
LGN and MGN RNA samples were harvested as described above and used for single-strand cDNA synthesis from oligoDT primers. Primers amplifying a 150–200 bp amplicon of each gene in enriched sets were designed by using the Whitehead Institute Primer3 program (for primer sequences, see supplemental Table 1, available at www.jneurosci.org as supplemental material). Primer efficiencies were calculated using a standard Ct curve on serial dilutions of cDNA template amplified with SYBR Green PCR mix (Qiagen) in the DNA Engine Opticon 2 System. PCRs were performed in triplicate for each sample and relative quantitation was calculated using the Pfaffl formula with Gapdh as reference: E (efficiency) = 10(−1/slope), fold change = [EtargetΔCt target (sample1−sample2)/EreferenceΔCt reference (sample1−sample2)].
In situ hybridization.
Digoxigenin (DIG)-labeled antisense and sense riboprobes (500–600 bp; for primer sequences, see supplemental Table 2, available at www.jneurosci.org as supplemental material) were synthesized from sample cDNA using a T7 reverse transcriptase in vitro kit with DIG-labeled ribonucleotides (Invitrogen). Riboprobes were purified on Micro Bio-spin Columns (Biorad) and quantified by UV spectrophotometry. Brains at P0 and P5 were flash frozen in isopentane, stored at −80°C and sectioned at 18 μm at −20°C on a cryostat (n = 3 per gene of interest at P0, n = 2 at P5). Sections were processed using an in situ hybridization protocol (Braissant and Wahli, 1998) and analyzed using a Zeiss Axioskop 2 microscope. Zic4 gradients were measured using a line capture tool along the nasal–temporal axis of the retina (4 sections of 1 animal) and the dorsolateral–ventromedial axis (n = 5 sections of 1 animal) of the LGN in ImageJ. Signal was normalized to the maximum value and averaged across 10 bins.
Gene set enrichment analysis.
Gene set enrichment analysis (GSEA) was performed on the following comparisons: P0 LGN versus P0 MGN, P5 LGN versus P5 MGN, and P5 MGN versus rwP5 MGN. Gene sets from the c2 (curated gene sets) collection of the Broad Institute Molecular Signatures Database (http://www.broad.mit.edu/gsea/index.jsp) were used. For a given probe i, the signal-to-noise ratio (SNR) between the mean expression level (Sμi) across samples of two conditions (s = P0 LGN, P0 MGN, etc.) was defined as follows: SNRi = (LGNμi − MGNμi)/(LGNσi + MGNσi), where Sσi = standard deviation across samples.
The probes were then ranked by SNR in an ordered list L. Given a set G containing NG probes, two cumulative distribution functions were compared: Phit(i) = proportion of genes in G with rank <i and Pmiss(i) = proportion of genes outside G that show a rank <i with the running enrichment score defined as RES(i) = Phit(i) − Pmiss(i). The peak enrichment score, ES, refers to the maximum deviation of RES(i) from 0. Statistical significance of ES was evaluated compared with a null distribution from 1000 permutations obtained by randomly shuffling the condition labels for each probe. Multiple comparisons were controlled for using the familywise error rate and enrichment scores were normalized across gene sets by centering and scaling: Phit(i) = #[g(j≤i) ∈ G]/NG, Pmiss(i) = #[g(j≤i) ∉ G]/[N − NG], L = {g1…gN}.
Anatomical tracing and retinogeniculate projection analysis.
Retinogeniculate projections were traced using intraocular injections of Alexa Fluor 488- and 594-conjugated cholera toxin-B (CTB, Invitrogen). Paired littermates of wild-type and Zic4-null mice (P30–P48) were injected using a Hamilton syringe with 1–2 μl of 488-CTB and 594-CTB in each eye. Mice were allowed to recover for 48 h to complete tracing and killed.
Retinogeniculate tracings were imaged from 50 μm brain sections using a Zeiss Axioskop 2 microscope with 5× lens objective lens and 1.25 eyepiece magnification (1 pixel = 1.88 μm). LGN images were divided into anterior, middle, and posterior groups. For each animal, two sections from each hemisphere in anterior, middle, and posterior groups were imaged and analyzed (n = 4 WT, n = 4 KO). Axiovision LE Rel. 4.4 software was used to measure the dorsomedial and dorsoventral length of the LGN and ipsilateral projection, as well as to count the number of ipsilateral clusters in each section. A Matlab script was used to measure the overlap between contralateral and ipsilateral terminals, the total number of LGN pixels, and the scatter of ipsilateral terminals (measured as the difference in intensity between a pixel and the average of its five nearest neighbors).
Activity-dependent induction of gene expression.
Littermate C57BL/6 mice at age P18 were placed in a dark, sound-proof room for 48 h and then either killed in the control group or treated with ramped exposure to 90 dB of white noise for 90 min and then killed. The white noise generator was an ASR-PRO1 acoustic startle reflex test apparatus (Med Associates), with intensity controlled using Startle Reflex software (Med Associates). The background noise of the room in which we performed acoustic stimulation was 55 dB. Mice were held in the apparatus in an acrylic cage that contained numerous holes to allow for the passage of sound. To minimize the risk of audiogenic seizures, mice were placed in the apparatus and exposed to background noise for 10 min and then to white noise at 60 dB for 10 min, 70 dB for 10 min, 80 dB for 10 min, and finally 90 dB for 90 min. Thalamic and cortical tissue was dissected bilaterally from n = 4 animals for both the experimental and control groups, flash frozen, and then homogenized with a RIPA buffer/proteinase inhibitor cocktail (Roche) with sodium orthovanadate (1 mm final concentration) and centrifuged. After supernatant protein concentration was quantified with a Bradford 595 Assay, 15 μg of cortical samples and 30 μg of thalamic samples were run at 200 V for 35 min on NuPAGE 4–12% Bis-Tris Gel (Invitrogen), transferred at 35 mV for 70 min to a PVDF membrane (Invitrogen), and stained with Foxp2 (1:500, Abcam), cFos (1:500, Santa Cruz Biotechnology), and GAPDH (1:500, Abcam) antibodies followed by 1:500 peroxidase-conjugated anti-rabbit secondary antibody (The Jackson Laboratory) and development with ECL Western blotting detection reagents (GE Healthcare) on Kodak film. Foxp2 and cFos signal was quantified and normalized to GAPDH signal in ImageJ. Data from three technical replicates was averaged and used for statistical analysis.
Visual rewiring of the MGN.
Neonatal P0 mice were anesthetized on ice (hypothermia) for 20 min. The scalp was opened and IC ablated bilaterally using a Bovie Cautery High Temperature Loop Tip (Aaron Medical). The incision was glued with Vetbond and animals were allowed to recover. At P5, thalamic tissue was dissected as described above. Intraocular CTB injections were also performed as described above.
Results
Gene microarrays identify patterns of differential gene expression between the perinatal LGN and MGN
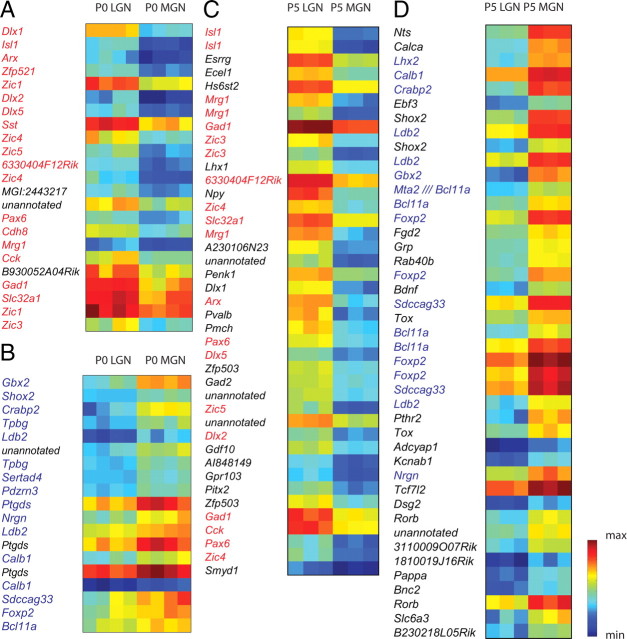
The positions of LGN and MGN are exposed at the lateral edges of the thalamus after cortex removal and were dissected under a light microscope (Fig. 1A–D). Of ∼45,000 probes screened on the Affymetrix 430 2.0 microarray chip, a SAM analysis identified 23 probes, corresponding to 20 unique genes, as enriched in the P0 LGN relative to the P0 MGN with a fold change (FC) >2 and FDR <0.01% (n = 15–20 per replicate, 4 replicates per group, delta value = 0.831) (Fig. 2A). Nineteen probes, corresponding to 13 genes, were enriched in P0 MGN relative to the P0 LGN (Fig. 2B). In the P5 LGN versus MGN screen, 125 probes, corresponding to 90 unique genes, were enriched in P5 LGN and 128 probes, corresponding to 74 unique genes, in P5 MGN using FC >2 and FDR <0.01% (n = 15–20 per replicate, 3 replicates per group, delta value = 1.441). Seventeen of twenty (85%) of the P0 LGN genes (Fig. 2A, red type) were present in the P5 LGN group and twelve of thirteen (92%) of the P0 MGN genes (Fig. 2B, blue type) were present in the P5 MGN group, demonstrating the presence of genes that are always different between LGN and MGN during a perinatal window (P0–P5). To reduce the P5 candidate genes to a more manageable number, we screened further using FC >3, yielding 41 probes, representing 31 unique genes, as enriched in P5 LGN (Fig. 2C) and 43 probes, representing 30 unique genes, in P5 MGN (Fig. 2D). Thirteen of thirty (43%) of these P5 LGN-enriched genes were present in the P0 LGN group (Fig. 2C, red type) and ten of thirty (33.3%) P5 MGN-enriched genes in the P0 MGN group (Fig. 2D, blue type).
Figure 2.
SAM identifies LGN- and MGN-enriched groups at P0 and P5. A, B, A group of 20 unique genes enriched in P0 LGN (A) and a group of 13 unique genes enriched in P0 MGN (B), determined in SAM using a fold change (FC) >2 and a delta value adjustment to 0.831 to yield a FDR < 0.01%. Seventeen of twenty (85%) of the P0 LGN genes (red type) were present in the P5 LGN group using the same SAM criteria and twelve of thirteen (92%) of the P0 MGN genes (blue type) were present in the P5 MGN group. Each P0 group has four replicates with n = 15–20 each. C, Thirty-one unique genes were enriched in P5 LGN with FC >3, delta = 1.4 for FDR <0.01%. Thirteen of thirty (43%) of these genes were present in the P0 LGN set (red type). D, Thirty unique genes were enriched in P5 MGN with these criteria; 10/30 (33.3%) were present in the P0 MGN set (blue type). Each P5 group has three replicates with n = 15–20 each. All genes are ranked by SAM d-score. Color bar, Blue represents minimum intensity value within the gene group on the microarray; red represents maximum intensity.
To control for potential differences in maturational state, we tested whether LGN and MGN undergo similar global processes of differentiation from P0 to P5 by using SAM analysis to compare LGN at P0 versus P5 and MGN at P0 versus P5 (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). In the LGN comparison, 12,712 probes were upregulated at P5 and 1655 were downregulated using FC >2 and FDR <0.01% (delta value = 0.596), while in the MGN comparison, using the same criteria, 13,266 were upregulated at P5 and 2846 were downregulated (delta value = 0.604). Since nearly a third of all the probes screened showed significant change between P0 and P5, we compared the most highly ranked genes. Among the top 115 probes for both the LGN and MGN age-enriched groups, 91/115 (79%) were identical, suggesting that LGN and MGN undergo common processes of maturation between P0 and P5 [supplemental Fig. 1 (green type), available at www.jneurosci.org as supplemental material].
Of the genes enriched in the LGN with respect to MGN, many were transcription factors, including those implicated in neuronal migration (Arx and Dlx1), cellular differentiation (Zic1, Zic3, Zic4, and Zic5), and areal patterning (Pax6). Non-transcription factor genes in LGN included cell-adhesion molecules (Cad8), neurotransmitter synthesis enzymes (Gad1), and cellular signals (Sst, Cck, Npy). Transcription factors were also overwhelmingly present in the genes enriched in the MGN with respect to LGN, including a previously identified dorsal thalamic marker, Gbx2. Another gene, Foxp2, a transcriptional repressor implicated in speech and language development in humans (Vargha-Khadem et al., 2005), was highly enriched in MGN. Non-transcription factor genes included those involved in retinoic acid signaling (Crabp2), Wnt signaling (Ck2-α), and Ca2+ signaling (Calb1).
To isolate genes with a potential role in sensory-specific processes of neural circuit formation, we retrieved P5 LGN and P5 MGN expression data for a list of genes with a demonstrated role in axon guidance compiled from the literature. Screening 118 unique genes represented by 309 probes (supplemental Table 3, available at www.jneurosci.org as supplemental material), we found the following factors enriched in P5 LGN with respect to P5 MGN: EphA7, ephrinA5, Fgfr1, Wnt4/5a, Sema5b/6a/6d, Slit2, Unc5b, NCAM1, Sfrp2, Neuropilin1/2, Ntrk2/3, and Slitrk5 (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). In turn, the following factors were enriched in P5 MGN with respect to P5 LGN: Bdnf, EphA1, EphB2, ephrinA2/A3, Frizzled1/3, netrinG1, and Slitrk6 (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material), suggesting that distinct groups of axon guidance factors contribute to neural connectivity and circuit formation in the two nuclei.
Real-time PCR and in situ hybridization confirm differential gene expression from LGN- and MGN-enriched sets
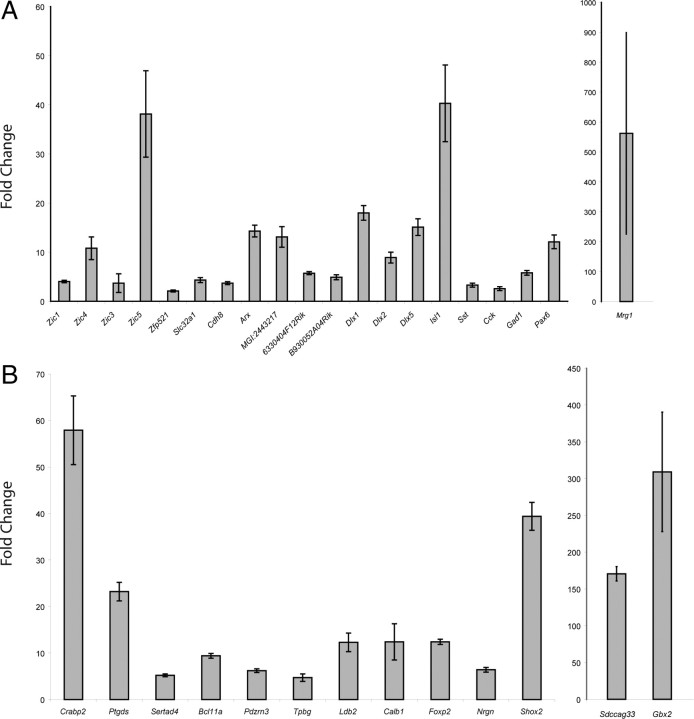
Differential gene expression levels between LGN and MGN were confirmed using quantitative real time PCR (qRT-PCR) on all genes of the P0 LGN and P0 MGN sets. All 20 of the P0 LGN-enriched genes were confirmed with qRT-PCR, with significant upregulation (p < 0.05) and FC >2 in LGN samples vs MGN samples (Fig. 3A). All 13 of the P0 MGN-enriched genes were also confirmed with qRT-PCR with significant upregulation (p < 0.05) and FC >4 (Fig. 3B).
Figure 3.
Quantitative RT-PCR of LGN- and MGN-specific genes confirms differential gene expression at P0. A, P0 LGN-enriched genes are all significantly upregulated (20/20, p < 0.05, FC >2 in LGN samples vs MGN samples). B, P0 MGN-enriched genes are all significantly upregulated (13/13, p < 0.05, FC >4) as well.
Top ranking genes for both the P0 and P5 LGN and MGN groups were selected for in situ hybridization to document anatomical expression patterns in more detail. At both P0 and P5, Zic4 and Zic1, two members of the Zic family of transcription factors, exhibited stronger expression in neonatal LGN than MGN (Fig. 4A,B,A′,B′), with no signal from sense probes (data not shown). Zic4 exhibited a graded expression pattern both in the dorsal subdivision of the LGN (LGd) with high levels dorsolaterally and in the retina with high levels on the temporal side (Fig. 4B; supplemental Fig. 3, available at www.jneurosci.org as supplemental material), suggesting a possible role in retinotopic patterning of retinogeniculate projections. At both P0 and P5, Crabp2, Foxp2, and Sdccag33, demonstrated stronger expression in neonatal MGN than LGN (Fig. 4C–E,C′–E′) and no signal with sense probes (data not shown). Foxp2 showed strong expression in both dorsal and ventral subdivisions of the MGN (MGd, MGv), while Crabp2 was localized primarily to the MGd and Sdccag33 localized exclusively to the MGv.
Figure 4.
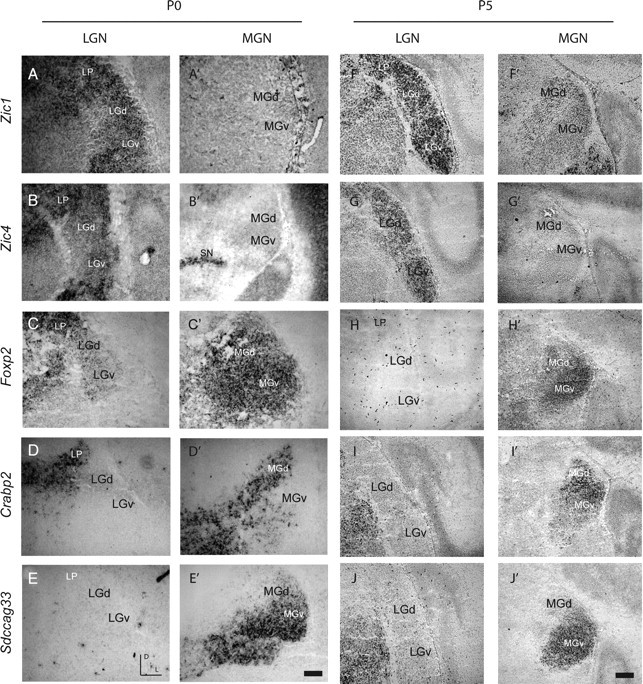
In situ hybridizations at P0 (A–E, A′–E′) and P5 (F–J, F′–J′) confirm the differential expression of LGN- and MGN-enriched genes. Zic1 (A, A′, F, F′) and Zic4 (B, B′, G, G′) are expressed in LGd (A, B, F, G) but not MGN (A′, B′, F′, G′), while Foxp2 (C, C′, H, H′), Crabp2 (D, D′, I, I′), and Sdccag33 (E, E′, J, J′) are expressed in MGN (C′–E′, H′–J′) but not in LGd (C–E, H–J). Sense probes were tested for all genes and yielded no signal (data not shown). LGd, Dorsal lateral geniculate nucleus; MGd, dorsal body of the medial geniculate nucleus; MGv, ventral division of the medial geniculate nucleus; LP, lateral posterior thalamic nucleus; SN, substantia nigra; LGv, ventral lateral geniculate nucleus. Scale bar: 150 μm for P0, 300 μm for P5.
Gene set enrichment analysis identifies functional pathways within LGN- and MGN-enriched sets
The analyses above are based on the study of individual genes. It is possible to screen for groups of genes that share similar functions, structure, biochemical pathways, and chromosomal location. Even if individual genes within a gene group show only minor changes in expression, group analysis can uncover large changes for the group as a whole. To identify functionally related groups of genes with collective enrichment in the LGN or MGN, GSEA using the c2 (curated gene sets) collection of the Broad Institute Molecular Signatures Database (http://www.broad.mit.edu/gsea/index.jsp) was performed (Subramanian et al., 2005). These pathways may represent developmental programs in the functional specification of the perinatal LGN and MGN. Eighteen of 1892 screened gene sets were significantly enriched in P0 LGN at p < 0.01 (supplemental Table 4, available at www.jneurosci.org as supplemental material). Among the top 18 pathways were the cardiac EGFP pathway, phosphatidylinositol signaling system, p53 upregulated genes, CD40 downregulated genes, and the IL-2 receptor pathway, the last two suggesting a role for cytokine signaling in cellular differentiation within the LGN.
One hundred eleven of 1892 gene sets had significant enrichment at p < 0.01 in P0 MGN (supplemental Table 5, available at www.jneurosci.org as supplemental material). The top pathway was the Alzheimer's disease set; additional pathways in the top 20 included the GATA3 gene set, upregulation with loss of Mecp2 set and the Hoxc8 pathway.
GSEA performed on the P5 data revealed 193/1892 gene sets enriched in the P5 LGN at p < 0.01 (supplemental Table 6, available at www.jneurosci.org as supplemental material). Pathways ranked in the top 20 included a gene set upregulated by TNFα signaling, both PI3K and mTOR signaling cascades, and the VEGF pathway. All four of these pathways have been previously suggested to play a role in the maintenance and plasticity of the developing and adult visual pathway (see Discussion), and their presence here suggests an additional role in early retinothalamic development. In the P5 MGN, 32/1892 gene sets were significantly enriched at p < 0.01 (supplemental Table 7, available at www.jneurosci.org as supplemental material). The most significant set was the Wnt signaling pathway, which has been implicated in diencephalic patterning with a Wnt source emanating from the alar plate (Braun et al., 2003; Zhou et al., 2004). Other top pathways included the NFAT pathway, Hox patterning genes, an additional Wnt signaling set, and the CCR3 pathway.
Zic4 contributes to the patterning of retinogeniculate projections
The role of Zic4 in visual pathway development was explored, because of its strong enrichment in the LGd and previous work identifying the role of gene family member, Zic2, in ipsilateral steering of retinal ganglion cell axons at the optic chiasm (Pak et al., 2004). While Zic2 expression is not appreciably detected with in situ hybridization in the LGd, Zic4 is strongly expressed in a gradient along the dorsolateral axis, as well as in a high temporal to low nasal gradient in retinal ganglion cells (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). To test whether Zic4 is necessary for normal retinogeniculate patterning, intraocular injections of CTB were performed in littermate control and Zic4-null mice at an age when retinogeniculate terminals have matured to their adult form (P30–P48) (Guido, 2008).
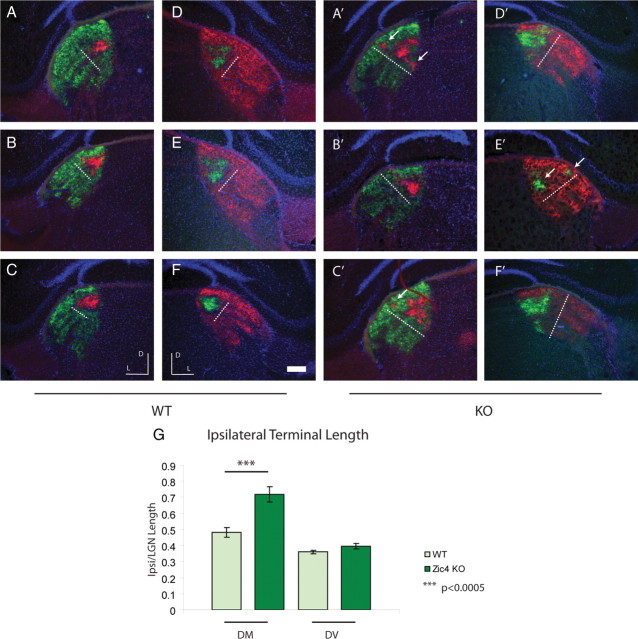
No changes in the size of LGd were observed (Fig. 5), suggesting that nuclear development and gross targeting were intact. However, a number of defects were detected in the rostral third portion of LGd, where Zic4-null mice exhibited the following: (1) a marked dorsomedial expansion of the ipsilateral termination zone (71.9 ± 4.8% of the dorsomedial axis in KOs vs 48.2 ± 3.3% in wild-type, p < 0.0005, t test, n = 4 each group) (Fig. 5A–F,A′–F′, dotted line, G), (2) increases in the number of ipsilateral terminal clusters (mean no. of clusters = 1.62 ± 0.17 vs 1.12 ± 0.08, KO vs control, p < 0.05, t test, n = 4 each group), and (3) an increase in the percentage of LGd filled by ipsilateral projections (21.18 ± 2.5% KO vs 14.49 ± 1.7% control, t test, p < 0.05, n = 4 each group). There was no statistically significant difference in the amount of overlap between contralateral and ipsilateral terminals (number of overlap pixels/total LGN pixels), although the Zic4-null mouse showed a trend of an increase in overlap (1.74 ± 0.26% KO vs 0.98 ± 0.34% control, p = 0.08, t test) suggesting that activity-dependent processes of eye-specific segregation were not largely disrupted, though they may have been slightly attenuated, in the absence of Zic4. Furthermore, no difference in ipsilateral spread along the dorsoventral axis was found, indicating that altered ipsilateral terminal spread occurs only along the dorsomedial axis in Zic4-null mice (Fig. 5G). Finally, despite an increased number of clusters in the knock-outs, there was no difference in the scatter of ipsilateral terminals, suggesting that ipsilateral zones are compact and segregated from contralateral terminals. Middle and posterior third portions of the LGd showed no difference between knock-outs and wild type (data not shown).
Figure 5.
Ipsilateral projections are disrupted in the rostral LGd of Zic4 KOs. Representative samples of intraocular 488- and 594-CTB tracings to the LGd in control (left, A–C; right, D–F) and Zic4 KO (left, A′–C′; right, D′–F′). Dotted white line, Length of ipsilateral zone along the dorsomedial axis; white arrows, multiple ipsilateral terminals in KO; D, dorsal, L, lateral. Scale bar, 300 μm. G, Ipsilateral terminal length along the dorsomedial (DM) and dorsoventral (DV) axes, expressed as a fraction of the total LGd terminal length.
Foxp2 is regulated by activity in the auditory thalamus
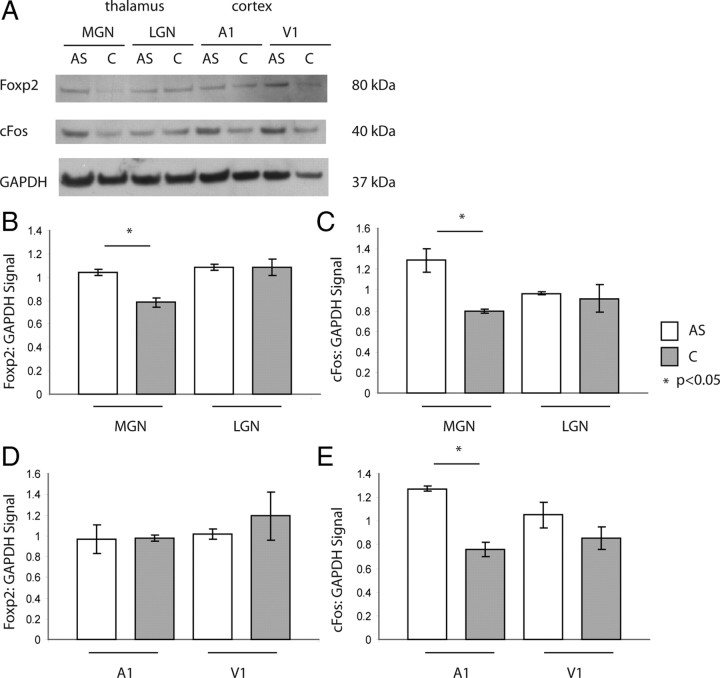
Foxp2 is a transcriptional repressor necessary for proper human speech and language development (for review, see Vargha-Khadem et al., 2005), but whose role in the auditory pathway has not yet been explored. Because Foxp2 expression has been shown to be dynamically regulated in songbirds during periods of plasticity (Teramitsu and White, 2006), we tested whether Foxp2 levels are modulated by auditory activity in vivo. Using Western blots, we found that Foxp2 protein was increased significantly in the MGN of P18 wild-type C57BL/6 animals after 120 min of high volume (90 dB) white noise stimulation compared with untreated controls (1.04 ± 0.025 vs 0.78 ± 0.037, p < 0.05, t test, n = 4 pooled for each group, technical replicate n = 3 for each group) (Fig. 6A,B,D). cFos expression was used as a positive control for early activity-dependent regulation of gene expression in the auditory thalamus and cortex (MGN: 1.29 ± 0.02 vs 0.78 ± 0.02, p < 0.05, A1: 1.27 ± 0.025 vs 0.76 ± 0.059, p < 0.05, t test) (Fig. 6A,C,E). In the LGN and V1, neither Foxp2 nor cFos levels were changed in experimental animals compared with control, suggesting that activity-dependent regulation is specific to the sensory pathway being stimulated (Foxp2 LGN: 1.09 ± 0.02 vs 1.08 ± 0.07, p = 0.927, t test; cFos LGN: 0.96 ± 0.02 vs 0.92 ± 0.12, p = 0.75, t test; Foxp2 V1: 1.01 ± 0.048 vs 1.19 ± 0.232, p = 0.54, t test; cFos V1: 1.05 ± 0.108 vs 0.85 ± 0.096, p = 0.786, t test) (Fig. 6B,C). Furthermore, Foxp2 expression did not change with auditory stimulation in A1, indicating that Foxp2 acts in an activity-dependent manner only in the thalamus (0.967 ± 0.14 vs 0.977 ± 0.03, p = 0.585, t test).
Figure 6.
Activity-dependent expression of Foxp2 protein in the auditory thalamus. A, Western blotting of Foxp2, cFos, and GAPDH in the MGN, LGN, A1, and V1 of P18 mice exposed to 2 h of ramped, high-decibel white noise versus no treatment. AS, Auditory stimulation, C, control. B, C, Quantitated signal of Foxp2 (B) and cFos (C) normalized to GAPDH levels shows an increase in Foxp2 expression in the MGN, but not LGN of mice undergoing auditory stimulation (Foxp2 MGN: 1.04 ± 0.025 vs 0.78 ± 0.037, p < 0.05, n = 4 pooled for each group, technical replicate n = 3 for each group; cFos MGN: 1.29 ± 0.02 vs 0.78 ± 0.02, p < 0.05). D, E, In the cortex, Foxp2 levels do not change in response to auditory stimulation (Foxp2 A1: 0.967 ± 0.14 vs 0.977 ± 0.03, p = 0.585; Foxp2 V1: 1.01 ± 0.048 vs 1.19 ± 0.232, p = 0.54), while cFos levels increase in both A1 and to a lesser extent, V1, of stimulated mice (A1: 1.27 ± 0.025 vs 0.76 ± 0.059, p < 0.05; cFos V1: 1.05 ± 0.108 vs 0.85 ± 0.096, p = 0.786).
Of note, we found that Foxp2 levels were higher in the normal LGN than the normal MGN at P18, a result consistent with expression results in the adult brain from the Allen Brain Atlas (http://www.brain-map.org/). This result suggests that dynamic levels of Foxp2 expression may be involved in later development of the visual pathway as well.
Visually rewired MGN exhibits LGN-like patterns of gene expression as well as novel patterns of plasticity-related genes
To clarify whether genetic programs of specification in the LGN and MGN are shaped by the identity of their inputs, we performed rewiring experiments in which retinal axons are rerouted to the MGN after ablation of the neonatal IC (Lyckman et al., 2001; Newton et al., 2004) and tested whether visual inputs to the MGN might induce LGN-like patterns of gene expression and/or downregulate genes typically enriched in the MGN (Fig. 7A–C). Screening 45,000 probes on the Affymetrix 430 2.0 microarray chip, a SAM analysis identified 14 probes, corresponding to 12 unique genes, as enriched and 1 probe representing 1 gene as downregulated in the P5 rwMGN with FC >2 and FDR <0.01% (n = 15–20 per replicate, 3 replicates per group, delta value = 0.489). To expand the candidate list, we loosened our criteria to FC >1.5 and FDR <3.37% (delta value = 0.666), yielding 44 probes, corresponding to 41 unique genes, as enriched (Fig. 7D) and 5 probes, corresponding to 5 unique genes, as downregulated in the rwMGN (Fig. 7E). Ten/41 (24.4%) of these enriched genes were present in the P5 LGN-enriched group, including Zic4, indicating that some LGN-specific patterns of gene expression are upregulated in the MGN after visual “rewiring” (Fig. 7D, red type). Zic4 upregulation in rwMGN was confirmed with in situ hybridization, as was a lack of change in MGN marker, Foxp2 (supplemental Fig. 6, available at www.jneurosci.org as supplemental material). The remaining 31 enriched genes potentially represent novel processes of plasticity and/or response to injury (Fig. 7D, black type). Only one of those genes, Pappa, was a gene enriched in the P5 MGN group compared with P5 LGN (Fig. 7E, blue type); this gene encodes for pregnancy-associated plasma protein A and has been implicated in positively regulating IGF-1 availability (Harrington et al., 2007).
Figure 7.
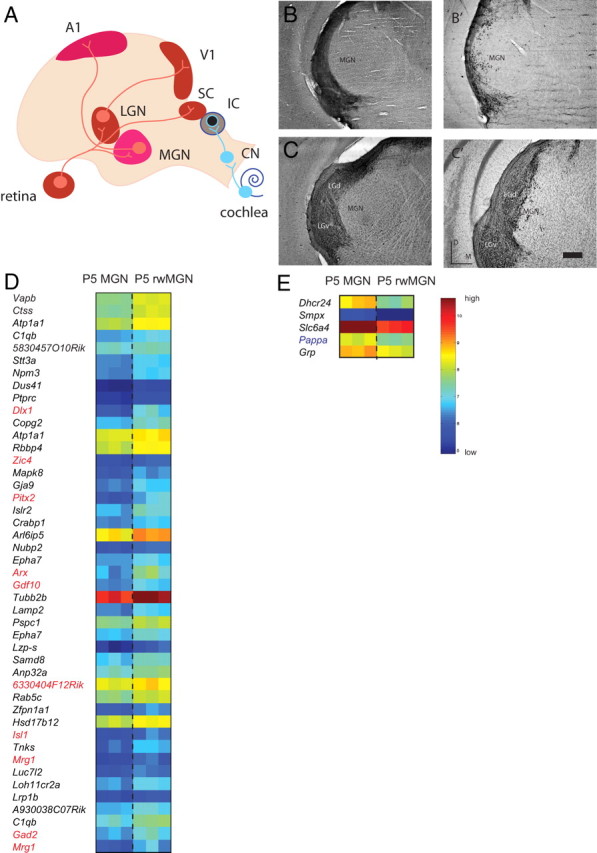
SAM identifies both novel and LGN-like patterns of gene expression in rwMGN at P5. A, Schematic of the rewired auditory pathway after IC ablation. MGN receives novel inputs from the retina, which drive the MGN and A1 to mediate visually driven responses. B, C, Intraocular injections of CTB project to the thalamus in normal (B, C) and visually rewired (B′, C′) mice. Coronal sections demonstrate ectopic retinogeniculate terminals in the rewired MGN (B′, C′) but not the normal MGN (B, C). D, Forty-four probes, corresponding to 41 unique genes, are enriched in the P5 rwMGN with a fold change (FC) >1.5 and false discovery rate <3.37% (n = 15–20 per replicate, 3 replicates per group, delta value = 0.666), and 10/41 (24.4%) of these enriched genes were present in the P5 LGN-enriched set (red type). E, Five probes, corresponding to five unique genes, were downregulated in the rwMGN, and only one was a gene enriched in the P5 MGN (blue type). Each group has three replicates with n = 15–20 each. All gene sets are ranked by SAM d-score. Color bar, Blue represents minimum intensity value within the gene set on the microarray; red represents maximum intensity.
Using the results of the SAM analysis of rwMGN and normal MGN, we searched for candidates with a potential role in guidance of retinal axons to their novel sensory target, rwMGN. To eliminate candidates with a nonspecific role in responsiveness to injury, we performed a control microarray on the P5 MGN from mice in which only the superior colliculus (SC) had been ablated at P0 (scMGN; data not shown). Ablating the SC, but not the IC, has been shown to induce ectopic targeting of retinal axons to the lateral posterior nucleus (LP) of the thalamus, but not the MGN, and therefore served as a control for changes in gene expression due to midbrain injury (Newton et al., 2004). One candidate, Slc6a4, a gene encoding a surface membrane serotonin transporter, was significantly downregulated in rwMGN and showed no difference between the normal MGN and scMGN (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). Because serotonin signaling has been shown to modulate target recognition of thalamocortical axons (Bonnin et al., 2007) and Slc6a4 has been shown to modulate eye-specific segregation of retinogeniculate terminals (Upton et al., 1999, 2002; Salichon et al., 2001), we hypothesized that this gene might play a role in axon targeting to the rwMGN. However, intraocular CTB injections of Slc6a4 knock-out mice revealed no ectopic projections to the MGN (n = 3 KO, n = 2 WT) (supplemental Fig. 4, available at www.jneurosci.org as supplemental material), suggesting that loss of Slc6a4 is not sufficient to induce rewiring of retinal axons to the MGN.
To identify additional candidates with a potential role in visual rewiring, we screened the P5 MGN and rwMGN data with our list of known axon guidance genes (supplemental Table 3, available at www.jneurosci.org as supplemental material). We found that EphA7, plexin-C1, RGMb, and Ntrk2 were upregulated in rwMGN, while semaphorin3a, Dcc, netrinG2, robo2, NCAM1, and slit2 were downregulated (supplemental Fig. 5, available at www.jneurosci.org as supplemental material), suggesting that a suite of axon guidance factors modulates targeting and topographic mapping of retinal ganglion cell axons into the rwMGN. Two upregulated factors in rwMGN, EphA7 and Ntrk2, were increased in P5 LGN compared with P5 MGN and possibly represent common mechanisms of retinogeniculate targeting and/or patterning in the LGN and rwMGN.
Finally, GSEA analysis identified 332/1892 gene sets significantly upregulated in the rwMGN relative to the P5 MGN at p < 0.01 (supplemental Table 8, available at www.jneurosci.org as supplemental material). Gene sets in the top 20 included a set of genes downregulated in response to rapamycin, inhibitor of the mTOR pathway, and the TNFα pathway, potentially reflecting cell death in response to deafferentation. Six/1892 gene sets were significantly downregulated at p < 0.01 (supplemental Table 9, available at www.jneurosci.org as supplemental material). Among these sets were the NF-κB pathway and Wnt signaling pathway, the latter of which was the most significantly enriched set of the P5 MGN.
Discussion
Using gene microarrays, we have identified groups of novel candidates with a potential role in LGN and MGN specification. At P0, a relatively small number of genes distinguish LGN from MGN, and a large proportion (85% and 92%, respectively) continue to show differential expression at P5, suggesting that a small number of programs orchestrate this postnatal differentiation. At P5, the number of differentially expressed genes increases about fivefold, reflecting a proliferation of secondary differentiation programs with age. Between P0 and P5, changes in expression are remarkably similar for the LGN and MGN (79% of the top differentially expressed probes are identical), suggesting that they undergo similar processes of maturation.
Among the candidates for LGN specification are four of the five members of the Zic family of transcription factors, which have been implicated in early patterning events (Aruga, 2004; Gaston-Massuet et al., 2005). Among the MGN candidates are three transcriptional modulators—Foxp2, a transcriptional repressor, Crabp2, a retinoic acid binding protein, and Sdccag33, a zinc-finger transcription factor. Crabp2 and Sdccag33 are enriched in the MGd and MGv, respectively, making them potentially useful MGN markers.
Functional significance of LGN- and MGN-enriched genes
Visual and auditory stimuli have distinct spatiotemporal characteristics and lead to different stimulus feature representations, frequency codes, and intra-areal circuitry (Sharma et al., 2000; Jones, 2007). Candidate genes enriched in the LGN and MGN potentially shape differences in circuit structure and function through axonal pathfinding (from E15 to P5), cellular differentiation, and synaptogenesis (Tuttle et al., 1998; Lyckman et al., 2001).
Differences in LGN and MGN expression may originate from differences in the position of precursor cells relative to local signaling centers (Vue et al., 2007; Szabó et al., 2009). The nature and identity of these signals are newly being discovered, including cues from the zona limitans intrathalamica (Shh), alar plate (Wnt), and border between the thalamus and pretectum (FGF8) (Kiecker and Lumsden, 2004; Kataoka and Shimogori, 2008; Vue et al., 2009). Our screen likely includes candidates that participate in these region- or lineage-specific programs of differentiation, and some previously published markers were confirmed: Dlx1 and Arx expression colocalizes with cells of the LGv (Kitamura et al., 1997) and selective expression of Isl1, Pax6 and Dlx1,2,5 in the LGN and Gbx2 in the MGN has been observed (Nakagawa and O'Leary, 2001; Kawasaki et al., 2004).
In addition to our screen for individual candidates, we used GSEA to search for functionally related groups of genes (Lyckman et al., 2008; Tilford and Siemers, 2009). Cytokine signaling pathways, such as the CD40-downregulated and IL-2 pathways, were identified in P0 LGN, while transcriptional networks, including MeCP2 and Hoxc8 pathways, were linked to P0 MGN. Intriguingly, several pathways in P5 LGN were implicated in visual plasticity: TNFα is involved in homeostatic increases of synaptic strength (Kaneko et al., 2008), as well as retinal ganglion cell (RGC) degeneration (Nakazawa et al., 2006); the mTOR pathway promotes adult RGC axon regeneration (Park et al., 2008); and the VEGF pathway supports RGC survival after injury (Kilic et al., 2006; Nishijima et al., 2007). Our results implicate these processes in retinogeniculate circuit formation of the perinatal LGN.
The advantage of a gene microarray approach is its capacity to identify previously unidentified candidates in processes of regional specification (Kawasaki et al., 2004; Sun et al., 2005; Murray et al., 2007, 2008; Leamey et al., 2008). Enriched groups provide the link between early patterning events and later functional processes of circuit formation and cellular differentiation. Limitations include the fact that the data reflect transcript, not protein levels, the measurement of which requires protein arrays. Dissections pooled structural subdivisions, as well as functional cell subpopulations, including specialized streams of visual information (Huberman et al., 2008; Kim et al., 2008). Laser dissection or fluorescence-activated cell sorting (FACS) techniques may be necessary to characterize the genetic identity of these subdivisions (Arlotta et al., 2005).
Zic4 exhibits a novel role in retinogeniculate mapping
Zic4 is a zinc-finger transcription factor with a role in cerebellar development, its heterozygous codeletion with Zic1 leading to cerebellar malformation in mice and Dandy-Walker malformation in humans (Grinberg et al., 2004). We provide the first report of Zic4's role in visual pathway patterning. While loss of family member, Zic2, leads to the failure of retinogeniculate fibers to project ipsilaterally (Pak et al., 2004), Zic4 loss produces the disordering of appropriately targeted ipsilateral fibers. Moreover, Zic2 is not detectably expressed in LGd, highlighting a divergent role for Zic4.
Increased clustering and disorganization of the ipsilateral terminals of Zic4-null mice resembles, but does not mimic, the phenotype of ephrin-A2/3/5-null mice (Pfeiffenberger et al., 2005). While both exhibit increased ipsilateral clustering, dorsomedial expansion of the ipsilateral projection in the Zic4 null is not present in the ephrin-A2/3/5 null. Interestingly, this dorsomedial expansion is the complement of the dorsoventral expansion of ipsilateral fibers in Ten_m3-null mouse (Leamey et al., 2008) and whether these molecules interact to establish proper ipsilateral retinotopic projections warrants future investigation.
An open question is whether it is loss of Zic4 in the projecting cells, or target cells, or both that leads to mismapping of ipsilateral fibers. Loss of the temporal high Zic4 gradient in the retina might allow ipsilateral axons formerly enriched in Zic4 to terminate more dorsomedially in the LGd. Alternatively, loss of Zic4 in LGd target cells might downregulate a repulsive cue that permits ipsilateral expansion. It is also unclear whether Zic4 regulates the expression of downstream axon guidance cues or acts itself as a cue, as has been demonstrated for other transcriptional factors such as En-2 (Brunet et al., 2005). In vitro assays, as well as region-specific mutants of Zic4 or in utero electroporation targeting retinal or thalamic cells for Zic4 knock-down or overexpression, would be useful in addressing this question.
Foxp2 and activity-dependent regulation in the mammalian brain
Foxp2 is a transcriptional repressor implicated in orofacial dyspraxia and corticostriatal abnormalities in humans with a heterozygous loss-of-function mutation (Vernes et al., 2006). It is necessary for ultrasonic vocalizations in perinatal mice (Shu et al., 2005) and song learning in adult canaries (Haesler et al., 2007), and its expression is dynamically regulated in songbirds with activity or conditions of learning (Haesler et al., 2004; Teramitsu and White, 2006).
We report here for the first time in mammals, that Foxp2 is positively regulated with auditory stimulation, suggesting an activity-dependent role in synaptic plasticity of the auditory pathway. Although a role in axonal pathfinding was not explored, Foxp2 may additionally contribute to mapping of auditory projections, as Foxp1 has been shown to regulate motoneuron differentiation and pathfinding (Rousso et al., 2008). A role for Foxp2 in early stages of auditory processing may contribute to higher level deficits of speech and language in humans (Vargha-Khadem et al., 2005).
Mechanisms of cross-modal plasticity in response to early injury
In the cortex, areal specification is influenced by intrinsic factors, including positional and lineage-derived gene programs (Miyashita-Lin et al., 1999) and extrinsic factors, such as the nature and identity of inputs (Dehay et al., 1993, 2001; Lukaszewicz et al., 2005). Similarly, we show that the functional differentiation of thalamic nuclei is influenced by its inputs. A quarter of genes and several gene pathways, including the mTOR-related pathway, that were upregulated in the rwMGN are also present in the LGN. As it is unlikely that LGN-like patterns of gene expression are the “default state” of the MGN in the absence of IC input, we propose that retinal axons induce specific LGN-like patterns of gene expression through their frequency code and/or chemical signals. Nonetheless, there may be constraints on how the MGN interprets novel input, leading to additional “plasticity-related” expression.
The molecular mechanism of retinal targeting to the deafferented MGN is not understood. Patterns of rwMGN gene expression include axon guidance pathways potentially mediating retinal ingrowth. A promising candidate, the downregulated serotonin transporter, Slc6a4, has previously been shown to modulate eye-specific segregation and influence somatosensory barrel formation (Upton et al., 1999, 2002; Salichon et al., 2001). However, Slc6a4-null mice are not rewired, suggesting either that its loss is not sufficient or that its effects are compensated by other genes. It is possible that other genes alone or in addition to downregulated Slc6a4—e.g., upregulated Zic4 (supplemental Fig. 6, available at www.jneurosci.org as supplemental material) or C1qb (Stevens et al., 2007)—are required or that the physical ablation of IC is necessary.
Conclusion
In summary, we report here newly identified, enriched groups of candidate genes for perinatal murine LGN and MGN specification. These genes and gene sets potentially reflect distinct differentiation programs set by differences in patterning cues and neural inputs. We identify for the first time a role for Zic4 in ipsilateral retinotopic patterning to the thalamus, and an activity-dependent regulation of Foxp2 in response to auditory stimulation. Furthermore, abnormal visual inputs respecify the MGN with both LGN-like and novel patterns of gene expression. These data thus identify novel molecular mechanisms linking early patterning events to functional differentiation of the visual and auditory pathways.
Footnotes
This work was supported by National Institutes of Health Grants F30NS057899 (S.H.), and R01EY015068, R01EY007023, and the Simons Foundation (M.S.). D.P. was supported by funding from the Nancy Lurie Marks Family Foundation. We thank Azadeh Moini for RT-PCR data analysis, Cortina McCurry for Western blot assistance, Orsi Kuti for technical support, Dennis Murphy for generating the Slc6a4-null mouse, and Alvin Lyckman, Cathy Leamey, Morgan Sheng, Yasonuri Hayashi, and Elly Nedivi for helpful comments on the project.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Bengel D, Murphy DL, Andrews AM, Wichems CH, Feltner D, Heils A, Mössner R, Westphal H, Lesch KP. Altered brain serotonin homeostasis and locomotor insensitivity to 3, 4-methylenedioxymethamphetamine (“Ecstasy”) in serotonin transporter-deficient mice. Mol Pharmacol. 1998;53:649–655. doi: 10.1124/mol.53.4.649. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bonnin A, Torii M, Wang L, Rakic P, Levitt P. Serotonin modulates the response of embryonic thalamocortical axons to netrin-1. Nat Neurosci. 2007;10:588–597. doi: 10.1038/nn1896. [DOI] [PubMed] [Google Scholar]
- Braissant O, Wahli W. A simplified in situ hybridization protocol using non-radioactively labeled probes to detect abundant and rare mRNAs on tissue sections. Biochemica. 1998;1:10–16. [Google Scholar]
- Braun MM, Etheridge A, Bernard A, Robertson CP, Roelink H. Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development. 2003;130:5579–5587. doi: 10.1242/dev.00685. [DOI] [PubMed] [Google Scholar]
- Brunet I, Weinl C, Piper M, Trembleau A, Volovitch M, Harris W, Prochiantz A, Holt C. The transcription factor Engrailed-2 guides retinal axons. Nature. 2005;438:94–98. doi: 10.1038/nature04110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Giroud P, Berland M, Smart I, Kennedy H. Modulation of the cell cycle contributes to the parcellation of the primate visual cortex. Nature. 1993;366:464–466. doi: 10.1038/366464a0. [DOI] [PubMed] [Google Scholar]
- Dehay C, Savatier P, Cortay V, Kennedy H. Cell-cycle kinetics of neocortical precursors are influenced by embryonic thalamic axons. J Neurosci. 2001;21:201–214. doi: 10.1523/JNEUROSCI.21-01-00201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figdor MC, Stern CD. Segmental organization of embryonic diencephalon. Nature. 1993;363:630–634. doi: 10.1038/363630a0. [DOI] [PubMed] [Google Scholar]
- Gaston-Massuet C, Henderson DJ, Greene ND, Copp AJ. Zic4, a zinc-finger transcription factor, is expressed in the developing mouse nervous system. Dev Dyn. 2005;233:1110–1115. doi: 10.1002/dvdy.20417. [DOI] [PubMed] [Google Scholar]
- Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- Grove EA, Fukuchi-Shimogori T. Generating the cerebral cortical area map. Annu Rev Neurosci. 2003;26:355–380. doi: 10.1146/annurev.neuro.26.041002.131137. [DOI] [PubMed] [Google Scholar]
- Guido W. Refinement of the retinogeniculate pathway. J Physiol. 2008;586:4357–4362. doi: 10.1113/jphysiol.2008.157115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, Scharff C. FoxP2 expression in avian vocal learners and non-learners. J Neurosci. 2004;24:3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington SC, Simari RD, Conover CA. Genetic deletion of pregnancy-associated plasma protein-A is associated with resistance to atherosclerotic lesion development in apolipoprotein E-deficient mice challenged with a high-fat diet. Circ Res. 2007;100:1696–1702. doi: 10.1161/CIRCRESAHA.106.146183. [DOI] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, Barres BA. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59:425–438. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG. The thalamus. Ed 2. New York: Plenum; 2007. [Google Scholar]
- Kaneko M, Stellwagen D, Malenka RC, Stryker MP. Tumor necrosis factor-alpha mediates one component of competitive, experience-dependent plasticity in developing visual cortex. Neuron. 2008;58:673–680. doi: 10.1016/j.neuron.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka A, Shimogori T. Fgf8 controls regional identity in the developing thalamus. Development. 2008;135:2873–2881. doi: 10.1242/dev.021618. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Crowley JC, Livesey FJ, Katz LC. Molecular organization of the ferret visual thalamus. J Neurosci. 2004;24:9962–9970. doi: 10.1523/JNEUROSCI.2165-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 2004;7:1242–1249. doi: 10.1038/nn1338. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- Kilic U, Kilic E, Järve A, Guo Z, Spudich A, Bieber K, Barzena U, Bassetti CL, Marti HH, Hermann DM. Human vascular endothelial growth factor protects axotomized retinal ganglion cells in vivo by activating ERK-1/2 and Akt pathways. J Neurosci. 2006;26:12439–12446. doi: 10.1523/JNEUROSCI.0434-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Yamagata M, Meister M, Sanes JR. Molecular identification of a retinal cell type that responds to upward motion. Nature. 2008;452:478–482. doi: 10.1038/nature06739. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Miura H, Yanazawa M, Miyashita T, Kato K. Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech Dev. 1997;67:83–96. doi: 10.1016/s0925-4773(97)00110-x. [DOI] [PubMed] [Google Scholar]
- Leamey CA, Glendining KA, Kreiman G, Kang ND, Wang KH, Fassler R, Sawatari A, Tonegawa S, Sur M. Differential gene expression between sensory neocortical areas: potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb Cortex. 2008;18:53–66. doi: 10.1093/cercor/bhm031. [DOI] [PubMed] [Google Scholar]
- Lim Y, Golden JA. Patterning the developing diencephalon. Brain Res Rev. 2007;53:17–26. doi: 10.1016/j.brainresrev.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, Kennedy H, Dehay C. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47:353–364. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyckman AW, Jhaveri S, Feldheim DA, Vanderhaeghen P, Flanagan JG, Sur M. Enhanced plasticity of retinothalamic projections in an ephrin-A2/A5 double mutant. J Neurosci. 2001;21:7684–7690. doi: 10.1523/JNEUROSCI.21-19-07684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyckman AW, Horng S, Leamey CA, Tropea D, Watakabe A, Van Wart A, McCurry C, Yamamori T, Sur M. Gene expression patterns in visual cortex during the critical period: synaptic stabilization and reversal by visual deprivation. Proc Natl Acad Sci U S A. 2008;105:9409–9414. doi: 10.1073/pnas.0710172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, Rubenstein JL. Early neocortical regionalization in the absence of thalamic innervation. Science. 1999;285:906–909. doi: 10.1126/science.285.5429.906. [DOI] [PubMed] [Google Scholar]
- Murray KD, Choudary PV, Jones EG. Nucleus- and cell-specific gene expression in monkey thalamus. Proc Natl Acad Sci U S A. 2007;104:1989–1994. doi: 10.1073/pnas.0610742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray KD, Rubin CM, Jones EG, Chalupa LM. Molecular correlates of laminar differences in the macaque dorsal lateral geniculate nucleus. J Neurosci. 2008;28:12010–120122. doi: 10.1523/JNEUROSCI.3800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, O'Leary DD. Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci. 2001;21:2711–2725. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T, Nakazawa C, Matsubara A, Noda K, Hisatomi T, She H, Michaud N, Hafezi-Moghadam A, Miller JW, Benowitz LI. Tumor necrosis factor-alpha mediates oligodendrocyte death and delayed retinal ganglion cell loss in a mouse model of glaucoma. J Neurosci. 2006;26:12633–12641. doi: 10.1523/JNEUROSCI.2801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JR, Ellsworth C, Miyakawa T, Tonegawa S, Sur M. Acceleration of visually cued conditioned fear through the auditory pathway. Nat Neurosci. 2004;7:968–973. doi: 10.1038/nn1306. [DOI] [PubMed] [Google Scholar]
- Nishijima K, Ng YS, Zhong L, Bradley J, Schubert W, Jo N, Akita J, Samuelsson SJ, Robinson GS, Adamis AP, Shima DT. Vascular endothelial growth factor-A is a survival factor for retinal neurons and a critical neuroprotectant during the adaptive response to ischemic injury. Am J Pathol. 2007;171:53–67. doi: 10.2353/ajpath.2007.061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD, Nakagawa Y. Patterning centers, regulatory genes and extrinsic mechanisms controlling arealization of the neocortex. Curr Opin Neurobiol. 2002;12:14–25. doi: 10.1016/s0959-4388(02)00285-4. [DOI] [PubMed] [Google Scholar]
- Pak W, Hindges R, Lim YS, Pfaff SL, O'Leary DD. Magnitude of binocular vision controlled by islet-2 repression of a genetic program that specifies laterality of retinal axon pathfinding. Cell. 2004;119:567–578. doi: 10.1016/j.cell.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C, Cutforth T, Woods G, Yamada J, Rentería RC, Copenhagen DR, Flanagan JG, Feldheim DA. Ephrin-As and neural activity are required for eye-specific patterning during retinogeniculate mapping. Nat Neurosci. 2005;8:1022–1027. doi: 10.1038/nn1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale CW, Grove EA. Patterning the mammalian cerebral cortex. Curr Opin Neurobiol. 2001;11:50–58. doi: 10.1016/s0959-4388(00)00173-2. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichon N, Gaspar P, Upton AL, Picaud S, Hanoun N, Hamon M, De Maeyer E, Murphy DL, Mossner R, Lesch KP, Hen R, Seif I. Excessive activation of serotonin (5-HT) 1B receptors disrupts the formation of sensory maps in monoamine oxidase a and 5-ht transporter knock-out mice. J Neurosci. 2001;21:884–896. doi: 10.1523/JNEUROSCI.21-03-00884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider GE. Early lesions of superior colliculus: factors affecting the formation of abnormal retinal projections. Brain Behav Evol. 1973;8:73–109. doi: 10.1159/000124348. [DOI] [PubMed] [Google Scholar]
- Sharma J, Angelucci A, Sur M. Induction of visual orientation modules in auditory cortex. Nature. 2000;404:841–847. doi: 10.1038/35009043. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development. 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder GA, Schmeidler J, De Gasperi R, Sosa MA, Rabidou D, Santucci AC, Perl D, Morrisey E, Buxbaum JD. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci U S A. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW, Barres BA. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Sur M, Garraghty PE, Roe AW. Experimentally induced visual projections into auditory thalamus and cortex. Science. 1988;242:1437–1441. doi: 10.1126/science.2462279. [DOI] [PubMed] [Google Scholar]
- Szabó NE, Zhao T, Zhou X, Alvarez-Bolado G. The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J Neurosci. 2009;29:2453–2466. doi: 10.1523/JNEUROSCI.4524-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramitsu I, White SA. FoxP2 regulation during undirected singing in adult songbirds. J Neurosci. 2006;26:7390–7394. doi: 10.1523/JNEUROSCI.1662-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilford CA, Siemers NO. Gene set enrichment analysis. Methods Mol Biol. 2009;563:99–121. doi: 10.1007/978-1-60761-175-2_6. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle R, Braisted JE, Richards LJ, O'Leary DD. Retinal axon guidance by region-specific cues in diencephalons. Development. 1998;125:791–801. doi: 10.1242/dev.125.5.791. [DOI] [PubMed] [Google Scholar]
- Upton AL, Salichon N, Lebrand C, Ravary A, Blakely R, Seif I, Gaspar P. Excess of serotonin (5-HT) alters the segregation of ispilateral and contralateral retinal projections in monoamine oxidase A knock-out mice: possible role of 5-HT uptake in retinal ganglion cells during development. J Neurosci. 1999;19:7007–7024. doi: 10.1523/JNEUROSCI.19-16-07007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton AL, Ravary A, Salichon N, Moessner R, Lesch KP, Hen R, Seif I, Gaspar P. Lack of 5-HT(1B) receptor and of serotonin transporter have different effects on the segregation of retinal axons in the lateral geniculate nucleus compared to the superior colliculus. Neuroscience. 2002;111:597–610. doi: 10.1016/s0306-4522(01)00602-9. [DOI] [PubMed] [Google Scholar]
- Vargha-Khadem F, Gadian DG, Copp A, Mishkin M. FOXP2 and the neuroanatomy of speech and language. Nat Rev Neurosci. 2005;6:131–138. doi: 10.1038/nrn1605. [DOI] [PubMed] [Google Scholar]
- Vernes SC, Nicod J, Elahi FM, Coventry JA, Kenny N, Coupe AM, Bird LE, Davies KE, Fisher SE. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15:3154–3167. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- von Melchner L, Pallas SL, Sur M. Visual behaviour mediated by retinal projections directed to the auditory pathway. Nature. 2000;404:871–876. doi: 10.1038/35009102. [DOI] [PubMed] [Google Scholar]
- Vue TY, Aaker J, Taniguchi A, Kazemzadeh C, Skidmore JM, Martin DM, Martin JF, Treier M, Nakagawa Y. Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol. 2007;505:73–91. doi: 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- Vue TY, Bluske K, Alishahi A, Yang LL, Koyano-Nakagawa N, Novitch B, Nakagawa Y. Sonic hedgehog signaling controls thalamic progenitor identity and nuclei specification in mice. J Neurosci. 2009;29:4484–4497. doi: 10.1523/JNEUROSCI.0656-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CJ, Pinson KI, Pleasure SJ. Severe defects in dorsal thalamic development in low-density lipoprotein receptor-related protein-6 mutants. J Neurosci. 2004;24:7632–7639. doi: 10.1523/JNEUROSCI.2123-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]