Abstract
Polar transport of the plant hormone auxin is controlled by PIN- and ABCB/PGP-efflux catalysts. PIN polarity is regulated by the AGC protein kinase, PINOID (PID), while ABCB activity was shown to be dependent on interaction with the FKBP42, TWISTED DWARF1 (TWD1). Using co-immunoprecipitation (co-IP) and shotgun LC–MS/MS analysis, we identified PID as a valid partner in the interaction with TWD1. In-vitro and yeast expression analyses indicated that PID specifically modulates ABCB1-mediated auxin efflux in an action that is dependent on its kinase activity and that is reverted by quercetin binding and thus inhibition of PID autophosphorylation. Triple ABCB1/PID/TWD1 co-transfection in tobacco revealed that PID enhances ABCB1-mediated auxin efflux but blocks ABCB1 in the presence of TWD1. Phospho-proteomic analyses identified S634 as a key residue of the regulatory ABCB1 linker and a very likely target of PID phosphorylation that determines both transporter drug binding and activity. In summary, we provide evidence that PID phosphorylation has a dual, counter-active impact on ABCB1 activity that is coordinated by TWD1–PID interaction.
Keywords: ABCB, PINOID, polar auxin transport, quercetin, TWISTED DWARF1
Introduction
Plant development and physiology depends on a unique, plant-specific process, the cell-to-cell or polar transport of auxin (PAT). PAT is controlled by efflux provided by members of the PIN-(PIN-FORMED) and B subfamily of ABC transporters (ABCBs), formerly called PGPs/MDRs (P-GLYCOPROTEIN, MULTIDRUG-RESISTANCE).
PIN-efflux carriers show mainly polar locations in PAT tissues and are thought to be the determinants of a ‘reflux loop’ in the root apex; their loss-of-function mutants are therefore characterized by strong developmental phenotypes (Blilou et al, 2005; Vieten et al, 2007). ABCB isoforms have been identified as primary, active (ATP-dependent) auxin pumps showing late developmental loss-of-function phenotypes (Geisler et al, 2005; Blakeslee et al, 2007; Mravec et al, 2008). Despite their predominnatly apolar locations, they have been demonstrated to contribute to PAT and long-range auxin transport (Geisler et al, 2003; Bouchard et al, 2006; Bailly et al, 2008). ABCB- and PIN-mediated auxin efflux can function independently and play identical cellular but separate developmental roles (Mravec et al, 2008). The current picture that emerges is that multilaterally expressed ABCBs minimize apoplastic reflux (Bailly et al, 2012a), while polar PINs provide a specific, vectorial auxin stream (Mravec et al, 2008). However, ABCBs and PINs are also capable of interactive and coordinated transport of auxin (Blakeslee et al, 2007).
On the posttranscriptional level, PAT has been shown to be controlled by protein–protein interaction, modulatory drugs and protein phosphorylation. The immunophilin-like FKBP42, TWISTED DWARF1 (TWD1), has been characterized as a central regulator of ABCB-mediated auxin transport by means of protein–protein interaction (Bailly et al, 2006). Positive regulation of ABCB1/PGP1- and ABCB19/PGP19/MDR1-mediated auxin transport (referred to as ABCBs hereafter) accounts for overlapping phenotypes between twd1 and abcb1 abcb19 (Bouchard et al, 2006; Bailly et al, 2008). ABCB1 and ABCB19 have been identified recently as binding proteins of the synthetic auxin-efflux inhibitor, 1-N-Naphtylphthalamic acid (NPA) (Murphy et al, 2002; Geisler et al, 2005; Rojas-Pierce et al, 2007; Nagashima et al, 2008; Kim et al, 2010). In addition, TWD1 binds to NPA and NPA binding disrupts TWD1–ABCB1 interaction (Murphy et al, 2002; Bailly et al, 2008). This leads to disruption of ABCB1 activity, suggesting that TWD1 and ABCB1 represent essential components of the NPA-sensitive-efflux complex (Bailly et al, 2008). On the contrary, several lines of evidence suggest that PIN proteins do not themselves act as direct targets of NPA (Lomax et al, 1995; Luschnig, 2001; Kim et al, 2010).
The serine–threonine kinase PINOID (PID) and the trimeric serine–threonine protein phosphatase 2A (PP2A) direct the polar targeting of PIN proteins (Friml et al, 2004; Michniewicz et al, 2007). The current model suggests that PID and PP2A antagonistically determine the fate of PIN cargoes for trafficking to the appropriate membrane by (de)phosphorylating conserved motifs of the hydrophilic loop of PIN proteins (Kleine-Vehn et al, 2009; Dhonukshe et al, 2010; Huang et al, 2010; Ding et al, 2011).
The regulatory A subunit, PP2AA1, called ROOTS CURL IN NPA1 (RCN1), is a negative regulator of basipetal transport in the root and as a consequence rcn1 roots exhibit a significant delay in gravitropism, consistent with an increased basipetal auxin transport (Sukumar et al, 2009; Rashotte et al, 2001). Importantly, the rcn1 gravitropic phenotype can be rescued by low concentrations of NPA, a concentration that is sufficient to block gravitropism in wild-type seedlings (Muday and DeLong). On the other hand, acropetal auxin transport is unaffected in rcn1, but shows a dramatic loss of NPA inhibition. Interestingly, rcn1 pin2 double-mutant analyses indicate that elevated basipetal transport in rcn1 does not require PIN2, leading to the suggestion that an NPA-binding protein is involved in this process (Rashotte et al, 2001).
PID belongs to the AGC family of serine/threonine kinases, and forms—together with AGC3-4/PID2, WAG1 and WAG2—the clade AGC3 (Galvan-Ampudia and Offringa, 2007). PID loss- or gain-of-function changes the apical (shoot-wards) or basal (root-wards) cellular localization of PIN proteins influencing the direction of the auxin movement (Friml et al, 2004). As a consequence, in the pid mutant, PIN1 localizes to the basal membrane of epidermal cells, which in turn redirects auxin away from the meristem and prevents the initiation of new lateral organs. This results in a pin-shaped inflorescence (Christensen et al, 2000). On the other hand, PID overexpression leads to a basal-to-apical switch of PIN1, PIN2 and PIN4 in root cortex and lateral root cap cells, and finally to a collapse of the root meristem probably due to auxin depletion (Michniewicz et al, 2007).
WAG1WAG2 loss-of-function mutations show an auxin-dependent root waving phenotype, and root curling is more resistant to NPA (Santner and Watson, 2006). Recently, PID, WAG1 and WAG2 were shown to phosphorylate PIN carriers at a conserved TPRXS(N/S) motif in the central hydrophilic loop leading to PIN recruitment into the apical recycling pathway (Dhonukshe et al, 2010; Huang et al, 2010). Moreover, disruption of PID and its three closest homologues completely abolishes the formation of cotyledons (Cheng et al, 2008). These findings, together with the fact that WAG1 and WAG2 are apolar and plasma-membrane-associated, suggested that AGC3 kinases act in the same or in a parallel regulatory pathway of PAT (Santner and Watson, 2006). Very recently, photoreceptor AGC4 kinase PHOTOTROPIN1 (phot1) was shown to phosphorylate nucleotide-binding domains (NBDs) of ABCB19 inhibiting its efflux activity (Christie et al, 2011).
Genetic and pharmacological analyses revealed that PID and PP2A antagonistically regulate basipetal auxin transport and gravitropic response in the root tip (Sukumar et al, 2009). Loss of PID activity alters the PIN2-mediated basipetal auxin transport and impedes the gravitropic response, without causing an obvious change in PIN2 cellular polarity. This finding indicates that PID promotes and enhances root gravitropism, but is not absolutely required. Furthermore, PID appears to have a specific regulatory effect on the basipetal transport machinery in the root, since the acropetal transport is unaffected in pid.
All these data are widely consistent with the concept of PID as a positive regulator of IAA efflux (Lee and Cho, 2006). The direct effect of PID on individual transporter activities, however, has not yet been addressed. Here, we identify and characterize PID as a relevant partner of the TWD1/ABCB subcomplex. Our data suggest that PID, besides its function as a molecular switch of PIN polarity, has a direct, dual impact on ABCB-mediated auxin-efflux activity. Transporter regulation is reversed by binding of the protein-kinase inhibitor, quercetin, a modulator of auxin transport.
Results
Identification of PINOID as a partner in the ABCB–TWD1 auxin-efflux complex
With the aim of identifying novel components of the auxin-efflux complex, characterized by ABCB1 and TWD1, we employed an IP approach followed by shotgun mass spectrometry analysis of TAPa-tagged TWD1 (TAPa-TWD1). Originally, we chose second-generation TAPa tagging, thus offering the possibility of IgG-BD-, 6xHIS- and 9xMYC epitope purification, that had been optimized for the identification of Arabidopsis protein complexes by two-step affinity purification. However, as IP using the HIS-tag only gave poor protein retention and the IgG-BD interfered with the MS analysis, we employed an one-step MYC IP. As starting material, we used total microsomes of 9-dag (days after germination) Arabidopsis roots, thus reducing typical nonspecific contaminants (like ribosomal proteins, chaperons and Rubisco; Supplementary Table S1). To gain further specificity of TWD1-interacting proteins, we subtracted identified vector control (35S:TAPa) proteins from pulled-down TAPa-TWD1 proteins, allowing elimination of proteins binding to the TAPa-tag alone (Supplementary Table S1). Finally, proteins with a score above 30 were considered as significant partners (Figure 1A). This procedure was repeated with each of the two independent transformants resulting in essentially the same TWD1 partners.
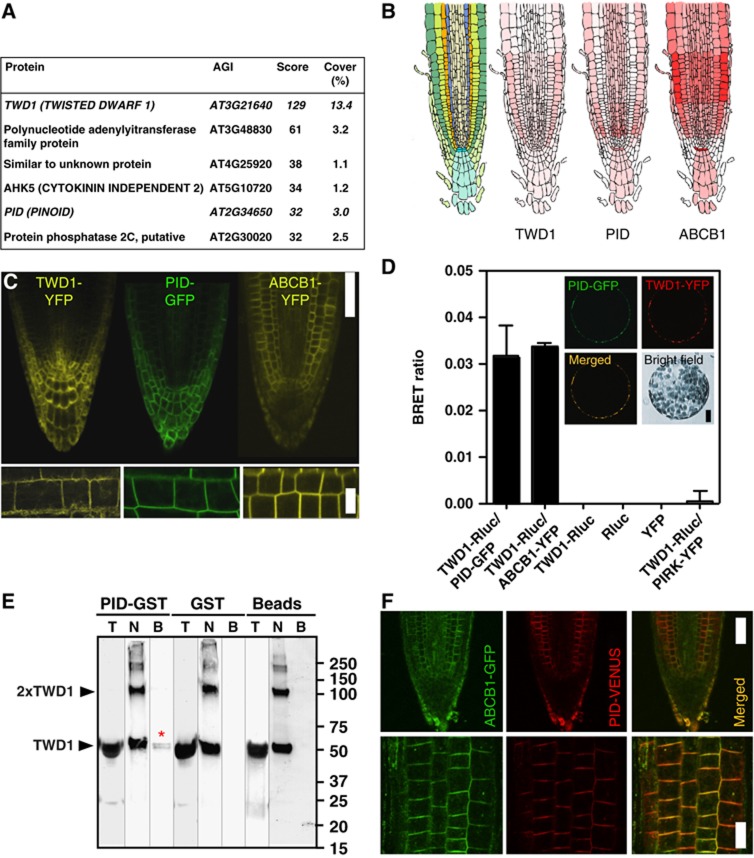
Figure 1.
PINOID physically interacts with the TWD1 immuno-complex. (A) TWD1 partners identified by co-IP using TAPa-TWD1 followed by shotgun MS/MS analysis. MASCOT-identified vector control (35S-TAPa) proteins were subtracted manually from TAPa-TWD1 proteins, and proteins with a score above 30 were considered as significant partners (see Supplementary Table S1 for complete listing and Supplementary Figure S1 for PID peptide sequences). (B, C) Expression of TWD1, PID and ABCB1 proteins overlap in primary root tips based on Arabidopsis gene Expression Database (AREXDB; B) and on confocal microscopy analysis of 35S:TWD1-YFP, 35S:PID-GFP and 35S:ABCB1-YFP (C). Upper row, confocal mid-plane of root tips (scale bar: 50 μm); lower row, close-up of cortical cells (bar, 20 μm). (D) BRET analysis of microsomes prepared from N. benthamiana leaves co-transfected with TWD1-Rluc (35S:TWD1-Rluc) and PID-GFP (35S:PID-GFP). Both TWD1 (TWD1-YFP) and PID (PID-GFP) colocalize on the plasma membrane of transfected protoplasts (inset). Positive BRET ratios that are comparable with established TWD1-Rluc/ABCB1-YFP interaction (Bailly et al, 2012a) are not found with negative plasma-membrane controls, such as PIRK-YFP, suggesting physical interaction in planta. (E) Immobilized PID-GST but not GST or glutathion-sepharose beads are able to pull-down significant amounts of purified TWD11-337in vitro indicated by an asterisk (T, total; N, non-bound; B, bound fractions). (F) ABCB1-GFP (ABCB1:ABCB1-GFP) colocalizes with PID-VENUS (PID:PID-VENUS) in epidermal layers of the root tip (upper row, confocal mid-plane of root tips (scale bar, 50 μm); lower row, epidermal planes (bar, 20 μm). Figure source data can be found with the Supplementary data.
Besides TWD1 as an obvious dominant pulled-down protein (protein score of 129, 13.4% coverage; Figure 1A), we found a so-far uncharacterized polynucleotide adenylyltransferase-like protein (At3G48830) and an unknown protein (At4G25920). In addition, two protein kinases, the histidine-like kinase AHK5/CYTOKININ INDEPENDENT2 (At5G10720) that regulates root elongation (Iwama et al, 2007) and the ACG kinase PINOID (At2G34650), and the catalytic domain of the PP2C-type protein phosphatase, AP2C1 (At2G30020), were also identified. AP2C1 is known to be involved in innate immunity responses by the negative regulation of the map kinases MPK4 and MPK6 (Schweighofer et al, 2007).
PINOID (PID) is a well-known key player in polar auxin transport regulation (Galvan-Ampudia and Offringa, 2007) and was therefore chosen for further analysis. TWD1/ABCB1 and PID show overlapping locations, mostly in epidermal and cortical cell layers, which is illustrated by confocal microscope analysis of PID-GFP, TWD1-YFP and ABCB1-YFP lines (Figure 1C). Although expressed under the control of the strong constitive 35SCaMV promoter, locations widely match expression profile predictions, such as those from the Arabidopsis gene Expression Database (AREXDB; Figure 1B), suggesting posttranscriptional modifications. TWD1-PID and ABCB1-PID colocations on the plasma membrane were further substantiated by co-expression in tobacco protoplasts (Figures 1D and 3B, Supplementary Figure S3).
TWD1–PID interaction was substantiated by in-planta bioluminescence resonance energy transfer (BRET) measurements (Bailly et al, 2008, 2012a) after co-expressing Renilla luciferase- and GFP-tagged versions of TWD1 (TWD1-Rluc) and PID (PID-GFP), respectively, in Nicotiana. benthamiana leaves. TWD1-Rluc, like Rluc-TWD1 (Bailly et al, 2012a), was shown to be functional by complementation of twd1-3 (Supplementary Figure S1E). Co-transfection of TWD1-Rluc and PID-GFP, widely colocalizing on the plasma membrane (Figure 1D inset), resulted in significant BRET ratios that are comparable to those found for the established TWD1–ABCB interaction (Bailly et al, 2012a), an indication for a physical proximity of <100 Å, and thus interaction. This interaction is specific since single expression of TWD1-Rluc, Rluc or YFP alone or TWD1-Rluc in combination with the non-related, plasma-membrane-bound protein kinase, PIRK (PIRK-YFP), only resulted in negligible BRET ratios.
Identification of PID as a TAPa-TWD1 partner does not necessarily imply direct physical interaction as the PID–TWD1 interaction might also be mediated via a third TWD1-interacting protein. Moreover, only one PID peptide with a MASCOT score of 32 was found in the MS analysis (Supplementary Figure S1). Therefore, in order to verify the IP data and to test a direct mode of interaction, we performed in vitro pull-down experiments using recombinant PID-GST (Christensen et al, 2000) and TWD11-337 protein. TWD11-337 purified as described (Kamphausen et al, 2002) contains all functional domains, such as the FKBD, the CaM-binding and TPR domain, except the C-terminal hydrophobic in-plane membrane anchor. Indeed, PID-GST, but not GST alone nor the empty-beads control, was able to pull-down small but significant amounts of TWD11-337 (Figure 1E).
In summary, our results demonstrate the utility of the IP approach employed for discovering valid protein–protein interactions of auxin transport complexes in Arabidopsis and suggest a relevant TWD1–PID interaction in planta.
PID has a dual impact on ABCB1-mediated auxin efflux
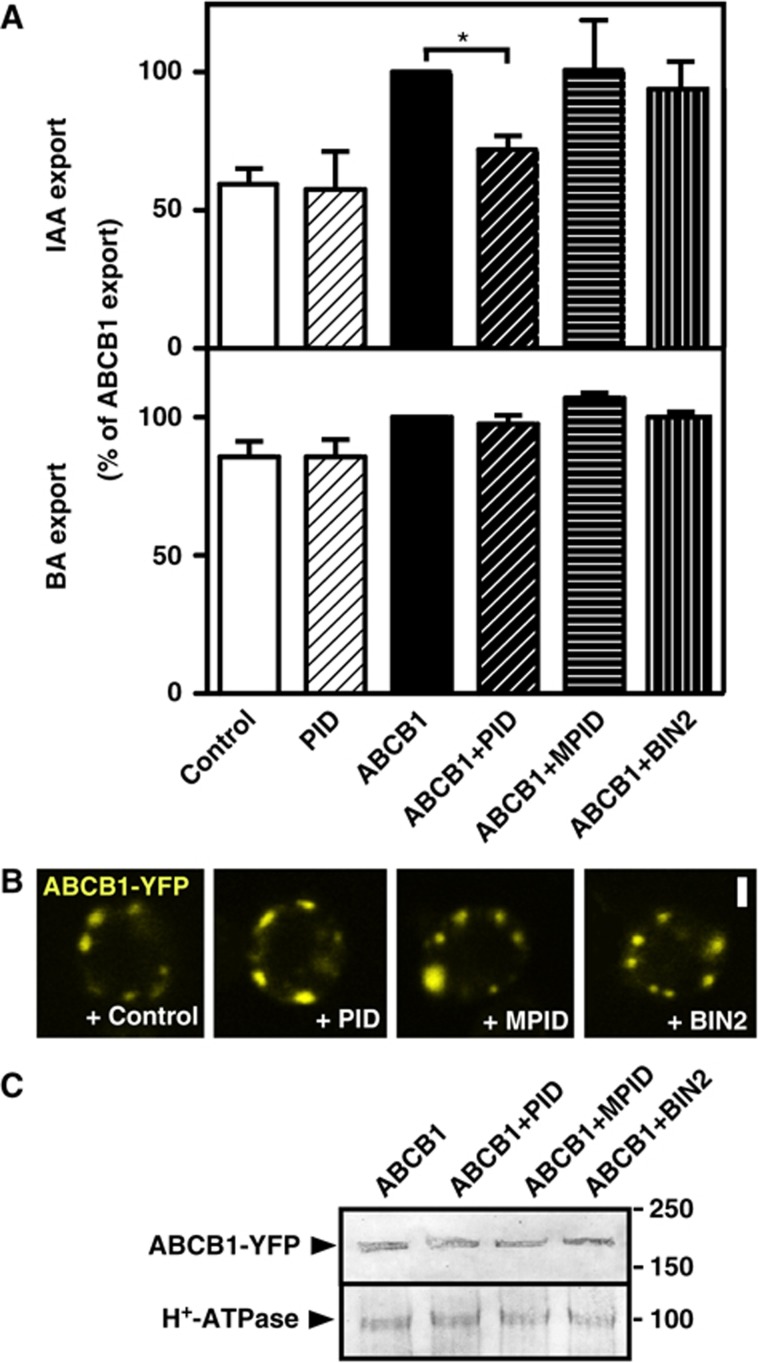
PID defines polar PIN locations and thus the directionality of auxin streams by direct PIN phosphorylation (Dhonukshe et al, 2010; Huang et al, 2010). A comparable mechanism has not so far been demonstrated for mainly nonpolar ABCBs or for TWD1, which functions as a regulator of ABCB1-efflux activity (Bouchard et al, 2006; Bailly et al, 2008). Therefore, we quantified ABCB1-mediated auxin efflux in the presence and absence of MYC-tagged PID in yeast. Surprisingly, PID significantly reduced ABCB1, but not vector control (background), IAA efflux by roughly 30% (Figure 2A). This inhibition corresponds to a 70% inhibition of ABCB1-specific efflux and is comparable to that found for ABCB1/TWD1 co-expression in yeast (Bailly et al, 2008). This inhibitory effect, caused by PID, was specific, as it was not found with the mutated, kinase-negative MPID (Christensen et al, 2000) or the non-related GSK-3-like kinase BRASSINOSTEROID-INSENSITIVE2 (BIN2), a negative regulator of brassinosteroid (BR) signalling and cell elongation (Vert, 2008). The specificity of PID-induced inhibition of IAA transport was further underlined by the fact that no effect was found for the unspecific, diffusion control, benzoic acid (BA) (Figure 2A lower panel).
Figure 2.
PID modulates ABCB1-mediated auxin efflux in yeast. (A) PID specifically inhibits ABCB1-mediated IAA export, while a mutated, inactive PID, MPID or unrelated protein kinase, BIN2, has no significant effect. Reduction of auxin retention (export) was calculated as relative export of initial export where ABCB1 was set to 100% (mean±s.e.; n=4–10). Significant differences (unpaired t-test with Welch’s correction, P<0.05) between –PID controls are indicated by asterisks. (B, C) Co-expression with PID, MPID or BIN2 does not significantly alter location (B) and expression (C) of ABCB1-YFP as revealed by confocal microscopy (B) and western analysis (C). Each 20 μg of protein was subjected to PAGE and western analysis using anti-GFP and plasma-membrane marker, anti-PMA1 (H+-ATPase). ABCB1-YFP localizes primarily to raft-like structures and the plasma membrane (see Supplementary Figure S2; Bailly et al, 2008). Bar, 2 μm.
Moreover, PID or MPID co-expression did not substantially alter ABCB1-plasma- membrane expression or localization in yeast (Figure 2B and C, Supplementary Figure S2A). ABCB1-YFP localizes primarily to raft-like structures at the boundaries of the plasma membrane as previously shown (Figure 2B; Bailly et al, 2008) and to the plasma membrane as demonstrated by comparison of anti-GFP immune-positive fractions in comparison with the plasma-membrane H+-ATPase, PMA1 (Supplementary Figure S2).
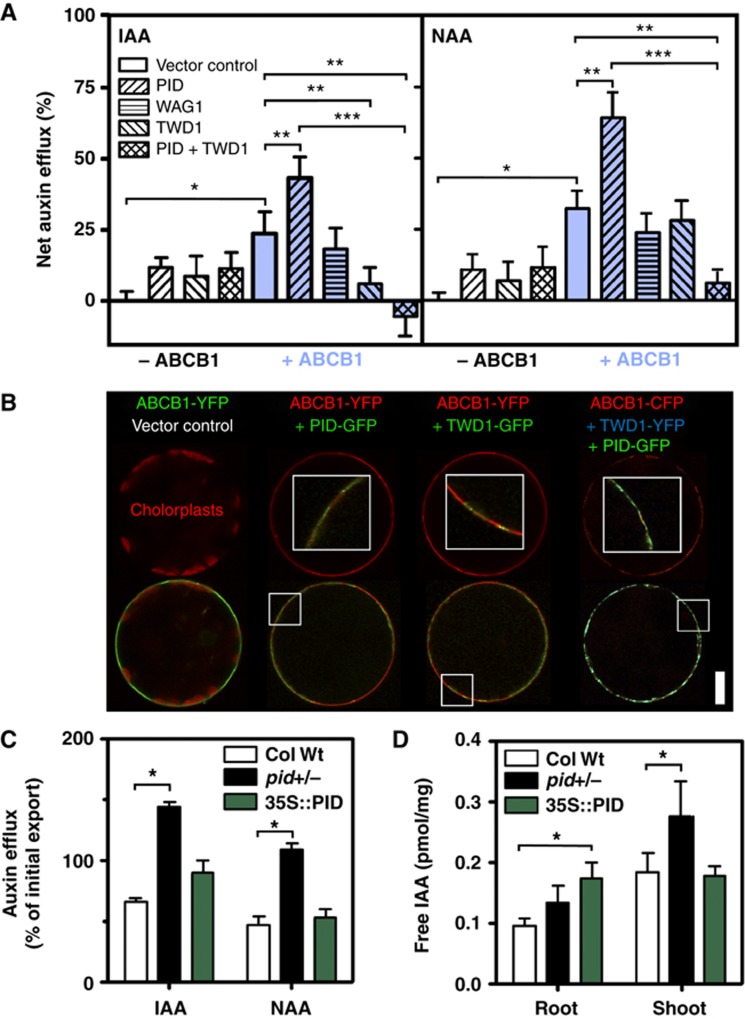
To further substantiate our yeast-generated results, we established a novel heterologous plant transport system that allowed quantification of auxin efflux of ABCB1, PID and TWD1 combinations from mesophyll protoplasts that were isolated from co-transfected N. benthamiana leaves. In agreement with described roles for PID on auxin-efflux activity (Lee and Cho, 2006), PID strongly activated ABCB1-catalysed IAA and NAA efflux. This action is specific as PID expression alone enhanced vector control IAA and NAA efflux only slightly, probably by activation of endogenous tobacco ABCB-type auxin exporters. Moreover, closely related AGC3 kinase, WAG1 sharing overlapping functionality and 39% protein sequence identity with PID (Dhonukshe et al, 2010; Huang et al, 2010), had no significant effect on ABCB1.
In order to address the role of TWD1 in PID-mediated activation of ABCB1, we quantified auxin efflux from triple-transfected ABCB1/PID/TWD1 protoplasts. Surprisingly, co-expression of ABCB1/PID/TWD1 entirely abolished ABCB1 auxin efflux. Co-expression of ABCB1/TWD1 revealed that at least for IAA a portion of this inhibitory effect is caused by TWD1 itself. However, NAA efflux analysed in parallel clearly demonstrated that PID, in the presence of TWD1, has a significant inhibitory effect on ABCB1 auxin efflux (Figure 3A). The finding that TWD1 only affected IAA but not NAA export, as previously reported for Arabidopsis (Bouchard et al, 2006) was also found for yeast, where TWD1 as reported here has an inhibitory role on ABCB1 (Bouchard et al, 2006; Bailly et al, 2008, 2012a), suggests that TWD1 besides its role as a regulator of activity also has an impact on ABCB1 specificity.
Figure 3.
PID negatively regulates ABCB1-mediated auxin efflux in planta. (A) Co-transfection of N. benthamiana protoplasts with PID specifically enhances ABCB1-mediated auxin (IAA and NAA) efflux in the absence of TWD1 but triple ABCB1/PID/TWD1 transfection strongly blocks ABCB1 activity (mean±s.e.; n=4). Significant differences (unpaired t-test with Welch’s correction, P<0.05) to vector control, ABCB1 and ABCB1/PID are indicated by one, two or three asterisks, respectively. (B) Co-transfection of PID (PID-GFP) and TWD1 (TWD1-GFP or TWD1-YFP) in N. benthamiana protoplasts does not significantly alter ABCB1 (ABCB1-YFP or ABCB1-CFP) location and expression in comparison to vector control co-expression (upper row). Note the colocalization of ABCB1, TWD1 and PID on the plasma membrane of tobacco protoplasts (lower row; insets in upper row show indicated details at higher magnification). For single and double co-expression controls, see Supplementary Figure S3H. Bar, 20 μm. (C) Efflux of native (IAA) and synthetic (NAA) auxin from Arabidopsis PID gain- (35S:PID) and loss-of-function (pid) protoplasts (means±s.e.; n=4; see Supplementary Figure S3C and D for time kinetics). (D) Free IAA levels determined by GC–MS are significantly elevated in the root of PID gain-of-function (35S:PID) and shoot of PID loss-of-function lines (pid+/−), respectively. Data are mean±s.e. (n=4 with each 40–50 seedlings). Note that material for C–D was prepared from heterozygous pid (pid+/−) plants since, due to technical limitations, a determination of homozygosity by shoot phenotyping or genotyping was not possible. Significant differences (unpaired t-test with Welch’s correction, P<0.05) between wild-type and mutant alleles/lines are indicated by asterisks.
Confocal microscopy analyses revealed that GFP-tagged PID and TWD1 as well as YFP-tagged ABCB1 and WAG1 all reside on the plasma membrane (Figure 3B, Supplementary Figure S3), where PID and TWD1 colocalize with ABCB1 (Figure 3B). TWD1 and PID, and to a lesser extend also ABCB1, revealed in some cases additionally to the plasma-membrane signals some intracellular signals (Figure 3, Supplementary Figure S3) that might represent artefacts caused by the constitutive, strong overexpression used in these assays. Importantly, and in analogy to the yeast system, co-expression of PID and TWD1 did not alter expression of ABCB1 nor its location on the plasma membrane as monitored microscopically (Figure 3B) and by western detection of ABCB1-MYC when compared to the plasma-membrane marker H+-ATPase, AHA2 (Supplementary Figure S3A).
In order to address the in-planta role of PID and to clarify its apparent dual role on ABCB1 activity, we quantified auxin efflux from Arabidopsis PID gain- and loss-of-function alleles. While 35S:PID mesophyll protoplasts did not show significant differences to the wild type, auxin efflux of both native and synthetic auxin from pid protoplasts was greatly enhanced (Figure 3C, Supplementary Figure S3) suggesting a negative impact on ABCB1 auxin export as found for ABCB1/PID/TWD1 co-expression in tobacco.
In summary, these results provide evidence that PID, dependent on the presence of TWD1, positively and negatively regulates ABCB1-mediated auxin efflux in an action that requires its kinase activity. However, based on our shoot-derived Arabidopsis model system, PID acts as a negative regulator in planta.
The protein-kinase inhibitor, quercetin, reverts PID-mediated ABCB1 modulation
Application of low concentrations of staurosporine, a well-known kinase inhibitor, to wild-type seedlings gives rise to phenotypes with the reduced basipetal auxin transport and delayed gravitropic response of pid suggesting PID as a primary target (Sukumar et al, 2009).
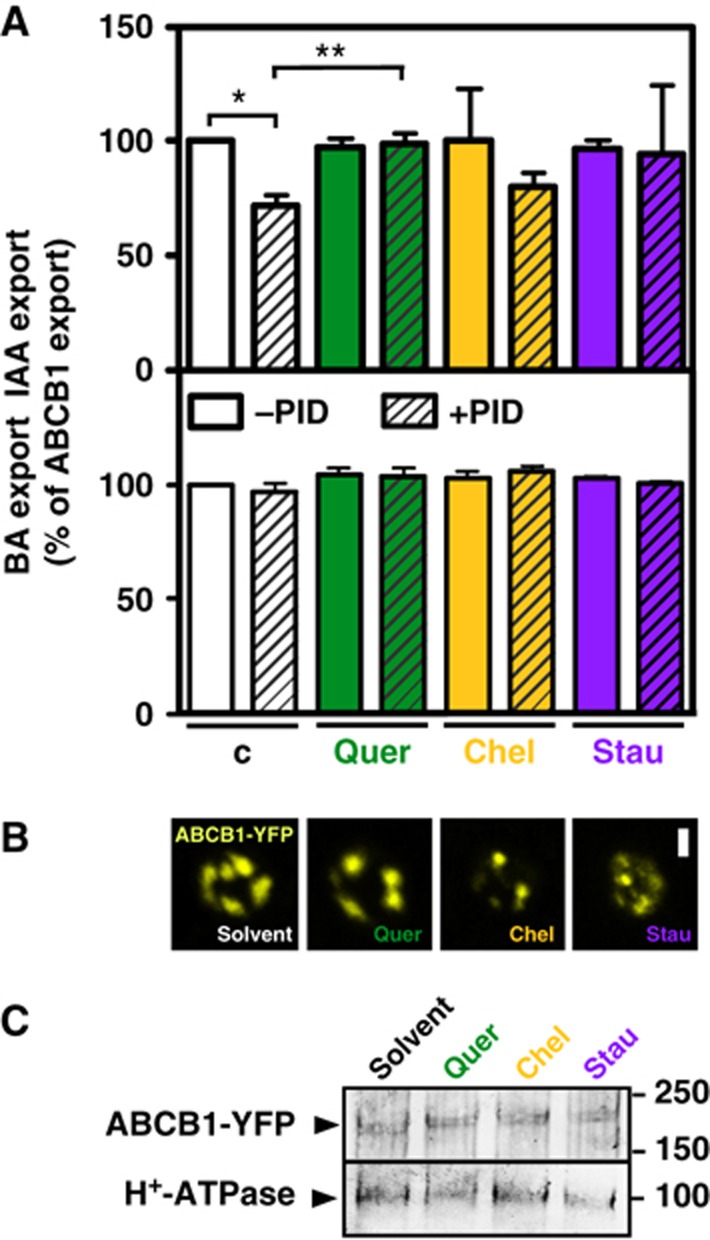
To further substantiate the mechanism underlying PID-mediated regulation of ABCB1, we therefore tested the effect of different protein-kinase inhibitors, like chelerythrine, staurosporine and quercetin, on regulation of PID in yeast. Chelerythrine is a potent and selective inhibitor of protein kinase C (PKC; IC50=0.7 μM), while staurosporine is known to be less specific but more potent (IC50 (PKC)=30 nM). The flavonoid quercetin is thought to act as a modulator of PAT and was shown to inhibit ABCB auxin transport (Geisler et al, 2005; Terasaka et al, 2005; Bouchard et al, 2006) and to disrupt ABCB1–TWD1 interaction (Bailly et al, 2008).
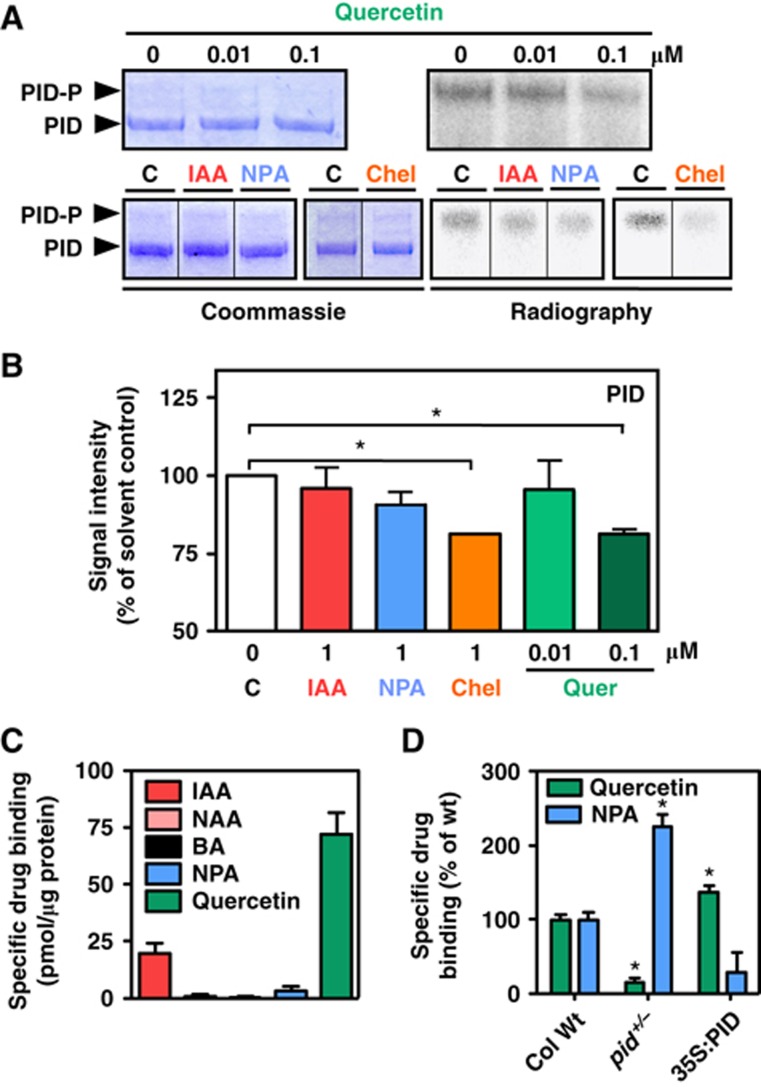
Quantification of ABCB1-catalysed auxin efflux in the presence and absence of PID revealed that only quercetin, and to a less significant extent staurosporine, was able to efficiently revert PID-mediated inactivation of ABCB1, while the mammalian protein-kinase inhibitor chelerythrine only had a mild effect (Figure 4). Again, reversal of drug inhibition was specific for IAA as the unspecific transport control, BA, assayed in parallel was not significantly affected. Moreover, confocal and western analyses revealed widely unchanged expression and location for ABCB1 in yeast upon treatment with kinase inhibitor (Figure 4B and C).
Figure 4.
The protein kinase and auxin transport inhibitor, quercetin, reverts PID-mediated ABCB1 inhibition in yeast. (A) Reduction of auxin (IAA) and BA retention (export) in the presence or absence of inhibitors (C, solvent control) is presented as relative export of initial export where ABCB1 solvent control was set to 100% (mean±s.e.; n=4–10). Significant differences (unpaired t-test with Welch’s correction, P<0.05) to −PID solvent control (one asterisk) or +PID solvent control (two asterisks) are indicated. Note that chelerythrine (chel; each 1 μM) unlike quercetin (quer) and staurosporine (stau) led to strong activation of vector control (backround) auxin efflux in the presence and absence of PID (not shown) requiring a relative presentation of activities. (B, C) Drug treatment (each 1 μM) does not significantly alter location (B) and expression (C) of ABCB1-YFP as revealed by confocal microscopy and western analysis. About 20 μg of each protein was subjected to PAGE and western analysis using anti-GFP and plasma-membrane marker, anti-PMA1 (H+-ATPase). ABCB1-YFP localizes primarily to raft-like structures and the plasma membrane (see Supplementary Figure S2; Bailly et al, 2008). Bar, 2 μm.
Quercetin binds to and inhibits PID kinase activity
Data, upon mutational and pharmacological treatments during yeast transport experiments, suggested that regulation of ABCB1 transport activity by PID is coupled to its function as a kinase. To verify this speculation, we quantified PID autophosphorylation and trans-phosphorylation of the standard kinase substrate, myelin basic protein (MBP), in the presence and absence of protein-kinase inhibitors and regulators of auxin transport. Quercetin blocked PID autophosphorylation (Figure 5A) as well as MBP trans-phosphorylation (Supplementary Figure S4) quantified by comparison of Coomassie stains (left panels) and the corresponding autoradiographs of phosphorylated PID (PID-P; Figure 5) and MBP bands (Supplementary Figure S4). In contrast to yeast transport experiments, a similar magnitude of quercetin inhibition of PID autophosphorylation (but not MBP trans-phosphorylation) was also found for chelerythrine (each 1 μM), while IAA and the synthetic auxin-efflux inhibitor, NPA, showed no significant effects.
Figure 5.
Quercetin binding blocks PID kinase activity. (A, B) In vitro autophosphorylation of PID-GST is inhibited by quercetin and chelerythrine while IAA and NPA have only slight effects. Coomassie stains (left panels) of non-phosphorylated PID (PID) was used as loading control (A). Autoradiographies (right panels) of autophosphorylated PID (PID-P), represented by the upper band in the Coomassie stain (Christensen et al, 2000), were quantified (B) and signal intensities were plotted against solvent controls (C; lower panel; means±s.e.; n=3). Significant differences (unpaired t-test with Welch’s correction, P<0.05) to solvent controls are indicated by asterisks. (C) PID-GST binds specifically quercetin and to a lesser amount as well IAA (mean±s.e.; n=4). Background drug binding to column material or GST alone (background) was corrected by subtracting specific binding to column-bound GST from column-bound PID-GST. (D) Microsomes prepared from PID loss- and gain-of-function lines show reduced and enhanced specific quercetin binding, respectively, but reciprocal specific NPA binding (mean±s.d.; n=4). Note that material was prepared from heterozygous pid (pid+/−) plants since, due to technical limitations, a determination of homozygosity by shoot phenotyping or genotyping was not possible. Reported values (C, D) are the means of specific radiolabelled drug bound in the absence of cold drug (total) minus radiolabelled drug bound in the presence of cold drug (unspecific). Significant differences (unpaired t-test with Welch’s correction, P<0.05) between GST alone (background; A) or wild-type and mutant microsomes (B) are indicated by asterisks. Figure source data can be found with the Supplementary data.
Chelerythrine and staurosporine have been shown to block PKC action by interacting with its catalytic domain. In order to investigate whether a similar mechanism does also account for PID inhibition, we measured the binding of radiolabelled quercetin to purified PID-GST and GST alone. Analysis of specific PID binding (binding to PID-GST minus binding to GST alone) showed significant quercetin binding (72.9±9.4 pmol/μg protein), while binding of NPA, NAA and BA was negligible (Figure 5C). Interestingly, despite its ineffectiveness in altering PID autophosphorylation, small but significant amounts of IAA were also bound to PID (20.2±4.2 pmol/μg). This phenomenon is currently under further investigation.
Direct binding of quercetin to PID was further supported by the fact that microsomes from PID loss-of-function alleles (pid+/−) showed drastically reduced quercetin binding (16.1±14.8% of wild-type), while gain-of-function lines (35S:PID) showed a significantly higher signal (137.9±20.4% of wild-type; Figure 5D). Surprisingly, interfering with PID expression had an inverse effect on NPA binding compared to quercetin: while NPA binding was enhanced in pid+/− by about a factor of 2, it was reduced to one-third in 35S:PID. This implies that PID, because it apparently does not bind NPA itself (Figure 5C), alters NPA-binding capacities of third-party NPA-binding proteins, such as ABCB1 or TWD1.
In summary, these data support the concept that PID is an in vivo target of quercetin that negatively regulates PID activity by direct drug binding.
The ABCB1 linker is a target of protein phosphorylation
The non-plant ABCB linker region of about 60 amino acids was identified to be subject of PKA (protein kinase A) or PKC phosphorylation altering its transport and associated ATPase activity (Chambers et al, 1994; Castro et al, 1999; Conseil et al, 2001). Proteomics approaches predicted three clusters of phosphorylation sites in the linker of Arabidopsis— ABCB1, ABCB4, ABCB11 and ABCB21 (Nuhse et al, 2004)—but the impact of these events on their activity is not entirely clear. Interestingly, most of the phosphorylation sites identified by in-silico prediction, experimentally or by phospho-proteomics, are well conserved among plant or animal orthologues, respectively, but are normally not shared among kingdoms (Figure 6C).
Figure 6.
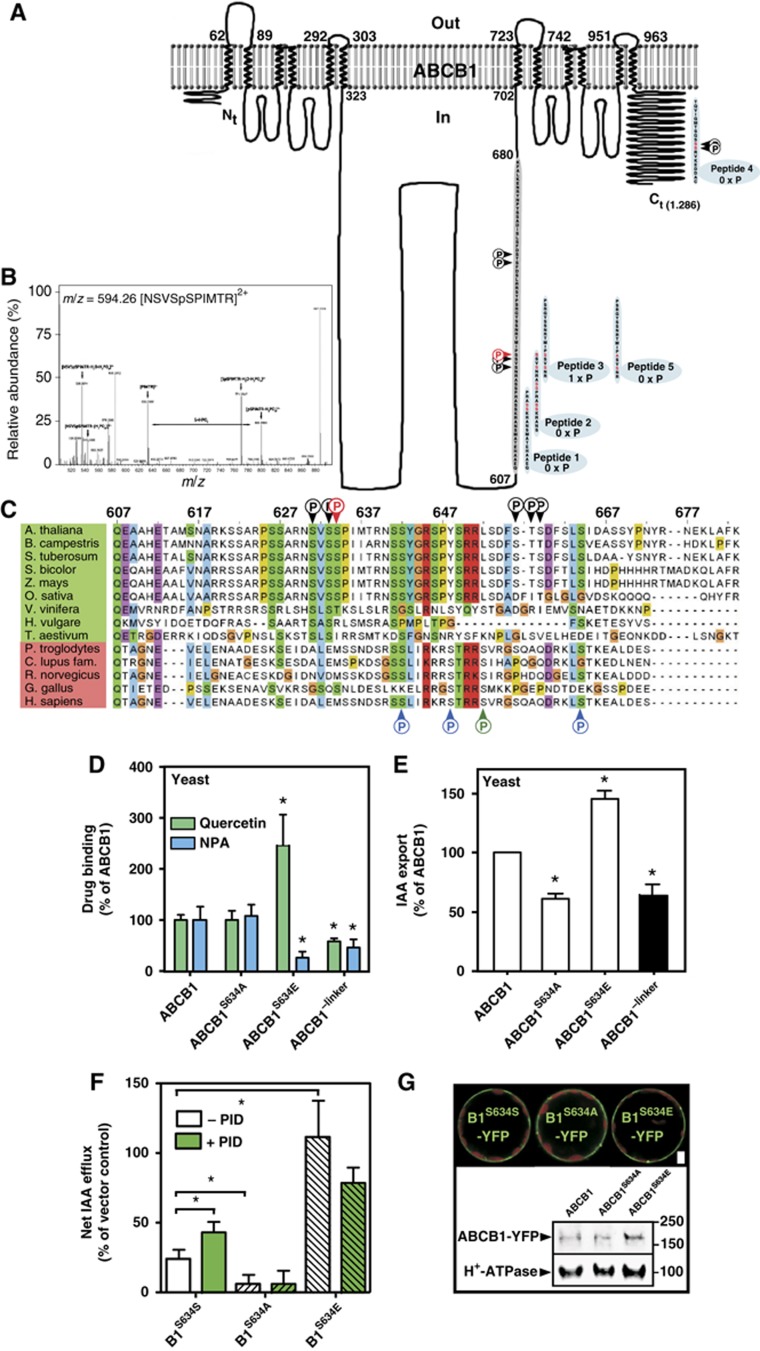
The ABCB1 linker domain is a target of protein phosphorylation. (A) Overview of ABCB1 linker (amino acids 607–680, grey background) phosphorylation sites either predicted in silico using NetPhos (red letters) or identified by phosphoproteomics (black P, Benschop et al, 2007; Nuhse et al, 2004; red P, this study). In vitro verification of linker phosphorylation by PID using synthetic peptides (blue background); number of PID phosphorylation sites (X × P) identified by MS/MS are indicated. Note that absence of peptide 2 phosphorylation is probably due to inefficient PID phosphorylation of S634 at the very C-terminus. (B) MS/MS spectrum and sequence of phosphorylated peptides derived from the ABCB1 linker after co-transfection of ABCB1 and PID in tobacco protoplast. Analysis of the data by detailed MASCOT search identified S634 as the only phosphorylation site in ABCB1. Fragmentation of the doubly charged parent peptide of 594.26 leads to the following results: The parent peptide has a mass of 594.26 Da; the five major peaks used for judging Ser634 phosphorylation are labelled by arrows (see text for details). The horizontal arrow illustrates the mass differences of a phosphorylated serine between y5 and y6 peptides (PIMTR (633.2988 Da) and SpSPIMTR (800.3568 Da)). A peak at 526.36 Da corresponds to the parent peptide losing a phosphate group (−98 Da) and a water molecule (−18 Da). As a second example, a peak at 545.43 Da corresponds to a peptide with neutral loss of a single phosphate group. The y5 fragment of parent peptide was detected at the correct size of 633.29 Da. After loss of a phosphate group and a water molecule the y7 of the parent peptide shows a peak at 771.20 Da. Finally, the peak at 800.35 Da represents the y6 parent peptide with neutral loss of a single phosphate group. (C) Relevant ABCB1 linker phosphorylation sites identified either by phosphoproteomics (black P, Nuhse et al, 2004; Benschop et al, 2007; red P, this study) or experimentally to be target of human PKC (blue P) or PKA (green P) phosphorylation are not conserved among plant and non-plant ABCB1 orthologues. (D) Microsomes prepared from yeast expressing a mutated version of ABCB1 mimicking S634 phosphorylation (ABCB1S634E) show reduced NPA but strongly enhanced quercetin binding, respectively, while deletion of the ABCB1 linker (ABCB1-linker) results in reduced NPA and quercetin binding (mean±s.e.; n=4). (E) Yeast expressing a mutated version of ABCB1 lacking S634 phosphorylation either by neutralizing (ABCB1S634A) or by linker deletion (ABCB1−linker) show reduced auxin export, while mimicking S634 phosphorylation (ABCB1S634E) has the opposite effect (mean±s.e.; n=4). Significant differences (unpaired t-test with Welch’s correction, P<0.05) to wild-type ABCB1 are indicated by asterisks. (F) Tobacco (N. benthamiana) protoplasts transfected with mutated versions of ABCB1 neutralizing (ABCB1S634A) or mimicking S634 phosphorylation (ABCB1S634E) show reduced and enhanced IAA export (mean±s.e.; n=4), respectively, which is not significantly altered by PID co-expression. Significant differences (unpaired t-test with Welch’s correction, P<0.05) to wild-type ABCB1 are indicated by an asterisk. (G) Mutagenesis of S634 in the ABCB (ABCB1-YFP) linker does not significantly alter ABCB1 expression and locations in N. benthamiana protoplasts as revealed by confocal microscopy (upper part) and western analysis (lower part). About 10 μg of each protein was subjected to western analysis using anti-GFP and plasma-membrane marker, anti-AHA2 (H+-ATPase); bar, 5 μm.
In order to explore whether the Arabidopis ABCB1 linker is indeed a target of protein-kinase phosphorylation under our experimental conditions, we analysed ABCB1 phosphorylation by LC—ESI—MS/MS after co-transfection with PID in tobacco leaves. The annotated MS spectra reported in Figure 6B shows the identified serine 634 (S634), detected previously by phosphoproteomics (Nuhse et al, 2004) as the only phosphorylation site in ABCB1 under our experimental settings.
In order to demonstrate that S634 identified in planta is indeed phosphorylated by PID and not via other kinases being themselves regulated by PID, we performed a PID in vitro phosphorylation analysis using overlapping peptides (peptide 1–3) covering the first half of the linker (Figure 6A). As a control, we used a peptide covering part of the ABCB1 C-terminus that has been shown to be phosphorylated upon early elicitor signalling (peptide 4; Benschop et al, 2007). MS analysis revealed that neither peptide 1, 2 nor 4 but only peptide 3 was phosphorylated by PID, leaving S633 and S634 as PID targets. S634 phosphorylation could be, however, excluded by using peptide 5 that contained an S634A exchange in comparison to peptide 3, suggesting that, in combination with previous phosphoproteomics (Nuhse et al, 2004), S634 is indeed a relevant PID phosphorylation site.
Interestingly, the phosphorylated peptide covering S634 (NSVSSPIMTR) showed no obvious sequence homology to the TPRXS(N/S) motif of PIN proteins recently shown to be phosphorylated by PID (Dhonukshe et al, 2010; Huang et al, 2010) suggesting a specific mode of PID action on auxin exporter classes. Moreover, repeating the same assay using human PKC resulted in phosphorylation of peptides 2–5 (Supplementary Figure S5) indicating a wider substrate specificity for human PKC in comparison to the plant PID kinase.
In summary, several lines of our own and previous analyses indicate that the ABCB1 linker is a target of protein phosphorylation with S634 being phosphorylated under our experimental conditions, which does, however, not strictly exclude that ABCB1 might be additionally phosphorylated at other sites when co-expressed in tobacco.
Quercetin and chelerythrine block root PAT but rescue the pin2 agravitropic phenotype
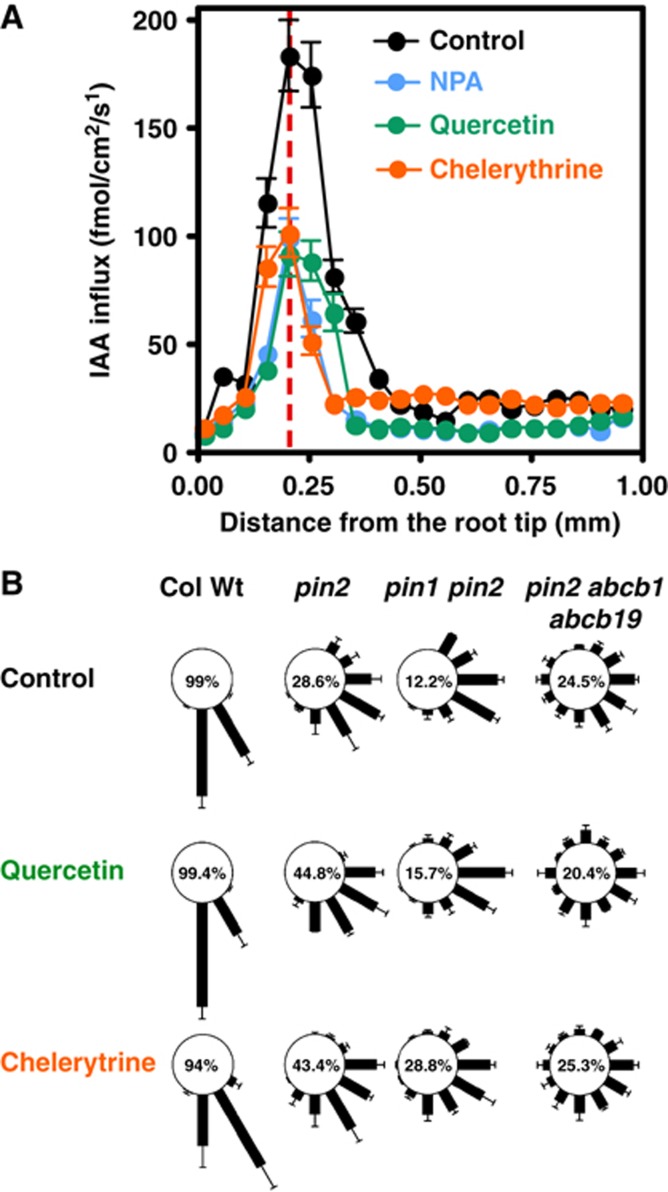
To test the physiological relevance of kinase inhibitors affecting the PID kinase activity as elaborated before in yeast and plant expression systems, we measured polar auxin transport in the root tip in the presence of quercetin and chelerythrine by using an auxin-specific auxin electrode (Mancuso et al, 2005). The underlying method for measurements of root IAA influxes has, besides the indirect visualization of expression of the auxin-responsive element DR5, become a well-established and reliable tool to quantify auxin fluxes in the root (Santelia et al, 2005; Bouchard et al, 2006; Bailly et al, 2008). Application of 5 μM chelerythrine and quercetin resulted in a strong inhibition of the maximal influx peaks at 200 μm from the root tip (Figure 7A). This reduction is similar to NPA treatments of equal concentrations (Bailly et al, 2008) and results in the same phenotype as the genetical loss of ABCB1/ABCB19 function alleles (Bouchard et al, 2006). Moreover, these data are consistent with a recent report where staurosporine was found to reduce basipetal IAA transport and gravity response (Sukumar et al, 2009). Interestingly, quercetin and chelerythrine—unlike NPA treatments—lead to basal and apical shifts of influx maxima, respectively. Moreover, IAA influx in distal regions (between 0.3 and 1 mm from the root tip) is insensitive to chelerythrine treatment but sensitive to NPA and quercetin. This suggests that overlapping targets are responsible for maximum influx peaks but distinct target spectra for distal root regions, as reported for abcb1 b19 and twd1 loss-of-function roots (Blakeslee et al, 2007; Bailly et al, 2008).
Figure 7.
Protein-kinase inhibitors, like quercetin and chelerythrine, block root PAT but rescue partially the pin2 agravitropic phenotype. (A) IAA influx profiles along wild-type roots in the presence of inhibitors (5 μM) measured using an IAA-specific microelectrode. Positive fluxes represent a net IAA influx. Data are means±s.e. (n=12). Note that chelerythrine and quercetin causes reduced influx peak at ca. 200 μm (dashed line) from the root tip comparable to NPA treatments. However, influx maxima are shifted apically and basipetally by chelerythrine and quercetin, respectively, but not by NPA (red line). (B) Quercetin- and chelerythrine-dependent rescue of pin2 agravitropic root phenotype requires ABCB1 and ABCB19. The length of each bar represents the mean percent angles±s.d. of seedlings showing the same direction of root growth of at least three independent experiments; numbers correspond to the mean per cent occurrence of 60° and 90° bending (sum of 60° and 90° sectors).
Recently, quercetin (like kaempferol) was shown to partially complement gravitropic-bending defects of pin2 (eir1-4) roots in a PIN1-dependent fashion and thus not found for pin1 pin2 alleles (Figure 7B; Santelia et al, 2008). Not surprisingly, this rescue was also found for chelerythrine treatments (mean per cent occurrence of 60 and 90° bending of 43.4% compared to 28.6% for the solvent control) at concentrations that do not inhibit root bending in wild-type seedlings. This supports the idea of a function for quercetin as an endogenous kinase inhibitor. In agreement, phorbole ester, a potent PKC activator, shows a slight inhibitory effect on pin2 gravitropic response (21.4%; Supplementary Figure S6), supporting the assumption that protein phosphorylation events trigger gravitropic root bending (Muday and DeLong). Surprisingly, staurosporine (26.4% compared to 28.6%) was less effective during pin2 rescue, indicating alternative targets or mode of actions for the different kinase inhibitors. Unlike quercetin (Santelia et al, 2008) and chelerythrine, which upregulates PIN1 expression at PIN2 domains (Supplementary Figure S7), staurosporine showed no influence on PIN1 expression or polar localization (Supplementary Figure S7; (Sukumar et al, 2009). The fact that the rescue of pin2 agravitropism was slightly enhanced in the presence of a combined quercetin and chelerythrine treatment (48.8% compared to 44.8 and 43.8%) argues for additive actions and independent pathways (Supplementary Figure S6). Partial rescue by chelerythrine in pin1 pin2 showed that the rescue by chelerythrine was, in contrast to what is found for flavonols, not PIN1-dependent (Figure 7B).
Remarkably, both quercetin and chelerythrine rescues were dependent on ABCB1 and ABCB19 functions, which became obvious by pin2 abcb1 abcb19 triple-mutant analysis (Figure 7B), suggesting ABCB1 and ABCB19 as direct chelerythrine/phosporylation targets. In summary, these physiological data support the idea that phosphorylation events trigger gravitropic root responses that depended on ABCB1 and ABCB19 phosphorylation.
Discussion
Identification of PID as a regulatory component of the TWD1/ABCB auxin-efflux complex
In this study, we have identified the AGC3 kinase PID by means of IP/MS analysis as a physical and functional partner of the auxin-efflux complex characterized by immunophilin-like TWD1 (Figure 1), a regulator of ABCB-mediated auxin efflux. A typical limitation of this approach is that fusion proteins may not be fully functional or capable of interaction, presumably due to steric hindrance from the epitope tag. Therefore, and in analogy to our previous yeast BRET system where an N-terminal fusion was proven functional (Bailly et al, 2008), we chose an N-terminal TAPa-tagged TWD1 as bait. We verified the functionality of TAPa-tagged TWD1 by genetic complementation test of the twd1-3 allele by TAPa-tagged TWD1. In all of the eight lines tested, TAPa-TWD1 complemented the ‘twisted syndrome’ in nearly all respects; only two complemented alleles had a slightly reduced growth compared to wild-type (Supplementary Figure S1A). Biochemical analysis revealed that auxin-efflux capacities were restored to wild-type level by TAPa-TWD1 complementation (Supplementary Figure S1C).
TWD1–PID interaction was verified by BRET analysis in planta and in vitro pull-downs. Assuming an equal PID–TWD1 interaction stoichiometry, pulled-down TWD1 was relatively low. This suggests that a portion of interacting proteins was eventually misfolded or that additive stabilizing or bridging factors were obviously absent in the in vitro assay. Relevant candidates might be ABCB1 itself, TOUCH3 (TCH3) or PINOID-BINDING PROTEIN1 (PBP1), which bind PID in a calcium-dependent manner, thus positively or negatively regulating its kinase activity, respectively (Benjamins et al, 2003). Of special interest is TCH3, a calmodulin-related protein (Benjamins et al, 2003), since calmodulin binding to the TWD1 CaM-binding domain has been shown previously (Kamphausen et al, 2002; Geisler et al, 2003).
The low protein coverage after co-IP is in agreement with a weak or transient mode of protein interaction and kinase action, and might explain also why we were unable to detect TWD1 in a reciprocal co-IP approach. Alternatively, the C-terminal GFP fusion employed in these studies might have resulted in a masked epitope or simply prevented protein interaction. Finally, expression of both TWD1 and PID is very low (Figure 1B) and microscopical analyses in Arabidopsis roots (Figure 1C and F) and tobacco protoplasts (Figure 3B, Supplementary Figure S3) suggest that PID-TWD1 (as well as PID-ABCB1) co-locations are limited to certain clusters on the plasma membrane further leading to a low ratio for the protein coverage.
Finally, interaction with TWD1 as a central part of the auxin-efflux complex provides a plausible ratio for a soluble kinase that has no obvious (plasma) membrane association motifs (Galvan-Ampudia and Offringa, 2007) to be plasma membrane bound (Figures 1 and 3; Friml et al, 2004; Michniewicz et al, 2007).
PID-mediated phosphorylation has a dual impact on ABCB1 activity
A plasma-membrane location for PID as shown here (Figures 1 and 3) and in other studies (Friml et al, 2004; Michniewicz et al, 2007) as well as interaction with TWD1 suggested a direct regulatory impact of PID on ABCB efflux activities. This was demonstrated by functional co-expression in yeast where PID was specifically blocking auxin efflux activities of ABCB1 (Figure 2). A negative impact of PID on ABCB1 in yeast was verified in planta where genetic deletion highly enhanced auxin efflux from pid mesophyll protoplasts (Figure 3).
Direct regulation of ABCB1 by PID was specific since it was found for effluxed auxin but not for the diffusion control, BA, and not with the closely related AGC3 kinase, WAG1 (Figure 3), or the unrelated GSK-3-like kinase, BIN2 (Figure 2). Moreover, PID regulation of ABCB1 was dependent on its PID kinase activity as kinase-deficient MPID had no effect and PID action was reverted by kinase inhibitors quercetin and staurosporine (Figure 4). This is in agreement with the data demonstrating pid to be staurosporine-insensitive providing evidence that PID is a direct target of staurosporine (Sukumar et al, 2009). Interestingly, both quercetin and staurosporine were shown to block PAT (Figure 7; Peer et al, 2004), gravitropism (Sukumar et al, 2009) and ABCB activity (Conseil et al, 1998; Geisler et al, 2005; Terasaka et al, 2005; Blakeslee et al, 2007; Sukumar et al, 2009).
However, use of N. benthamina protoplasts as a novel heterologous in-planta transport system revealed that ABCB1/PID co-expression resulted in enhanced auxin efflux (Figure 3). These findings are in agreement with recent data describing enhanced efflux from tobacco BY-2 cells upon PID overexpression (Lee and Cho, 2006) but contrast at first hand with yeast and Arabidopsis transport data (Figures 2 and 3). This discrepancy could be solved by the finding that triple co-expression of ABCB1/PID/TWD1 resulted in entire loss of auxin efflux, suggesting that the presence of TWD1 defines the positive or negative regulatory impact of PID on ABCB1. ABCB1/PID interference in the absence of TWD1 is supported by overlapping expression in certain tissue, such as the root stele (Figure 1B), and plasma membrane colocalizations in epidermal cell files (Figure 1F).
The obvious question that now arises is why PID has a negative impact on ABCB1 in yeast in the absence of TWD1. The most likely explanation is that ScFKBP12 is able to functionally complement TWD1 in yeast as has been suggested for TWD1 modulation of ABCB1 (Bouchard et al, 2006; Bailly et al, 2008). This is supported by the findings that ABCB1-mediated auxin efflux from yeast is strongly reduced in an fkbp12 strain (Bouchard et al, 2006) in analogy to mammalian MDR3 that was shown to be dependent on ScFKBP12 (Hemenway and Heitman, 1996). Finally, ScFKBP12 is able to widely complement twd1 loss-of-function alleles (unpublished data), which is slightly surprising as ScFKBP12 lacks functional TPR and calmodulin-binding domains as well as the C-terminal membrane anchor.
PID, like closely related AGC3 kinases, WAG1 and WAG2, phosphorylates the middle serines of cytoplasmic loops of PIN proteins in three conserved TPRXS(N/S) motifs (Michniewicz et al, 2007; Dhonukshe et al, 2010; Huang et al, 2010). Despite the fact that PID recognition motifs in the ABCB1 linker are distinct, three lines of evidence support an analogous event for the ABCB1 linker: First, PID co-expression regulates ABCB activity in an action that is dependent on its kinase activity as shown by mutational and pharmacological inhibition of PID kinase activity (Figures 2, 3, 4). Second, S634 of the ABCB1 linker is a target of kinase phosphorylation as shown by MS/MS analysis of ABCB1 co-expressed with PID in tobacco and PID in vitro peptide phosphorylation. And, third, mutational analyses of S634 alter ABCB1 activity and NPA-binding capacity expressed in yeast and tobacco in a manner that is in agreement with ABCB-PID co-expression (Figure 6).
Alanine neutralization of S634 (ABCB1S634A) as well as linker deletion strongly reduced auxin export to vector control level. On the contrary, phospho-mimicry (ABCB1S634E) of linker phosphorylation strongly enhanced ABCB1-mediated export in yeast and tobacco (Figure 6E and F) overcompensating ABCB1/PID co-expression (Figure 6F). As shown for yeast, mutation of the ABCB1 linker in the tobacco system also does not alter significantly ABCB1 expression or location (Figure 6G). However, the finding that co-expression of mutated ABCB1S634A and ABCB1S634E with PID had no significant influence on ABCB1 activity strongly supports the concept that PID phosphorylates this residue in the absence of TWD1. This would obviously require a functional ABCB1–PID interaction, which is supported by co-locations in Arabidopsis (Figure 1F) and tobacco (Figure 3B, Supplementary Figure S3).
Our data from mutational analyses (Figure 6) are best in agreement with a model in which PID, in the absence of TWD1, does phosphorylate S634, resulting in ABCB1 activation (Supplementary Figure S8A). On the other hand, negative ABCB1 regulation in the presence of TWD1 argues together with in-planta measurements of auxin transport (Figure 3) for a second, PID-specific ABCB1 phosphorylation site that does not essentially need to be part of the linker. This aspect is currently under investigation.
However, structure modelling of the Arabidopsis ABCB1 on the inward-facing crystal structure of mouse ABCB1/PGP1 (Aller et al, 2009) illustrates that S634 is a central residue of the linker domain connecting both NBDs (Supplementary Figure S8B) that themselves fuel transport by ATP hydrolysis. In order to test how the linker mechanistically might alter ABCB functionality, we computed electrostatic surface potentials in ABCB1 with and without linker (Bailly et al, 2012b). In agreement with our transport studies (Figure 6), these results indicate that removal of the linker significantly ameliorates the surface potential of neighbouring transmembrane domains (TMDs) suggested to be responsible for substrate binding and gating (Supplementary Figure S9C). Alternatively, phosphorylation of the linker that is in direct connection to the N-terminal nucleotide-binding fold (see Figure 6) might also alter ATP binding to these ATP pockets.
In summary, our data support PID-mediated ABCB1 linker phosphorylation as a novel mode of plant ABCB activity regulation in analogy to mammalian ABCBs shown to be phosphorylated by PKC and PKA (Chambers et al, 1994). In analogy, plant ABCBs are also obviously regulated by multiple (linker) phosphorylation events that result in inverse regulatory effects (Figure 6, Supplementary Figure S8) as found for mammalian ABCBs, that seem to be fine-tuned via their linker phosphorylation status (Goodfellow et al, 1996; Castro et al, 1999; Conseil et al, 2001).
The kinase and auxin transport inhibitor, quercetin, blocks PID activity by drug binding
Inhibitor treatment of PID auto- and MBP trans-phosphorylation (Figure 5, Supplementary Figure S4), transport assays (Figure 4) and non-invasive quantification of PAT (Figure 7) suggested that protein-kinase inhibitors, chelerythrine, staurosporine and quercetin, block PID by inhibiting its kinase activity. These data are consistent with a recent report suggesting PID as a primary target of staurosporine (Sukumar et al, 2009). Staurosporine had no significant effect on transporter locations and only mildly upregulated ABCB19 expression (Supplementary Figure S7), indicating a direct effect on transporter activity as shown for mammalian ABCBs (Conseil et al, 1998; Castro et al, 1999)
Of special interest was quercetin, a well-known clinical kinase inhibitor (Gschwendt et al, 1983) and modulator of auxin transport (Peer and Murphy, 2007). Quercetin efficiently blocked PID action at nM concentrations (Figure 5) and was specifically shown to bind to recombinant PID and in planta (Figures 5 and 7). This suggests a novel facet of auxin transport regulation where quercetin would block PID activity and thereby phosphorylation-dependent (in)activation of individual transporters by direct drug binding. Interestingly, inactivation of auxin transport by quercetin was recently also described for TWD1-dependent ABCB1 activation by disruption of protein–protein interaction (Bailly et al, 2008).
PID is not a direct target of NPA
Currently, PID is seen as a positive regulator of NPA-sensitive PAT, which is based on the correlation of the following findings: First, the pid mutant shoot phenotype can—in analogy to the more drastic one of pin1(Palme and Galweiler, 1999)—be widely phenocopied by NPA treatment (Wisniewska et al, 2006). Second, pid shoots (Bennett et al, 1995) and roots (Sukumar et al, 2009) show reductions of acropetal and basipetal PAT, respectively. And third, root defects of 35S:PID alleles can be rescued by NPA treatment (Christensen et al, 2000; Benjamins et al, 2001).
However, here we show that NPA has only a slight, nonsignificant inhibitory effect on PID kinase activity and does not bind to PID (Figure 5). Surprisingly, although PID itself does not bind NPA, PID loss- or gain-of-function does obviously inversely alter NPA-binding capacities of NPA-binding proteins. As PIN proteins do obviously not bind NPA (Rojas-Pierce et al, 2007; Kim et al, 2010), one plausible explanation is that PID phosphorylation of the ABCB1 linker might not only modulate ABCB1 activity but also NPA-binding capacities. As a proof-of-concept, NPA (quercetin) binding is significantly reduced (elevated) in yeast ABCB1S634E microsomes (Figure 6D), mimicking ABCB1 phosphorylation. While alanine neutralization had no significant effect, probably because yeast lacks a PID AGC3 kinase orthologue, deletion of the linker abolished both NPA and quercetin binding. This implies that enhanced (reduced) NPA (quercetin) binding to PID gain-of-function microsomes (Figure 5D) might be a direct result of altered ABCB1 phosphorylation at S634 by PID.
These findings, however, also suggest that the pinoid phenotype and repression of 35S:PID defects by NPA are at least to a certain magnitude taken over by PIN-independent transport mechanisms, such as ABCBs. This is also supported by additive, drastic developmental defects of pin1 pid alleles (Furutani et al, 2007). NPA action might be therefore mediated by closely related AGC3 kinases, like PID2 or WAG1/WAG2, that have been shown to share the regulation of identical NPA-sensitive PAT pathways (Santner and Watson, 2006; Dhonukshe et al, 2010). Obviously, this role might be shared also by other protein kinases, like co-purified putative TWD1 interactor, AHK5, that has been localized also to the plasma membrane (Desikan et al, 2008).
Is PID a negative or a positive regulator of auxin transport?
Previous results from different labs have created the somewhat confusing picture that depending on the test system or on the examined tissue, PID either functions as positive or negative regulator of PAT. Here, we provide a molecular rationale for these discrepancies by verifying the ABCB1 linker as a putative PID target and TWD1 as PID interactor deciding for the regulatory impact of PID phosphorylation on ABCB1 activity: First, as discussed above, comparison of transport analyses obtained with heterologous yeast and tobacco and Arabidopsis systems unambiguously suggest that PID has a negative or positive impact on ABCB1 activity depending on the presence or absence of TWD1 or functional TWD1 orthologues, such as FKBP12 in yeast. We also provide evidence that positive and negative regulation is encoded most likely by distinct ABCB1 phosphorylation sites (Supplementary Figure S8).
Second, and as a direct consequence of the above, depending on the plant origin of the test system and therefore depending on its molecular environment, PID regulation might result in positive or negative net fluxes. This is illustrated by a negative impact of PID phosphorylation using shoot (Figures 3 and 6) or root transport systems (Figure 7). Although pharmacological studies obviously do have their pitfalls, our non-invasive measurements of root IAA fluxes in the presence of protein-kinase inhibitors support a positive PID regulation (Figure 7). As TWD1 is low but expressed throughout the plant body (Bailly et al, 2012a), this implies that the regulatory impact of TWD1 on ABCB1 phosphorylation by PID might be regulated by ABCB1–TWD1 interaction (Bouchard et al, 2006; Bailly et al, 2008, 2012a) that itself is under the control of the PAT modulator, quercetin (Bailly et al, 2008). This overall concept is in agreement with previous data that show PID to have specific, dose-dependent and inverse regulatory roles in the root and shoot (Friml et al, 2004; Sukumar et al, 2009). As a result, free IAA is elevated in 35S:PID roots and pid shoots (Figure 3D), which is in agreement with reduced IAA levels in the tips but enhanced signals in distal parts with emerging lateral roots (Friml et al, 2004). These also obviously match the findings that pid roots (unlike pid shoots) show only a mild phenotype while the opposite holds true for 35S:PID alleles (Michniewicz et al, 2007).
The situation is even more complicated by the fact that PIN–ABCB interactions have been shown to be synergistic (PIN1-ABCB1) and antagonistic (PIN2-ABCB1) on one hand (Blakeslee et al, 2007) and that PID controls PIN polarity (Michniewicz et al, 2007) on the other.
In summary, our data suggest that PID, besides its function as a molecular switch of PIN polarity, has a direct impact on auxin-efflux transporter activity. This is in principle in analogy to the recently suggested model of ABCB19 regulation by photoreceptor kinase, phot1 (Christie et al, 2011). Moreover, also for PINs a direct regulation by D6 protein kinases has been suggested (Zourelidou et al, 2009).
Our findings suggest an attractive scenario where TWD1 functions in recruiting PID for ABCB phosphorylation (Supplementary Figure S8A) and as such defines the impact of ABCB1 phosphorylation and regulation. ABCB1 activity regulation by PID is reverted by binding of quercetin, an inhibitor of auxin transport and protein kinases.
MS/MS and mutational analyses indicate phosphorylation of the ABCB1 linker at S634 as a key event in ABCB1 regulation. This is of relevance as the mode of ABCB regulation by TWD1 is unknown, but was initially thought to be dependent on the TWD1 PPIase/rotamase activity (Geisler et al, 2003; Bouchard et al, 2006; Bailly et al, 2008; Kim et al, 2010). However, all attempts to demonstrate such an enzymatic activity for TWD1 failed until now. Alternatively, ABCB1 linker phosphorylation might also alter ABCB1–TWD1 interaction, which is currently under investigation. Importantly, both modes of ABCB1 regulation, directly via TWD1 interaction and PID phosphorylation, might also take place in parallel or in competition, resulting in fine-tuning of ABCB activity as reported for mammalian ABCBs. As such, PID would require TWD1 as a shuttle to find and dock to ABCB targets.
Materials and methods
Construction of TAPa-TWD1 gain-of-function alleles
TWD1 cDNA (At3G21640) was amplified by PCR, inserted BamHI/NotI into pENTR-3C (Invitrogen) and transferred by Gateway recombination into pBIN20-N-TAPa resulting in an N-terminal TAPa fusion (35S:TAPa-TWD1). pBIN20-N-TAPa was constructed by transferring the 35S:N-TAPa:Term cassette from pN-TAPa (Rubio et al, 2005) into pBIN20. Columbia wild type and twd1-3 were transformed with 35S:TAPa-TWD1 and 35S:TAPa (vector control); positive transformants were selected by resistance to kanamycin and verified by western analysis using anti-MYC and anti-TWD1 (see below). Homozygous lines were selected by progeny analysis and used for tandem affinity purification.
One-step affinity purification of TAPa-TWD1-interacting proteins
About 0.3 g of 9-dag Arabidopsis roots from 35S:TAPa-TWD1 or pBIN20-N-TAPa (vector control) seedlings grown vertically on ½ MS plates at 16-h light (100 μE) was homogenized with 0.3 ml lysis buffer (50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100) and the homogenate was centrifuged at 1000 g at 4°C for 5 min. The supernatant was centrifuged at 8000 g at 4°C for 10 min and used for IP using anti-MYC MicroBeads (Miltenyi Biotec, Germany) according to the manufacturer’s instructions. Elutes were precipitated using 10% TCA/acetone, washed twice with acetone, and then pellets were dissolved in 6 M Urea/100 mM ammonium bicarbonate and digested with 0.01 mg/ml trypsin (sequence grade; Promega) and 50 mM ammonium bicarbonate at 37°C for 16 h.
Shotgun mass spectrometric analysis and database searching
Trypsin-digested peptides were analysed by LC–MS/MS using an LTQ-Orbitrap XL-HTC-PAL system (Thermo Fisher Scientific, Bremen, Germany) as described in Fukao et al (2011) with the following modifications: the elution gradient was 5–45% (v/v) acetonitrile in 0.1% (v/v) formic acid over 70 min and the range of MS scan was m/z 450–1500.
MS/MS spectra were analysed using the MASCOT server (version 2.2.) searching the TAIR8 database (The Arabidopsis Information Resource). The MASCOT search parameters were as follows: set-off threshold at 0.05 in the expectation value cutoff, peptide tolerance at 10 p.p.m., MS/MS tolerance at ±0.8 Da, peptide charge of 2+ or 3+, trypsin as enzyme allowing up to one missed cleavage, carboxymethylation on cysteines as a fixed modification and oxidation on methionine as a variable modification. MASCOT-identified vector control proteins were subtracted manually from TAPa-TWD1 co-purified proteins, and proteins with a score above 30 were considered as significant partners (Figure 1 and Supplementary Figure S1).
Interaction analyses
PID-GST was expressed from pGEX4T-1-PID and purified as described in Christensen et al (2000). Ca. 1 μg of PID-GST bound to glutathione sepharose was incubated with a 20-fold access of purified TWD11-337 protein (Bailly et al, 2008) and pull-down assays were performed as in Geisler et al (2003). Equal amounts of loading control, non-bound material and elutes were separated by PAGE, and detected using anti-TWD11075-1078.
For BRET analyses, TWD1 cDNAs were inserted by PCR into pCR8-TOPO (Invitrogen) and transferred into compatible BRET destination vectors, pPZP-35S:attR-hRluc (AY995143; Subramanian et al, 2006) by Gateway recombination (Invitrogen). Microsomes from N. benthamiana leaves co-infiltrated with agrobacteria (GV3101) containing 35S:PID-GFP (pGREEN0179-PID-GFP), 35S:TWD1-Rluc, 35S:PIRK-YFP or corresponding empty vector controls) using standard protocols were prepared 4 days after infiltration (dai). BRET signals were recorded from microsomes (each ca. 10 μg) in the presence of 5 μM coelenterazine (Biotium Inc.) and BRET ratios were calculated as described previously (Bailly et al, 2008, 2012a). Results are the average of 10 readings collected every minute; values presented are averages from three independent experiments (independent agrobacterium infiltrations) each with four replica.
Auxin transport assays
ABCB1 was expressed from pNEV-PGP1/ABCB1 (Geisler et al, 2005) and pNEV-ABCB1-YFP (Bouchard et al, 2006). PID (At2g34650) or BIN2 cDNA (At4g18710) were PCR amplified from pGEX4T-1-PID (Christensen et al, 2000) or pUC8-BIN-GFP and inserted BamHI/SalI into pRS314CUP, resulting in pRS314CUP-PID or pRS314CUP-BIN2. The inactive pRS314CUP-MPID and pNEV-ABCB1S634A/E/D were constructed by introducing D205A and S634A/D exchanges by site-directed mutagenesis (QuikChange, Stratagene). Yeast IAA transport was assayed with the unspecific BA as control in parallel and performed as in Bailly et al (2008). Relative IAA/BA export was calculated from retained radioactivity as follows: (radioactivity in the yeast at time t=10 min)−(radioactivity in the yeast at time t=0))*(100%)/(radioactivity in the yeast at t=0 min).
Simultaneous IAA and NAA export from Arabidopsis mesophyll protoplasts was analysed as in Geisler et al (2005). Simultaneous IAA and NAA export and confocal microscopy analyses from N. benthamiana mesophyll protoplasts was analysed 4 dai by agrobacterium-mediated co-transfection of combinations of 35S:ABCB1-MYC, 35S:ABCB1-YFP, ABCB1:ABCB1-CFP, 35S:PID-FLAG, 35S:PID-GFP, 35S:WAG1-YFP, 35S:TWD1-GFP and 35S:TWD1-YFP from pBI121-PGP1 (Sidler et al, 1998), pBIN19-ABCB1-YFP (Bailly et al, 2012a), pMOA37-ABCB1-CFP (Bailly et al, 2012a), pMOA34-PID-FLAG (see below; Dhonukshe et al, 2010), pGREEN0179-WAG1-YFP-HA (Dhonukshe et al, 2010), pMDC85-TWD1-GFP (see below) and pBIN19-TWD1-YFP (Bailly et al, 2012a), respectively. pMOA34-PID-FLAG and pCAMBIA1300-TWD1-GFP were constructed by subcloning the 35S:PID-FLAG-TERM cassette from pART7-PID-FLAG (Michniewicz et al, 2007) into binary plasmid pMO34 and gateway recombination of TWD1 cDNA from pDONR207-TWD1 (Bailly et al, 2012a) into pMDC85. Tobacco protoplast preparation and transport assays were identical to the Arabidopsis protocol (Geisler et al, 2005) except that a 25% Percoll gradient was used. Relative IAA/NAA export was calculated from effluxed radioactivity as follows: ((radioactivity in the medium at time t)−(radioactivity in the medium at time t=0))*(100%)/(radioactivity in the medium at t=0).
Average values are presented from 6 to 8 independent experiments (yeast: independent transformations; protoplasts: infiltrations of independent agrobacterium transformants).
Phosphorylation assays
PID-GST autophosphorylation and PID-dependent phosphorylation of myelin-binding protein (MBP) was assayed as described in Christensen et al (2000). Assays were carried out in the presence of indicated drugs or solvent control, and signal intensities of radiolabelled PID or MBP bands were quantified after PAGE using a Cyclone Phosphoimager and Scion Image software 1.63 (Scion Corporate). Average values are presented from two independent PID-GST preparations each with 3–8 experiments.
In vitro kinase assays each using 5-μg synthetic ABCB1 peptides (Peptide 2.0) were performed as described above each either in the presence of 50 ng human PKCα (Sigma-Aldrich) or 1 μg purified GST-PID (Christensen et al, 2000). Peptides were Ziptip purified (Millipore), dried and analysed by MS/MS. Sequences are as follows: peptide 1: Q(607)EAAHETAMSNARKSSARPC, peptide 2: S(616)NARKSSARPSSARNSVSSC, peptide 3: R(629)NSVSSPIMTRNSSYGRSPC, peptide 4: T(1268)QVIGMTSGSSSRVKEDDAC and peptide 5: R(629)NSVSAPIMTRNSSYGRSPC.
For in-planta phosphorylation analyses, 35S:ABCB1-MYC was co-transfectedin N. benthamiana leaves with 35S:PID-FLAG as described above. Total microsomes were isolated, proteins were trypsin-digested, enriched by immobilized metal-ion affinity chromatography (IMAC)/titanium dioxide affinity chromatography and analysed by LC–ESI–MS/MS on a LTQ-Orbitrap XL mass spectrometer as described recently (Endler et al, 2009). The MS/MS spectra were acquired using both collision energy dissociation (CID) and electron transfer dissociation (ETD) fragmentations techniques. The MS/MS data were searched using MASCOT version 2.1.0.4. (Matrix Science, London, UK) and phosphopeptide identifications were accepted with a minimal MASCOT ion score of 25 and a MASCOT expect value of ≤0.05. The spectra relative to phosphopeptides were further manually validated by verifying the neutral loss of H3PO4 (98 Da).
Analysis of PID, TWD1 and ABCB1 expression
PID:PID-VENUS (Michniewicz et al, 2007) or 35S:PID-GFP (pGREEN0179-PID-GFP) were introduced into homozygous ABCB1:ABCB1-GFP (Mravec et al, 2008) or Wt (all in ecotype Columbia). 35S:TWD1-YFP was introduced into twd1-3, and 35S:ABCB1-YFP was introduced into abcb1-1 (Bailly et al, 2012a), using agrobacterium-mediated transformations. Homozygous T3 seedlings were grown vertically for 5 dag as described above and analysed by confocal laser scanning microscopy (Leica TCS SP5). Sequential scans were used to record the emission of GFP (excitation 488 nm, emission 498–510 nm), VENUS (excitation 514 nm, emission 524–580 nm), CFP (excitation 458 nm, emission 468–500 nm) or YFP (excitation 514 nm, emission 524–550 nm). Images were electronically coloured and merged using Photoshop@ 10.0.1. software (Adobe Systems, Mountain View, CA). Relative transcript levels were extracted from the Arabidopsis gene Expression Database (AREXDB).
Drug-binding studies
Drug-binding assays using Arabidopsis or yeast microsomes or PID-GST were performed by vacuum filtration as described elsewhere (Bailly et al, 2008). [3H](G)quercetin (10 Ci/mmol; 1.0 mCi/ml) was custom-synthesized by ARC Inc. (St. Louis, USA). In short, for determining specific drug binding, four replicates of each 20 μg of protein or 1 μg of column-bound PID-GST (or GST alone) were incubated with 10 nM radiolabelled drugs (30–60 Ci/mmol) in the presence and absence of the corresponding 10 μM non-radiolabelled drug. Reported values are the means of specific radiolabelled drug bound in the absence of cold drug (total) minus radiolabelled drug bound in the presence of cold drug (unspecific) from at least three independent experiments each with four replicates. Background drug binding to column material or GST alone (background) was corrected by subtracting specific binding to column-bound GST from column-bound PID-GST.
In-planta analysis of IAA contents and fluxes
Endogenous free IAA was quantified from shoot and root segments of MeOH-extracted seedlings by using gas chromatography–mass spectrometry (GC–MS) as described in Bouchard et al (2006). Data are means of four independent lots of 30–50 seedlings each.
A platinum microelectrode was used to monitor IAA fluxes in Arabidopsis roots as described previously (Santelia et al, 2005; Bouchard et al, 2006; Bailly et al, 2008; Kim et al, 2010). For measurements, Columbia wild-type plants were grown in hydroponic cultures and used at 5 dag. Differential currents were recorded in the absence and presence of 5 μM NPA (data taken from Bailly et al, 2008), quercetin or chelerythrine.
Quantitative analysis of root gravitropism
Root gravitropism in the dark of wild-type and pin1 (AT1G73590), pin2 (AT5G57090), abcb1/pgp1 (AT2G36910) and abcb19/pgp19 (AT3G28860) mutant combinations (all ecotype Columbia (Col Wt)) in the presence of protein-kinase inhibitors (100 nM quercetin, 400 nM chelerythrine, 50 nM staurosporine and 10 μM phorbole ester) was performed as described previously (Santelia et al, 2008). Helical wheels were plotted using PolarBar software.
Data analysis
Data were analysed using Prism 4.0b (GraphPad Software, San Diego, CA). The ABCB1 2D structure was created by TMRPres2D software (http://bioinformatics.biol.uoa.gr/TMRPres2D) and TMD predictions were performed using HMMTOP (http://www.enzim.hu/hmmtop/index.php) and PRED-TMR (http://athina.biol.uoa.gr/PRED-TMR). ABCB linker alignments were conducted using CLUSTAL X. Putative phosphorylation sites in ABCB1 were predicted using NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos). Structure modelling was performed as in Bailly et al (2012b) and surface electrostatic potentials were computed using the PyMol APBS Tools2 plugin with default settings in the presence or absence of the linker domain.
Supplementary Material
Acknowledgments
We thank V Vincenzetti for the outstanding technical support, I Hwang for BIN2 cDNA, R Offringa for pART7-PID-FLAG, pGREEN0179-PID-GFP and pGREEN0179-WAG1-YFP-HA, M Heisler for PID:PID-VENUS lines, S Christensen for pGEX4T-PID, T Kato for pBIN20-N-TAPa, A Fuglsang for anti-PMA1 and PIRK-YFP, A von Arnim for the donation of BRET vectors and help with the construction, A Buchala for comments on the text and E Martinoia for continuous support and mentorship. This work was supported by grants from the Novartis Foundation (to MG), from the Pool de Recherche of the University of Fribourg (to MG), from the Danish Research School for Biotechnology, FOBI (to MG and AS) and from the Swiss National Funds (to MG).
Author contributions: SH and MG designed the research; SH performed auxin transport, root gravitropism, PIN1,2-GFP and ABCB1,19-GFP and drug-binding analyses; ML and MW purified TWD11-337 protein; SH, AE and PN analysed ABCB1 phosphorylation; BW analysed BRET interaction and performed confocal imaging; SM analysed root IAA fluxes; SP analysed free auxins; JZ, AB, BW and SCO conducted site-directed mutagenesis and transport assays; AB did structure modelling and analyses; YF performed IP and MS analysis; and MG performed PID-GST pull-down, drug-binding, linker in vitro phosphorylation assays and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323: 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A, Sovero V, Geisler M (2006) The TWISTED DWARF’s ABC: how immunophilins regulate auxin transport. Plant Signaling Behav 1: 277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M (2008) Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J Biol Chem 283: 21817–21826 [DOI] [PubMed] [Google Scholar]
- Bailly A, Wang B, Zwiewka M, Vincenzetti V, Henrichs S, Sovero V, Pollmann S, Schenck D, Lüthen H, Azzarello E, Mancuso S, Maeshima M, Friml J, Schulz A, Geisler M (2012a) TWISTED DWARF1 controls cell elongation by modulating ABCB-mediated auxin export. Plant Cell (under review) [Google Scholar]
- Bailly A, Yang Y, Martinoia E, Geisler M, Murphy A (2012b) Plant lessons: understanding substrate specificity through structural modeling of ABCBs. Front Plant Sci 2: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Ampudia CS, Hooykaas PJ, Offringa R (2003) PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol 132: 1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Bennett S, Alvarez J, Bossinger G, Smyth D (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Development 128: 4057–4067 [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, Geisler M, Nagashima A, Sakai T, Martinoia E, Friml J, Peer WA, Murphy AS (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, Schulz B, Geisler M (2006) Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem 281: 30603–30612 [DOI] [PubMed] [Google Scholar]
- Castro AF, Horton JK, Vanoye CG, Altenberg GA (1999) Mechanism of inhibition of P-glycoprotein-mediated drug transport by protein kinase C blockers. Biochem Pharmacol 58: 1723–1733 [DOI] [PubMed] [Google Scholar]
- Chambers TC, Pohl J, Glass DB, Kuo JF (1994) Phosphorylation by protein kinase C and cyclic AMP-dependent protein kinase of synthetic peptides derived from the linker region of human P-glycoprotein. Biochem J 299Part 1: 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y (2008) NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci USA 105: 21017–21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D (2000) Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Christie JM, Yang H, Richter GL, Sullivan S, Thomson CE, Lin J, Titapiwatanakun B, Ennis M, Kaiserli E, Lee OR, Adamec J, Peer WA, Murphy AS (2011) phot1 Inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol 9: e1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conseil G, Baubichon-Cortay H, Dayan G, Jault JM, Barron D, Di Pietro A (1998) Flavonoids: a class of modulators with bifunctional interactions at vicinal ATP- and steroid-binding sites on mouse P-glycoprotein. Proc Natl Acad Sci USA 95: 9831–9836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conseil G, Perez-Victoria JM, Jault JM, Gamarro F, Goffeau A, Hofmann J, Di Pietro A (2001) Protein kinase C effectors bind to multidrug ABC transporters and inhibit their activity. Biochemistry 40: 2564–2571 [DOI] [PubMed] [Google Scholar]
- Desikan R, Horak J, Chaban C, Mira-Rodado V, Witthoft J, Elgass K, Grefen C, Cheung MK, Meixner AJ, Hooley R, Neill SJ, Hancock JT, Harter K (2008) The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS One 3: e2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mahonen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, Offringa R (2010) Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, Friml J (2011) Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Endler A, Reiland S, Gerrits B, Schmidt UG, Baginsky S, Martinoia E (2009) In vivo phosphorylation sites of barley tonoplast proteins identified by a phosphoproteomic approach. Proteomics 9: 310–321 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, Hooykaas PJ, Palme K, Offringa R (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Fukao Y, Ferjani A, Tomioka R, Nagasaki N, Kurata R, Nishimori Y, Fujiwara M, Maeshima M (2011) iTRAQ analysis reveals mechanisms of growth defects due to excess zinc in Arabidopsis. Plant Physiol 155: 1893–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Kajiwara T, Kato T, Treml BS, Stockum C, Torres-Ruiz RA, Tasaka M (2007) The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development 134: 3849–3859 [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Offringa R (2007) Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci 12: 541–547 [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, Ejendal KF, Smith AP, Baroux C, Grossniklaus U, Muller A, Hrycyna CA, Dudler R, Murphy AS, Martinoia E (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-Kalman Z, Koncz C, Dudler R, Blakeslee JJ, Murphy AS, Martinoia E, Schulz B (2003) TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol Biol Cell 14: 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow HR, Sardini A, Ruetz S, Callaghan R, Gros P, McNaughton PA, Higgins CF (1996) Protein kinase C-mediated phosphorylation does not regulate drug transport by the human multidrug resistance P-glycoprotein. J Biol Chem 271: 13668–13674 [DOI] [PubMed] [Google Scholar]
- Gschwendt M, Horn F, Kittstein W, Marks F (1983) Inhibition of the calcium- and phospholipid-dependent protein kinase activity from mouse brain cytosol by quercetin. Biochem Biophys Res Commun 117: 444–447 [DOI] [PubMed] [Google Scholar]
- Hemenway CS, Heitman J (1996) Immunosuppressant target protein FKBP12 is required for P-glycoprotein function in yeast. J Biol Chem 271: 18527–18534 [DOI] [PubMed] [Google Scholar]
- Huang F, Zago MK, Abas L, van Marion A, Galvan-Ampudia CS, Offringa R (2010) Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama A, Yamashino T, Tanaka Y, Sakakibara H, Kakimoto T, Sato S, Kato T, Tabata S, Nagatani A, Mizuno T (2007) AHK5 histidine kinase regulates root elongation through an ETR1-dependent abscisic acid and ethylene signaling pathway in Arabidopsis thaliana. Plant Cell Physiol 48: 375–380 [DOI] [PubMed] [Google Scholar]
- Kamphausen T, Fanghanel J, Neumann D, Schulz B, Rahfeld JU (2002) Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J 32: 263–276 [DOI] [PubMed] [Google Scholar]
- Kim JY, Henrichs S, Bailly A, Vincenzetti V, Sovero V, Mancuso S, Pollmann S, Kim D, Geisler M, Nam HG (2010) Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. J Biol Chem 285: 23309–23317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J (2009) PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Cho HT (2006) PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18: 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax TL, Muday GK, Rubery PH (1995) Auxin transport. InPlant Hormones: Physiology, Biochemistry and Molecular Biology Davies PJ (ed.) pp509–530Dordrecht, Netherlands: Kluwer, [Google Scholar]
- Luschnig C (2001) Auxin transport: why plants like to think BIG. Curr Biol 11: R831–R833 [DOI] [PubMed] [Google Scholar]
- Mancuso S, Marras AM, Magnus V, Baluska F (2005) Noninvasive and continuous recordings of auxin fluxes in intact root apex with a carbon nanotube-modified and self-referencing microelectrode. Anal Biochem 341: 344–351 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, Weigel D, Meyerowitz EM, Luschnig C, Offringa R, Friml J (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J (2008) Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135: 3345–3354 [DOI] [PubMed] [Google Scholar]
- Muday GK, DeLong A (2001) Polar auxin transport: controlling where and how much. Trends Plant Sci 6: 535–542 [DOI] [PubMed] [Google Scholar]
- Murphy AS, Hoogner KR, Peer WA, Taiz L (2002) Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol 128: 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Suzuki G, Uehara Y, Saji K, Furukawa T, Koshiba T, Sekimoto M, Fujioka S, Kuroha T, Kojima M, Sakakibara H, Fujisawa N, Okada K, Sakai T (2008) Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. Plant J 53: 516–529 [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Stensballe A, Jensen ON, Peck SC (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palme K, Galweiler L (1999) PIN-pointing the molecular basis of auxin transport. Curr Opin Plant Biol 2: 375–381 [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, DeLong A, Muday GK (2001) Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13: 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Pierce M, Titapiwatanakun B, Sohn EJ, Fang F, Larive CK, Blakeslee J, Cheng Y, Cutler SR, Peer WA, Murphy AS, Raikhel NV (2007) Arabidopsis P-glycoprotein19 participates in the inhibition of gravitropism by gravacin. Chem Biol 14: 1366–1376 [DOI] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, Martinoia E (2008) Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem 283: 31218–31226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Vincenzetti V, Azzarello E, Bovet L, Fukao Y, Duchtig P, Mancuso S, Martinoia E, Geisler M (2005) MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett 579: 5399–5406 [DOI] [PubMed] [Google Scholar]
- Santner AA, Watson JC (2006) The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J 45: 752–764 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, Buchala A, Cardinale F, Meskiene I (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R (1998) Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10: 1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Woo J, Cai X, Xu X, Servick S, Johnson CH, Nebenfuhr A, von Arnim AG (2006) A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J 48: 138–152 [DOI] [PubMed] [Google Scholar]
- Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK (2009) PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol 150: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, Yazaki K (2005) PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17: 2922–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G (2008) Plant signaling: brassinosteroids, immunity and effectors are BAK!. Curr Biol 18: R963–R965 [DOI] [PubMed] [Google Scholar]
- Vieten A, Sauer M, Brewer PB, Friml J (2007) Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci 12: 160–168 [DOI] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Zourelidou M, Muller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C (2009) The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136: 627–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.