Abstract
Through targeted inactivation of the ssrA and smpB genes, we establish that the trans-translation process is necessary for normal growth, adaptation to cellular stress, and virulence by the bacterial pathogen Francisella tularensis. The mutant bacteria grow slower, have reduced resistance to heat and cold shocks, and are more sensitive to oxidative stress and sub-lethal concentrations of antibiotics. Modifications of the tmRNA tag and use of higher resolution mass spectrometry approaches enabled the identification of a large number of native tmRNA substrates. Of particular significance to understanding the mechanism of trans-translation, we report the discovery of an extended tmRNA tag and extensive ladder-like pattern of endogenous protein tagging events in F. tularensis that are likely to be a universal feature of tmRNA activity in eubacteria. Furthermore, the structural integrity and the proteolytic function of the tmRNA tag are both crucial for normal growth and virulence of F. tularensis. Significantly, trans-translation mutants of F. tularensis are impaired in replication within macrophages and are avirulent in mouse models of tularemia. By exploiting these attenuated phenotypes, we find that the mutant strains provide effective immune protection in mice against lethal intradermal, peritoneal, and intranasal challenges with the fully virulent parental strain.
Keywords: smpB, ssrA, tmRNA, trans-translation, F. tularensis
INTRODUCTION
Trans-translation is an evolutionarily conserved mechanism for post-transcriptional quality control in bacteria that is activated in response to stalling of ribosomes on defective messenger RNAs. The specialized hybrid transfer-messenger RNA (tmRNA), in association with its protein co-factor, SmpB (Karzai et al., 1999), orchestrates this unique process (Dulebohn et al., 2007, Karzai et al., 2000, Keiler, 2008). The primary event leading to the commencement of trans-translation is stalling of ribosomes at the 3′ end of nonstop mRNAs (mRNAs lacking in frame termination signals). Such defective mRNAs may arise as a consequence of endonucleolytic cleavage, 3′-5′ exonucleolytic activity, premature transcription termination, point mutations, a string of rare codons, or decoding of specific stall sequences (Abo et al., 2000, Hayes et al., 2002a, Hayes et al., 2002b, Inada & Aiba, 2005, Okan et al., 2006, Li et al., 2008, Mehta et al., 2006, Richards et al., 2006, Roche & Sauer, 1999, Roche & Sauer, 2001). During trans-translation, alanine-charged tmRNA, in a quaternary complex with SmpB and the GTP-bound form of elongation factor Tu (EF-Tu-GTP), engages the stalled ribosome and acts as a tRNA to get accommodated into the empty ribosomal A-site. Accommodation is followed by transpeptidation of the incomplete nascent peptide to the alanine charge of tmRNA and resumption of translation, in trans, on a short mRNA open reading frame encoded by tmRNA (tmRNA ORF). Translation terminates on a stop codon provided by the tmRNA ORF, thus permitting normal recycling of the translation machinery. The nascent polypeptide chain is thus appended with the tmRNA peptide tag, targeting it for degradation by C-terminal specific cellular proteases. The tmRNA tag sequence contains sequence signals for recognition by the ClpXP, its co-factor SspB (also known as MglB in F. tularensis), ClpAP, and Lon proteases (Flynn et al., 2001, Ge & Karzai, 2009, Gottesman et al., 1998, Gur & Sauer, 2008, Karzai et al., 1999, Keiler et al., 1996, Levchenko et al., 2005, Levchenko et al., 2003, Levchenko et al., 2000, Lies & Maurizi, 2008, Baron & Nano, 1998, Choy et al., 2007).
Impairment of trans-translation leads to a wide variety of phenotypes in different bacterial species, ranging from very subtle growth defects to lethality. The most common of these phenotypes indicate that this system enables the bacterium to withstand various cellular stresses (Roche & Sauer, 1999, Karzai et al., 1999, Komine et al., 1994, Muto et al., 2000, Shin & Price, 2007, Corvaisier et al., 2003, De la Cruz & Vioque, 2001, Konno et al., 2004, Okan et al., 2006, Okan et al., 2010). Other possible roles, which are thought to be specific to particular gene circuits, include the ability to support the growth of certain bacteriophage in Escherichia coli (Karzai et al., 1999, Retallack et al., 1994), induction of virulence factors in Yersinia (Okan et al., 2006, Okan et al., 2010), and cell cycle regulation in Caulobacter crescentus (Keiler & Shapiro, 2003).
Although the smpB and ssrA genes are conserved in eubacteria, a number of key questions regarding the utility and necessity for this conservation remain unanswered. For instance, it is not clear whether this system is uniformly functional in eubacteria, whether it plays a role in virulence of all pathogenic bacteria, or whether similar substrates are tagged by this system in all bacteria. Additionally, it is unclear why this system is essential for survival of some bacterial species while dispensable for growth and viability of others. It has been suggested that a redundant, or partially redundant, system might be operational in larger-genome bacteria that has been either lost or inactivated in small-genome bacteria. Indeed, recent studies have uncovered two gene products in E. coli that could provide alternative ribosome rescue functions (Chadani et al., 2011, Chadani et al., 2010, Garza-Sanchez et al., 2011).
In the present study, we have conducted a comprehensive evaluation of the hitherto untested function of the trans-translation system (ssrA and smpB genes) in the small-genome bacteria F. tularensis. This gram-negative pathogen is the etiological agent of the zoonotic disease tularemia (Ellis et al., 2002). There are four subspecies of F. tularensis: F. tularensis subsp. tularensis, F. tularensis subsp. holarctica, F. tularensis subsp. mediasiatica, and F. tularensis subsp. novicida (Oyston et al., 2004). F. tularensis subsp. tularensis (the type A biovar) causes the most severe form of tularemia, while F. tularensis subsp. holarctica (the type B biovar) is less virulent and causes milder symptoms in healthy humans. A modified F. tularensis subsp. holarctica, which was developed as a live vaccine strain (LVS) in the former Soviet Union (Dennis et al., 2001), is currently extensively used as a model system to study tularemia, as it causes a lethal infection in mice that closely resembles the human disease (Anthony & Kongshavn, 1987).
A highly intriguing feature of the tmRNA system is that its function has been adapted to fulfill the specific physiological needs of various bacterial species. Studies of tmRNA in several distinct bacterial species (Withey & Friedman, 1999, Keiler & Shapiro, 2003, Shin & Price, 2007, Abe et al., 2008, Keiler, 2008, Mikulik et al., 2008, Thibonnier et al., 2008, Ujiie et al., 2009, Okan et al., 2010) have yielded new and unique insights into the structure, function, and diverse physiological roles played by this system. Our studies of the tmRNA system in F. tularensis demonstrate that this small genome bacterium has an active trans-translation system that is important for normal growth, resistance to a variety of stress conditions, and virulence of the pathogen in a mouse model of tularemia. We have uncovered the identity of an uncommonly long tmRNA open reading frame (ORF) in Francisella and show that it encodes an extended peptide tag. Further more, we have discovered an unusual ladder pattern-like of tagging in a number of tmRNA substrates and demonstrate that the tagging for proteolysis function of the tmRNA system is essential for virulence of this pathogen. We have also explored the possibilities of manipulating the F. tularensis trans-translation system for pathogen attenuation and development of a potential live vaccine strain. The results presented herein expand our understanding of the roles and mechanism of trans-translation in bacteria, the unusual nature and identity of the F. tularensis peptide tag, as well as our knowledge of the human pathogen F. tularensis.
RESULTS
Inactivation of ssrA and smpB genes of F. tularensis
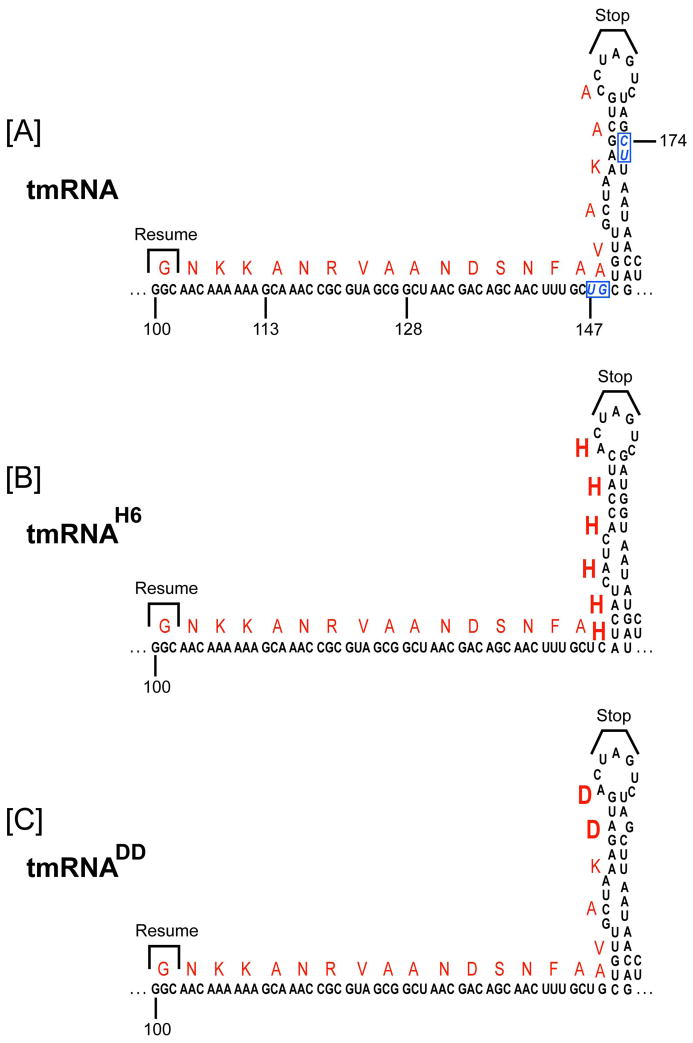
To address the question of whether trans-translation plays a role in the viability of the small-genome (~1.9 Mb) bacterial pathogen F. tularensis, we endeavored to create gene disruption mutants of the smpB and ssrA genes in the wild-type (WT) F. tularensis LVS. The genome of F. tularensis has a putative single-piece ssrA gene encoding a tmRNA that contains a number of predicted conserved structural elements (helices and pseudoknots), which are similar in overall architecture to the well-studied E. coli tmRNA (Dulebohn et al., 2007, Karzai et al., 2000, Keiler, 2008). However, bioinformatics analysis of the mRNA-like domain of F. tularensis tmRNA highlights a potential ambiguity in the resume signal, size, and identity of the tmRNA-encoded peptide tag (see below). Unlike in E. coli, Salmonella, and Yersinia, the gene for the Francisella tmRNA cofactor, smpB, is not located immediately upstream of the ssrA gene; rather, it is present at a different locus in the F. tularensis genome. Our initial attempts to inactivate the smpB and ssrA genes by allelic exchange were not successful. However, we were able to obtain several independent inactivating ssrA and smpB gene mutations in WT F. tularensis LVS via a recently developed Lactobacillus intron (LtrB) insertion gene targeting strategy, adapted for Francisella (Rodriguez et al., 2009). We obtained two ssrA gene disruption strains using this gene targeting strategy. In the first strain, the LtrB intron was chromosomally inserted at base pair position 147 of the ssrA gene (ssrA::LtrB-bp147), creating strain A147, in which the tmRNA gene was disrupted within the tmRNA open reading frame (Fig. 1A). In the second strain, the LtrB intron was chromosomally inserted at base pair position 174 of the ssrA gene (ssrA::LtrB-bp174), creating strain A174, in which the tmRNA gene was disrupted within the putative helix-5 of the tmRNA gene (Fig. 1A). Using the LtrB gene targeting method, we were also able to generate a targeted smpB gene disruption mutant of F. tularensis LVS. In this strain, B255 (smpB::LtrB-bp255), the LtrB intron disrupts the 474 base pairs-long F. tularensis smpB gene approximately in the middle, after nucleotide 255 of the SmpB coding sequence. All LtrB intron insertions were confirmed by PCR and DNA sequencing, and were stable over many generations of growth. We interpret these results to indicate that the trans-translation process is not essential for F. tularensis viability.
Figure 1.
[A]. A schematic representation of the predicted secondary structure of the F. tularensis tmRNA ORF and helix-5. The mRNA ORF sequence is shown, highlighting the translation resume codon, termination signal, and the encoded amino acid residues (in red letters). Nucleotide positions of this segment of tmRNA, indicated by numbers below the sequence, are shown with respect to the start of the ssrA gene. LtrB intron insertions sites (pos. 147 and pos. 174 of the tmRNA encoding ssrA gene) are shown in boxed blue italic letters. [B]. The diagram depicts the tmRNAH6 variant used in this study, highlighting the encoded six histidines and compensatory changes introduced to preserve the stem-loop structure of helix-5. [C]. The diagram depicts the tmRNADD variant used in this study, highlighting the altered sequence and the encoding aspartic acid residues.
Inactivation of trans-translation in F. tularensis leads to increased sensitivity to stress conditions
To assess the potential growth defects of these smpB and ssrA mutants, we examined their abilities to grow in various media and withstand a range of stress conditions. All three strains (ssrA A147, ssrA A174, and the smpB B255) had decreased growth rates on solid medium at 37°C, as exhibited by smaller colony sizes in comparison to the parental strain (Fig. 2A). This phenotype was observed when the mutant strains were streaked either from frozen stocks or when plated from fresh liquid cultures, with the B255 strain exhibiting the slowest growth rate. The same three mutant strains (A147, A174 and B255) exhibited decreased growth rates in rich liquid media (Mueller-Hinton and brain heart infusion broths) and in the Chamberlain chemically-defined minimal medium formulations (Fig. 2B, and Fig. S1). Interestingly, the magnitude of the growth defect in liquid media varied among trans-translation mutants and depended on the type of growth medium. Strain A147 (tmRNA tag ORF disruption) was the least affected in all media tested, while the ssrA A174 (helix 5 disrupted) and smpB B255 mutant strains were more severely affected (Fig. 2).
Figure 2.
Growth phenotypes of the F. tularensis ssrA and smpB mutants. [A]. The indicated smpB and ssrA mutants, the complemented mutants, and the WT strain were grown in liquid medium to early stationary phase, diluted, and spread on Mueller Hinton Chocolate Agar plates to obtain single colonies. Photographs were taken after 3–4 days incubation at 37°C. [B]. The smpB and ssrA mutants, the complemented mutants, and the WT strain were inoculated in MH broth from overnight cultures, and growth was monitored by taking OD600 measurements at the indicated time points. [C]. Growth complementation of ssrA mutants with plasmids expressing proteolysis-resistant variants (ssrADD and ssrAHis6) of F. tularensis tmRNA. Complemented ssrA mutants and the WT strain were grown in MH broth, and their growth was monitored by taking OD600 measurements at the indicated time points. Results shown are the means and standard deviations from four independent experiments. Error bars show standard deviations.
To ensure that the observed growth defects were not due to polar effects, or the presence of unintended spurious mutations, we performed genetic complementation assays by introducing the respective ssrA or smpB gene expression cassettes on a plasmid vector under the control of their native promoters, or by targeted insertion at a different chromosomal locus. Complementation studies showed that introduction of the ssrA or smpB genes into the respective mutant strains substantially improved or restored their growth deficiencies to levels comparable to the parental strain (Fig. 2B), thus confirming that the observed growth defects were due to loss of smpB and ssrA gene functions.
We next evaluated the sensitivity of the ssrA and smpB mutants to various stress conditions. Growth of the mutant strains on solid medium at 40°C was only slightly impaired in comparison to their growth at 37°C (Fig. S2). The mutants were considerably more sensitive to heat shock at 50°C in liquid cultures (Fig. 3A). The smpB mutant strain was also considerably more sensitive to oxidative stress, induced by inclusion of hydrogen peroxide in liquid culture or in filter disk assays (Table I, and Fig. 3B). F. tularensis ssrA and smpB mutant strains were also more sensitive to cold shock, as applied by placing liquid cultures on ice for 4 h. The mutant bacteria did recover from the effects of this treatment but grew much slower on plates. The increased heat and cold sensitivity of the mutants was compensated by genetic complementation (Fig. 3, and Fig. S2). Furthermore, we evaluated the sensitivity of the F. tularensis smpB and ssrA mutants to various antibiotics by filter-disk growth-inhibition assays. The mutant strains exhibited increased sensitivity, compared to the parental strain, to the presence of translation-specific antibiotics, including kanamycin, chloramphenicol, streptomycin, gentamycin, spectinomycin, and tetracycline, with the smpB mutant exhibiting the highest level of sensitivity (Table I). Plasmid-borne copies of the WT ssrA or smpB genes complemented the respective defects of the mutant strains (Table I). Among other antibiotic agents, the transcription inhibitor rifampicin, DNA replication inhibitors novobiocin and nalidixic acid, and the nucleic acid intercalating agent ethidium bromide also caused notably increased growth inhibition in the B255 strain. In contrast, the ssrA and smpB mutant strains, like the isogenic parental strain, were fully resistant to the cell wall biosynthesis inhibitor ampicillin (due to the presence of a beta-lactamase gene in the genome of F. tularensis LVS), and the membrane permeabilizing agent polymixin B, to which LVS exhibits naturally high resistance. Similarly, application of osmotic shock with 2% sodium chloride, and acid shock (at pH 4) did not uncover any increased sensitivity in the mutant strains (data not shown). These data suggest that the observed enhanced sensitivity to antibiotics is unlikely to be caused by a general defect in drug efflux systems or alteration in membrane permeability.
Figure 3.
ssrA and smpB mutant strains of F. tularensis are more sensitive to heat shock and oxidative stress. [A]. The smpB and ssrA mutants, the complemented mutants, and the WT strain were grown overnight in MH broth, diluted to matching optical densities with fresh medium, and incubated at 50°C. Viable bacteria were enumerated at the indicated time points using CFU assays. [B]. The smpB mutant, the smpB complemented mutant, and the WT strain were grown overnight in MH broth, diluted to matching optical densities with fresh medium, and grown to OD600 of 0.5. Hydrogen peroxide was added to a final concentration of 0.002% and viable bacteria were enumerated at the indicated time points using CFU assays. Results shown are the means and standard deviations from three independent experiments. Error bars show standard deviations.
Table 1.
smpB and ssrA mutants of F. tularensis are more sensitive to drugs.
| Drug Concentration (μg) | WT | B255 | B255/psmpB | A174 | A174/pssrA |
|---|---|---|---|---|---|
| Streptomycin (10) | 23±1a | 32±1 | 24±1 | 30±1 | 24±1 |
| Gentamycin (10) | 30±1 | 40±1 | 32±1 | 40±1 | 32±1 |
| Kanamycin (5) | 21±1 | 28±1 | 23±1 | 30±1 | 24±1 |
| Tetracycline (5) | 31±1 | 42±1 | 33±1 | 42±1 | 31±1 |
| Chloramphenicol (5) | 34±1 | 53±1 | 36±1 | 45±1 | 35±1 |
| Spectinomycin (10) | 16±1 | 36±1 | 20±1 | 20±1 | 16±1 |
| Novobiocin (30) | 22±1 | 30±1 | 24±1 | 30±1 | 24±1 |
| Rifampicin (10) | 20±1 | 28±1 | 22±1 | 24±1 | 20±1 |
| Nalidixic acid (5) | 32±1 | 35±1 | 32±1 | 32±1 | 32±1 |
| Polymixin B (100) | 6±0 | 6±0 | 6±0 | 6±0 | 6±0 |
| Ampicillin (10) | 6±0 | 6±0 | 6±0 | 6±0 | 6±0 |
| EtBr (5) | 21±1 | 28±1 | 22±1 | 22±1 | 21±1 |
| H2O2 (30) | 23±1 | 28±1 | 23±1 | 28±1 | 23±1 |
Average diameter of the zone of inhibition (including filter disk) in millimeters, with standard deviation obtained from three independent experiments. The diameter of the filter disk is 6 mm.
To assess the importance of tmRNA-mediated degradation for the observed growth defects in F. tularensis, we created two known proteolysis-resistant tmRNA variants, tmRNAH6 (Fig. 1B), with six histidines replacing the last six native residues of tmRNA tag, and tmRNADD (Fig. 1C), with two aspartic residues replacing the two native C-terminal alanine residues of the tmRNA tag. These mutants are functional in trans-translation; however, cellular proteins tagged by the tmRNADD variant are highly resistant to proteolysis, while substrates tagged by the tmRNAH6 variant are partially resistant to proteolysis by C-terminal specific proteases (Choy et al., 2007, Ge & Karzai, 2009, Gottesman et al., 1998, Keiler et al., 1996). Creation of a functional tmRNAH6 variant required the retention of the stem-loop structure in helix-5 of tmRNA, which was accomplished by introduction of compensating base pair partnering residues (Fig. 1B). Complementation studies showed that the tmRNADD variant was less efficient in compensating the growth defects of ssrA mutant, compared to complementation by the WT ssrA gene (Figure 2C). The tmRNAH6 variant, with the compensatory substitutions in helix-5, was functional in an endogenous tagging assay (see below) and complemented the growth defect of the ssrA mutant strain, albeit not as well as the WT ssrA gene (Fig. 2C). The tmRNAH6 variant with uncompensated helix-5 mutations, on the other hand, did not complement the growth defects of the ssrA mutant strains. These results underscore the importance of the degradative potential of the tmRNA tag and support the presence and functional importance of helix-5 for F. tularensis tmRNA activity.
F. tularensis tmRNA tag sequence and identification of tagged substrates
In light of the aforementioned findings, we sought to determine the actual sequence of the Francisella tmRNA peptide tag. In many bacterial species, the predicted tmRNA tag sequence is highly conserved and begins with an alanine (GCX) codon –the E. coli tmRNA tag sequence, ANDENYALAA, conforms to this rule. The tag peptide contains signal sequences at both its N- and C-terminal ends for recognition by various proteases and their cofactors. Based on comparative tag sequence analysis, there was some ambiguity as to the resume codon of the F. tularensis tmRNA tag. There are three potential translation resume sites: the tag sequence could start with a GCG (Ala) codon beginning at nucleotide 124 (Fig. 1A), resulting in the AANDSNFAAVAKAA peptide tag, which is very similar to the E. coli tag (ANDENYALAA), with an N-terminal proximal signal for SspB/MglB (AAND) and a C-terminal signal for ClpXP (KAA). The second possibility was that the tag sequence could start with a GCA (Ala) codon located four codons upstream, at nucleotide 113, resulting in the ANRVAANDSNFAAVAKAA peptide tag (Fig. 1A). Alternatively, the tag sequence could begin with a GGC (Gly) codon, a further four codons upstream at nucleotide 100, resulting in the GNKKANRVAANDSNFAAVAKAA peptide tag, which is substantially longer than the canonical E. coli tag (Fig. 1A). In an attempt to determine the identity of the tmRNA tag and discover native substrates of the F. tularensis tmRNA system, we introduced the tmRNAH6 expression vector into an ssrA mutant strain. The tmRNAH6 variant was functional in this background and tagged a range of cellular proteins (Fig. 4A), as measured by endogenous tagging assays (Roche & Sauer, 2001, Sundermeier et al., 2008, Sundermeier et al., 2005). Our initial mass spectrometry (LC-MS/MS) analysis of these tagged proteins showed an assortment of endogenously tagged substrates carrying the (A)-GNKK trypsin digested segment of the tag –the first alanine (A) comes from the aminoacylated tRNA-like domain of tmRNA. These data suggested that the F. tularensis tmRNA ORF began with a GGC (Gly) resume codon at nucleotide 100, yielding the 22 amino acid encoded tag [(A)-G1-N2-K3-K4-A5-N6-R7-V8-A9-A10-N11-D12-S13-N14-F15-A16-A17-V18-A19-K20-A21-A22] (Fig. 1A).
Figure 4.
[A]. Analysis of proteins that are endogenously tagged by the F. tularensis tmRNAH6 variant. Cultures of the ssrA mutant strain A147 (control), or the pssrAA5EH6 variant complemented strain (right lane) were grown to stationary phase, and tmRNAH6-tagged endogenous proteins were purified by Ni-NTA affinity chromatography. H6-tag containing proteins were resolved by electrophoresis on 15% Tris-Tricine gels and examined by western blot analysis with anti-H6 antibodies. [B]. Mass-spectrometric analysis of endogenous tmRNA-tagging in F. tularensis reveals the sequence of the tmRNA-encoded peptide tag and ladder-like pattern of tagging sites. A schematic representation of this pattern is provided for one of the tmRNA-tagged proteins, F. tularensis GroEL, highlighting the junction site and addition of the tmRNA tag to consecutive amino acids on a single site of the GroEL protein. The junction site alanine residue, in parenthesis, is derived from the tmRNA charge. The WT sequence of the tmRNA tag is shown below the schematic for comparison. [C]. An example is shown of MS/MS fragmentation patterns of tmRNA tag-containing junction peptides from F. tularensis catalase protein (KatG), forming a ladder-like tagging pattern. The four spectra show the catalase peptide TVALIAGGH (in black) with the added F. tularensis tmRNA-tag fragment AGNK (in red), and of three junction peptides, where the same tmRNA-tag fragment is incorporated at C-terminally adjacent position of KatG protein. These spectra show the y-ions produced during fragmentation of the peptides inside the mass spectrometer. Due to the nature of the mass-spectrum, the direction of the peptide sequences shown is from right (N-terminus) to left (C-terminus). The amino acid sequence of the four junction peptides is shown below the MS-MS spectra.
In an effort to confirm these observations and further facilitate the identification of tag-junction peptides (the exact site of tmRNA tag addition), we constructed tmRNAAla5Glu-H6. In this variant, the native alanine residue at position 5 of the F. tularensis tmRNA tag was substituted with glutamic acid to permit subsequent controlled cleavage of the tmRNA-tagged peptides with an additional endopeptidase, Glu-C, prior to mass-spectrometric analysis. For more detailed analysis of tagged proteins, we transformed the F. tularensis ssrA strain with a vector carrying the tmRNAAla5Glu-H6 variant and grew the cultures to saturation. We subsequently performed affinity purification of tmRNA tagged proteins, from total cell lysates under denaturing conditions, by capturing H6-tagged protein on a Ni-NTA column. The affinity-purified protein pool was then subjected to controlled proteolysis by trypsin, endopeptidase Glu-C, or both enzymes, followed by high-resolution mass-spectrometric analysis of the peptide digests by the MudPIT (Multidimensional Protein Identification Technology) method. This analysis revealed the identity of multiple junction peptides, each carrying the tmRNA tag (Table S1 and Table S2). The tmRNA-encoded part of all junction peptides revealed the sequence (A)-G1N2K3, consistent with our earlier finding that the Francisella tmRNA-tag ORF begins with the GGC (Gly) codon at position 100 (Fig. 1A and Table S2).
Using this purification and proteomics approach, we analyzed over 20,000 putatively tagged peptides, among which greater than 1,350 had clear and discernable tag junction peptides, representing over 200 distinct cellular proteins (Table S1 and Table S2). Surprisingly, the majority of the tmRNA-tagged proteins were appended at internal sites. Only a few received C-terminal tags, i.e. tags added after the terminal residues of the protein. Among these substrates, we did not encounter tagging due to predicted sequence motifs, previously shown to promote ribosome stalling and trans-translation. For instance, we did not observe any tags added after the putative NPP-weak stop signal, originally identified in E. coli (Hayes et al., 2002a, Hayes et al., 2002b, Roche & Sauer, 2001). Most interestingly, our analysis revealed a ladder-like pattern of tmRNA-tag addition, where the peptide tag is appended after consecutive adjacent residues within a segment of the protein (Fig. 4B and 4C, and Table S2). The junction peptides were clearly discernable from these ladders, with some having up to ten consecutive amino acids being tagged (Fig. 4B and Table S2). A few of the substrates, e.g., catalase KatG, (Fig. 4C) contained tmRNA tagging loci along their entire length, while others contained only a limited number or a single tagging locus.
ssrA and smpB mutants of F. tularensis are impaired in replication within macrophages
SmpB and tmRNA are known to contribute to the virulence of a number of pathogenic bacteria (Hutchison et al., 1999, Julio et al., 2000, Okan et al., 2006, Okan et al., 2010, Thibonnier et al., 2008). Therefore, we assessed the potential importance of the smpB and ssrA genes for F. tularensis virulence and pathogenesis. F. tularensis is known to replicate inside host cells, including macrophages, and mutants defective in intracellular replication (such as mglA and mglB) are avirulent (Barker & Klose, 2007, Baron & Nano, 1998). To examine the role of the trans-translation system in F. tularensis pathogenesis, we evaluated the ability of ssrA and smpB mutant strains to infect and replicate inside mouse bone marrow-derived macrophages (BMDM). Cultured primary BMDM were infected with identical multiplicity of infection (MOI) of the smpB, ssrA, or the WT strain. To ensure that we were monitoring intracellular bacterial growth exclusively, extracellular bacteria were killed by treatment with gentamicin. CFU assays were conducted to estimate the number of surviving intracellular bacteria. CFU assays performed at 1 h post-infection showed that ssrA mutant bacteria were internalized as efficiently as the parental strain. In sharp contrast, at 16 h post-infection the parental strain replicated to levels approximately 100-fold higher than the 1 h time point (Fig. 5A). The ssrA mutant A147, a partially functional mutant, was able to replicate only 10-fold during the same time period, while the ssrA A174 mutant strain showed no or minimal intracellular replication (Fig. 5A). The intracellular replication defect of the ssrA mutants was fully complemented by a plasmid-borne copy of the WT ssrA gene (Fig. 5A). Interestingly, complementation with the proteolysis-resistant tmRNADD variant did not restore the intracellular replication defect of the ssrA mutants, suggesting that the timely disposal of some tmRNA tagged proteins was important for intracellular replication (Fig. 5A).
Figure 5.
F. tularensis ssrA and smpB mutants exhibit intracellular growth defects. Mouse bone marrow-derived macrophages were infected with the indicated ssrA [A] or smpB [B] mutants, the complemented mutants, or the WT strain at an MOI of 10 for 2 h. After killing of extracellular bacteria by addition of gentamicin, the host cells were lysed at 16 h [A] or 24 h [B] post-infection. Serial dilutions of the lysates were spread on MH Chocolate Agar plates for CFU analysis. The graphs show the means from three independent experiments and the error bars show standard deviations. The differences in the intracellular replication levels between the mutant and the WT strain and the mutant and the complemented strain were statistically significant (p<0.005).
The smpB B255 mutant was similarly defective in intracellular replication (Fig. 5B). The intracellular replication defect was not due to a delay in replication, as later time points did not yield higher CFU of mutant bacteria. To independently confirm these observations, we performed macrophage infection assays, using an smpB mutant and a WT strain carrying an expression plasmid for the green fluorescent protein. Consistent with our earlier findings, we observed far fewer fluorescent bacteria at later time points as compared to the parental strain (Fig. S3). These results demonstrate that the SmpB-tmRNA mediated trans-translation process in general and the proteolytic function in particular are required for intracellular replication of F. tularensis within host macrophages. These findings are also consistent with the conclusion that the mutant bacteria are defective in their ability to replicate within primary macrophages.
smpB mutants of F. tularensis are defective in phagosomal escape
Francisella is a facultative intracellular pathogen. It gains entry into the host cell by the process of looping phagocytosis (Clemens et al., 2005). The phagosomal membrane surrounding Francisella restricts the initial access of the engulfed bacteria to the cytoplasm, the ultimate site of intracellular bacterial replication. The WT bacteria have evolved mechanisms to rapidly escape the phagosomal compartment, which enable the released bacteria to replicate to high numbers inside the cytoplasm of infected cell (Clemens et al., 2004, Golovliov et al., 2003).
One possibility for the observed inability of the Francisella smpB mutant to replicate within host macrophages was that it lacked the ability to escape from the phagosomal compartment. To assess this possibility, we examined the capacity of the mutant bacteria to escape from the phagosomes of primary BMDM. To this end, we performed phagosomal integrity assays by infecting BMDM with WT and smpB mutant bacteria and examined phagosomal escape at 1 h post infection. The 1 h time point was selected as it permitted sufficient time for a large number of WT bacteria to escape the phagosomal compartment (see below), but it was decidedly insufficient for any significant increases in cell number due to bacterial replication. The phagosome integrity assay we employed uses a 2-step permeabilization protocol. In the first step, the membrane of the infected cells is permeabilized by digitonin treatment, which permits staining of bacteria that have escaped to the cytoplasm. In the second step, all cellular membranes are permeabilized by a triton X-100 treatment. Total intracellular bacteria are then labeled (Fig. 6). The differential staining of cytoplasmic (yellow) and phagosomal (green) bacteria facilitates a reliable measure of phagosomal escape (Fig. 6A). For the WT strain, we observed 82±4% of the total bacteria in cytoplasm of the infected cells at 1 h post infection. This level of escape is similar to what has been reported in previous studies (Checroun et al., 2006). Compared to the WT strain, we found the smpB mutant to have greater than 2.5-fold defect in phagosomal escape at 1 h post infection, with only 32±2% of the total bacteria in the cytoplasm of the infected cells (Fig. 6B).
Figure 6.
F. tularensis smpB mutant is defective in phagosomal escape within bone marrow-derived macrophages. [A] Representative results for phagosomal integrity assay. Mouse bone marrow-derived macrophages were infected with the WT or the smpB mutant strain at MOI of 25. The infected macrophages were permeabilized with digitonin, which enables staining of only cytoplasmic bacteria by mouse monoclonal anti-LPS antibody (red). The cells were subsequently treated with triton X-100, which permits staining of total intracellular bacteria by rabbit polyclonal anti-LVS antibody (green). [B]. Quantification of the ability of the of WT and smpB mutant strains to escape from the phagosomal compartment into the cytoplasm at 1 h post infection. Phagosomal escape is represented as percentage of cytoplasmic bacteria (yellow). In each experiment digitonin-permeabilized cells were stained for C-terminal cytoplasmic tail of calnexin (ER membrane protein) as a control. The data presented are from three independent experiments, each performed in triplicates. For each strain, over 7000 bacteria are counted to estimate the phagosomal escape. [C]. Quantification of the ability of the of WT and smpB mutant strains to escape from the phagosomal compartment into the cytoplasm at 4 h post infection. Phagosomal escape is represented as percentage of cytoplasmic bacteria. In each experiment digitonin-permeabilized cells were stained for C-terminal cytoplasmic tail of calnexin as a control. The data presented are from three independent experiments, each performed in triplicates. For each strain, over 7000 bacteria were counted to estimate the phagosomal escape. (*) The difference in phagosomal escape between WT and mutant strains is statistically significant (P<0.0001).
To ascertain whether the smpB mutant exhibited a delay in phagosomal escape we performed phagosomal integrity assays, by infecting BMDM with WT and smpB mutant bacteria, and examined phagosomal escape at 4 h post infection. This analysis revealed that the SmpB mutant still had a 2-fold defect in phagosomal escape at 4 h post infection, with only 42±2% of the total bacteria in the cytoplasm of the infected cells (Fig. 6C). This indicates that the loss of smpB gene function results in compromised ability of this strain to modify the phagosomal compartment. Under our experimental conditions, the percentage of escaped bacteria is independent of the growth rate of the bacteria. These findings suggest that the overall failure of the smpB mutant to replicate within macrophages is due in part to its inability to escape from the phagosomal compartment.
The smpB mutant of F. tularensis is avirulent in mouse models of tularemia
The observation that the smpB mutant was unable to replicate within macrophages hinted at a potential role for the SmpB-tmRNA system in F. tularensis virulence. To investigate this possibility, we evaluated the ssrA and smpB mutant strains in mouse infection models of tularemia. First, we infected groups of C3H/HeN mice via the intradermal route with either WT or the mutant strains. We chose an infective dose (107 CFU per animal) that is considered lethal in this mouse model. Indeed, all mice infected with the parental F. tularensis strain succumbed to this treatment within ten days of infection (Fig. 7). In sharp contrast, although mice infected with the same dose of the ssrA (Fig. 7A) or smpB mutants (Fig. 7B) exhibited some of the initial symptoms of the infection (malaise, ruffled fur, hunched posture, decreased mobility, and weight loss), they all fully recovered and survived the infection. The virulence defect of the ssrA mutants was compensated by complementation with a plasmid-borne copy of WT ssrA (Fig. 7A), and that of the smpB mutant with a chromosomally integrated copy of WT smpB (Fig. 7B). Two orders of magnitude more cells (109 CFU) of the ssrA A174 strain were required to cause lethality via the intradermal route. The smpB B255 mutant strain did not cause lethality by this route even at the highest dose (5×109 CFU).
Figure 7.
F. tularensis ssrA and smpB mutants do not cause lethal disease in mouse models of tularemia. [A]. C3H/HeN mice were infected intradermally with 107 CFU of F. tularensis ssrA mutant A147, the complemented mutant containing a plasmid-borne copy of ssrA, or with the WT strain. Mice were monitored for survival for at least three weeks, and survival curves were plotted. [B]. Intradermal infections and survival curves for mice infected, at the indicated CFU, with F. tularensis smpB mutant B255, the mutant complemented with a chromosomal copy of smpB (B255 bla::smpB), or the WT strain were performed as described in panel [A]. [C]. Intraperitoneal infections and survival curves for mice infected, at the indicated CFU, with F. tularensis smpB mutant B255 or the WT were performed as described in panel [A]. [D]. Intranasal infections and survival curves for mice infected, at the indicated CFU, with F. tularensis smpB mutant B255 or the WT strain were performed as described in panel [A]. Two independent experiments with 5 mice per bacterial strain were performed, and the data were combined. The smpB mutant was significantly attenuated compared to the parental LVS (P < 0.0001), and complementation restored virulence to the smpB mutant (P < 0.01).
To assess the extent of the virulence defect of the smpB strain, we evaluated additional routes of infection. To this end, we examined infections via the intraperitoneal and intranasal routes. We found that only 1–2 CFU of the parental LVS strain were required to cause mortality via the intraperitoneal route. To achieve the same level of mortality via the intraperitoneal route, 106 CFU or higher doses of the smpB mutant strain were required (Fig. 7C). Examination of infections via the intranasal route revealed that 104 to 105 CFU of WT were sufficient to cause mortality. Infection with the smpB mutant strain by intranasal route, while causing disease at higher doses (108 –109 CFU), did not cause lethality at doses up to 5×109 CFU (Fig. 7D). These data demonstrate that a functional SmpB-tmRNA mediated trans-translation system is required for virulence and pathogenesis of F. tularensis.
To further dissect the observed in vivo virulence defects, we performed bacterial organ burden assays on mice infected with either the smpB mutant or with the control parental LVS strain via the intradermal, intraperitoneal, or intranasal routes. In agreement with the results of the survival assays, significantly higher doses of the mutant bacteria were necessary to recover similar levels of the bacteria from the internal organs of the infected mice (Fig. 8, and data not shown). At the highest infective doses tested, and at the early time points post-infection (day 1 to day 5), the mutant bacteria were found in the spleen, liver, and lungs of the infected mice, indicating that at these higher doses the mutant bacteria were capable of disseminating from the primary site of infection (Fig. 8). In contrast to the parental strain, however, these initial pools of infecting mutant bacteria did not persist, and the bacterial burden declined substantially in all infected organs by the end of the first week, leading to full recovery of the infected mice.
Figure 8.
Organ burden analysis of mice infected with F. tularensis smpB or ssrA mutants. C3H/HeN mice were infected by intranasal route with 108 CFU of the F. tularensis smpB B255 mutant or with 105 CFU of the WT strain. The infected mice were sacrificed at the indicated post-infection time points, and their organs (lung [A], liver [B], or spleen [C]) were collected. The organs were homogenized, and serial dilutions of the resident bacteria were spread on MH Chocolate Agar plates for CFU determination. Two independent experiments with 5 mice per time point per bacterial strain were performed, and the data were combined. The bars indicate mean CFU values. The difference between mutant and WT strains was significant for all organs at day 9 post-infection (p<0.01).
Immunization with the live smpB mutant strain protects mice against subsequent challenge with a lethal dose of virulent F. tularensis
The mouse infection and organ burden assays suggested that the smpB-ssrA mutants of F. tularensis were highly attenuated, and infection of mice with these strains, even at substantially higher doses, was not associated with signs of severe disease and mortality. These phenotypes are desired characteristics for an effective vaccine strain. Therefore, we sought to investigate the capacity of the smpB mutant strain to induce protective immunity against subsequent lethal infection by virulent F. tularensis LVS. Groups of five C3H/HeN mice were inoculated via the intradermal route with sub-lethal doses (105 CFU) of WT, 106 CFU of the smpB mutant, or an equal volume of PBS. One month later, all immunized mice were challenged via the intradermal route with 108 CFU of F. tularensis LVS, equal to 10 lethal doses for the C3H/HeN mouse model. All control mice, in the PBS-treated group, succumbed to the disease by day 5 post-infection (Table 2). In sharp contrast, all of the smpB-vaccinated mice were fully protected against this lethal dose of infection and survived the challenge with little signs of disease. We also utilized a similar strategy for intraperitoneal vaccination of mice with the smpB mutant. Intraperitoneal vaccination with F. tularensis LVS is difficult to achieve due to the low lethal dose (1–2 CFU). As a consequence, some of the LVS vaccinated mice succumbed to the infection prior to challenge, while others did not reach immune state and died after the challenge, presumably because they did not receive any inoculating bacteria (Table 2). In contrast, we were able to vaccinate mice intraperitoneally with 105 CFU of the smpB strain without any ill effects. Most interestingly, all of the smpB-vaccinated mice were fully protected against a lethal intraperitoneal challenge with a high lethal dose (103 CFU) of F. tularensis LVS. Finally, we tested the smpB mutant strain for its ability to confer protection against the pneumonic form of tularemia in a mouse intranasal model of infection. To this end, we inoculated mice via the intranasal route with either 105 CFU of the smpB mutant strain, 103 CFU of the LVS, or PBS alone. The intranasal vaccination approach with the smpB mutant conferred complete protection against a lethal intranasal challenge with 105 CFU of WT, while all the mock-treated mice succumbed to the challenge (Table 2). Taken together, these finding suggest that the F. tularensis smpB mutant strain has the potential for developing effective live cell vaccine for tularemia.
Table 2.
Survival of immunized mice that were challenged via the indicated routes with lethal doses of F. tularensis LVS
| Immunization Route | PBS control
|
F. tularensis B255 (smpB)
|
F. tularensis LVS
|
||||
|---|---|---|---|---|---|---|---|
| Intradermal | intraperitoneal | intranasal | Intradermal | intraperitoneal | intranasal | ||
| Immunization Dose (CFU) | - | 106 | 105 | 105 | 105 | 1 | 103 |
| Challenge Dose (CFU) | (108, 103, 105) a | 108 | 103 | 105 | 108 | 103 | 105 |
| # Survivors/Challenged c | 0/15 | 5/5 | 5/5 | 5/5 | 5/5 | 2/4 b | 5/5 |
Mice in each PBS control group were challenged via the appropriate routes with the indicated CFU of F. tularensis (LVS)
One mouse in this group did not survive the vaccination
The vaccination experiment was performed once with five mice per group
DISCUSSION
In this study, we have demonstrated that F. tularensis maintains an active SmpB-tmRNA-mediated trans-translation system and relies on it for adaptation to a variety of environmental conditions. Our findings clearly show that insertional inactivation of the smpB and ssrA genes results in significant changes in cellular physiology and lead to the profound inability of the bacterium to withstand a variety of physiological and environmental stresses. The ability to adapt to different environmental conditions is particularly important for pathogenic bacteria, since they must deal with a variety of host defense mechanisms. SmpB and tmRNA deficient strains of F. tularensis, unlike their E. coli and Yersinia counterparts, exhibit a slower in vitro growth phenotype, which might contribute to their observed intracellular growth defects. However, our studies also demonstrate that F. tularensis smpB and ssrA mutants are unable to replicate within macrophages, and this defect is due in part to a reduction in the capacity of the mutant bacteria to escape the phagosomal compartment, even at early time points post infection where cell growth and replication are not dominant factors (Fig. 6). Therefore, since the capability of F. tularensis to replicate within host cells appears to be necessary for its virulence, and the smpB and ssrA mutants are impaired in their ability to grow inside macrophages, we postulated that the smpB and ssrA mutants of F. tularensis might be avirulent and unable to cause disease in mouse models of tularemia. Indeed, smpB and ssrA mutants of F. tularensis suffered severe defects in their ability to cause lethal disease in mice. Most strikingly, studies of infections via the intraperitoneal route revealed that while only 1–2 CFU of the LVS were sufficient to cause mortality, over 106 smpB mutant bacteria were required to achieve the same outcome. Thus, the SmpB-tmRNA mediated tagging and ribosome rescue system plays an important role in F. tularensis virulence.
F. tularensis mutants with impaired trans-translation capacity exhibited another attenuated phenotype that makes them potential candidates for developing live cell tularemia vaccines. This attenuated phenotype was revealed by organ burden analyses of mice infected with high doses of the mutant bacteria, showing that while the mutant bacteria were disseminated to various target organs (spleen, liver, and lungs), they were cleared much more rapidly than the WT bacteria and the infected mice made full recovery. This led us to examine the possibility that perhaps the host immune system had been sufficiently educated by its encounters with the mutant bacteria to enable protection of the host against subsequent challenges with F. tularensis LVS. Indeed, direct examination of this possibility demonstrated that immunization of mice with the smpB mutant strain provided full protection against subsequent lethal intradermal, intraperitoneal, and intranasal challenges with the WT strain. Our immunization experiments, conducted with the LVS strain, which is fully virulent in mice, hold promise for use of trans-translation mutants as the basis for human vaccine development. Although our current studies look extremely promising, comprehensive future studies with the type A Schu S4 strain will be required to establish the practicality of trans-translation mutants as the basis for development of tularemia vaccine.
Studies in E. coli and other bacterial species have demonstrated that the tmRNA tag sequence contains multiple signal sequences for recognition by various proteases and their co-factors (Flynn et al., 2001, Levchenko et al., 2000, Ge & Karzai, 2009, Choy et al., 2007, Okan et al., 2006). Our proteomic studies permitted the conclusive determination of the F. tularensis tmRNA-encoded peptide tag, demonstrating that translation of the tmRNA ORF resumes with a GGC (gly) codon, and that the encoded 22 amino acid tag is longer in comparison to tmRNA tags from other well-studied bacterial species. Interestingly, the F. tularensis tag contains signals for ClpXP and its cofactor, SspB/MglB. However, the SspB recognition signal sequence (Ala-Asn-Asp) is located in the middle of the tag, while in many other bacterial species this sequence is located at the N-terminus of the tag. It is unclear why the F. tularensis tmRNA tag is longer. Studies of the Mycoplasma pneumonia tmRNA tag, which is 27 amino acid residues in length, have shown that parts of this extended tag have evolved to accommodate additional recognition signals for the Lon protease (Ge & Karzai, 2009). Further studies are required to decipher why the F. tularensis peptide tag is longer or what additional signal motifs it might contain.
Consistent with previously reported observations (Okan et al., 2010, Okan et al., 2006, Mehta et al., 2006), the tmRNA structure and proteolytic targeting function of the encoded peptide tag appear to be important in F. tularensis. Complementation studies with the proteolysis-resistant tmRNADD variant showed decreased efficiency in compensating the growth and virulence defects of the F. tularensis ssrA mutants, suggesting that tmRNA-tag dependent proteolysis of target proteins is essential for F. tularensis growth and adaptation to adverse environmental conditions. Similarly, our gene disruption and mutational studies pointed to the importance of maintaining the structural integrity of tmRNA for its function. In particular, preserving the integrity of helix-5 appears to be of functional consequence. Helix-5 is formed in part by the tmRNA ORF and the adjacent 3′ segment. Disruptions by either intron insertion (as with the ssrA A174 mutant) or nucleotide substitutions that weaken the base-pairing propensity of this helix (as in the uncompensated tmRNAH6 variant) resulted in severe impairment of tmRNA activity, confirming the significance of this structure to tmRNA function. We observed some functional differences between ssrA mutant strains. We attribute these differences to the degree of tmRNA inactivation, i.e., to the retention of some degree of partial trans-translation related functions (ribosome rescue or tagging for proteolysis) in the ssrA A147 strain. Indeed, mutations of E. coli ssrA at positions following the tmRNA ORF have been shown to retain some of the functions of the native molecule, including the ribosome rescue function (Nameki et al., 2000, Wower et al., 2004). The smpB mutant, as judged by the magnitude of its growth and stress-related defects, is likely a fully inactivating mutant, lacking all trans-translation related functions, similar to what has been observed for smpB mutants in other bacterial species (Baumler et al., 1994, Karzai et al., 1999, Hong et al., 2005, Okan et al., 2006, Abe et al., 2008, Mikulik et al., 2008, Thibonnier et al., 2008, Okan et al., 2010).
Our efforts to determine the nature and identity of the F. tularensis tmRNA tag necessitated the construction of tmRNAH6 variants and the use of a much more sensitive mass spectrometry (MudPIT-LC-MS-MS) approach. Such high-resolution approaches had not been previously employed to either examine endogenous tmRNA substrates or determine the exact junction site of the peptide tag. Application of this proteomics approach to tmRNA-mediated endogenous tagging in F. tularensis uncovered a high number of hitherto unknown native protein substrates of trans-translation. It also enabled the unambiguous determination of the tmRNA tag peptide junction sites for close to two hundred proteins (Table S1). These trans-translation substrate proteins appear to be functionally diverse. One well-represented group contains proteins with translation related functions. These include proteins of both large and small ribosomal subunits, translation elongation and initiation factors, and protein folding chaperones. The frequency of trans-translation mediated tagging in this case may simply be due to high-level synthesis of these proteins, necessary for both normal cell growth and adaptations to physiological stresses. Alternatively, tagging could simply serve to remove unwanted and surplus proteins from the cell and provide nutritional building blocks for synthesis of new and essential proteins. Another well-represented group includes components of the outer membrane. We also found several known F. tularensis virulence factors, including intracellular growth locus C (IglC) and macrophage infectivity factor (Mif), which are necessary for full virulence and intracellular replication in mammalian cells (Lai et al., 2004, Lauriano et al., 2004, Lindgren et al., 2004, Santic et al., 2005).
The unprecedented observation of a frequent ladder-like pattern of tmRNA-tagging sites, across many endogenous substrates, is quite intriguing, and could provide mechanistic insights into the tagging process and the trans-translation system in general. The existence of this ladder-like pattern of tagging by the tmRNA system may have eluded the attention of prior investigations of endogenously tagged substrates because high-resolution mass spectrometric methods for the capture and detection of the tagged peptide were not employed. The existence of this pattern of tagging is not unique to F. tularensis. Using this exact approach, we have detected the same ladder-like pattern of tagging in E. coli and Y. pseudotuberculosis (AS and WK, unpublished data). The mechanism by which this pattern of tagging arises is of great interest to a more complete understanding of the trans-translation process and will require further investigation.
Experimental Procedures
Bacterial strains and culture conditions
F. tularensis LVS (ATCC 29684), was a gift from Dr. Karen Elkins (Center for Biologics Evaluation and Research, U.S. Food and Drug Administration, Rockville, MD). Frozen stocks of F. tularensis were prepared from liquid cultures by addition of 20% glycerol and stored at −80°C. F. tularensis was grown on ready-made plates of Mueller-Hinton (MH) agar supplemented with 1% bovine hemoglobin and 2% IsoVitaleX Enrichment (BD Biosciences, Lincoln Park, NJ), or on the same formulation with 2.5% defibrinated sheep blood substituting for hemoglobin, and with an additional 1% proteose peptone and 0.1% glucose (LoVullo et al., 2006). Tryptic soy broth or tryptic soy agar with addition of 0.1% cysteine and 25μgml −1 each of sodium pyruvate, ferrous sulfate and sodium metabisulfite was used for growth of bacteria after electroporation (Rodriguez et al., 2009). F. tularensis plates were incubated for 2–3 days at 37°C in incubators infused with 5% carbon dioxide. Liquid rich medium for F. tularensis were either MH broth (cation-adjusted, BD Biosciences) supplemented with 2% IsoVitaleX Enrichment, 0.1% glucose and 335μM ferric pyrophosphate; brain heart infusion broth (BHI) titrated to pH 6.8, or BHI supplemented with 0.1% cysteine. Liquid minimal medium for Francisella (Chamberlain medium) was prepared as previously described (Chamberlain, 1965). F. tularensis culture media were supplemented, when indicated, with the following concentrations of antibiotics: kanamycin, 5 μg ml−1, hygromycin B, 200 μg ml−1, or ampicillin, 50 μg ml−1. Sucrose selection was performed by inclusion of 5% sucrose in solid growth media.
Construction of F. tularensis mutants and complemented strains
All oligonucleotide primers used in this study are listed in Table S3. F. tularensis LVS tmRNA (ssrA) and smpB targeted mutation strains were constructed using TargeTron technology adapted for use in Francisella (Rodriguez et al., 2009). The respective gene sequences were used as inputs for Sigma-Aldrich’s computer-based algorithm, according to the manufacturer’s specifications. The following output primers were obtained for targeting these genes with the LtrB intron: ssrA-147s-IBS, ssrA-147s-EBS1d and ssrA-147s-EBS2 for LtrB insertion at base pair (bp) position 147 of ssrA; ssrA-174s-IBS, ssrA-174s-EBS1d, and ssrA-174s-EBS2 for insertion at bp174 of ssrA; and smpB-255s-IBS, smpB-255s-EBS1d and smpB-255s-EBS2 for insertion at bp 255 of smpB. The universal LtrB intron-specific primer EBS-Universal was used for all target genes. The mutations were confirmed by PCR with both intron-specific primers and gene-specific primers external to the insertion sites. Sigma-Aldrich’s TargeTron kit was used to obtain the targeting constructs based on plasmid pKEK1140 [kindly provided by Dr. Karl Klose (Rodriguez et al., 2009)]. The parental F. tularensis LVS cells were electroporated with the respective constructs and the transformants were grown on kanamycin-containing plates. Mutant clones were selected by PCR. DNA sequencing of the PCR products was used to confirm the LtrB intron insertion sites. The targeting vector was cured from mutant clones by growth on medium without antibiotics at 37°C, and the vector loss was confirmed by testing for kanamycin sensitivity.
Francisella shuttle vector pMP633 was used for gene-complementation experiments (LoVullo et al., 2009b, LoVullo et al., 2006). For complementation with ssrA, the gene with its putative promoter region of 100 bp upstream of the first nucleotide and an additional 20 bp downstream of the last nucleotide of ssrA was PCR amplified from a WT colony with primers ssrA-UPfw-MluI and ssrA-DNrev-MluI and sub-cloned into the MluI site of the pMP633 vector to create pMP633-ssrA. For complementation with the smpB gene, including its putative promoter region of 135 bp upstream from the start codon, was PCR-amplified with primers smpB-UPfw-MluI and smpB-DNrev-MluI and sub-cloned into the MluI site of the pMP633 to create pMP633-smpB. We also employed an allelic exchange strategy (LoVullo et al., 2009a) for complementation with smpB by inserting the gene into the chromosomal blaB locus of F. tularensis, which encodes β-lactamase and confers ampicillin resistance to the WT bacteria. The smpB gene with 200 bp of upstream and downstream flanking sequence was PCR amplified with primers smpB-UP200 and smpB-DN200 and sub-cloned between the EcoRV and BamHI sites of the allelic exchange vector pMP815 (a kind gift from Dr. Pavelka, University of Rochester), creating pMP815-smpB. The smpB mutant strain B255 was transformed with pMP815-smpB, primary recombinants were selected on kanamycin, and the recombinants were confirmed for sucrose sensitivity and kanamycin resistance by re-streaking on the respective selective media. Primary recombinants were then grown on MH plates with sucrose to select for a secondary recombination event resulting in replacement of the WT copy of blaB with the smpB sequence. Secondary recombinants were then confirmed for ampicillin and kanamycin sensitivity and sucrose resistance. Insertion of smpB into the blaB locus was confirmed by PCR with primers blaBseq-FW and blaBseq-REV, external to the insertion site.
The ssrAH6, ssrAA5EH6, and ssrADD constructs were created by site-directed mutagenesis using pMP633ssrA DNA as the substrate. Primer pairs for the mutagenesis reactions were: ssrAH6-mutF and ssrAH6mutR, ssrA-Ala5Glu-mutF and ssrA-Ala5Glu-mutR, and ssrADD-mutF and ssrADD-mutR. Both ssrAH6 constructs were also made to contain a similar-length helix-5 stem-loop structure downstream of the ssrA tag open reading frame. Primers for the stem-loop creation by site-directed mutagenesis were ssrAH6stem-mutF and ssrAH6stem-mutR.
Growth rate, stress resistance and antibiotics sensitivity assays
For growth on solid medium, F. tularensis strains were grown on MH agar plates for 2–3 days. Single colonies were inoculated into 5 ml of MH broth and grown overnight (16 h) at 37°C, with shaking at 220 rpm. Fresh cultures were inoculated from overnight cultures and grown to an optical density at 600 nm (OD600) of 0.6–0.7. Aliquots containing equal number of OD600 units were diluted in fresh MH medium to obtain single colonies, spread on MH agar plates, and photographed 3–4 days later. For the heat sensitivity assay, 10-fold serial dilutions of bacterial cultures obtained as above were plated on MH agar and kept at 37°C or 40°C for several days. For heat shock experiments, 0.25 ml aliquots of OD600-normalized bacterial suspensions were placed in microcentrifuge tubes and incubated at 37°C or 50°C in a dry-heat incubator. Serial dilutions of these cultures were plated for CFU counts at indicated time points. Cold-shock was performed by incubating aliquots of the culture on ice for 4 h. For liquid growth assays, bacteria were grown from frozen stocks on MH agar plates, inoculated and grown in appropriate liquid growth medium overnight, and diluted into fresh media to an OD600 of 0.05. Liquid cultures were then grown at 37°C, aliquots were removed at different time points and their optical densities measured and plotted. For oxidative stress assays, hydrogen peroxide was added to MH broth cultures to a final concentration of 0.002% (higher hydrogen peroxide concentrations tested resulted in complete absence of growth of the WT bacteria). Antibiotic sensitivity assays were performed using filter paper disks of 6 mm diameter (Becton-Dickinson). Bacteria were grown in MH broth, diluted to the same OD600 values, and equal volumes were spread on MH agar plates using cotton-tipped applicators to achieve bacterial lawns. Filter disks were then placed on the surface of the streaked plates, and 5μl solutions of the appropriate antibiotic were applied to each filter disk. At least three disks were used per condition, and each experiment was repeated at least two times.
Infection of bone marrow-derived macrophages (BMDM)
Mouse macrophages were derived from the bone marrows of C57BL/6 mice (Pujol & Bliska, 2003) and seeded as a monolayer of 1.5×105 cells per well in 24-well tissue culture plates. Francisella strains were grown to mid-exponential growth phase in brain heart infusion (BHI) broth, washed and suspended in phosphate-buffered saline (PBS), and used for infection at a multiplicity of infection (MOI) of ten bacterial cells per macrophage. Plating serial dilutions of the same sample on Chocolate agar and colony forming units (CFU) counting was used to control for the number of bacteria. For fluorescence detection, bacteria were transformed with the pFNLTP6-groE-GFP plasmid, described previously (Maier et al., 2004). To facilitate contact of the bacteria with the host cells, the co-culture plates were centrifuged for 5 min at 800 × g. Infection was permitted to proceed for 2 h at 37°C, after which the medium was aspirated and the cells washed twice with PBS. The extracellular bacteria were then killed by addition of cell culture medium containing 100 3g of gentamicin for 1 h. At the indicated post-infection time points, the BMDM were washed three times with PBS, lysed by addition of 1% saponin (Sigma) in PBS, and collected. Serial dilutions were prepared in PBS and spread on Chocolate agar plates. CFU were determined after 3–4 days of growth. Each experimental condition was performed in duplicate and the experiments were repeated three times.
Phagosome Integrity Assay
The phagosome integrity assay was modified from Chong A et al (Chong et al., 2008). Mouse macrophages were derived from the bone marrows of C57BL/6 mice and seeded as a monolayer of 1.5×105 cells per well in 24-well tissue culture plates on glass coverslips. Francisella strains were grown to mid-exponential growth phase in brain heart infusion (BHI) broth, washed and suspended in phosphate-buffered saline (PBS), and used for infection at a multiplicity of infection of twenty five bacterial cells per macrophage. At 1 h post infection, the cells were washed twice with KHM buffer (110 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2, pH 7.3) and permeabilized with 50 3g ml−1 digitonin (Sigma) in KHM buffer for 1 min at room temperature. Cells were then washed immediately with KHM buffer and blocked with 2% BSA in PBS. Mouse monoclonal antibodies against Francisella LPS were delivered to stain the cytoplasmic bacteria. The cells were then fixed in 3% paraformaldehyde and treated with 50mM NH4Cl and subsequently permeabilized with 0.5% Triton X-100 (Sigma) for 5 minutes at room temperature and blocked with 2% BSA. Rabbit polyclonal antibodies against heat killed LVS were delivered to stain total cellular bacteria. Alexa Fluor-594 (red) conjugated goat anti-mouse (Invitrogen) and Alexa Fluor-488 (green) conjugated goat anti-rabbit (Invitrogen) antibodies were added to detect cytoplasmic and total cellular bacteria respectively. The coverslips were mounted on microscope slides with ProLong Gold Antifade Reagent (Invitrogen) and visualized by fluorescence microscopy using a Zeiss Axioplan2 microscope equipped with a 40X objective. A Spot camera (Dignostic Instruments) was used to sequentially capture Alexa Fluor 594 (red) and Alexa Fluor 488 (green) signals in 10 fields from each coverslip. The red and green images were overlaid using Adobe Photoshop and the percent escape was quantified by counting cytoplasmic bacteria (yellow) and dividing this number by total bacteria (green). For each strain more than 7000 bacteria were counted. Digitonin-permeabilized cells were stained for C-terminal cytoplasmic tail of calnexin (ER membrane protein) as a control. The results are from more than three independent experiments.
Mouse infections and bacterial organ burden assays
All animal experiments were performed in accordance with the protocol approved by the Institutional Animal Care and Use Committee of Stony Brook University. Female mice of C3H/HeN strain at 6–8 weeks of age were obtained from Charles River Laboratories. The bacteria for all routes of infection were grown to mid-exponential phase in BHI media containing cysteine, washed and suspended in PBS prior to infection. Retrospective counts were performed to determine actual CFU doses. For intradermal infections, indicated doses of the bacteria were suspended in 100 μl of PBS and injected at the base of the tail in groups of five or six animals. The same volume (100 μl) of PBS was used for mock infection controls. For intraperitoneal infections, bacteria were prepared similarly and injected into the abdominal cavity. For intranasal infection, mice were first anesthetized with a mixture of ketamine HCl (Fort Dodge Animal Health Laboratories) (100 mg ml −1) and xylazine HCl (Ben Venue Laboratories) (20 mg ml −1) mixed 2:1, which was further diluted 3:20 in PBS before a dose of 6 ml kg−1 of body weight was delivered by intraperitoneal injection. A 20 μl suspension of bacterial in PBS was delivered through a micro-pipet tip into mouse nares (10 μl each), immediately followed by an additional 20 μl of sterile PBS. The mice were monitored for survival twice daily for a period of at least 21 days. Animals in the terminal stage of disease were euthanized. For bacterial organ burden assays, groups of mice (at least ten per group) were infected, as above, and euthanized with carbon dioxide at indicated post-infection times. Their organs were aseptically removed and homogenized in 1 ml of PBS. Serial dilutions of the homogenates were spread on MH agar plates. CFU counts were performed 3–5 days later. Limit of detection for bacterial burden assay was 100 CFU per organ.
Purification and analysis of endogenous tmRNA-tagging substrates
For determination of endogenous tmRNA tagging substrates in F. tularensis, ssrA mutant strains were transformed with constructs pMP633-ssrAH6 or pMP633ssrAA5EH6 (the latter strain was used in all mass-spectrometry experiments reported here). The strains were grown overnight in BHI medium or CDM with addition of hygromycin. For large-scale purification, 1L cultures were inoculated from overnight cultures to a starting OD600 of 0.1. The cultures were then grown to late stationary phase at 37°C and harvested by centrifugation at 4000 × g. Bacterial pellets were suspended in 30 ml of lysis buffer [8 M urea, 100 mM sodium biphosphate, 10 mM tris-(hydroxymethyl)-aminomethane (Tris) at pH 8, 20 mM imidazole and 2 mM β-mercaptoethanol in distilled water]. Cell lysis was allowed to proceed for 1 h at room temperature with rotation. The lysates were sonicated with three 1 min pulses on ice. The lysates were then spun for 30 min at 30000xg, to remove the insoluble material. Aliquots of the whole-cell lysate were saved for western blot analysis. The rest of the soluble sample was mixed with 1 ml Ni-NTA slurry, pre-equilibrated in lysis buffer, and the mixture was rotated for 1 h to allow binding of the H6-tagged proteins. The lysate-resin mix was then loaded into an empty gravity-flow column and washed 5 times with 20 ml of lysis buffer, and once with the same buffer without urea. Bound proteins were eluted by addition of 5 ml of elution buffer (250 mM imidazole, 100 mM sodium biphosphate, 10 mM Tris pH 8, 1 mM beta-mercaptoethanol) in 1 ml fractions. Eluted proteins were precipitated by addition of 10% trichloroacetic acid (TCA). The precipitated material was collected by centrifugation at 30000 × g for 30 min, and washed with acetone. For western blot analysis of the tmRNA-tagged proteins, aliquots of the acetone-washed sample were resuspended and boiled in SDS–sample buffer containing 4 M urea, resolved by electrophoresis on 15% Tris-tricine polyacrylamide gels, and transferred to PVDF membranes. Membrane-bound proteins were detected with a mouse monoclonal antibody to the H6-epitope tag, a secondary goat anti-mouse alkaline phosphatase-conjugated antibody, and the colorimetric BCIP/NBT substrate.
Sample preparation for proteomics analysis
TCA/acetone pellets were resuspended in a buffer containing 8 M urea, and then diluted to 2 M final urea concentrations with a 0.1 M ammonium bicarbonate buffer. After the sample was reduced with 0.1 M DTT and alkylated with 0.2 M iodoacetamide, it was split in two and treated with either trypsin or Glu-C. After incubation for 16 h, the samples were split in two again, and trypsin was added to the Glu-C digest and Glu-C was added to the trypsin digest. After incubation of the double digests, the samples were combined, acidified with formic acid (5% final concentration) and used for mass spectrometric analysis.
Mass Spectrometry
The samples were analyzed for protein content using a modification of the multidimensional protein identification technology (MudPIT) method (Washburn et al., 2001). Samples were pressure-vessel loaded onto a column, which was packed with 3 cm of a strong cation-exchange matrix (Partisphere SCX, 5 μm, Whatman) and 3 cm of C18 matrix (Magic, Michrom Bioresources, 5 μm). Following sample loading, the column was washed for 10 min with Buffer A [2% Acetonitrile (ACN), 0.1% Formic Acid (FA)] at ~200–300 nl min-1. The MudPIT column was connected to a C18 separation column (100 μm) which was pulled using a P-2000 CO2 laser puller (Sutter Instruments, Novato, CA) to a 5 μm tip and packed with 10 cm of 5 μm Magic C18 material (Agilent, Santa Clara, CA), using a pressure vessel, and subsequently equilibrated in buffer A.
The dual-column construct was placed in line with an Eksigent NanoLC-2D HPLC pump. The HPLC separation, at 300 nl min−1, was provided by a three-component gradient, implemented in 13 steps. Each step consisted of the following sub-steps: 5 min wash with 100% Buffer A; 5 min wash with a fixed percentage of Buffer C (0.5 M ammonium acetate, in Buffer A); 10 min wash with 100% Buffer A; 60 min gradient of 0% to 40% Buffer B (90% ACN, 0.1% FA); 30 min wash, and 100% Buffer A. The 13 steps varied the fixed Buffer C from 0 to 100 %. The application of a 1.8 kV distal voltage electro-sprayed the eluted peptides directly into a Thermo Fisher Scientific LTQ Orbitrap XL ETD ion trap mass spectrometer equipped with a nanoLC electrospray ionization source (ThermoFinnigan, San Jose, CA). Full masses (MS/MS) spectra were recorded on the peptides over a 400–2000 m/z range at 60,000 resolution, followed by five tandem mass (MS/MS) events, sequentially generated in a data-dependent manner on the first, second, third, fourth and fifth most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer-scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan, San Jose, CA).
MS/MS spectra were extracted from the RAW file with Readw.exe (http://sourceforge.net/projects/sashimi). The MS/MS data were searched with Inspect (Tanner et al., 2005) against a F. tularensis LVS database with no enzyme restrictions and optional modifications: +15.9994 on methionine, +57.0214 on cysteine, and +627.3340, 498.2914, 370.1964 on the C-terminus of peptides, which corresponds to the amino acids left over from the addition of the tag. The results were p-valued to 0.1, and a new database was constructed where all potentially tagged peptides have the entire tmRNA tag added to their C-termini. The MS/MS data were again searched against the new database, with no enzyme restrictions and optional modifications of +15.9994 on methionine and +57.0214 on cysteine. Only peptides with at least a p-value of 0.01 and complete protease cleavage sites (either K, R, E, or D) were analyzed further. The proteins identified in proteomic screens were assigned to functional categories based on the F. tularensis genome database of University of Washington (http://www.francisella.org).
Statistical Analysis
The results of the organ burden assays were compared using the Mann-Whitney test for nonparametric data with one-tailed probability (P) values. The mouse survival curves were compared using the log-rank test. Results were analyzed for significance using data obtained from three independent experiments with multiple replicates. P values were calculated by one-way analysis of variance and Tukey’s multiple-comparison posttest. Statistical calculations were performed using Prism 4.0 (GraphPad Software). P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank members of the Karzai lab for critical reading of the manuscript and for helpful discussions and suggestions. We thank Drs. Karl Klose and Martin Pavelka for bacterial strains and plasmids. We thank Galina Romanov for preparation of mouse bone marrow-derived macrophages. This work was supported by the NIH grants RO1-GM-065319 and PO1-AI-055621.
References
- Abe T, Sakaki K, Fujihara A, Ujiie H, Ushida C, Himeno H, Sato T, Muto A. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Mol Microbiol. 2008;69:1491–1498. doi: 10.1111/j.1365-2958.2008.06381.x. [DOI] [PubMed] [Google Scholar]
- Abo T, Inada T, Ogawa K, Aiba H. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J. 2000;19:3762–3769. doi: 10.1093/emboj/19.14.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LS, Kongshavn PA. Experimental murine tularemia caused by Francisella tularensis, live vaccine strain: a model of acquired cellular resistance. Microb Pathog. 1987;2:3–14. doi: 10.1016/0882-4010(87)90110-0. [DOI] [PubMed] [Google Scholar]
- Barker JR, Klose KE. Molecular and genetic basis of pathogenesis in Francisella tularensis. Annals of the New York Academy of Sciences. 2007;1105:138–159. doi: 10.1196/annals.1409.010. [DOI] [PubMed] [Google Scholar]
- Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol. 1998;29:247–259. doi: 10.1046/j.1365-2958.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Kutsukake K, Abo T. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA-and ArfA-mediated pathways. Mol Microbiol. 2011;80:772–785. doi: 10.1111/j.1365-2958.2011.07607.x. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol. 2010;78:796–808. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- Chamberlain RE. Evaluation of Live Tularemia Vaccine Prepared in a Chemically Defined Medium. Appl Microbiol. 1965;13:232–235. doi: 10.1128/am.13.2.232-235.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Aung LL, Karzai AW. Lon protease degrades transfer-messenger RNA-tagged proteins. J Bacteriol. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect Immun. 2005;73:5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvaisier S, Bordeau V, Felden B. Inhibition of transfer messenger RNA aminoacylation and trans-translation by aminoglycoside antibiotics. J Biol Chem. 2003;278:14788–14797. doi: 10.1074/jbc.M212830200. [DOI] [PubMed] [Google Scholar]
- De la Cruz J, Vioque A. Increased sensitivity to protein synthesis inhibitors in cells lacking tmRNA. RNA. 2001;7:1708–1716. [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Tonat K. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sanchez F, Schaub RE, Janssen BD, Hayes CS. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol Microbiol. 2011;80:1204–1219. doi: 10.1111/j.1365-2958.2011.07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Karzai AW. Co-evolution of multipartite interactions between an extended tmRNA tag and a robust Lon protease in Mycoplasma. Mol Microbiol. 2009;74:1083–1099. doi: 10.1111/j.1365-2958.2009.06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjostedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003;71:5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease. Proc Natl Acad Sci U S A. 2008;105:16113–16118. doi: 10.1073/pnas.0808802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002a;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci U S A. 2002b;99:3440–3445. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Tran QA, Keiler KC. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol Microbiol. 2005;57:565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) Plays a Role in Salmonella enterica Serovar Typhimurium Pathogenesis. J Bacteriol. 2000;182:1558–1563. doi: 10.1128/jb.182.6.1558-1563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-Translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L. TmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J Bacteriol. 2003;185:573–580. doi: 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T, Kurita D, Takahashi T, Muto A, Himeno H. Initiation-shift of trans-translation by aminoglycosides. Nucleic acids symposium series. 2004:299–300. doi: 10.1093/nass/48.1.299. [DOI] [PubMed] [Google Scholar]
- Lai XH, Golovliov I, Sjostedt A. Expression of IglC is necessary for intracellular growth and induction of apoptosis in murine macrophages by Francisella tularensis. Microb Pathog. 2004;37:225–230. doi: 10.1016/j.micpath.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Grant RA, Flynn JM, Sauer RT, Baker TA. Versatile modes of peptide recognition by the AAA+ adaptor protein SspB. Nat Struct & Mol Biol. 2005;12:520–525. doi: 10.1038/nsmb934. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Grant RA, Wah DA, Sauer RT, Baker TA. Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag. Mol Cell. 2003;12:365–372. doi: 10.1016/j.molcel.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Li X, Yagi M, Morita T, Aiba H. Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:462–473. doi: 10.1111/j.1365-2958.2008.06167.x. [DOI] [PubMed] [Google Scholar]
- Lies M, Maurizi MR. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J Biol Chem. 2008;283:22918–22929. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H, Golovliov I, Baranov V, Ernst RK, Telepnev M, Sjostedt A. Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol. 2004;53:953–958. doi: 10.1099/jmm.0.45685-0. [DOI] [PubMed] [Google Scholar]
- LoVullo ED, Molins-Schneekloth CR, Schweizer HP, Pavelka MS., Jr Single-copy chromosomal integration systems for Francisella tularensis. Microbiology. 2009a;155:1152–1163. doi: 10.1099/mic.0.022491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoVullo ED, Sherrill LA, Pavelka MS., Jr Improved shuttle vectors for Francisella tularensis genetics. FEMS Microbiol Lett. 2009b;291:95–102. doi: 10.1111/j.1574-6968.2008.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoVullo ED, Sherrill LA, Perez LL, Pavelka MS., Jr Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology. 2006;152:3425–3435. doi: 10.1099/mic.0.29121-0. [DOI] [PubMed] [Google Scholar]
- Maier TM, Havig A, Casey M, Nano FE, Frank DW, Zahrt TC. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol. 2004;70:7511–7519. doi: 10.1128/AEM.70.12.7511-7519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Richards J, Karzai AW. tmRNA determinants required for facilitating nonstop mRNA decay. RNA. 2006;12:2187–2198. doi: 10.1261/rna.247706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulik K, Paleckova P, Felsberg J, Bobek J, Zidkova J, Halada P. SsrA genes of streptomycetes and association of proteins to the tmRNA during development and cellular differentiation. Proteomics. 2008;8:1429–1441. doi: 10.1002/pmic.200700560. [DOI] [PubMed] [Google Scholar]
- Muto A, Fujihara A, Ito KI, Matsuno J, Ushida C, Himeno H. Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes Cells. 2000;5:627–635. doi: 10.1046/j.1365-2443.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- Nameki N, Tadaki T, Himeno H, Muto A. Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett. 2000;470:345–349. doi: 10.1016/s0014-5793(00)01349-1. [DOI] [PubMed] [Google Scholar]
- Okan NA, Bliska JB, Karzai AW. A Role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2006;2:e6. doi: 10.1371/journal.ppat.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan NA, Mena P, Benach JL, Bliska JB, Karzai AW. smpB-ssrA Mutant of Yersinia pestis Functions As a Live Attenuated Vaccine to Protect Mice Against Pulmonary Plague Infection. Infect Immun. 2010;78:1284–93. doi: 10.1128/IAI.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- Pujol C, Bliska JB. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun. 2003;71:5892–5899. doi: 10.1128/IAI.71.10.5892-5899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallack DM, Johnson LL, Friedman DI. Role for 10Sa RNA in the growth of lambda-P22 hybrid phage. J Bacteriol. 1994;176:2082–2089. doi: 10.1128/jb.176.7.2082-2089.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J, Mehta P, Karzai AW. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- Rodriguez SA, Davis G, Klose KE. Targeted gene disruption in Francisella tularensis by group II introns. Methods. 2009;49:270–274. doi: 10.1016/j.ymeth.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 2005;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- Shin JH, Price CW. The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures. J Bacteriol. 2007;189:3729–3737. doi: 10.1128/JB.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier T, Ge Z, Richards J, Dulebohn D, Karzai AW. Studying tmRNA-mediated surveillance and nonstop mRNA decay. Methods in Enzymology. 2008;447:329–358. doi: 10.1016/S0076-6879(08)02217-9. [DOI] [PubMed] [Google Scholar]
- Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci U S A. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Analytical Chemistry. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- Thibonnier M, Thiberge JM, De Reuse H. Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS ONE. 2008;3:e3810. doi: 10.1371/journal.pone.0003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujiie H, Matsutani T, Tomatsu H, Fujihara A, Ushida C, Miwa Y, Fujita Y, Himeno H, Muto A. Trans-translation is involved in the CcpA-dependent tagging and degradation of TreP in Bacillus subtilis. J Biochem. 2009;145:59–66. doi: 10.1093/jb/mvn143. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Withey J, Friedman D. Analysis of the role of trans-translation in the requirement of tmRNA for lambdaimmP22 growth in Escherichia coli. J Bacteriol. 1999;181:2148–2157. doi: 10.1128/jb.181.7.2148-2157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower IK, Zwieb C, Wower J. Contributions of Pseudoknots and Protein SmpB to the Structure and Function of tmRNA in trans-Translation. J Biol Chem. 2004;279:54202–54209. doi: 10.1074/jbc.M410488200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.