Abstract
Radiation-induced bystander effects have been extensively studied at low doses, since evidence of bystander induced cell killing and other effects on unirradiated cells were found to be predominant at doses up to 0.5 Gy. Therefore, few studies have examined bystander effects induced by exposure to higher doses of radiation, such as spatially fractionated radiation (GRID) treatment. In the present study, we evaluate the ability of GRID treatment to induce changes in GRID adjacent (bystander) regions, in two different murine carcinoma cell lines following exposure to a single irradiation dose of 10 Gy. Murine SCK mammary carcinoma cells and SCCVII squamous carcinoma cells were irradiated using a brass collimator to create a GRID pattern of nine circular fields 12 mm in diameter with a center-to-center distance of 18 mm. Similar to the typical clinical implementation of GRID, this is approximately a 50:50 ratio of direct and bystander exposure. We also performed experiments by irradiating separate cultures and transferring the medium to unirradiated bystander cultures. Clonogenic survival was evaluated in both cell lines to determine the occurrence of radiation-induced bystander effects. For the purpose of our study, we have defined bystander cells as GRID adjacent cells that received approximately 1 Gy scatter dose or unirradiated cells receiving conditioned medium from irradiated cells. We observed significant bystander killing of cells adjacent to the GRID irradiated regions compared to sham treated controls. We also observed bystander killing of SCK and SCCVII cells cultured in conditioned medium obtained from cells irradiated with 10 Gy. Therefore, our results confirm the occurrence of bystander effects following exposure to a high-dose of radiation and suggest that cell-to-cell contact is not required for these effects. In addition, the gene expression profile for DNA damage and cellular stress response signaling in SCCVII cells after GRID exposure was studied. The occurrence of GRID-induced bystander gene expression changes in significant numbers of DNA damage and cellular stress response signaling genes, providing molecular evidence for possible mechanisms of bystander cell killing.
INTRODUCTION
A long-standing concept in radiobiology has been that cells must be directly exposed to ionizing radiation for DNA damage, and possible cell death, to occur. However, considerable evidence now exists that challenges this belief. In fact, as early as the 1940s, published literature provided evidence for DNA damage brought about by targeting not only the cells or tissues, but also the surrounding medium (1, 2). The term “radiation-induced bystander effects” refers to effects seen in cells that have not directly been exposed to ionizing radiation. Kotval and Gray (3) demonstrated that α particles passing close to, but not through, chromatin threads produced chromosomal breaks and exchanges. An increase in sister chromatid exchanges in cells that were not directly exposed to ionizing radiation, but were in the vicinity of directly irradiated cells (4), provided further proof of bystander effects. Among the numerous studies, each with its own nuances and protocols now accumulated in the literature, radiation-induced bystander effects can be broadly classified into two types: those mediated by gap-junctions and requiring cell-to-cell communication and those brought about by the presence of medium secreted factors, which do not require cell-to-cell contact (5–10). There are also reports in the literature where evidence of bystander effects from irradiation was not found, especially when low-LET radiation sources were used. Therefore, the phenomenon appears to have specific requirements to occur.
Spatially fractionated radiation therapy (sometimes referred to as “GRID” therapy) refers to the delivery of a single radiation fraction (10–20 Gy peak doses) by dividing a radiation field into smaller segments interspersed with segments receiving no (or very low doses) direct irradiation. This approach has been shown to have potent palliative benefits without increasing toxicity (11, 12). Although originally designed mostly to avoid normal tissue toxicity, over the last 100 years, this approach has been sporadically, and in general successfully, used to improve treatment of bulky and deep-seated tumors. Our recent clinical experience along with that of others suggests that GRID may be combined with traditional dose/time fractionated radiation therapy or used along with other treatment modalities, including chemotherapy, to achieve better control of bulky tumors while minimally extending the treatment course (11, 13). Although GRID dose distribution is nonuniform, regression of the tumor mass receiving GRID has exhibited uniform regression clinically (11, 13). One plausible explanation might be the enhanced reoxygenation of the tumor following GRID, which would be expected to improve the effect of subsequently applied radiation or chemotherapy. We have recently observed evidence of reoxygenation of tumors after spatially fractionated micro-beam radiation (61). Induction of tumor necrosis factor α and ceramide, as well as down regulation of transforming growth factor β1, have also been observed following GRID (14, 15). Increased cytokine production has resulted in broad systemic effects as well (11). It also is possible that bystander effects might play a role in killing adjacent nonirradiated or partially irradiated cells.
In the present study, we sought to evaluate the possible contribution of bystander effects to the overall therapeutic effect of spatially fractionated (GRID) radiation therapy. In our experiments, we considered the regions that were exposed to 10 Gy of radiation as “directly irradiated”. The cells adjacent to the direct irradiation fields that did not receive direct irradiation, but were exposed to indirect radiation (i.e., scattering and some degree of a very steep dose gradient), which amounted to a valley dose of approximately 1 Gy, were considered as “bystander cells”. Clonogenic survival was used to determine bystander cell killing in two different murine carcinoma cell lines following exposure to a single dose of 10 Gy using either confluent cultures and selectively harvesting bystander cells or by performing medium exchange experiments. In addition, we evaluated the expression profiles of genes involved in DNA damage signaling and stress response signaling in murine cells.
MATERIALS AND METHODS
Cell Culture
Two murine tumor cell lines (passage 3–5) were used in these experiments. SCK is a mouse mammary carcinoma cell line derived from A/J mice (16, 17). SCK cells are considered to be radiation resistant (18). We chose these cells to determine the occurrence of bystander effects in response to spatially fractionated radiation therapy at clinically relevant doses. The cells were grown in RPMI 1640 culture medium, with 2 mM L-glutamine (Cellgro, Manassas, VA) supplemented with 10% bovine serum albumin (Hyclone, Logan, UT) and 1% penicillin-streptomycin (10,000 units/ml penicillin G sodium, 10,000 μg/ml streptomycin in 0.85% saline) (Hyclone). SCCVII is a mouse squamous cell carcinoma cell line. We used these cells in our experiments, since GRID therapy has been used at our institution and others to treat mainly head and neck cancers. SCCVII cells were cultured in D-MEM culture medium with 4.5 g/L glucose, sodium pyruvate and 2 mM (concentration) L-glutamine (Cellgro, Manassas, VA), supplemented with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO) and 1% penicillin-streptomycin (10,000 units/ml penicillin G sodium, 10,000 μg/ml streptomycin in 0.85% saline) (Hyclone). SCK and SCCVII cells were sub-cultured by seeding at a concentration of 2.0 × 105 cells in 25-cm2 culture flasks (Corning, NY), and were grown in a fully humidified incubator with 5% CO2 at 37°C. The doubling time of these cell lines is approximately 24 h.
Radiation Survival Studies
SCK and SCCVII cells were sparsely plated (200–250 cells) in 25-cm2 culture flasks containing 5 ml culture medium. The irradiation was performed approximately 16–20 h later using a CP-160 cabinet X-radiator system (Faxitron, Lincolnshire, IL) at room temperature. The cells were exposed to 0, 1 and 2 Gy X rays at a dose rate of 1 Gy per min at 150 kV and 6.6 mA. The medium was changed immediately after irradiation and the flasks were returned to the incubator for 8 days, after which the colonies (>50 cells) were stained using crystal violet and counted to determine colony forming efficiency. Colony forming efficiency was defined as the ratio of the number of colonies to the number of plated cells. In all cases, the clonogenic survival was normalized to the cloning efficiency of appropriately sham treated and time-matched incubations.
Small Animal Conformal Radiation Research System (SACRRS) Used for GRID Irradiation
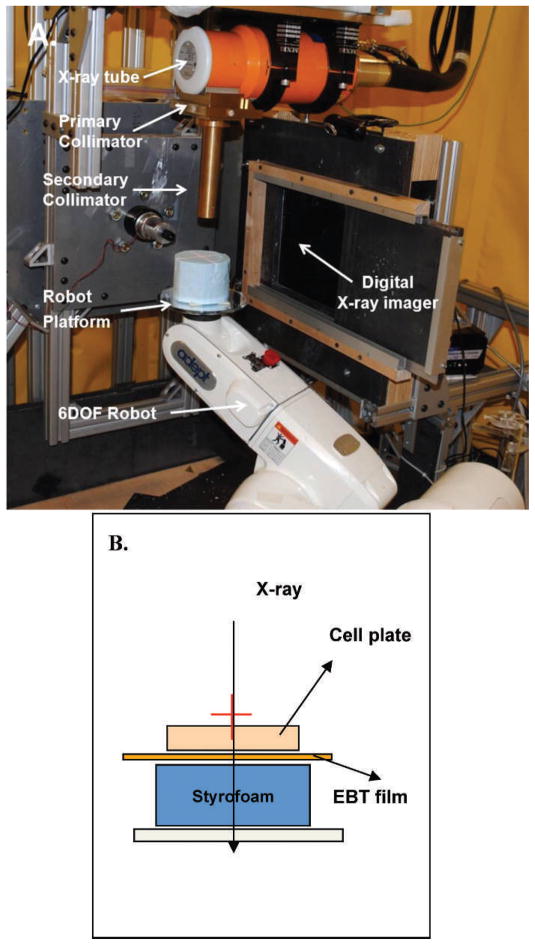
The irradiation system has a 225kVp X-ray tube with focal spots of 0.4 mm (used for imaging) and 3 mm (used for therapy). The precise positioning of the target/beam for both therapy and imaging is achieved by a programmable robotic arm (Adept Viper S650, Adept Technology Inc., Pleasanton, CA). The system is equipped with a flat-panel amorphous silicon digital X-ray imager (XRD 0820 CM3, Perking Elmer Inc., San Jose, CA), which captures 1024 × 1024 pixel image (200 μm pixel size) at a frame rate of 7.5 Hz. The system can deliver an accurate and quantifiable conformal radiation dose to selected targets in single or multiple fractions. In addition, the system is also equipped with a variable aperture collimation system mounted at 20 cm from the X-ray source that can produce field size up to 20 cm2 at the isocenter. These specially designed brass collimators are mounted on two sliders that move in opposite direction from the center using a uni-axial motor (Velmex Inc.) to make square fields. During the GRID irradiation, cells are placed on the “palm” of the robot and aligned with the X-ray beam. The GRID pattern of irradiation is then created by programming the robot platform to move normal to X-ray beam direction (Fig. 1A). The cells were then irradiated to create a pattern of nearly 50:50 direct and bystander exposure pattern of 9 circular fields, 12 mm in diameter with a center-to-center distance of 18 mm. The cells were irradiated with 10 Gy using GRID, at a dose rate of 1.9 Gy per min at 255 kV and 13 mA.
FIG. 1.
Schematic diagram of set-up used in GRID irradiation experiments. Panel A: The Small Animal Conformal Radiation Research System (SACCRS) used in GRID irradiation experiments. Panel B: A Gafchromic EBT-2 film was attached to the bottom of a 100 mm Petri dish containing cells. The bottom of the dish was placed on top a 3-inch thick styrofoam board, and the entire set-up was placed on a robot platform and irradiated in a GRID pattern.
A Gafchromic EBT-2 film (International Specialty Products, Wayne, NJ) was used for dosimetry. A calibration curve was plotted using the pixel values against the doses by irradiating the EBT-2 film under a Cobalt-60 beam. Subsequently, the EBT-2 films were irradiated at 1 cm below the isocenter, at 225 kVp energy, with 12 mm brass collimator duplicating the conditions under which the murine cells were irradiated (Fig. 1B). The films were exposed at different times and pixel values were recorded. The dose rate at 1 cm depth was then calculated.
Bystander Survival Studies
Medium transfer based studies
SCK and SCCVII cells were plated at confluent density in 25-cm2 culture flasks containing 5 ml culture medium and were irradiated 16–20 h later with 10 Gy. Flasks were returned to the incubator and cells were harvested by scraping at 0, 24, 48 and 168 h after irradiation. The 168 h (1 week) time point was included to determine the long-term effect of direct or bystander radiation exposure. Any clumps of cells were manually disrupted using a 500-μl pipette tip and the cells were then re-plated sparsely in 6-well cell culture dishes (Greiner Bio-One, Germany) and stained for colony-forming efficiency following 8 days to determine the effect of direct irradiation on these cells. Donor cell cultures for providing the conditioned medium were plated with 2 million confluent cells in 5 ml culture medium in 25-cm2 flasks and were exposed to 10 Gy of radiation after 16–20 h. Medium from sham-exposed cells was used for the controls. Medium transfer was performed 4 h after irradiation by collecting all medium from donor flasks and passing it through a 0.22 μm filter (Corning, NY) to ensure that no cells were present in the medium, but that soluble proteins or other factors could be retained. This filtrate was considered as the conditioned medium. The original medium was aspirated from bystander recipient cells and was replaced with filtered conditioned medium. Cells were harvested by scraping at 0, 24, 48 and 168 h after medium transfer and were re-plated (200–250 cells) in 6-well cell culture dishes and stained for clonogenic survival after 8 days.
GRID based bystander studies
SCK and SCCVII cells were plated at confluent density in 100 mm cell culture dishes (Greiner Bio-One, Germany) a day prior to irradiation. In our experiments, we considered the regions that were exposed to 10 Gy of radiation as “directly irradiated” and the adjacent cells which did not receive direct irradiation, but were exposed to indirect radiation (i.e., scattering) which amounted to a valley dose of approximately 1 Gy, were considered as “bystander cells”. The irradiated and adjacent bystander cells were demarcated using a waterproof histological marker, Pap pen (Research Products International Corp, Mt. Prospect, IL) to separate the irradiated and bystander fields using a small square of Gafchromic film that was attached to the bottom of the flask during irradiation as a guide and separately isolated. Cells were harvested at 0, 4, 24 and 48 h after GRID irradiation. Clumps were manually disrupted using a 500-μl pipette tip and cells were then sparsely re-plated (200–250 cells) in fresh medium, in 6-well culture plates to determine clonogenic survival.
RNA Isolation and Reverse Transcription
SCCVII cells were plated at confluent density in 100 mm culture dishes a day prior to irradiation. Twenty-four hours later, cells were exposed to 10 Gy of radiation in a GRID pattern using SACRRS. The directly irradiated cells, as well as bystander cells, were harvested at different times to determine the effects of GRID exposure (direct or indirect) as a function of time. Cells were harvested at 0, 4 and 24 h after GRID exposure by separately scraping the irradiated and bystander cells into 5 ml of culture medium. Cultures were then centrifuged at 300g for 5 min. Cell pellets were flash-frozen using dry ice and stored at −80°C for subsequent use. Total RNA was isolated from the cell pellet using the RNeasy Mini RNA isolation kit (Qiagen, Valencia, CA). Total RNA purity and yield was determined by measuring the absorbance at 260 nm and 280 nm using a NanoDrop (ThermoFisher, Rockford, IL), and the RNA integrity number (RIN) was determined using a Bioanalyzer (Agilent, Santa Clara, CA). Five micrograms of total RNA was then reverse transcribed with random hexamer primers using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) as per the manufacturer’s protocol.
Gene Expression Analysis
Gene expression was quantified using the RT2 Profiler PCR Array Mouse DNA Damage Signaling Pathway and Mouse Stress Response to Cellular Damage Pathway (SABiosciences, Frederick, MD). Reactions were performed as per the manufacturer’s protocol, using a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) and results were analyzed using the 7900 HT Fast System SDS Software. Threshold cycle (Ct) values obtained from real-time PCR were used in our calculations. The 2−ΔΔCt method (19) was used to calculate the normalized Ct values. The formula used can be represented as:
where GOI represents the Ct value for the gene of interest, RG represents the reference (housekeeping) gene Ct value, ‘exp’ represents the exposed (direct or bystander) cells and ‘cntr’ represents the sham-exposed controls. The 84 genes within each pathway were normalized against the average of 2 reference genes: Gapdh and Actb. Ct values greater than 32.0 were considered to be beyond the limit of detection
Statistical Analyses
Measurements of clonogenic survival are presented as mean values with ±SE of 5 independent experiments, each performed in duplicate. Student’s paired t test was used to calculate the significance of the difference irradiated (direct or bystander) and sham-exposed survival values. The O’Brien’s OLS statistic was used for gene expression testing due to sample-size constraints. The t test is also used for gene-specific testing. The level of significance was set at a P value of 0.05.
The patterns of gene expression were then compared between the different times and treatment groups. If a gene exhibited a fold-change greater than 2, it was considered as showing an “effect” and was selected for further analysis. If no gene in a particular functional group exhibited an effect, the group was considered as showing “no effect”. The number of effect genes were then counted and compared to determine whether more genes showed an increase or a decrease in expression. In the case that equal number of genes showed both up and down regulation, the effect was considered “unknown”.
RESULTS
GRID Film Dosimetry
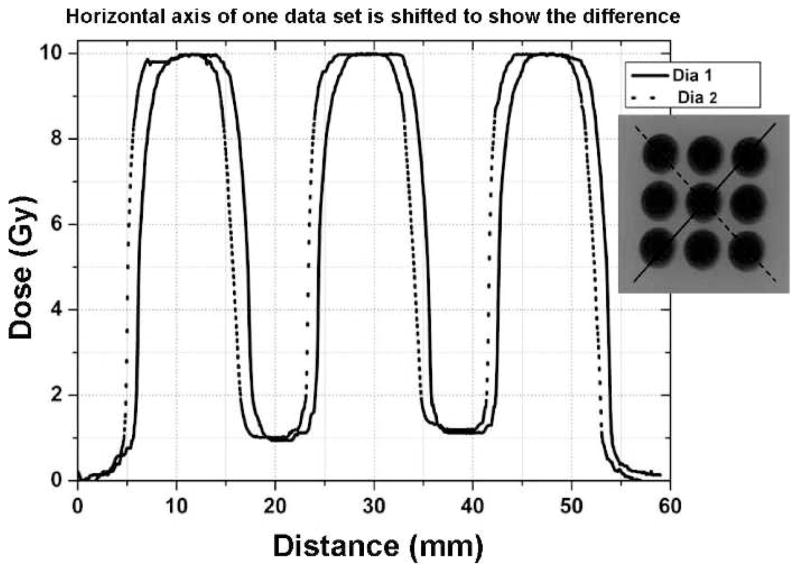
In the case of GRID based cell irradiation experiments, a Gafchromic EBT-2 film was attached to the bottom of a 100 mm Petri dish containing cultured cells, and the irradiation was performed with a 12 mm diameter brass collimator and the 225 kVp SACRRS beam. Cells were placed on the top of the robot platform while on top of a 3-inch thick Styrofoam board. The robot was programmed to create a GRID pattern of 9 circular areas and center-center distances of each irradiated beam center was 18 mm in horizontal and vertical directions. For each irradiation point, the cell layer was positioned 1 cm below the isocenter and was irradiated with 10 Gy. The same batch of EBT-2 films was used in all measurements and irradiated films were marked for their orientation and scanned at the same area of the scanner. The films were scanned 24 h after irradiation using the Epson perfection V700 scanner and were analyzed using ImageJ. In vivo film measurements showed that cells growing on the plate surface were irradiated with a dose of 9.81 ± 0.26 Gy within the peak region and cells in the valley region (20) received 0.91 ± 0.26 Gy on average (Fig. 2).
FIG. 2.
Cells were irradiated using spatially fractionated radiation to evaluate bystander effects. Cells were irradiated at a peak dose of 10 Gy using a brass collimator to create a GRID pattern of 9 open circular areas, 12 mm in diameter with a center-to-center distance of 18 mm. The bystander cells were harvested from the valley dose region along the diagonal lines illustrated, which represents about 10% of the total radiation.
Cell Survival
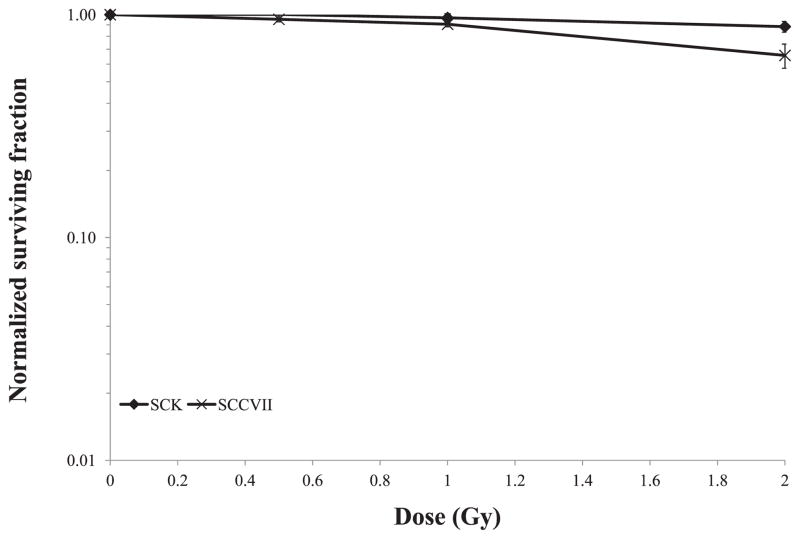
Traditional radiation survival studies were used to determine cell survival after exposure to about 1 Gy, since that was the observed background dose in the bystander (valley) region (Fig. 3). We observed a negligible decrease in survival in confluent SCK or SCCVII cells exposed to 1 Gy irradiation (data not shown). We observed on average a 3–10% decrease in survival in sparsely plated SCK or SCVII cells exposed to 1 Gy irradiation (Fig. 3). Survival of confluent SCK and SCCVII cells after direct exposure to 10 Gy of radiation is illustrated in Fig. 4. SCK and SCCVII cells exhibited an average of 10% survival after exposure to 10 Gy of radiation using either the Faxitron (Fig. 4A) or the GRID system (Fig. 4B). The decrease in survival of directly irradiated cells was found to be statistically significant (P < 0.05) at all times evaluated compared to the sham treated controls.
FIG. 3.
Clonogenic survival of murine mammary carcinoma (SCK) and head and neck cancer (SCCVII) cells after exposure to various doses of X irradiation using a cabinet Faxitron system. Vertical bars represent the standard error of the measurements: n = 5 observations per data point. For some data points, the error bars are too small to be visible. We observed 3 to 10% decrease in survival in cells exposed to 1 Gy, which is the approximate valley dose region in the GRID bystander studies. The clonogenic survival was normalized to the cloning efficiency of the control cells.
FIG. 4.
Clonogenic survival in directly irradiated mouse mammary carcinoma (SCK) and head and neck cancer (SCCVII) cells. SCK and SCCVII cells were exposed to 10 Gy of radiation. Panel A: A cabinet Faxitron system and panel B: spatially fractionated radiation (GRID). Cells were harvested by scraping at the various times after irradiation and were re-plated for clonogenic survival. Clonogenic survival was normalized to the cloning efficiency of the control cells. The x-axis represents the time after irradiation that cells were re-plated for clonogenic study. Vertical bars represent the standard error of the measurements. For some data points, the error bars are too small to be visible.
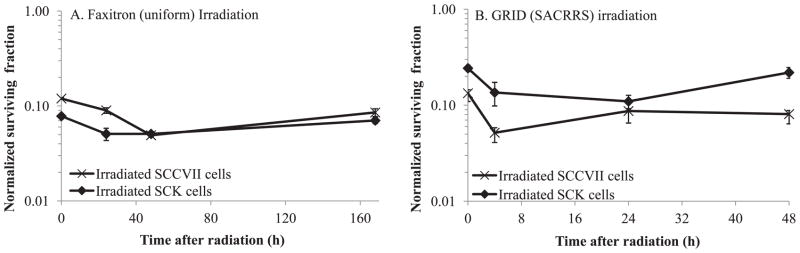
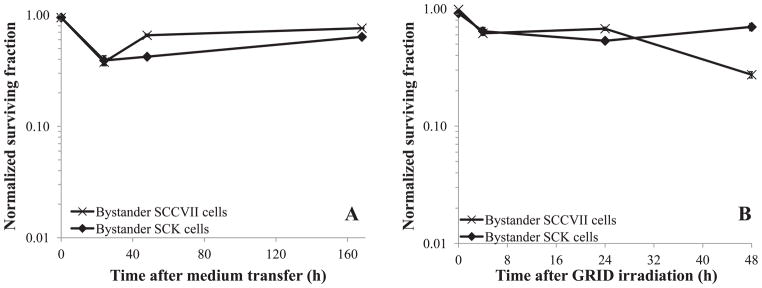
When medium from irradiated cells was transferred to unirradiated cultures bystander killing was observed (Fig. 5A). We also observed significant (P < 0.05) bystander killing above and beyond what could be expected by background/scatter irradiation (3 to 10% cell killing noted above for 1 Gy in our cells) after GRID irradiation. Therefore, clonogenic cell survival was reduced in bystander cells after a medium exchange that was nearly identical to that obtained within the bystander cell contact (GRID) experiments (Fig. 5B). Both SCK and SCCVII bystander clonogenic cell survival was reduced to nearly 50% compared to sham treated cells. Bystander SCK and SCCVII cell survival was significantly different (P < 0.05) from the respective controls when assessed 4 and 24 h after irradiation. However, when cells were harvested immediately after medium transfer or GRID exposure (0 h), there was little to no bystander killing observed. Survival values were corrected for multiplicity by normalizing to sham treated controls, which were handled with the exact same time course as the treated cells and would account for any degree of multiplicity in the plating efficiency obtained.
FIG. 5.
Clonogenic survival in bystander mouse mammary carcinoma (SCK) and head and neck cancer (SCCVII) cells. SCK and SCCVII cells were irradiated in two different systems. Panel A: Cells were irradiated in a cabinet Faxitron system and subsequently medium from irradiated cells was transferred 4 h later to bystander cells. Panel B: Cells were irradiated using a conformal X-ray system (SACRRS) and bystander cells in the diagonal region (received a background dose of 1 Gy, which resulted in 3 to 10% decrease in survival) of a GRID irradiated pattern were selectively isolated at various times and the clonogenic survival was determined. The clonogenic survival was normalized to the cloning efficiency of the control cells. The x-axis represents the time following medium transfer (panel A) or GRID irradiation (panel B) when cells were re-plated for clonogenic study. N = 3–5 experiments for all data points. Vertical bars represent the standard error of the measurements. For some data points, the error bars are too small to be visible.
Gene Expression Changes in Bystander Cells
We proceeded to evaluate the effects of direct and bystander exposure to a GRID treatment of 10 Gy on SCCVII cells using the SYBR green based real-time PCR technology at 0, 4 and 24 h after exposure. Real-time PCR arrays specific for mouse DNA damage and cellular stress response pathways were studied in our experiments. GRID-induced bystander gene expression changes were observed in several groups of genes involved in the repair/response to DNA damage and stress response signaling genes, thus providing molecular evidence for possible mechanisms of cell killing in nonirradiated cells.
DNA damage response genes
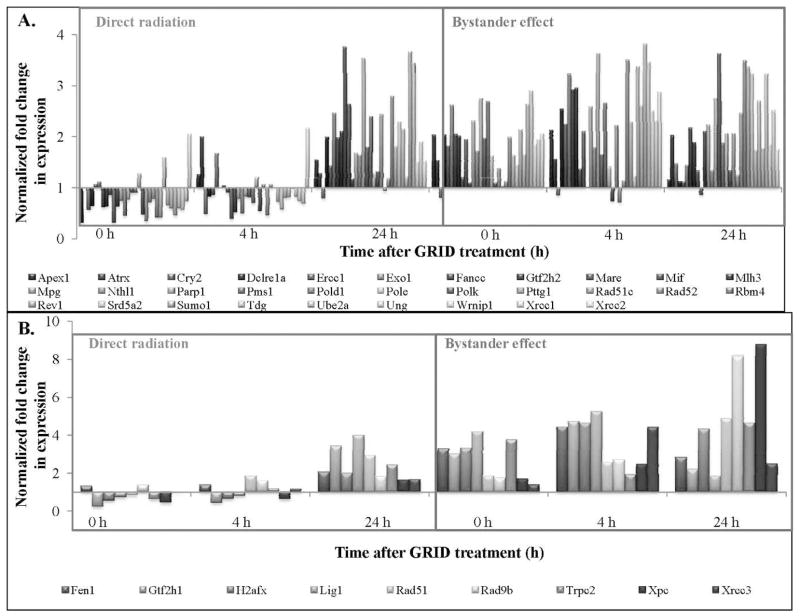
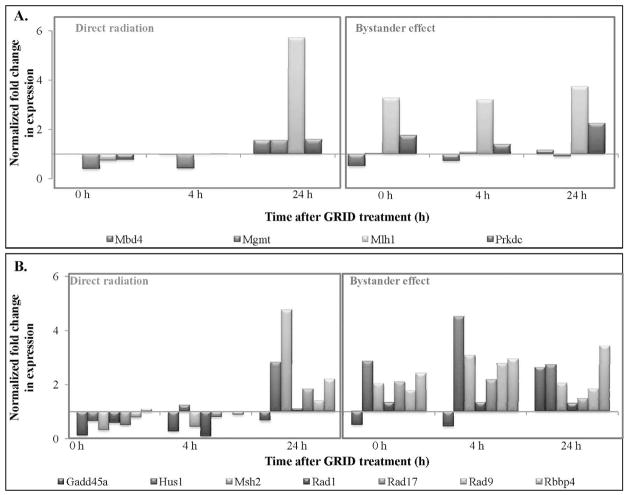
Figure 6A and B illustrate the expression of genes involved in DNA damage repair that exhibited a greater than 2- and 4-fold change, respectively, in directly irradiated and bystander SCCVII cells. DNA damage response genes exhibiting 2-fold or more change in expression in directly irradiated and bystander cells are presented in Table 1. Table 2 illustrates the statistical analysis of genes involved in the DNA damage signaling pathway. The fold-change values were compared between bystander and directly irradiated cells at each time, to determine the effect of exposure type (direct or bystander) on gene expression. Most DNA repair genes demonstrated an increase in fold-change expression in bystander cells compared to directly irradiated cells (Table 2). Expression of DNA repair genes was found to be significantly (P < 0.03) higher in bystander cells compared to GRID exposed cells at the times evaluated (0–24 h). Genes involved in damaged DNA binding, including Rad51c, Xpc and Xrcc1 (P = 0.002, 0.04 and 0.05, respectively), showed higher expression in the bystander cells. Base excision repair genes, such as Mpg, Nthl1 and Parp1 (P = 0.02, 0.01 and 0.009, respectively), were to be significantly higher. Dclre1a (nucleotide excision repair) and Xrcc6 and H2afx1 (double-strand break repair) all exhibited significant changes in bystander expression (P < 0.01). In the “mismatch repair” and “other genes related to DNA repair” subgroups, Mlh1, Mlh3, Apex1, Chaf1a, Fen1, Gtf2h1, Gtf2h2, Lig1, Pold1, Rbm4, Rev1, Sumo1, Tdg and Ube2a all exhibited higher (P < 0.05) expression in bystander cells than in directly irradiated cells. Genes involved in apoptosis (Fig. 7A) exhibited a change in expression in bystander cells compared to directly irradiated cells. Expression of genes involved in apoptosis was found to be significantly different in bystander cells compared to directly irradiated cells at 4 h after exposure (P = 0.014). Mgmt (P = 0.03) and Rad21 (P = 0.009) were expressed significantly higher in bystander cells compared to directly irradiated cells. Cell cycle genes also exhibited a significant difference in expression between the two exposure groups (Fig. 7B). Directly irradiated cells showed a decrease in expression at 0 and 4 h after GRID treatment, whereas we observed an increase in expression in bystander cells. Cell cycle arrest genes, such as Hus1 and Msh2 (P = 0.03), cell cycle checkpoint genes like Rad9 and Rad17 and Rad21 (P = 0.02, 0.02 and 0.09, respectively), all exhibited significantly (P < 0.03) higher expression in cells harvested from the bystander region compared to GRID-irradiated cells.
FIG. 6.
Expression patterns of genes involved in DNA damage repair. Mouse head and neck carcinoma (SCCVII) cells were exposed to a single fraction of 10 Gy using GRID. Cells from the directly irradiated and bystander regions were selectively and gene expression changes were evaluated using quantitative real-time PCR. Panel A: DNA damage repair genes exhibiting 2- to 4-fold change in expression. Panel B: DNA damage repair genes exhibiting greater than a 4-fold change in expression.
TABLE 1.
Fold-Change Values of DNA Damage Response Genes Exhibiting 2-Fold or More Change at one or More Timepoints Studied in Either Directly Irradiated or Bystander Cells, Compared to Sham Treated Controls
| Gene symbol | Description | Fold change values
|
|||||
|---|---|---|---|---|---|---|---|
| Directly
|
Bystander cells
|
||||||
| 0 Fold change | 4 h Fold change | 24 h Fold change | 0 Fold change | 4 h Fold change | 24 h Fold change | ||
| Apex1 | Apurinic/apyrimidinic endonuclease 1 | 0.30 | 1.25 | 1.54 | 2.03 | 2.12 | 1.16 |
| Atrx | Alpha thalassemia/mental retardation syndrome X-linked homolog (human) | 1.00 | 1.99 | 1.28 | 1.52 | 1.55 | 2.01 |
| Dclre1a | DNA cross-link repair 1A, PSO2 homolog (S. cerevisiae) | 0.64 | 0.83 | 1.99 | 2.03 | 2.53 | 1.13 |
| Ercc1 | Excision repair cross-complementing rodent repair deficiency, complementation group 1 | 1.06 | 0.85 | 1.42 | 1.82 | 2.24 | 1.12 |
| Exo1 | Exonuclease 1 | 1.12 | 1.68 | 2.45 | 2.61 | 3.22 | 1.43 |
| Fancc | Fanconi anemia, complementation group C | 0.61 | 0.99 | 1.97 | 2.04 | 2.92 | 2.16 |
| Fen1 | Flap structure specific endonuclease 1 | 1.31 | 1.38 | 2.04 | 3.28 | 4.42 | 2.83 |
| Gadd45a | Growth arrest and DNA-damage-inducible 45 alpha | 0.12 | 0.27 | 0.65 | 0.49 | 0.45 | 2.65 |
| Gtf2h1 | General transcription factor II H, polypeptide 1 | 0.21 | 0.43 | 3.42 | 3.02 | 4.72 | 2.20 |
| Gtf2h2 | General transcription factor II H, polypeptide 2 | 0.63 | 1.04 | 2.10 | 2.01 | 2.94 | 1.89 |
| H2afx | H2A histone family, member X | 0.52 | 0.64 | 1.98 | 3.31 | 4.64 | 4.32 |
| Hus1 | Hus1 homolog (S. pombe) | 0.65 | 1.25 | 2.83 | 2.88 | 4.52 | 2.73 |
| Lig1 | Ligase I, DNA, ATP-dependent | 0.73 | 0.78 | 3.99 | 4.17 | 5.25 | 1.85 |
| Mare | Alpha globin regulatory element containing gene | 0.85 | 0.91 | 3.75 | 1.19 | 1.36 | 1.33 |
| Mbd4 | Methyl-CpG binding domain protein 4 | 0.99 | 0.97 | 1.56 | 0.49 | 0.72 | 1.16 |
| Mgmt | O-6-methylguanine-DNA methyltransferase | 0.38 | 0.41 | 1.55 | 1.03 | 1.08 | 0.89 |
| Mif | Macrophage migration inhibitory factor | 0.31 | 0.38 | 2.63 | 1.94 | 2.09 | 0.86 |
| Mlh1 | MutL homolog 1 (E. coli) | 0.73 | 0.98 | 5.71 | 3.28 | 3.19 | 3.72 |
| Mlh3 | MutL homolog 3 (E coli) | 0.63 | 0.51 | 1.17 | 1.10 | 1.01 | 2.09 |
| Mpg | N-methylpurine-DNA glycosylase | 0.74 | 0.77 | 1.68 | 2.30 | 2.58 | 2.22 |
| Msh2 | MutS homolog 2 (E. coli) | 0.31 | 0.44 | 4.76 | 2.04 | 3.09 | 2.05 |
| Nthl1 | Nth (endonuclease III)-like 1 (E. coli) | 0.45 | 0.49 | 1.63 | 1.71 | 1.77 | 1.33 |
| Parp1 | Poly (ADP-ribose) polymerase family, member 1 | 0.76 | 0.83 | 3.53 | 2.75 | 3.63 | 2.74 |
| Pms1 | Postmeiotic segregation increased 1 (S. cerevisiae) | 0.90 | 0.81 | 1.80 | 1.96 | 1.65 | 3.63 |
| Pold1 | Polymerase (DNA directed), delta 1, catalytic subunit | 0.90 | 0.69 | 2.38 | 2.68 | 2.64 | 1.86 |
| Pole | Polymerase (DNA directed), epsilon | 1.27 | 1.19 | 1.27 | 1.62 | 1.40 | 2.06 |
| Polk | Polymerase (DNA directed), kappa | 0.47 | 0.54 | 1.31 | 1.09 | 0.73 | 1.33 |
| Prkdc | Protein kinase, DNA activated, catalytic polypeptide | 0.76 | 1.03 | 1.59 | 1.74 | 1.41 | 2.24 |
| Pttg1 | Pituitary tumor-transforming gene 1 | 0.33 | 1.07 | 2.43 | 1.37 | 2.21 | 2.05 |
| Rad1 | RAD1 homolog (S. pombe) | 0.57 | 0.07 | 1.10 | 1.34 | 1.35 | 1.29 |
| Rad17 | RAD17 homolog (S. pombe) | 0.49 | 0.79 | 1.83 | 2.10 | 2.18 | 1.47 |
| Rad51 | RAD51 homolog (S. cerevisiae) | 0.82 | 1.87 | 2.94 | 1.87 | 2.58 | 4.88 |
| Rad51c | Rad51 homolog c (S. cerevisiae) | 0.71 | 0.45 | 0.93 | 1.03 | 0.71 | 1.24 |
| Rad52 | RAD52 homolog (S. cerevisiae) | 0.78 | 1.06 | 1.18 | 1.12 | 1.15 | 2.45 |
| Rad9 | RAD9 homolog (S. pombe) | 0.79 | 0.98 | 1.40 | 1.79 | 2.78 | 1.83 |
| Rad9b | RAD9 homolog B (S. cerevisiae) | 1.38 | 1.61 | 1.82 | 1.74 | 2.71 | 8.16 |
| Rbbp4 | Retinoblastoma binding protein 4 | 1.11 | 0.88 | 2.20 | 2.42 | 2.95 | 3.45 |
| Rbm4 | RNA binding motif protein 4 | 0.41 | 0.99 | 2.78 | 1.98 | 3.50 | 3.48 |
| Rev1 | REV1 homolog (S. cerevisiae) | 0.42 | 0.72 | 1.80 | 1.63 | 2.27 | 3.36 |
| Srd5a2 | Steroid 5 alpha-reductase 2 | 1.58 | 0.57 | 2.29 | 1.44 | 1.22 | 3.22 |
| Sumo1 | SMT3 suppressor of mif two 3 homolog 1 (yeast) | 0.64 | 0.80 | 2.14 | 2.14 | 3.37 | 1.73 |
| Tdg | Thymine DNA glycosylase | 0.59 | 0.82 | 1.20 | 1.64 | 2.60 | 2.69 |
| Trpc2 | Transient receptor potential cation channel, subfamily C, member 2 | 0.61 | 1.18 | 2.44 | 3.75 | 1.93 | 4.64 |
| Ube2a | Ubiquitin-conjugating enzyme E2A, RAD6 homolog (S. cerevisiae) | 0.45 | 0.97 | 3.67 | 2.62 | 3.82 | 1.77 |
| Ung | Uracil DNA glycosylase | 0.59 | 0.82 | 3.44 | 2.89 | 3.46 | 3.22 |
| Wrnip1 | Werner helicase interacting protein 1 | 0.56 | 0.74 | 1.51 | 1.84 | 2.50 | 1.84 |
| Xpc | Xeroderma pigmentosum, complementation group C | 0.44 | 0.62 | 1.64 | 1.70 | 2.47 | 8.77 |
| Xrcc1 | X-ray repair complementing defective repair in Chinese hamster cells 1 | 0.73 | 0.69 | 1.89 | 1.94 | 2.31 | 2.50 |
| Xrcc2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | 2.05 | 2.16 | 1.52 | 2.05 | 2.86 | 1.75 |
| Xrcc3 | X-ray repair complementing defective repair in Chinese hamster cells 3 | 0.95 | 1.18 | 1.68 | 1.39 | 4.41 | 2.50 |
TABLE 2.
P Values and Patterns for DNA Damage Response Genes
| 0 h
|
4 h
|
24 h
|
||||
|---|---|---|---|---|---|---|
| P | Pattern | P | Pattern | P | Pattern | |
| DNA repair | ||||||
| Damaged DNA binding | 0.014 | Increase | 0.023 | Increase | 0.271 | Increase |
| Base-excision repair | 0.073 | Increase | 0.004 | Increase | 0.287 | Decrease |
| Nucleotide-excision repair | 0.017 | Increase | 0.004 | Increase | 0.35 | Increase |
| Double-strand break repair | 0.051 | Increase | 0.007 | Increase | 0.2 | Increase |
| Mismatch repair | 0.034 | Increase | 0.004 | Increase | 0.435 | Unknown |
| Other genes related to DNA Repair | 0.006 | Increase | 0.002 | Increase | 0.433 | Decrease |
| Apoptosis | 0.106 | Increase | 0.014 | Increase | 0.443 | No effect |
| Cell cycle | ||||||
| Cell cycle arrest | 0.024 | Increase | 0.007 | Increase | 0.46 | Unknown |
| Cell cycle checkpoint | 0.065 | Increase | 0.119 | Increase | 0.235 | No effect |
| Other genes related to the cell cycle | 0.035 | Increase | 0.01 | Increase | 0.423 | No effect |
Notes. The fold-change expression values were compared between direct and bystander groups using the O’Brien’s OLS statistic. P < 0.05 were considered significant. The expression patterns for the different treatment groups indicate expression in bystander cells compared to directly irradiated cells at the various times.
FIG. 7.
Expression patterns of genes involved in apoptosis (panel A) and cell cycle exhibiting (panel B) greater than 2-fold change in expression analyzed by quantitative real-time PCR.
Cellular stress response
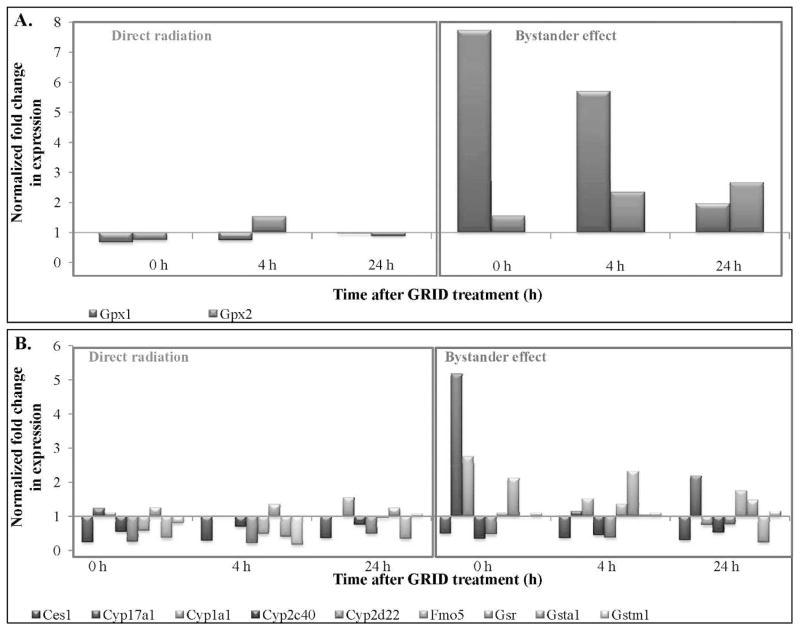
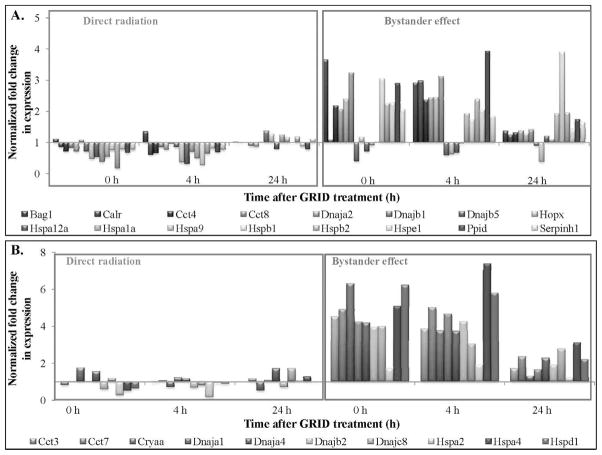
The values of cellular stress response genes exhibiting 2-fold or more change in directly irradiated and bystander cells are presented in Table 3. Antioxidant and pro-oxidant genes demonstrated an increase in fold-change expression in bystander cells, while directly irradiated cells showed a decrease (Fig. 8A). Several genes involved in xenobiotic metabolism (Fig. 8B) exhibited an increase in expression in bystander cells at all times evaluated. Figure 9A and B illustrate the expression of molecular chaperone genes that exhibited a greater than 2- and 4-fold change, respectively, in directly irradiated and bystander SCCVII cells. We observed an increase in expression in bystander cells at all times evaluated compared to directly irradiated cells. Table 4 illustrates the statistical analysis of the genes involved in the cellular stress signaling pathway. The fold-change values were compared between bystander and directly irradiated cells at each time, to determine the effect of exposure type (direct or bystander) on gene expression. Expression of genes coding for antioxidant and pro-oxidant enzymes, as well as molecular chaperones, including heat shock proteins, were found to be significantly different in bystander cells compared to directly irradiated cells at 0–24 h after exposure (P = 0.05). Genes coding for antioxidant and pro-oxidant enzymes, such as Gpx1, Sod1 and Xdh were expressed at significantly higher levels (P < 0.01) in bystander cells compared to directly irradiated cells at 0–24 h after exposure. Heat shock proteins, such as Dnaja1, Dnajb1, Hspa4, Hspd1 and Hspe1 all exhibited significantly (P < 0.05) different expression in bystander cells at all times evaluated. Cct7 (P < 0.02), a molecular chaperone and Gsta1 (P < 0.05), a gene involved in response to xenobiotic metabolism (detoxification) both demonstrated changes in bystander expression compared to expression in directly irradiated cells.
TABLE 3.
Fold-Change Values of Cellular Stress Response Genes Exhibiting 2-Fold or More Change in Directly Irradiated and Bystander Cells, Compared to Sham Treated Controls
| Gene symbol | Description | Fold-change values
|
|||||
|---|---|---|---|---|---|---|---|
| Directly irradiated cells
|
Bystander cells
|
||||||
| 0 h Fold change | 4 h Fold change | 24 h Fold change | 0 h Fold change | 4 h Fold change | 24 h Fold change | ||
| Bag1 | Bcl2-associated athanogene 1 | 1.10 | 1.35 | 1.02 | 3.64 | 2.90 | 1.37 |
| Calr | Calreticulin | 0.85 | 0.60 | 0.98 | 1.07 | 2.97 | 1.24 |
| Cct3 | Chaperonin containing Tcp1, subunit 3 (gamma) | 0.81 | 0.95 | 1.00 | 4.52 | 3.86 | 1.71 |
| Cct4 | Chaperonin containing Tcp1, subunit 4 (delta) | 0.70 | 0.64 | 0.99 | 2.17 | 2.35 | 1.32 |
| Cct7 | Chaperonin containing Tcp1, subunit 7 (eta) | 0.98 | 1.06 | 1.14 | 4.93 | 5.02 | 2.36 |
| Cct8 | Chaperonin containing Tcp1, subunit 8 (theta) | 0.83 | 0.84 | 0.89 | 2.05 | 2.43 | 1.36 |
| Ces1 | Carboxylesterase 1 | 0.23 | 0.28 | 0.35 | 0.48 | 0.35 | 0.29 |
| Cryaa | Crystallin, alpha A | 1.74 | 0.69 | 0.50 | 6.33 | 3.74 | 1.31 |
| Cyp17a1 | Cytochrome P450, family 17, subfamily a, polypeptide 1 | 1.24 | 1.02 | 1.01 | 5.17 | 1.13 | 2.20 |
| Cyp1a1 | Cytochrome P450, family 1, subfamily a, polypeptide 1 | 1.11 | 1.01 | 1.55 | 2.74 | 1.51 | 0.74 |
| Cyp2c40 | Cytochrome P450, family 2, subfamily c, polypeptide 40 | 0.54 | 0.69 | 0.75 | 0.33 | 0.43 | 0.51 |
| Cyp2d22 | Cytochrome P450, family 2, subfamily d, polypeptide 22 | 0.24 | 0.20 | 0.47 | 0.45 | 0.36 | 0.76 |
| Dnaja1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | 0.98 | 1.21 | 1.09 | 4.24 | 4.67 | 1.64 |
| Dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 0.70 | 0.77 | 0.86 | 2.38 | 2.44 | 1.29 |
| Dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 0.70 | 0.77 | 0.86 | 2.38 | 2.44 | 1.29 |
| Dnajb1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 1.08 | 0.96 | 1.02 | 3.22 | 3.11 | 1.40 |
| Dnajb5 | DnaJ (Hsp40) homolog, subfamily B, member 5 | 0.70 | 0.85 | 1.37 | 0.39 | 0.58 | 0.89 |
| Dnajc8 | DnaJ (Hsp40) homolog, subfamily C, member 8 | 1.17 | 0.78 | 1.72 | 4.00 | 3.03 | 2.77 |
| Fmo5 | Flavin containing monooxygenase 5 | 0.58 | 0.48 | 0.93 | 1.08 | 1.35 | 1.75 |
| Gpx1 | Glutathione peroxidase 1 | 0.66 | 0.73 | 0.95 | 7.73 | 5.70 | 1.96 |
| Gpx2 | Glutathione peroxidase 2 | 0.74 | 1.52 | 0.88 | 1.55 | 2.35 | 2.67 |
| Gsr | Glutathione reductase | 1.26 | 1.35 | 1.24 | 2.12 | 2.32 | 1.47 |
| Gsta1 | Glutathione S-transferase, alpha 1 (Ya) | 0.36 | 0.40 | 0.33 | 0.97 | 1.04 | 0.23 |
| Gstm1 | Glutathione S-transferase, mu 1 | 0.80 | 0.16 | 1.06 | 1.08 | 1.08 | 1.15 |
| Hopx | HOP homeobox | 0.47 | 0.35 | 1.27 | 1.17 | 0.62 | 0.37 |
| Hspa12a | Heat shock protein 12A | 0.52 | 0.30 | 0.79 | 0.70 | 0.66 | 1.20 |
| Hspa1a | Heat shock protein 1A | 0.37 | 0.70 | 1.24 | 0.91 | 0.97 | 1.10 |
| Hspa2 | Heat shock protein 2 | 0.26 | 0.14 | 1.04 | 1.71 | 1.91 | 1.21 |
| Hspa4 | Heat shock protein 4 | 0.49 | 0.92 | 1.26 | 5.09 | 7.38 | 3.12 |
| Hspa9 | Heat shock protein 9 | 0.53 | 0.49 | 1.16 | 1.02 | 1.93 | 1.92 |
| Hspb1 | Heat shock protein 1 | 0.74 | 0.27 | 0.99 | 3.04 | 1.73 | 3.90 |
| Hspb2 | Heat shock protein 2 | 0.16 | 0.62 | 1.18 | 2.23 | 2.38 | 1.95 |
| Hspd1 | Heat shock protein 1 (chaperonin) | 0.63 | 0.87 | 0.96 | 6.23 | 5.82 | 2.21 |
| Hspe1 | Heat shock protein 1 (chaperonin 10) | 0.77 | 0.80 | 0.87 | 2.26 | 2.03 | 1.42 |
| Ppid | Peptidylprolyl isomerase D (cyclophilin D) | 0.65 | 0.68 | 0.77 | 2.89 | 3.94 | 1.73 |
| Serpinh1 | Serine (or cysteine) peptidase inhibitor, clade H, member 1 | 0.76 | 0.76 | 1.13 | 2.05 | 1.83 | 1.63 |
FIG. 8.
Expression patterns of antioxidant (panel A) and xenobiotic metabolism genes (panel B) exhibiting greater than 2-fold change in expression, harvested from directly irradiated and bystander cells after exposure to 10 Gy using GRID in SCCVII cells.
FIG. 9.
Expression patterns of molecular chaperone genes exhibiting (panel A) 2- to 4-fold change in expression and (panel B) greater than 4-fold change in expression after direct or bystander exposure to 10 Gy GRID in SCCVII cells.
TABLE 4.
P Values and Patterns for Cellular Stress Response Genes
| 0 h
|
4 h
|
24 h
|
||||
|---|---|---|---|---|---|---|
| P | Pattern | P | Pattern | P | Pattern | |
| Antioxidant and Pro-oxidant enzymes | 0.008 | Increase | 0.004 | Increase | 0.084 | Increase |
| Molecular Chaperones | ||||||
| Heat Shock Proteins | 0.003 | Increase | 0.0001 | Increase | 0.011 | Increase |
| Other Molecular Chaperones | 0.012 | Increase | 0.005 | Increase | 0.056 | Increase |
| Xenobiotic Metabolism | ||||||
| Cytochrome P450s | 0.136 | Increase | 0.144 | No effect | 0.15 | Decrease |
| Other Genes Related to Xenobiotic Metabolism | 0.187 | Increase | 0.184 | Increase | 0.345 | No effect |
Notes. The fold-change expression values were compared between direct and bystander groups using the O’Brien’s OLS statistic. P < 0.05 were considered significant. The expression patterns for the different treatment groups indicate expression in bystander cells compared to directly irradiated cells at the various times.
In summary, our results demonstrated higher expression of genes involved in DNA repair, cell cycle arrest and checkpoint, antioxidant and molecular chaperone pathways in the GRID irradiated and bystander cells compared to sham treated controls.
DISCUSSION
In the current study, we evaluated the ability of spatially fractionated radiation (GRID) to induce bystander effects in murine carcinoma cells after exposure to a single dose of 10 Gy, analogous to the high single doses (usually 10–20 Gy) used in spatial fractionation. A significant decrease in clonogenic survival was observed in bystander cells after exposure to medium obtained from cells exposed to 10 Gy. We also observed a significant bystander killing in cells adjacent to irradiated regions of the same monolayer compared to the sham treated controls. The decrease in survival of cells in the adjacent regions was found to be more than that expected from exposure to only background “valley” or scatter doses, suggesting the existence of true cytotoxic bystander effects after GRID irradiation.
We observed greater than a 2-fold increase in expression of genes involved in DNA damage repair, cell cycle arrest and apoptosis in GRID irradiated cells 24 h after exposure. Bystander (GRID-adjacent) cells exhibited increased expression of genes involved in DNA repair, cell cycle arrest, and apoptosis immediately after exposure or 4 h after exposure. In some instances, the increase persisted up to 24 h after GRID irradiation. We observed increased expression of antioxidant, heat shock and chaperone genes immediately after irradiation or 4 h after GRID exposure in the bystander cells. In a few instances, we observed a persistent increase in expression up to 24 h after GRID exposure. We did not observe significant increase in expression of these genes in the GRID irradiated cells. In contrast, it has been reported that p53-related genes exhibited minimal activation in bystander cells, while genes involved in NFκB were activated to equal degrees in direct and bystander cells (21, 22) after α-particle irradiation of fibroblasts.
Of the many genes studied using the arrays we selected, we observed significantly higher levels of genes encoding Glutathione peroxidase, Gpx1 and superoxide dismutase, Sod1 in bystander cells compared to sham treated controls. These antioxidant enzymes have been known to be important in cellular defense to oxidative stress (23, 24). Factors responsible for transmitting the bystander signal or agents that cause cytotoxicity or cell stress from irradiated to naive bystander cells have been previously hypothesized to be proteins that can withstand freezing and thawing (25–29). In addition, certain studies have indicated the involvement of reactive oxygen species (26, 30, 31), growth factors and cytokines in the maintenance of the bystander signal (32). Our results suggest that secreted factors that lead to reactive oxygen species are very likely candidates for the effects observed. We observed the highest increase in expression of antioxidant genes in the bystander cells immediately after GRID treatment suggesting that oxidative stress was occurring in these cells.
An increase in expression of DNA damage response genes, such as those involved in apoptosis, cell cycle arrest and repair was also observed in bystander cells compared to sham treated controls in our experiments. In past studies, exposure of cells to low doses of α particles have resulted in increased expression of DNA damage response genes, such as TP53, CDKN1A, CDC2, CCN1 and RAD51 (33). Therefore, as might be expected, our results agree with those of others to suggest that a part of the killing mechanism involved in bystander effects is through DNA damage, which is something that can be induced by a variety of reactive oxygen species.
Overall, radiation-induced bystander effects have been demonstrated using a wide variety of end points, including decreased cell survival (34, 35), apoptosis (36–40), increased chromosomal damage (4, 41–44) and DNA double-strand breaks (6, 45, 46). Changes in expression of mitogen (commonly referred to as stress) activated protein kinases (31, 47, 48) have been observed after exposure to medium obtained from cells exposed to ionizing radiation. Differential expression of genes involved in maintaining cell structure and motility, signal transduction and cell-to-cell communication was observed in one study in bystander cells by 30 min after treatment (22). Gene ontology studies revealed that the majority of these genes were responsible for maintaining cell-to-cell signaling, including adhesion, activation and stimulation of normal cells (22). The AKT-GSK3β-β catenin signaling pathway has been suspected to play a role in maintaining and developing prolonged bystander states (22). In addition, the expression of genes involved in the NFκB signaling pathway exhibited differential expression in bystander cells at 4 h after treatment as shown in another study (21).
Bystander effects are not necessarily restricted to the exposure of neighboring cells to ionizing radiation. Recently, the ability of genotoxic stress producing agents, other than ionizing radiation, to induce bystander effects has been reported. We and others have found that chemotherapeutic drugs, such as chloroethylnitrosurea (49), paclitaxel (20), mitomycin C (41, 50) and phleomycin (41) can induce bystander effects through secretion of medium soluble factors. In addition, photosensitizers (51), heat (52) and photodynamic stress (53) agents have all been reported to cause some type of bystander effect. Therefore, the true nature of bystander effects appears to be a cell stress related phenomenon and not necessarily a unique by-product of radiation damage or a specific type of treatment.
In our studies, bystander cells exhibited approximately a 50% decrease in cell survival, whereas directly irradiated cells exhibited a >90% decrease in survival compared to sham treated controls. Bystander cells exhibited changes in expression of greater numbers of genes involved in DNA damage repair and cell cycle arrest compared to directly irradiated cells at the times studied. Bystander cells received a low (scatter/valley) dose of 1 Gy compared to the 10 Gy directly irradiated cells. The massive damage and cell death occurring in the directly irradiated cells is most likely the reason that gene expression levels became suspended to an extent, while the lower amounts of cell death and damage inflicted in the ‘radiation-adjacent’ cells promoted more detectable changes in gene expression patterns. Interestingly, survival of directly irradiated cells over time also did not exhibit evidence of potentially lethal damage repair, which would normally be expected since they were nearly confluent at the time of irradiation. It is possible that the bystander cells themselves secreted a death-inducing factor that was able to affect the irradiated cell viability. Further study of 10 Gy direct irradiation without bystander influences would be needed to clarify this intriguing possibility.
A few studies have attempted to understand what may be involved in the mechanism(s) by which GRID therapy at doses as high as 20 Gy improves tumor response and whether it is simply a product of increased cell kill or something more. Induction of tumor necrosis factor-α (TNF-α) and ceramide, as well as down regulation of transforming growth factor β1 have been observed in the serum of patients after GRID (14, 15). In general, and as might be expected after a large cytotoxic event, GRID therapy appears to lead to increased cytokine production, resulting in broad systemic effects (11). Due to the on/off nature within GRID fields, we hypothesized that bystander effects might play a role in killing adjacent nonirradiated or partially irradiated cells. To our knowledge, study of the type of intercellular communication that might exist between adjacent cells in the open and closed areas of GRID has not been approached to date and may provide valuable information on the mechanisms involved in the promising clinical results that have been observed at our institution (11, 13) and in the various other studies cited above.
Interestingly, the bulk of literature on radiation-induced bystander effects has focused on low doses (generally below 5 Gy). It has been demonstrated that the cell killing in bystander cells is most clearly detected at doses below 0.5 Gy (54). The role of LET on bystander effects has also been studied by numerous groups (21, 22). These studies observed that there appears to be a greater incidence of bystander effect with high-LET irradiation when equal doses are compared to low-LET.
It has also been suggested that bystander effects plateau at low doses and further increases in the radiation dose has no effect on bystander response (5). Gow et al. observed a decrease in surviving fraction of bystander HPV-G cells receiving medium from 0.5 Gy or 5 Gy irradiated cells, as well as recipients of medium from 10 Gy of radiation when plated densely. However, the surviving fractions returned to near control levels when sparsely plated recipient cells received medium from cells that had been exposed to 10 Gy (55). In our study, using a dose of 10 Gy, both medium transfer and GRID irradiation assays reduced bystander cell viability and, at least in the case of the GRID exposures, the bystander cells were densely plated which would agree with the previous study results. In another study, cells receiving medium from cells irradiated with fractionated doses (i.e., 2 Gy per fraction), rather surprisingly exhibited decreased survival fractions compared to cells that directly received the fractionated irradiation (i.e., bystander cells receiving medium from fractionated radiation exhibited more cell killing than cells that were directly exposed to fractionated doses of radiation) (56, 57). In terms of radiation treatment, this suggests that although the direct effect of fractionated radiation doses is hoped to be normal tissue sparing, the occurrence of bystander effects coming from the irradiated tumor or normal cells after each fraction of radiation may increase the overall tumor and normal tissue responses.
Recently, intensity modulated radiation therapy (IMRT), with doses ranging from 3 to 20 Gy, was used to study cellular response to radiation (58). The authors observed three distinct types of bystander effects. Type 1 was a decrease in cell survival observed in unirradiated cells adjacent to cells irradiated with a moderate dose (3 Gy). Type 2 was an increase in survival in cells adjacent to cells receiving a lethal dose (20 Gy) of radiation. Type 3 was an increased survival of cells receiving a high dose of radiation, when they were adjacent to cells receiving a low dose, possibly due to the generation of unspecified mechanisms of survival response in the low-dose cells, which were conducive to colony formation. In our study, we observed decrease in survival of cells adjacent to cells exposed to 10 Gy irradiation. Other studies involving IMRT determined the effect of modulated fields of irradiation on cell survival. Radiation-resistant and radiation-sensitive cells were exposed to modulated and nonmodulated irradiation up to 8 Gy using a multi-leaf collimator. No significant difference was found in survival of cells exposed to modulated radiation or nonmodulated fields of irradiation, suggesting the occurrence of bystander effects in modulated fields (59). In addition, survival of cells that were in the same flask but not exposed to radiation (out-of-field) was found to be less than that expected solely due to radiation scattering (60). Inhibition of intercellular communication using nitric oxide synthase inhibitor increased cell survival, suggesting the occurrence of bystander effects in the out-of-field cellular responses.
In our radiation therapy clinic, treatment of head and neck cancer usually involves GRID therapy followed by a conventional course of 2 Gy fractionated radiation therapy and chemotherapy. In addition to bystander killing by the GRID dose, the occurrence of sub-lethal bystander effects after GRID therapy might make these cells more susceptible to the subsequent chemotherapy and radiation treatment, further contributing to the overall tumor response. A detailed understanding of possible bystander effects involved in low or high dose or spatially fractionated radiation therapy, and the factors that mediate such effects may uncover molecular mechanisms involved and suggest new therapeutic targets to improve treatment outcomes, either by increasing tumor cell killing or enhancing normal cell protection. Studies designed in vivo will need to take into consideration possible contributions of tissue and tumor effects, including hypoxia, on bystander effects. Understanding the involvement of bystander effects within various dose gradients present in state-of-the art radiation techniques may enable us to optimize therapies and design potent combination treatment approaches.
Acknowledgments
This work was supported by Central Arkansas Radiation Therapy Institute (CARTI) and NIH grant CA44114. The authors thank N. Koonce and A. Jamshidi-Parsian, for their input and guidance.
References
- 1.Dale WM. The effect of X-rays on enzymes. Biochem J. 1940;34:1367–73. doi: 10.1042/bj0341367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dale WM. The effect of X-rays on the conjugated protein d-aminoacid oxidase. Biochem J. 1942;36:80–5. doi: 10.1042/bj0360080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotval JP, Gray LH. Structural changes produced in microspores of Tradescantia by alpha-radiation. J Genet. 1947;48:135–54. 1947. doi: 10.1007/BF02989375. [DOI] [PubMed] [Google Scholar]
- 4.Nagasawa H, Little JB. Induction of sister chromatid exchanges by extremely low doses of alpha-particles. Cancer Res. 1992;52:6394–6. [PubMed] [Google Scholar]
- 5.Mothersill C, Seymour CB. Radiation-induced bystander effects–implications for cancer. Nat Rev Cancer. 2004;4:158–64. doi: 10.1038/nrc1277. [DOI] [PubMed] [Google Scholar]
- 6.Hu B, Wu L, Han W, Zhang L, Chen S, Xu A, Hei TK, Yu Z. The time and spatial effects of bystander response in mammalian cells induced by low dose radiation. Carcinogenesis. 2006;27:245–51. doi: 10.1093/carcin/bgi224. [DOI] [PubMed] [Google Scholar]
- 7.Azzam EI, de Toledo SM, Little JB. Oxidative metabolism, gap junctions and the ionizing radiation-induced bystander effect. Oncogene. 2003;22:7050–7. doi: 10.1038/sj.onc.1206961. [DOI] [PubMed] [Google Scholar]
- 8.Azzam EI, de Toledo SM, Little JB. Direct evidence for the participation of gap junction-mediated intercellular communication in the transmission of damage signals from alpha-particle irradiated to nonirradiated cells. Proc Natl Acad Sci USA. 2001;98:473–8. doi: 10.1073/pnas.011417098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzam EI, Little JB. The radiation-induced bystander effect: evidence and significance. Hum Exp Toxicol. 2004;23:61–5. doi: 10.1191/0960327104ht418oa. [DOI] [PubMed] [Google Scholar]
- 10.Mothersill C, Seymour C. Radiation-induced bystander effects: past history and future directions. Radiat Res. 2001;155:759–67. doi: 10.1667/0033-7587(2001)155[0759:ribeph]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Penagaricano JA, Moros EG, Ratanatharathorn V, Yan Y, Corry P. Evaluation of spatially fractionated radiotherapy (GRID) and definitive chemoradiotherapy with curative intent for locally advanced squamous cell carcinoma of the head and neck: initial response rates and toxicity. Int J Radiat Oncol Biol Phys. 2010;76:1369–75. doi: 10.1016/j.ijrobp.2009.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Penagaricano JA, Griffin R, Corry P, Moros E, Yan Y, Ratanatharathorn V. Spatially fractionated (GRID) therapy for large and bulky tumors. J Ark Med Soc. 2009;105:263–5. [PubMed] [Google Scholar]
- 13.Huhn JL, Regine WF, Valentino JP, Meigooni AS, Kudrimoti M, Mohiuddin M. Spatially fractionated GRID radiation treatment of advanced neck disease associated with head and neck cancer. Technol Cancer Res Treat. 2006;5:607–12. doi: 10.1177/153303460600500608. [DOI] [PubMed] [Google Scholar]
- 14.Sathishkumar S, Boyanovsky B, Karakashian AA, Rozenova K, Giltiay NV, Kudrimoti M, Mohiuddin M, Ahmed MM, Nikolova-Karakashian M. Elevated sphingomyelinase activity and ceramide concentration in serum of patients undergoing high dose spatially fractionated radiation treatment: implications for endothelial apoptosis. Cancer Biol Ther. 2005;4:979–86. doi: 10.4161/cbt.4.9.1915. [DOI] [PubMed] [Google Scholar]
- 15.Sathishkumar S, Dey S, Meigooni AS, Regine WF, Kudrimoti MS, Ahmed MM, Mohiuddin M. The impact of TNF-alpha induction on therapeutic efficacy following high dose spatially fractionated (GRID) radiation. Technol Cancer Res Treat. 2002;1:141–7. doi: 10.1177/153303460200100207. [DOI] [PubMed] [Google Scholar]
- 16.Burgher AH, Swanlund DJ, Griffin RJ, Song CW, Bischof JC, Roberts KP. Sensitization of thermotolerant SCK cells to hyperthermia and freezing with reduction of intracellular pH: implications for cryosurgery. J Surg Oncol. 2003;82:160–9. doi: 10.1002/jso.10214. [DOI] [PubMed] [Google Scholar]
- 17.Griffin RJ, Williams BW, Wild R, Cherrington JM, Park H, Song CW. Simultaneous inhibition of the receptor kinase activity of vascular endothelial, fibroblast, and platelet-derived growth factors suppresses tumor growth and enhances tumor radiation response. Cancer Res. 2002;62:1702–6. [PubMed] [Google Scholar]
- 18.Dings RP, Williams BW, Song CW, Griffioen AW, Mayo KH, Griffin RJ. Anginex synergizes with radiation therapy to inhibit tumor growth by radiosensitizing endothelial cells. Int J Cancer. 2005;115:312–9. doi: 10.1002/ijc.20850. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Alexandre J, Hu Y, Lu W, Pelicano H, Huang P. Novel action of paclitaxel against cancer cells: bystander effect mediated by reactive oxygen species. Cancer Res. 2007;67:3512–7. doi: 10.1158/0008-5472.CAN-06-3914. [DOI] [PubMed] [Google Scholar]
- 21.Ghandhi SA, Yaghoubian B, Amundson SA. Global gene expression analyses of bystander and alpha particle irradiated normal human lung fibroblasts: synchronous and differential responses. BMC Med Genomics. 2008;1:63. doi: 10.1186/1755-8794-1-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghandhi SA, Ming L, Ivanov VN, Hei TK, Amundson SA. Regulation of early signaling and gene expression in the alpha-particle and bystander response of IMR-90 human fibroblasts. BMC Med Genomics. 2010;3:31. doi: 10.1186/1755-8794-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell JB, Russo A. The role of glutathione in radiation and drug induced cytotoxicity. Br J Cancer Suppl. 1987;8:96–104. [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Z, Sarsour EH, Kalen AL, Li L, Kumar MG, Goswami PC. Late ROS accumulation and radiosensitivity in SOD1-overexpressing human glioma cells. Free Radic Biol Med. 2008;45:1501–9. doi: 10.1016/j.freeradbiomed.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayanan PK, Goodwin EH, Lehnert BE. Alpha particles initiate biological production of superoxide anions and hydrogen peroxide in human cells. Cancer Res. 1997;57:3963–71. [PubMed] [Google Scholar]
- 26.Lehnert BE, Goodwin EH. A new mechanism for DNA alterations induced by alpha particles such as those emitted by radon and radon progeny. Environ Health Perspect. 1997;5(105 Suppl):1095–101. doi: 10.1289/ehp.97105s51095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mothersill C, Seymour CB. Cell-cell contact during gamma irradiation is not required to induce a bystander effect in normal human keratinocytes: evidence for release during irradiation of a signal controlling survival into the medium. Radiat Res. 1998;149:256–62. [PubMed] [Google Scholar]
- 28.Iyer R, Lehnert BE, Svensson R. Factors underlying the cell growth-related bystander responses to alpha particles. Cancer Res. 2000;60:1290–8. [PubMed] [Google Scholar]
- 29.Shao C, Prise KM, Folkard M. Signaling factors for irradiated glioma cells induced bystander responses in fibroblasts. Mutat Res. 2008;638:139–45. doi: 10.1016/j.mrfmmm.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Konopacka M, Rzeszowska-Wolny J. The bystander effect-induced formation of micronucleated cells is inhibited by antioxidants, but the parallel induction of apoptosis and loss of viability are not affected. Mutat Res. 2006;593:32–8. doi: 10.1016/j.mrfmmm.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Lyng FM, Maguire P, McClean B, Seymour C, Mothersill C. The involvement of calcium and MAP kinase signaling pathways in the production of radiation-induced bystander effects. Radiat Res. 2006;165:400–9. doi: 10.1667/rr3527.1. [DOI] [PubMed] [Google Scholar]
- 32.Barcellos-Hoff MH, Brooks AL. Extracellular signaling through the microenvironment: a hypothesis relating carcinogenesis, bystander effects, and genomic instability. Radiat Res. 2001;156:618–27. doi: 10.1667/0033-7587(2001)156[0618:esttma]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 33.Azzam EI, de Toledo SM, Gooding T, Little JB. Intercellular communication is involved in the bystander regulation of gene expression in human cells exposed to very low fluences of alpha particles. Radiat Res. 1998;150:497–504. [PubMed] [Google Scholar]
- 34.Mothersill C, Seymour C. Medium from irradiated human epithelial cells but not human fibroblasts reduces the clonogenic survival of unirradiated cells. Int J Radiat Biol. 1997;71:421–7. doi: 10.1080/095530097144030. [DOI] [PubMed] [Google Scholar]
- 35.Sawant SG, Zheng W, Hopkins KM, Randers-Pehrson G, Lieberman HB, Hall EJ. The radiation-induced bystander effect for clonogenic survival. Radiat Res. 2002;157:361–4. doi: 10.1667/0033-7587(2002)157[0361:tribef]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Mothersill C, Seymour RJ, Seymour CB. Increased radiosensitivity in cells of two human cell lines treated with bystander medium from irradiated repair-deficient cells. Radiat Res. 2006;165:26–34. doi: 10.1667/rr3488.1. [DOI] [PubMed] [Google Scholar]
- 37.Belyakov OV, Folkard M, Mothersill C, Prise KM, Michael BD. Bystander-induced apoptosis and premature differentiation in primary urothelial explants after charged particle microbeam irradiation. Radiat Prot Dosimetry. 2002;99:249–51. doi: 10.1093/oxfordjournals.rpd.a006775. [DOI] [PubMed] [Google Scholar]
- 38.Belyakov OV, Folkard M, Mothersill C, Prise KM, Michael BD. Bystander-induced differentiation: a major response to targeted irradiation of a urothelial explant model. Mutat Res. 2006;597:43–9. doi: 10.1016/j.mrfmmm.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Belyakov OV, Malcolmson AM, Folkard M, Prise KM, Michael BD. Direct evidence for a bystander effect of ionizing radiation in primary human fibroblasts. Br J Cancer. 2001;84:674–9. doi: 10.1054/bjoc.2000.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyng FM, Seymour CB, Mothersill C. Production of a signal by irradiated cells which leads to a response in unirradiated cells characteristic of initiation of apoptosis. Br J Cancer. 2000;83:1223–30. doi: 10.1054/bjoc.2000.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asur RS, Thomas RA, Tucker JD. Chemical induction of the bystander effect in normal human lymphoblastoid cells. Mutat Res. 2009;676:11–6. doi: 10.1016/j.mrgentox.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Prise KM, Belyakov OV, Folkard M, Michael BD. Studies of bystander effects in human fibroblasts using a charged particle microbeam. Int J Radiat Biol. 1998;74:793–8. doi: 10.1080/095530098141087. [DOI] [PubMed] [Google Scholar]
- 43.Deshpande A, Goodwin EA, Bailey SM, Marrone BL, Lehnert BE. Alpha-particle-induced sister chromatid exchange in normal human lung fibroblasts: evidence for an extranuclear target. Radiat Res. 1996;145:260–7. [PubMed] [Google Scholar]
- 44.Lorimore SA, Kadhim MA, Pocock DA, Papworth D, Stevens DL, Goodhead DT, Wright EG. Chromosomal instability in the descendants of unirradiated surviving cells after alpha-particle irradiation. Proc Natl Acad Sci USA. 1998;95:5730–3. doi: 10.1073/pnas.95.10.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sedelnikova OA, Nakamura A, Kovalchuk O, Koturbash I, Mitchell SA, Marino SA, et al. DNA double-strand breaks form in bystander cells after microbeam irradiation of three-dimensional human tissue models. Cancer Res. 2007;67:4295–302. doi: 10.1158/0008-5472.CAN-06-4442. [DOI] [PubMed] [Google Scholar]
- 46.Sokolov MV, Smilenov LB, Hall EJ, Panyutin IG, Bonner WM, Sedelnikova OA. Ionizing radiation induces DNA double-strand breaks in bystander primary human fibroblasts. Oncogene. 2005;24:7257–65. doi: 10.1038/sj.onc.1208886. [DOI] [PubMed] [Google Scholar]
- 47.Asur R, Balasubramaniam M, Marples B, Thomas RA, Tucker JD. Bystander effects induced by chemicals and ionizing radiation: evaluation of changes in gene expression of downstream MAPK targets. Mutagenesis. 2010;25:271–9. doi: 10.1093/mutage/geq003. [DOI] [PubMed] [Google Scholar]
- 48.Azzam EI, De Toledo SM, Spitz DR, Little JB. Oxidative metabolism modulates signal transduction and micronucleus formation in bystander cells from alpha-particle-irradiated normal human fibroblast cultures. Cancer Res. 2002;62:5436–42. [PubMed] [Google Scholar]
- 49.Demidem A, Morvan D, Madelmont JC. Bystander effects are induced by CENU treatment and associated with altered protein secretory activity of treated tumor cells: a relay for chemotherapy? Int J Cancer. 2006;119:992–1004. doi: 10.1002/ijc.21761. [DOI] [PubMed] [Google Scholar]
- 50.Rugo RE, Almeida KH, Hendricks CA, Jonnalagadda VS, Engelward BP. A single acute exposure to a chemotherapeutic agent induces hyper-recombination in distantly descendant cells and in their neighbors. Oncogene. 2005;24:5016–25. doi: 10.1038/sj.onc.1208690. [DOI] [PubMed] [Google Scholar]
- 51.Dahle J, Angell-Petersen E, Steen HB, Moan J. Bystander effects in cell death induced by photodynamic treatment UVA radiation and inhibitors of ATP synthesis. Photochem Photobiol. 2001;73:378–87. doi: 10.1562/0031-8655(2001)073<0378:beicdi>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 52.Dabrowska A, Gos M, Janik P. “Bystander effect” induced by photodynamically or heat-injured ovarian carcinoma cells (OVP10) in vitro. Med Sci Monit. 2005;11:BR316–24. [PubMed] [Google Scholar]
- 53.Chakraborty A, Held KD, Prise KM, Liber HL, Redmond RW. Bystander effects induced by diffusing mediators after photodynamic stress. Radiat Res. 2009;172:74–81. doi: 10.1667/RR1669.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seymour CB, Mothersill C. Relative contribution of bystander and targeted cell killing to the low-dose region of the radiation dose-response curve. Radiat Res. 2000;153:508–1. doi: 10.1667/0033-7587(2000)153[0508:rcobat]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 55.Gow MD, Seymour CB, Byun SH, Mothersill CE. Effect of dose rate on the radiation-induced bystander response. Phys Med Biol. 2008;53:119–32. doi: 10.1088/0031-9155/53/1/008. [DOI] [PubMed] [Google Scholar]
- 56.Mothersill C, Seymour CB. Bystander and delayed effects after fractionated radiation exposure. Radiat Res. 2002;158:626–33. doi: 10.1667/0033-7587(2002)158[0626:badeaf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 57.Mothersill CE, Moriarty MJ, Seymour CB. Radiotherapy and the potential exploitation of bystander effects. Int J Radiat Oncol Biol Phys. 2004;58:575–9. doi: 10.1016/j.ijrobp.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 58.Mackonis EC, Suchowerska N, Zhang M, Ebert M, McKenzie DR, Jackson M. Cellular response to modulated radiation fields. Phys Med Biol. 2007;52:5469–82. doi: 10.1088/0031-9155/52/18/001. [DOI] [PubMed] [Google Scholar]
- 59.Butterworth KT, McGarry CK, O’Sullivan JM, Hounsell AR, Prise KM. A study of the biological effects of modulated 6 MV radiation fields. Phys Med Biol. 2010;55:1607–18. doi: 10.1088/0031-9155/55/6/005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butterworth KT, McGarry CK, Trainor C, O’Sullivan JM, Hounsell AR, Prise KM. Out-of-field cell survival following exposure to intensity-modulated radiation fields. Int J Radiat Oncol Biol Phys. 2011;9:1516–22. doi: 10.1016/j.ijrobp.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffin RJ, Koonce NA, Dings RPM, Siegel E, Moros EG, et al. Microbeam radiation therapy alters vascular architecture and tumor oxygenation and is enhanced by a galectin-1 targeted anti-angiogenic peptide. Radiat Res. 2012;177:804–12. doi: 10.1667/rr2784.1. [DOI] [PMC free article] [PubMed] [Google Scholar]