The SWI/SNF complex is a key element of the yeast CWI MAPK pathway, which mediates the chromatin remodeling necessary for an adequate transcriptional response to cell wall stress. The MAPK Slt2 mediates, through Rlm1, nucleosome rearrangements at cell wall stress–responsive genes by targeting the SWI/SNF complex.
Abstract
In Saccharomyces cerevisiae, the transcriptional program triggered by cell wall stress is coordinated by Slt2/Mpk1, the mitogen-activated protein kinase (MAPK) of the cell wall integrity (CWI) pathway, and is mostly mediated by the transcription factor Rlm1. Here we show that the SWI/SNF chromatin-remodeling complex plays a critical role in orchestrating the transcriptional response regulated by Rlm1. swi/snf mutants show drastically reduced expression of cell wall stress–responsive genes and hypersensitivity to cell wall–interfering compounds. On stress, binding of RNA Pol II to the promoters of these genes depends on Rlm1, Slt2, and SWI/SNF. Rlm1 physically interacts with SWI/SNF to direct its association to target promoters. Finally, we observe nucleosome displacement at the CWI-responsive gene MLP1/KDX1, which relies on the SWI/SNF complex. Taken together, our results identify the SWI/SNF complex as a key element of the CWI MAPK pathway that mediates the chromatin remodeling necessary for adequate transcriptional response to cell wall stress.
INTRODUCTION
The cell wall of Saccharomyces cerevisiae is an external envelope that protects yeast against extreme environmental conditions. This macromolecular complex is essential for maintaining cell shape and integrity (Lesage and Bussey, 2006). Stressful conditions that damage the fungal cell wall trigger cellular responses to guarantee cell survival through the remodeling of this extracellular matrix. The adaptive response of S. cerevisiae to cell wall stress is mainly mediated by the cell wall integrity pathway (CWI; Levin, 2011). A pair of membrane proteins, Mid2 and Wsc1, act as the main sensors of this pathway. Under activation conditions, these sensors interact with the guanine nucleotide exchange factor Rom2, activating the small GTPase Rho1, which then interacts with and activates Pkc1. The main role of activated Pkc1 is to trigger a mitogen-activated protein kinase (MAPK) module. Phosphorylation of the MAPK kinase kinase Bck1 activates a pair of redundant MAPK kinases (Mkk1 and Mkk2), which finally phosphorylate the MAPK Slt2/Mpk1 (Levin, 2011). The phosphorylated form of this protein acts mainly on two transcription factors: Rlm1 (Watanabe et al., 1997) and SBF (Baetz et al., 2001).
The final consequence of the activation of the CWI pathway by cell wall stress is the induction of an adaptive transcriptional program that has been extensively studied by genome-wide expression profiling (Lagorce et al., 2003; García et al., 2004, 2009). Recent work has established a noncatalytic mechanism by which Slt2 regulates the transcription of a small subset of CWI-responsive genes, including FKS2, through SBF (Kim et al., 2008; Kim and Levin, 2011). However, the transcriptional response triggered through the CWI pathway is largely dependent on Rlm1 (Jung and Levin, 1999; García et al., 2004). Rlm1 is a MADS-box transcription factor related to members of the mammalian MEF2 family of transcriptional regulators, and not only does it share sequence similarity with MEF2 isoforms, but it also has the same DNA-binding specificity in vitro (CTA[T/A]4TAG; Watanabe et al., 1995; Dodou and Treisman, 1997). Mutations in Rlm1 at putative Slt2 phosphorylation sites Ser-427 and Thr-439 are critical for stress-induced transcription of cell wall stress genes, suggesting that Slt2 regulates the transcriptional activity of Rlm1 by direct phosphorylation of these residues (Jung et al., 2002). The elements of the yeast transcriptional machinery working in concert with Rlm1 for the transcriptional activation upon cell wall stress and the molecular mechanisms involved in this process are completely unknown.
Promoter-chromatin disassembly is a potent mechanism of transcriptional activation (Adkins et al., 2007), playing a critical role in establishment and maintenance of transcriptional programs. Posttranslational modifications of individual histones by histone modifiers (reviewed in Wang et al., 2004) and disassembly and removal of nucleosomes by ATP-dependent chromatin-remodeling complexes (Clapier and Cairns, 2009) work in concert to regulate this process (Narlikar et al., 2002). The yeast SWI/SNF family, including SWI/SNF and RSC, are large chromatin-remodeling machines that can move or eject nucleosomes, facilitating transcription and other nuclear processes (Kasten et al., 2011). S. cerevisiae SWI/SNF consists of 11 subunits: Snf2, Swi3, Swi1, Snf5, Swp82, Snf12, Arp7, Arp9, Snf6, Snf11, and Taf14, with the Snf2 subunit serving as an ATPase that provides the energy for nucleosome remodeling (Côté et al., 1994; Smith et al., 2003). Because SWI/SNF levels within cells are very low and its interaction with nucleosomal DNA has no sequence specificity, the complex needs to be targeted to specific genes for nucleosome reorganization. Targeted recruitment of the SWI/SNF chromatin-remodeling complex can be achieved through direct interaction with gene-specific transcriptional activators (Cosma et al., 1999; Neely et al., 1999; Yudkovsky et al., 1999; Peterson and Workman, 2000). On recruitment, SWI/SNF locally alters nucleosome positioning at the promoter, facilitating the binding of transcription factors to their binding sites and then stimulating transcriptional initiation by Pol II (Neely et al., 1999; Yudkovsky et al., 1999).
The participation of the yeast SWI/SNF complex in the regulation of specific gene expression in response to glucose starvation, intracellular phosphate concentration, and heat shock is well documented (Adkins et al., 2007; Biddick et al., 2008; Erkina et al., 2008; Shivaswamy and Iyer, 2008). In the context of MAPK signaling, both RSC and SWI/SNF complexes are recruited to osmostress-responsive genes by Hog1 (Proft and Struhl, 2002; Mas et al., 2009), although RSC seems to be the key remodeling enzyme to promote chromatin reorganization under these conditions (Mas et al., 2009).
In this work, we identified for the first time the SWI/SNF ATP-dependent chromatin-remodeling complex as an essential element required for yeast transcriptional reprogramming upon conditions that affect cell wall integrity. We show that SWI/SNF is recruited to the promoters of cell wall stress–responsive genes under stress conditions through direct interaction with Rlm1, and SWI/SNF targeting is required for nucleosome rearrangements at cell wall stress–responsive genes. Therefore our results add novel insights on the mechanisms by which the yeast cell wall stress transcriptional program is regulated.
RESULTS
SWI/SNF is required for the transcriptional response triggered by cell wall stress through the CWI pathway
To identify novel elements required for proper gene expression under conditions affecting cellular integrity, we performed a large-scale screening using the whole collection of haploid deletion strains in all nonessential genes of S. cerevisiae transformed with the plasmid pJS05. This reporter system is based on the transcriptional fusion of the MLP1/KDX1(YKL161C) promoter to the coding sequence of the NAT1 gene, which confers resistance in yeast to the antibiotic nourseothricin (Rodríguez-Peña et al., 2008). MLP1 shows low basal gene expression levels, but it is highly expressed under cell wall stress, and this induction is largely dependent on Slt2 and Rlm1 (García et al., 2004). In previous work we used this collection of mutant strains to identify genes whose absence produces constitutive activation of the CWI pathway under vegetative (no stress) growth conditions (Arias et al., 2011). Here transformants were tested for their ability to grow under cell wall stress (1 U/ml zymolyase) in the presence of nourseothricin to identify genes required for transcriptional activation under these conditions. Those mutants unable to activate a correct adaptive response will not induce the reporter system and therefore will not be able to grow under the concentration of nourseothricin assayed.
A total of 159 mutants defective in the induction of MLP1 were identified (see Materials and Methods for details). Analysis of the complete gene data set using the Web-based tools GeneCodis (http://genecodis.dacya.ucm.es) and FunSpec (http://funspec.med.utoronto.ca) allowed us to establish transcription (GO:0006350) as the biological process with the highest statistical significance. Within this group, several protein complexes related to regulation of gene expression were identified (Table 1), including the SWI/SNF ATP-dependent chromatin-remodeling complex. Although this complex had not been previously associated with cell wall stress responses mediated by the CWI pathway, the identification of five members belonging to this complex (SNF2, SNF6, SWI3, SNF11, and TAF14) clearly suggested its participation.
TABLE 1:
Protein complexes related to genes identified in the screening belonging to the group of transcription.
| Category | p value | Number of genes | |||
|---|---|---|---|---|---|
| Input | Total | ||||
| SAGA complex | 1.89E-12 | 7 | 16 | ||
| SWI/SNF transcription activator complex | 1.55E-09 | 5 | 10 | ||
| SAGA-like complex (SLIK) | 2.04E-06 | 3 | 5 | ||
| RNA polymerase II | 2.60E-06 | 4 | 17 | ||
| ADA complex | 4.07E-06 | 3 | 6 | ||
| SBF complex | 3.58E-05 | 2 | 2 | ||
| RNA polymerase I | 7.16E-05 | 3 | 14 | ||
| Kornberg's mediator (SRB) complex | 0.000254 | 3 | 21 | ||
The Munich Information Center for Protein Sequences (Neuherberg, Germany) protein complexes (1142 categories). The analysis was performed using the bioinformatic tool FunSpec. The p values represent the probability that the intersection of a given list with any given functional category occurs by chance. Input, number of genes from the input cluster in a given category. Total, total number of genes included in each category.
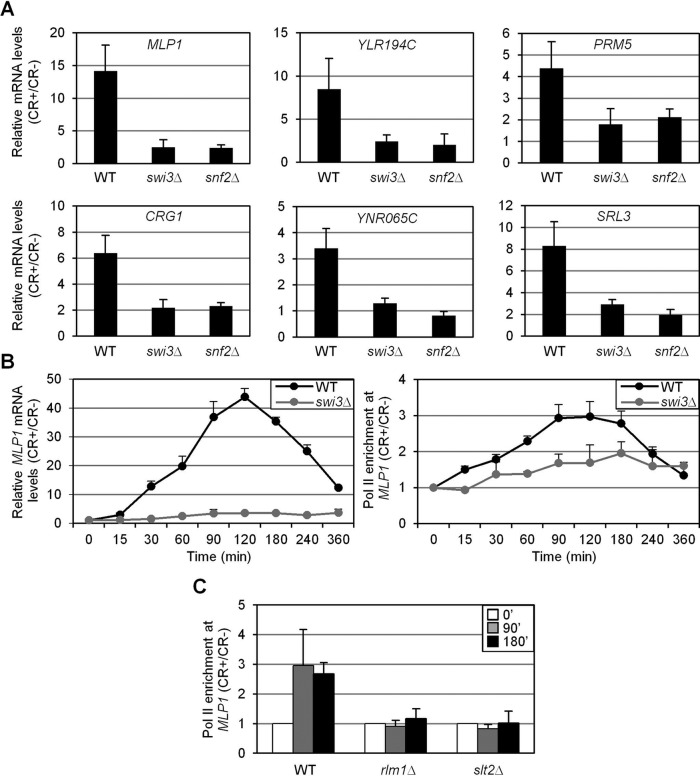
To study the relevance of SWI/SNF in CWI-mediated gene expression, we analyzed mRNA levels of several genes induced through the CWI pathway in response to stress mediated by Congo red (CR) in wild-type (WT), swi3Δ, and snf2Δ cells. As shown in Figure 1A, transcriptional activation of MLP1, YLR194C, CRG1, YNR065C, PRM5, and SRL3 upon stress was severely compromised in the absence of SWI/SNF. Moreover, characterization of the kinetics of MLP1 expression as a consequence of stress in WT and swi3Δ strains clearly indicated a dependence on the SWI/SNF complex for induction through the whole time course (Figure 1B, left). In agreement, stress treatment elicited a clear recruitment of RNA Pol II—characterized by chromatin immunoprecipitation (ChIP) analysis—to the MLP1 promoter, and this recruitment was largely dependent on Swi3 (Figure 1B, right). Furthermore, Pol II binding triggered by stress was completely blocked in slt2∆ and rlm1∆ strains (Figure 1C). Therefore cell wall stress transcriptional response involves the recruitment of RNA Pol II to the corresponding responsive genes, and binding of this complex requires the activator Rlm1, the MAPK Slt2, and the chromatin-remodeling enzyme SWI/SNF.
FIGURE 1:
The SWI/SNF remodeling complex is required for the transcriptional response triggered by cell wall stress. (A) mRNA levels of several CWI-responsive genes were analyzed by RT-qPCR in WT, swi3Δ, and snf2Δ strains after 2 h of CR treatment. Values represent the ratio between CR-treated and nontreated cells. (B) Left, kinetics of MLP1 gene expression analyzed by RT-qPCR in WT and swi3Δ strains at different times of CR treatment. Values represent the ratio between CR-treated and nontreated cells. Right, recruitment of RNA Pol II Rpb1 subunit to MLP1 at MLP1PRO2 region (−143 to +56) determined by ChIP analysis in WT and swi3Δ strains subjected to CR treatment at the times indicated. Results are shown as the fold induction of CR-treated against nontreated samples. (C) The entry of RNA Pol II at MLP1 under cell wall stress determined in WT, rlm1Δ, and slt2Δ strains as indicated in B.
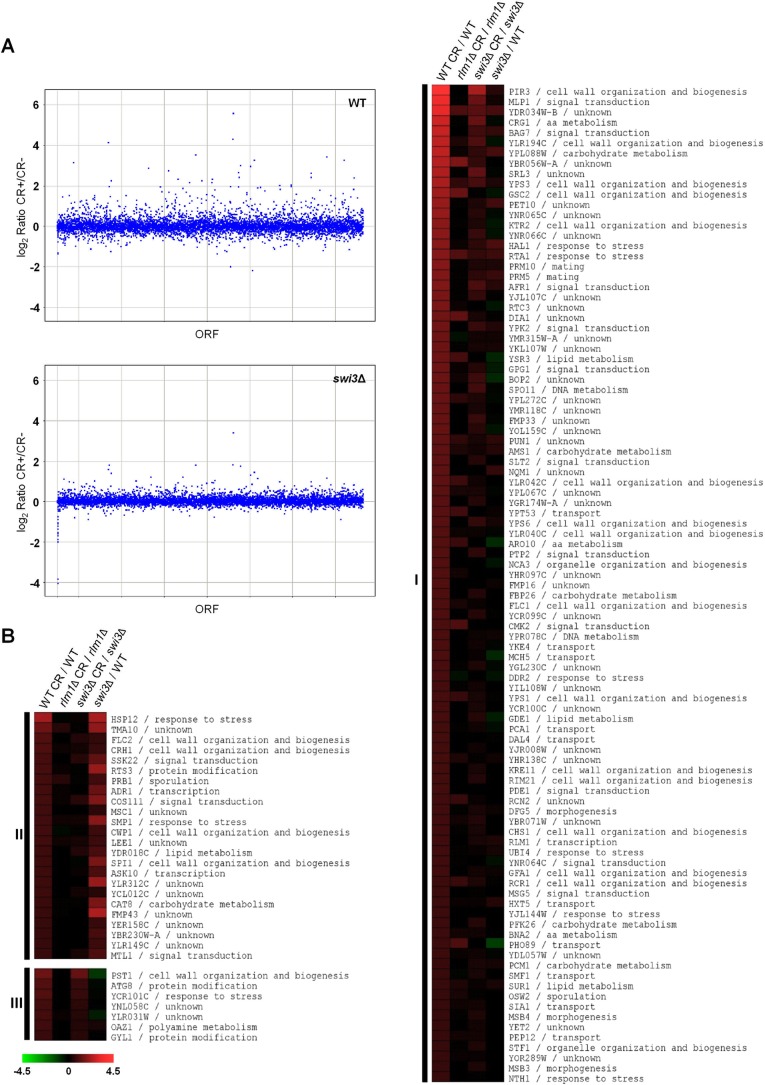
These results prompted us to characterize the genome-wide expression profiles of WT and swi3Δ strains challenged with cell wall stress using Affymetrix GeneChips. Microarray analyses revealed that 128 genes were induced (ratio of transcript levels ≥2), and 15 genes were repressed (ratio of transcript levels ≤0.5), respectively, upon treatment with CR in a WT strain. In contrast, only 14 genes were induced by cell wall stress in the swi3Δ mutant strain (Figure 2A), most of these genes showing a significant reduction in gene expression with respect to the WT strain. A more detailed comparison of the transcriptional induction profiles in WT and mutant cells (see Materials and Methods) uncovered three clusters of genes within the response with respect to the Swi3 dependence: 1) genes whose induction was dependent on Swi3 (97genes; 76%); 2) genes whose basal expression levels was dependent on Swi3 (24 genes; 19%); and 3) a minor group of 7 genes induced by cell wall stress independent of Swi3 (5%; Figure 2B). Moreover, stress induction of the majority of genes (∼90%) within groups 1–3 required the Rlm1 transcription factor, as deduced from the transcriptional profile of an rlm1Δ strain (Figure 2B).
FIGURE 2:
Genome-wide expression profiles of WT (BY4741), swi3Δ, and rlm1Δ strains challenged with CR. (A) Scatter plot of normalized gene expression data in WT and swi3Δ strains. Gene expression ratios (treated/untreated) were plotted against their ORFs using Spotfire software. (B) Heat map obtained by MeV 4.6 software shows gene expression ratios comparing the transcriptional response to CR (treated vs. untreated) in the WT, rlm1Δ, and swi3Δ strains respectively (three columns on the left). Gene expression ratios of a swi3Δ mutant vs. a WT strain in the absence of stress are shown in the fourth column. The genes included in the analysis were those up-regulated in the WT strain by CR treatment. Genes were grouped together (clusters 1–3) on the basis of their dependence on Swi3 for basal and CR-mediated activation (see the text for details). Gene functional categories were assigned according to the information from the Biobase BioKnowledge Library (BKL; http://rous.mit.edu/index.php/Biobase_BioKnowledge_Library). The degree of color saturation represents the expression log2 ratio value, as indicated by the scale bar.
Of importance, Slt2 phosphorylation levels upon cell wall stress in swi3Δ and snf2Δ mutants were similar to those in the WT (Supplemental Figure S1), indicating that the absence of SWI/SNF does not affect the functionality of the CWI pathway.
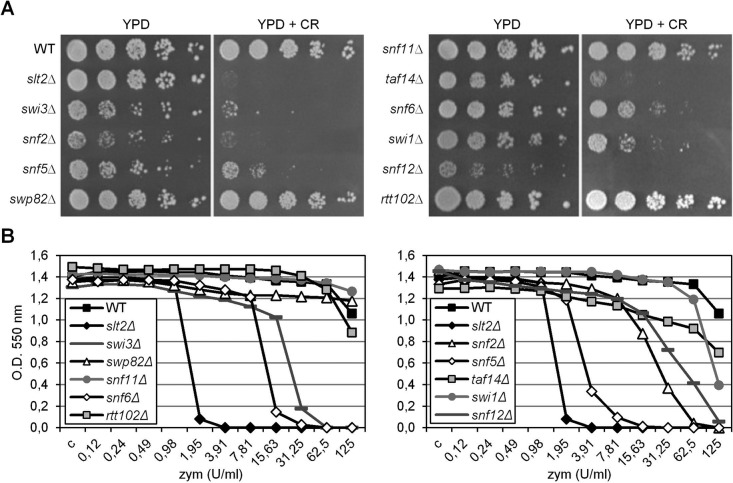
Phenotypic analysis of a full set of swi/snf mutants growing in the presence of the cell wall–interfering compounds CR and zymolyase revealed that deletion of SWI3, SNF2, SNF5, TAF14, SNF6, SWI1, and SNF12, although with some differences, impaired cell growth under cell wall stress conditions (Figure 3). Thus the SWI/SNF chromatin-remodeling complex is essential for eliciting gene expression in response to cell wall stress and therefore for cell survival under these conditions.
FIGURE 3:
Deletion of genes that encode SWI/SNF subunits renders cells sensitive to cell wall stress. (A) The indicated strains were spotted on YPD plates without or with 50 μg/ml CR, and plates were incubated for 72 h at 30°C. (B) Zymolyase sensitivity was performed in a 96-well microtiter plate assay with zymolyase 20T, giving concentrations ranging from 125 to 0.12 U/ml YPD. Each well was inoculated with ∼104 cells from an exponentially growing culture. Plates were incubated for 24–48 h at 30°C, and cell growth was determined by measuring absorbance at 550 nm.
Rlm1 and SWI/SNF are interdependently recruited to CWI-responsive genes under cell wall stress, and this requires activation of the MAPK Slt2
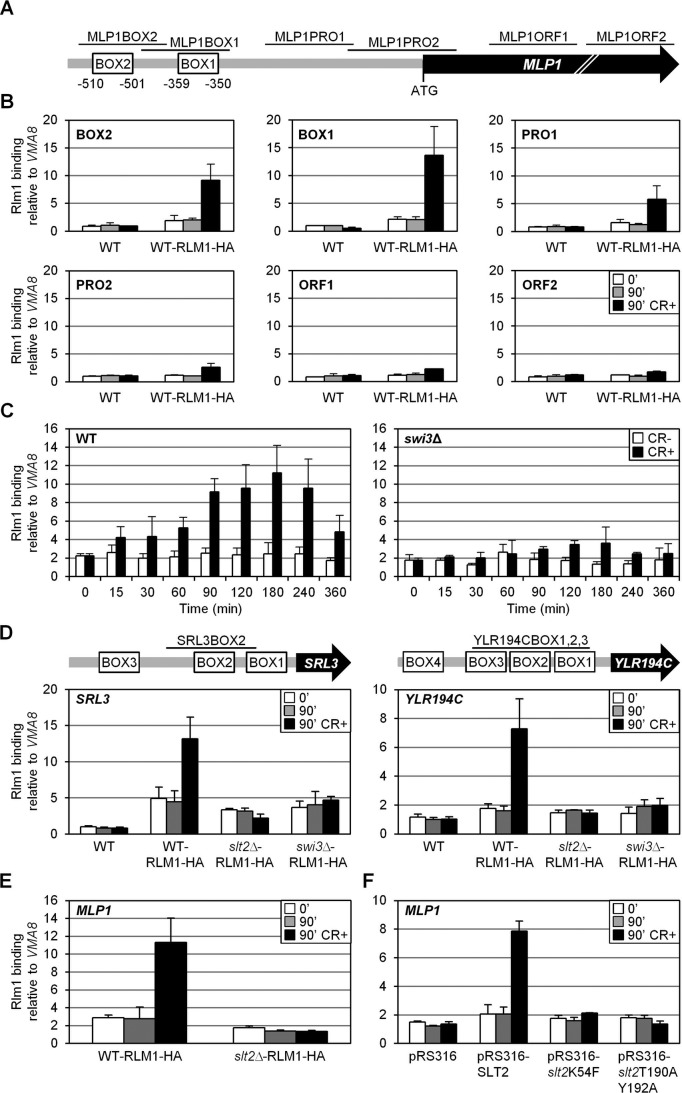
The mechanisms by which Rlm1 induces transcription of CWI-responsive genes are not completely understood. ChIP assays were used to follow the binding of Rlm1 at MLP1 before and during cell wall stress conditions. Chromatin from a WT strain expressing a functional hemagglutinin (HA) epitope–tagged Rlm1 was immunoprecipitated with anti-HA antibodies and analyzed by quantitative PCR to check occupation in different regions of the MLP1 gene (Figure 4A). As shown in Figure 4B, Rlm1 was present at the MLP1 promoter in the absence of stress, as deduced from the comparison in Rlm1 binding between tagged and untagged WT strains. Moreover, cell wall stress induced high levels of Rlm1 recruitment. This enrichment was found through the entire analyzed promoter region but was more pronounced in the region of Rlm1 putative binding sites (BOX1 and BOX2; Figure 4B). Of note, both sites are functional, since fusions of either BOX1 or BOX2 of MLP1 to a minimal CYC1 promoter-lacZ results in a reporter system that can be transcriptionally activated by cell wall stress (unpublished data).
FIGURE 4:
Rlm1 occupies the MLP1 promoter in vivo under basal conditions, and it is recruited under cell wall stress in a SWI3- and SLT2-dependent manner. (A) Schematic representation of the MLP1 gene. BOX1 and BOX2 mark the Rlm1-binding sites. Regions amplified by primers for ChIP experiments are shown as horizontal lines. (B) Rlm1-HA association with the MLP1 gene analyzed by ChIP in a WT strain expressing or lacking Rlm1-HA before and after CR addition. The regions of MLP1 analyzed are indicated in the upper left corner. (C) Kinetics of Rlm1-HA recruitment to the MLP1 promoter (MLP1BOX1 region) analyzed by ChIP in WT (left) and swi3Δ mutant (right) strains expressing Rlm1-HA in the absence and presence of CR at the indicated times. (D) Rlm1 recruitment to SRL3 (left) and YLR194C (right) promoters under cell wall stress (CR 30 μg/ml) and nonstress conditions analyzed by ChIP in WT-tagged (Rlm1-HA) and untagged strains, as well as in Rlm1-HA–tagged swi3Δ and slt2Δ strains. Schematic representation of the SRL3 and YLR194C genes, indicating putative Rlm1-binding sites, with the regions analyzed by ChIP (shown as horizontal lines) is included. (E) Rlm1 association to the MLP1BOX1 region of MLP1 analyzed by ChIP in WT and slt2∆ cells expressing Rlm1-HA before and after CR treatment. (F) Rlm1 binding analyzed, as in E, in slt2∆ cells expressing tagged Rlm1-HA transformed with plasmid pRS316-SLT2, pRS316-slt2K54F, or pRS316-slt2T190A/Y192A.
The kinetics of Rlm1 recruitment at the MLP1 promoter was further characterized at different times of CR treatment in WT and swi3Δ strains. In the WT strain, a peak of maximum occupancy between 90 and 180 min was found (Figure 4C, left), in agreement with the kinetics of MLP1 expression (Figure 1B). Furthermore, Rlm1 recruitment mediated by CR stress was largely dependent on Swi3 (Figure 4C, right). These results were further confirmed for two additional genes of the cell wall stress response, SRL3 and YLR194C (Figure 4D), clearly demonstrating the requirement of the SWI/SNF remodeling complex for the stress-dependent recruitment of Rlm1 to target genes.
Characterization of Rlm1 protein levels in the swi3∆ strain revealed a significant reduction with respect to the WT (Supplemental Figure S2A). Under stress conditions, this is probably in part related to the lack of a feedback transcriptional mechanism through Rlm1, whose expression is induced by cell wall stress. However, equalizing the levels of Rlm1 in both strains by overexpressing Rlm1 (Supplemental Figure S2A) did not rescue defects in cell wall stress–mediated Rlm1 recruitment (Supplemental Figure S2B), induction of CWI-responsive genes (Supplemental Figure S2C), and CR hypersensitivity due to the SWI3 deletion (Supplemental Figure S2D), clearly demonstrating the requirement of the SWI/SNF remodeling complex for the CWI transcriptional response.
Moreover, signaling through the CWI pathway is critical for Rlm1 recruitment to CWI-responsive genes following stress since this binding was completely dependent on the presence of Slt2 (Figure 4, D and E), and it required both the catalytic activity of the MAPK and phosphorylation of the MAPK by Mkk1/2, as deduced from the absence of Rlm1 binding to the MLP1 promoter in cells expressing versions of Slt2 carrying mutations within the ATP-binding site (pRS316-slt2K54F) or within the activation loop of the MAPK (pRS316-slt2T190A/Y192A; Figure 4F).
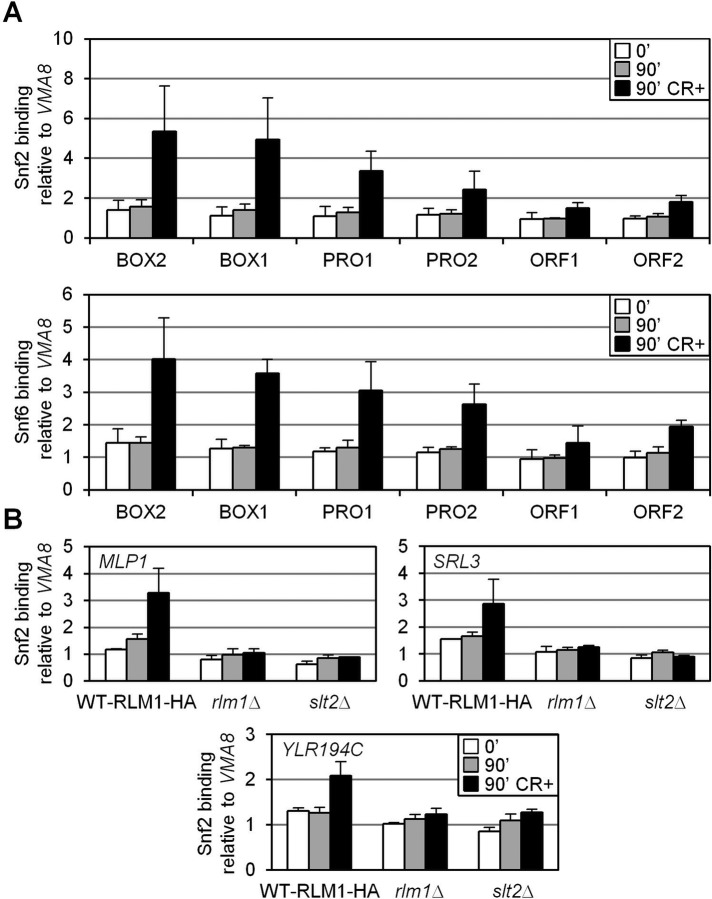
Binding of the Snf2 and Snf6 subunits to various regions of MLP1 gene was then determined by ChIP, using specific polyclonal antibodies against these proteins, in a WT strain before and after CR addition. In agreement with a key role for SWI/SNF in cell wall stress–dependent gene expression, the complex associated with MLP1 after 90 min of cell wall stress as determined by ChIP analysis. As shown in Figure 5A, Snf2 and Snf6 recruitment was detected primarily on regions of the MLP1 promoter, with maximal occupation at Rlm1-binding sites. On stress, Snf2 is also recruited to the promoters of SRL3 and YLR194C (Figure 5B). Furthermore, stress-mediated SWI/SNF recruitment depends on the CWI pathway, since Snf2 binding to MLP1, SRL3, and YLR194C promoters is blocked in both slt2∆ and rlm1∆ strains (Figure 5B).
FIGURE 5:
Snf2 and Snf6 are recruited to the activated MLP1 promoter in vivo in a CWI- dependent manner. (A) Association of Snf2 and Snf6 along the different regions of MLP1 (Figure 4A) determined by ChIP in a WT strain grown in the presence or absence of CR. (B) Recruitment of Snf2 at MLP1 (MLP1BOX1), SRL3 (−265 to −135) and YLR194C (−254 to −123) gene promoters analyzed by ChIP as in A in WT, rlm1∆, and slt2∆ strains.
Rlm1 physically interacts with the SWI/SNF complex in a cell wall stress–dependent manner
The interdependence of Rlm1 and SWI/SNF binding to the promoters of CWI pathway-responsive genes led us to examine whether the SWI/SNF complex physically associates with Rlm1. In vitro, purified SWI/SNF complex clearly interacted with Rlm1, and the DNA-binding domain of Rlm1 is not required for this association (Figure 6A). Moreover, interaction between SWI/SNF and Rlm1 was also detected in vivo by pull-down experiments with extracts from rlm1Δ snf2Δ cells expressing Myc-tagged Rlm1 and HA-tagged Snf2. As shown in Figure 6B, Rlm1 coprecipitated with Snf2 in a stress-dependent manner. Thus these data provide biochemical evidence that Rlm1 and the SWI/SNF complex interact after cell wall stress.
FIGURE 6:
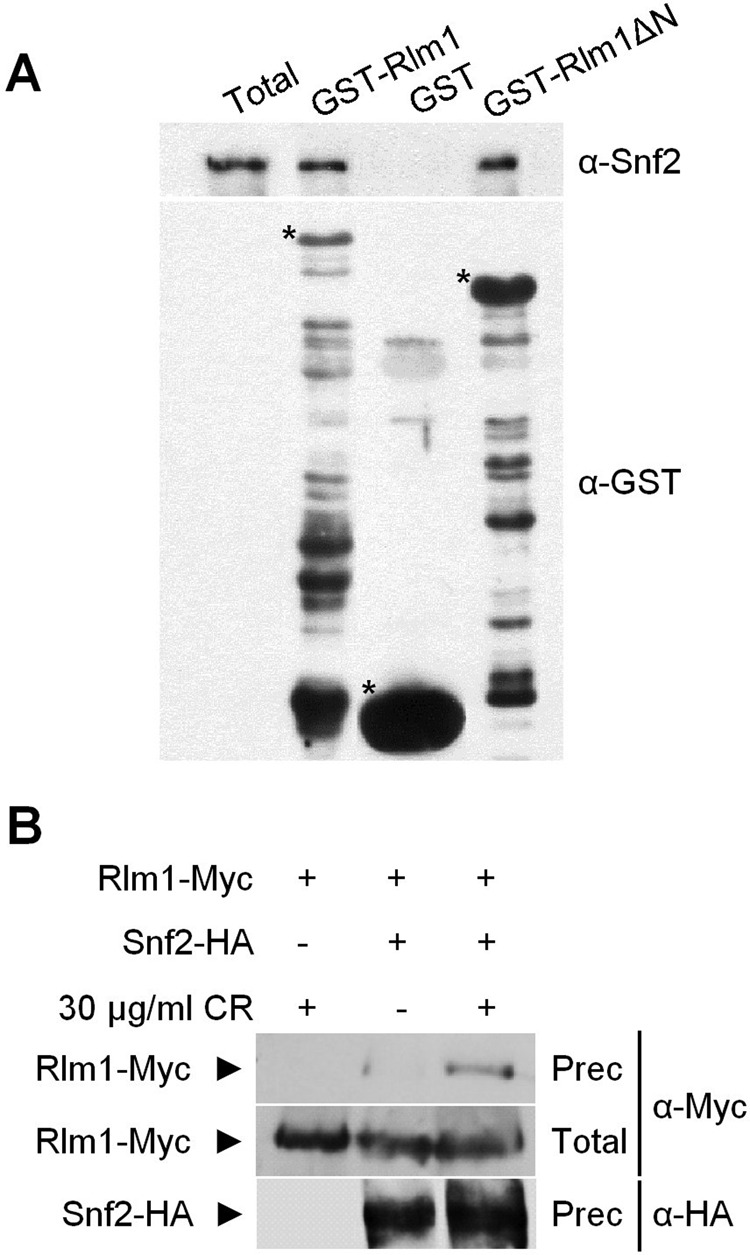
Rlm1 physically interacts with SWI/SNF. (A) Rlm1 associates directly with the SWI/SNF complex in vitro. GST, GST-Rlm1, and GST-Rlm1ΔN were expressed and purified from E. coli, and the SWI/SNF complex was purified from a yeast strain bearing TAP-tag Snf2. SWI/SNF complex was incubated with GST proteins bound to glutathione–Sepharose beads, and interacting proteins were probed using antibodies against GST and Snf2. Specific GST fusion proteins are labeled with an asterisk. Total represents 20% of total input protein. (B) In vivo binding of Rlm1 and SWI/SNF. An rlm1Δ snf2Δ strain was transformed with episomal plasmids expressing Rlm1-Myc and Snf2-HA under their native promoters. Samples were taken from yeast cultures growing in the presence or absence of CR (30 min). Snf2-HA was immunoprecipitated by monoclonal antibodies against HA. Snf2-HA and Rlm1-Myc proteins were detected by Western blotting against HA and Myc epitopes, respectively. Total extract represents 10% of the total input protein.
Cell wall stress directs nucleosome displacement at the MLP1 locus in a SWI/SNF-dependent manner
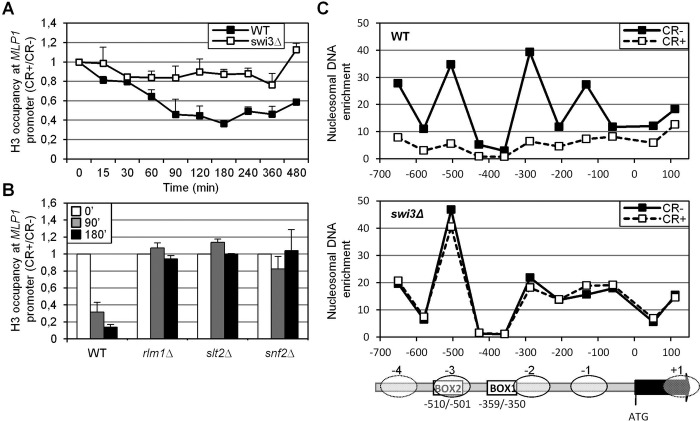
To examine the effect of SWI/SNF complex on chromatin organization and to check whether this complex was promoting nucleosome rearrangements, we first studied histone H3 occupancy at MLP1 in WT cells after CR addition. A decrease in H3 occupancy was observed at different regions of the MLP1 promoter but not at the coding region, with a peak of H3 eviction after 3 h of CR treatment (Supplemental Figure S3). This timing was coincident with the time course of Rlm1 occupation and was maximal at MLP1BOX1 and MLP1BOX2 regions, in agreement with the highest recruitment of SWI/SNF at these locations (Figure 5A). More important, H3 histone eviction in WT stressed cells was completely dependent on Swi3 (Figure 7A). Moreover, H3 displacement was also dependent on Snf2, Slt2, and Rlm1 (Figure 7B), supporting the idea that activation of the CWI pathway as a consequence of cell wall stress leads to chromatin remodeling at the CWI-responsive genes through the activity of the SWI/SNF complex.
FIGURE 7:
Lack of Swi3 abrogates chromatin remodeling at MLP1 locus. (A) Kinetics of histone H3 binding to MLP1 promoter (MLP1BOX1 region) in WT and swi3Δ strains under CR treatment at the indicated times. (B) Association of histone H3 to MLP1 promoter (MLP1BOX1) in WT, rlm1Δ, slt2Δ, and snf2Δ strains analyzed by ChIP before and after 90 and 180 min of CR addition. In both cases, results are shown as the ratio between the fold enrichment of the treated against the untreated culture. (C) Nucleosome-scanning analysis of MLP1 locus (from −699 to +161) in WT and swi3Δ strains under basal and cell wall stress conditions. Mononucleosomes were isolated from cultures growing in the absence and presence of CR (3 h). Purified DNA samples were analyzed by qPCR as detailed in Materials and Methods. Bottom, nucleosomes represented as ovals at the indicated position of the MLP1 locus.
To further support this conclusion, we analyzed nucleosome positioning at the MLP1 promoter before and after stress. Mononucleosomes were obtained by micrococcal nuclease digestion of chromatin, followed by nucleosome scanning analysis using overlapping primer pairs covering an 800–base pair region from −699 to +161 of MLP1. As shown in Figure 7C, in WT cells growing under normal conditions, DNA at the MLP1 promoter is packaged into an array of four positioned nucleosomes. After cell wall stress, the pattern of nucleosome organization was dramatically changed, with a complete loss of nucleosomes at the MLP1 promoter, consistent with their eviction. This chromatin-remodeling effect was not observed in a swi3Δ mutant (Figure 7C). Although slight differences in the nucleosome pattern of WT and swi3Δ cells without stress were observed, particularly at the nucleosomes −1 and −2, nucleosome positioning in the swi3Δ strain was independent of the presence of CR (Figure 7C). Therefore all these data demonstrate that SWI/SNF complex function mediates the chromatin-remodeling activity necessary for induction of CWI-responsive genes upon cell wall stress.
DISCUSSION
Yeast cells exposed to environmental stress respond through conserved MAPK pathways resulting in rapid transcriptional induction of stress-responsive genes. The regulation of gene transcription in eukaryotic cells involves a dynamic balance between packaging of regulatory sequences into chromatin and access of transcriptional regulators to these sequences (Cairns, 2009). Here we describe a novel connection between MAPK signaling and chromatin remodeling. The combination of gene expression analyses, ChIP, and chromatin-remodeling assays allowed us to establish a model for the sequence of events during transcriptional initiation upon cell wall stress (Figure 8). The MAPK Slt2 mediates, through Rlm1, nucleosome rearrangements at CWI-responsive genes by selective targeting of the SWI/SNF remodeling complex. This process is essential for assembly of the transcription preinitiation complex and proper orchestration of transcriptional adaptive responses.
FIGURE 8:
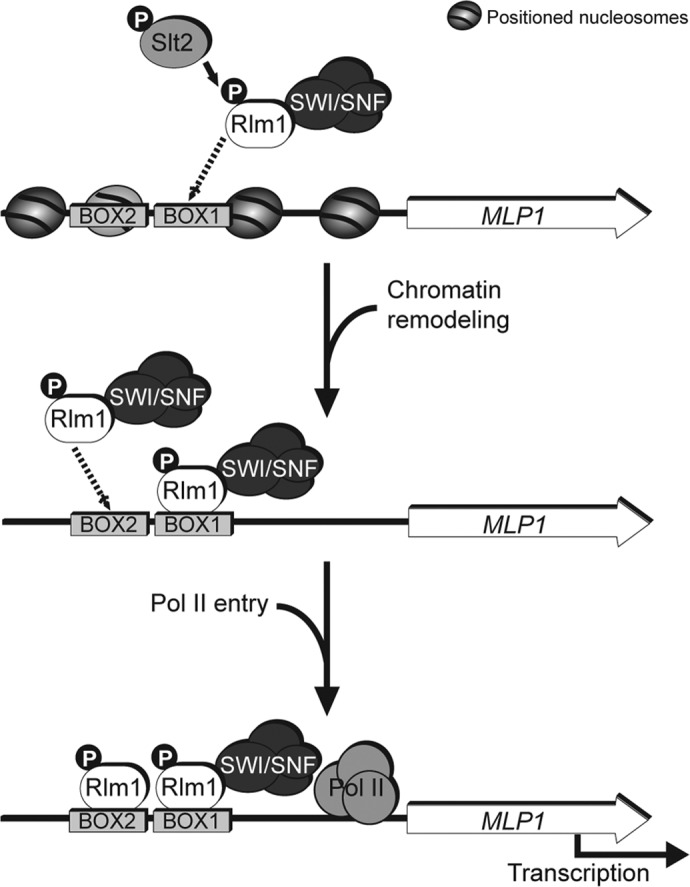
Model for transcriptional activation of CWI-responsive genes. In MLP1, those exposed Rlm1-binding sites would allow transcription factor access to the promoter, whereas occluded binding sites within nucleosomes would not be accessible. Under cell wall stress conditions, Slt2 phosphorylates Rlm1, and the SWI/SNF remodeling complex is targeted to the promoters of CWI-responsive genes via interaction with the activated transcription factor. On recruitment, SWI/SNF locally alters nucleosome positioning at the MLP1 promoter, facilitating the binding of Rlm1 to binding sites previously occluded by positioned nucleosomes in a two-step mechanism. Finally, the binding of Pol II stimulates transcription initiation.
Nucleosome positioning can influence the accessibility of the DNA-binding sites in the promoters to specific transcription factors, and consequently occupancy of such binding sites can be regulated by chromatin-remodeling factors (Li et al., 2007; Jansen and Verstrepen, 2011). The nucleosome-positioning pattern characterized here for MLP1 correlates well with data from multiple high-resolution genome-wide maps of in vivo nucleosomes across the S. cerevisiae genome (Jiang and Pugh, 2009). The MLP1 promoter presents two DNA-binding sites for Rlm1: BOX1 (−359/−350) and BOX2 (−510/−501). The former seems to be exposed in the linker DNA between nucleosomes −3 and −2, or partially exposed at the −2 nucleosome edge, whereas the latter would be completely occluded within nucleosome −3. The exposed site would allow the transcription factor Rlm1 access to the promoter in a first step, but chromatin remodeling would be required under stress to expose the additional nucleosomal site in a two-step model for activation (Figure 8). Such a requirement has also been established at the yeast PHO5 promoter (Fascher et al., 1990; Cairns, 2009). In our ChIP assays, Rlm1 is bound to the promoter of the cell wall–responsive genes in the absence of stress. This basal binding of Rlm1 is independent of Slt2 kinase activity and phosphorylation of the MAPK by Mkk1/Mkk2, suggesting that Rlm1 preloaded into CWI-responsive genes could be inactive. However, this binding seems to be also partially dependent on the presence of Slt2. Because there is some Slt2 MAPK activity under vegetative growth conditions, a small amount of active Rlm1 could be bound to the CWI gene promoters in the absence of stress. In agreement, a very small amount of SWI/SNF also resides on the MLP1 promoter under nonstress conditions, which is dependent on Slt2 and Rlm1.
On cell wall stress, Slt2 interacts with and phosphorylates Rlm1, regulating its transcriptional activation function (Watanabe et al., 1997; Jung et al., 2002). We show here that recruitment of Rlm1 to cell wall stress genes is highly induced by stress and it requires phosphorylation by Slt2 and the activity of SWI/SNF. Because SWI/SNF is targeted to the promoters of the cell stress genes and this recruitment leads to clear alterations of nucleosome architecture, our data favor a model in which chromatin remodeling by the SWI/SNF complex would facilitate the binding of the transcription factor and then stimulate transcriptional initiation by Pol II (Figure 8). Thus massive recruitment of Rlm1 to the MLP1 promoter under cell wall stress requires preliminary action by the SWI/SNF complex. In support of this model, it has been shown that the SWI/SNF complex can facilitate Gal4 binding to occluded sites to promote transcription in vivo by removing nucleosomes that occupy the Gal4-binding site (Burns and Peterson, 1997).
The absence of Rlm1 also abolished the binding of SWI/SNF to the cell wall stress gene promoters. This interdependence between Rlm1 and SWI/SNF indicates a close functional connection between both elements for promoter recruitment. How is the remodeling complex recruited to specific CWI gene promoters? The existence of a direct physical interaction shown here, both in vitro and in vivo, between Rlm1 and SWI/SNF, and the fact that recruitment of SWI/SNF depends on MAPK and Rlm1 favor a model in which the remodeling complex is targeted to the promoters via interaction with the transcription factor Rlm1 (Figure 8). Physical interaction with the specific transcription factor has been shown to be one of the main determinants for the recruitment of the SWI/SNF remodeling complex to different gene promoters (Cosma et al., 1999; Neely et al., 1999; Yudkovsky et al., 1999). The interaction of activated Rlm1 and the remodeling complex could take place in solution, and then those Rlm1 DNA-binding sites that are not occupied by the transcription factor under nonactivating conditions would direct the remodeling complex to the chromatin (Figure 8). Alternatively, Rlm1 prebound to the accessible DNA-binding sites within the promoter could be activated by Slt2, leading to the recruitment of SWI/SNF. The two alternatives do not exclude one another.
The mechanisms of regulation of gene expression proposed here are not restricted to MLP1 but they seem to represent a general mechanism for transcriptional activation of the CWI responsive–genes regulated by Rlm1. Supporting this, we show that 1) the global yeast gene expression profile in response to cell wall stress is severely affected in swi3Δ mutants, 2) the SWI/SNF complex is also recruited to other promoters of CWI-responsive genes, like SRL3 and YLR194C, in a Slt2- and Rlm1-dependent manner, 3) Rlm1 recruitment to these promoters is also dependent on SWI/SNF, and 4) deletion of SWI/SNF elements renders cells hypersensitive to cell wall stress.
As a conclusion, the results presented in this work characterize the SWI/SNF complex as a novel element of the CWI MAPK pathway. Recruitment of this complex to cell wall stress–responsive genes mediates the chromatin remodeling necessary for developing adequate transcriptional responses upon cell wall stress. In addition, several elements of the SAGA histone-modifying complex were also uncovered in the screening developed here. Future work is necessary to define the nature of possible cooperation between the two complexes in the regulation of these processes.
MATERIALS AND METHODS
Yeast strains and growth conditions
Experiments were performed with the S. cerevisiae strain BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and mutant derivatives provided by the European Saccharomyces cerevisiae Archive for Functional Analysis (EUROSCARF; Institute for Molecular Biosciences, Johann Wolfgang Goethe-University Frankfurt, Frankfurt, Germany). snf12Δ and swi1Δ mutants were obtained from the diploid heterozygous disruptants provided by EUROSCARF after sporulation and tetrad analyses using standard yeast genetic techniques. Double mutant snf2Δ rlm1Δ (BY4741, but snf2::KanR rlm1::HIS3) was obtained following a similar approach after the generation of the corresponding diploid heterozygous disruptant. The tagging strains in BY4741 background SJ01 (WT RLM1-3HA::LEU2), SJ02 (slt2Δ RLM1-3HA::LEU2), SJ03 (snf2Δ RLM1-3HA::LEU2), and SJ04 (swi3Δ RLM1-3HA::LEU2) were obtained by integrating RLM1-3xHA by homologous recombination into the RLM1 locus. To this end, the RLM1 open reading frame was PCR amplified and cloned into the integrative LEU2 vector pRS305 containing the 3xHA epitope (pRS305-3xHA; kindly provided by V. J. Cid, Universidad Complutense de Madrid). Integration of the HA-tagged version in the RLM1 locus was achieved by transforming the plasmid previously obtained (pRS305-Rlm1-3HA) linearized at the unique HindIII site. Correct integration was confirmed with a PCR-based strategy.
Cells were grown, depending on the experimental approaches, on YPD (1% yeast extract, 2% peptone, and 2% glucose) or SC medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose) supplemented with the required amino acids. Routinely, cells were grown overnight in liquid media (YPD or SD in the case of cells bearing plasmids) at 220 rpm and 24°C to an optical density of 0.8–1 (A600). The culture was refreshed to 0.2 (A600) in YPD, grown for an additional 2 h, and then divided into two parts. One part was allowed to continue growing under the same conditions (nontreated culture), and the other one was supplemented with CR (30 μg/ml; Merck, Darmstadt, Germany). Cells were collected at the indicated times and processed depending on the experimental approach.
Plasmids
The plasmid YEp352-RLM1 (episomal URA3) was obtained by cloning the insert from plasmid YEp181-RLM1 previously described (Watanabe et al., 1995), containing the RLM1 promoter and coding regions, into a YEp352 shuttle vector. To obtain plasmid YEp352-RLM1-3xHA, a 3xHA epitope from the pRS305-3xHA plasmid was cloned in-frame at the C-terminus of RLM1 present in the YEp352-RLM1 plasmid. A similar strategy was used to construct the episomal LEU2 plasmid YEp181-RLM1-6Myc (a 6xMyc epitope was cloned in-frame at the C-terminus of RLM1 into the plasmid YEp181-RLM1). Plasmid CP233 contains the SNF2-HA-6HIS fusion (Peterson et al., 1994) inserted into the YEp24 shuttle vector (episomal URA3). The complete open reading frame (ORF) of RLM1 or a variant lacking the sequence corresponding to the N-terminal 222 amino acids (RLM1ΔN) was fused to glutathione S-transferase (GST) under the control of the tac promoter in the pGEX-KG vector to obtain pGEX-KG-RLM1 and pGEX-KG-RLM1ΔN, respectively. Plasmids pRS316-SLT2, pRS316-slt2K54F, and pRS316-slt2T190A/Y192A were kindly provided by H. Martín (Universidad Complutense de Madrid).
Screening for yeast mutants defective in MLP1 induction
The screening was developed using the whole collection of haploid deletion strains in all nonessential genes of S. cerevisiae (provided by EUROSCARF). Plasmid pJS05 (Rodríguez-Peña et al., 2008) was transformed in the collection of mutant strains as previously described (Arias et al., 2011), and 5 μl of culture for each transformant was inoculated in 96-well microtiter plates (Nunclon; Nunc, Langenselbold, Germany) containing 95 μl of YPD plus 200 μg/ml nourseothricin (Werner BioAgents, Jena, Germany) and 1 U/ml zymolyase 20T (MP Biomedicals, Solon, OH) per well. These conditions allowed growth of the wild-type strain but inhibited the growth of mutant strains deleted in genes required for cell wall stress–mediated MLP1 induction, such as slt2Δ or sho1Δ. The plates were incubated in a static culture at 28°C for 48–72 h, and growth was determined by measuring absorbance at 550 nm in each well with a Model 680 microplate reader (Bio-Rad, Hercules, CA). Under these conditions, 347 mutants were initially scored in the screening for inhibited cell growth with respect to the WT strain. Positive mutants were transformed individually with the plasmid MLP1-LacZ carrying the promoter region of MLP1 fused to the lacZ gene and further confirmed by measurement of β-galactosidase activity as described (García et al., 2009). In total, 159 mutants were finally confirmed as defective in transcriptional activation of MLP1 by cell wall stress.
Phenotypic analyses
Yeast cells were grown at 24ºC in YPD to mid-log phase, and sensitivity assays to the cell wall perturbing agents CR and zymolyase were performed as previously described (Bermejo et al., 2008).
Microarray experiments
Total RNA isolation and purification was carried out as detailed elsewhere (García et al., 2004). Genome-wide transcriptional profiles were obtained using Affymetrix GeneChip Yeast Genome 2.0 arrays (Affymetrix, Santa Clara, CA). cDNA synthesis and chip hybridization, image analysis, data processing, and statistical analysis were basically carried out as described (Arroyo et al., 2011). The files generated from the scanning (.CEL) were converted to gene expression signals using the RMA algorithm included in Affymetrix Expression Console. For each experimental condition, three microarray experiments corresponding to three independent biological samples were processed and analyzed. Fold changes between experimental conditions under comparison were calculated as a quotient between the mean of the gene expression signals. The genes were considered to be up- or down-regulated when their expression ratio under the conditions tested was ≥2 or ≤0.5, respectively. Statistical analysis was performed with Student's t test. Those values with p < 0.05 were considered as significant and the corresponding genes considered for further analysis.
To determine whether the gene induction observed in the WT strain after CR treatment (30 μg/ml, 4 h) was significantly reduced in swi3Δ and rlm1Δ mutant strains, we used the relationship between the responses of each mutant (mutant ratio) versus those of the WT strain (WT ratio). Thus a value of mutant ratio/WT ratio of 0.65 was considered the threshold for defining a significant reduction in gene induction (García et al., 2009). In any case, genes whose mutant ratio was <1.6 were deemed as not being up-regulated. The same threshold for the relationship between mutant/WT versus WT ratio was used to determine genes whose basal expression levels was dependent on SWI3 and RLM1. Clustering analysis was performed using the MeV MultiExperiment Viewer, version 4.6 (Saeed et al., 2006).
The microarray data described here follow the minimum information about a microarray experiment recommendations and have been deposited at the National Center for Biotechnology Information gene expression and hybridization array data repository with accession number GSE31176.
Quantitative real-time PCR assays
Real-time quantitative RT-PCR (RT-qPCR) assays were performed as detailed in García et al. (2004). For quantification, the abundance of each gene was determined relative to the standard transcript of ACT1 for input cDNA normalization, and the final data on relative gene expression between the conditions tested were calculated following the 2−∆∆Ct method (Livak and Schmittgen, 2001). Primer sequences are available upon request.
Western blotting assays
The procedures used for immunoblot analyses, including cell collection and lysis, collection of proteins, fractionation by SDS–PAGE, and transfer to nitrocellulose membranes, have been described previously (Bermejo et al., 2008).
Chromatin immunoprecipitation assays
ChIP was performed as described elsewhere (Papamichos-Chronakis and Peterson, 2008). The antibodies used in these experiments were as follows: polyclonal anti-HA (ab9110; Abcam, Cambridge, MA), polyclonal anti-H3 (ab1791; Abcam), monoclonal anti-Pol II (8WG16; Covance, Berkeley, CA), and polyclonal anti-Snf2 and anti-Snf6 antibodies kindly provided by J. Reese (Pennsylvania State University, University Park, PA). The immunoprecipitated DNA was quantified by qPCR using the following primers, with locations indicated by the distance from the respective ATG initiation codon: MLP1BOX2 (RLM1-binding domain at the promoter), −548/−408; MLP1BOX1 (RLM1-binding domain at the promoter), −453/−313; MLP1PRO1 (promoter), −253/−98; MLP1PRO2 (promoter/ORF), −143/+56; MLP1ORF1 (ORF), +98/+250; MLP1ORF2 (ORF), +555/+687; YLR194C (promoter), −254/−123; SRL3 (promoter), −265/−135; and VMA8 (promoter), −321/−191. The fold enrichment (FE) at specific DNA regions was calculated using the comparative Ct method (Aparicio et al., 2004) and the promoter region of the VMA8 gene as sequence control. Thus the Ct of the input sample was subtracted from the Ct of the immunoprecipitated sample to calculate the ∆Ct, both in the control sequence (ΔCtcont) and in the target DNA (ΔCtexp), for each condition. Finally, FE was calculated by using the formula FE = 2−(∆Ctexp−∆Ctcont).Data represent the mean and SD of at least three independent experiments.
Nucleosome-scanning analysis
MNase digestion of chromatin, protein degradation, and DNA purification were basically performed as previously described (Liu et al., 2005). The amount of MNase to yield >80% mononucleosomal DNA was determined in initial titrations. Eventually, 40 and 20 U of micrococcal nuclease (Worthington Biochemical, Lakewood, NJ) were used for the WT and swi3Δ strains, respectively. Control samples of genomic DNA were obtained as described (Sekinger et al., 2005), but using 0, 1.2, 1.4, and 1.6 U of MNase. Genomic DNA samples were Qiagen column purified (Qiagen, Valencia, CA). Genomic DNA and MNase-treated chromatin were electrophoretically separated on a 1.5% agarose gel, and mononucleosome-sized (140–220 base pairs) fragments were excised from the gel and purified. Purified DNA samples were analyzed by qPCR using a set of 11 overlapping primer pairs, each of which generate 100 ± 8–base pair PCR products that overlap 30 ± 10 base pairs along the MLP1 promoter region (from −699 to +161). The nucleosomal DNA enrichment level of a given DNA region was calculated as the ratio between the amounts of PCR product obtained from the problem samples to that of the genomic DNA (control samples).
Purification of SWI/SNF complex and GST fusion proteins
Yeast SWI/SNF complex was purified as described (Yang et al., 2007). For GST-fusion protein purification, Escherichia coli BL21 (Rosetta) cells transformed with the plasmid pGEX-KG, pGEX-KG-RLM1, or pGEX-KG-RLM1ΔN were grown at 30°C in Luria Bertani (LB) medium supplemented with 0.2 mM isopropyl-β-d-thiogalactoside for 5 h to induce protein expression. Cells were collected by centrifugation, washed with cold phosphate-buffered saline (PBS), and resuspended in PBS supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). Then, cells were broken by sonication, and after centrifugation at 15,000 rpm for 1 h at 4°C, the supernatant was taken. Finally, 250 μl of glutathione–Sepharose 4B beads (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) were added to the supernatant and mixed for 2 h at 4°C and subsequently washed three times with cold PBS.
In vitro coprecipitation assay
A 0.5-pmol amount of the SWI/SNF complex was mixed with 20 μl of beads containing GST, GST-RLM1, or GST-RLM1ΔN fusion. Each reaction was carried out in 200 μl of binding buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 2 MgCl2, 10% glycerol, 100 μg/ml bovine serum albumin, and protease inhibitors [2 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, and 1 mM PMSF]). After incubation for 2 h at 4°C, beads were extensively washed with binding buffer and boiled in 2× SDS loading buffer. Bound proteins were analyzed by Western blotting using anti-GST (Z-5) sc-459 (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-Snf2 polyclonal antibodies.
In vivo coprecipitation assay
The double mutant rlm1Δ snf2Δ was cotransformed with episomal plasmids CP233 and YEp181-RLM1-6Myc expressing the HA- and Myc-tagged Snf2 and Rlm1 protein, respectively, or with YEp181-RLM1-6Myc and the empty vector YEp352. These cells were grown overnight at 24°C in SC-Ura-Leu medium. To study the in vivo interaction between Snf2 and Rlm1, we performed a formaldehyde-based cross-linking procedure as previously described (Hall and Struhl, 2002), but that cross-linking step was quenched by addition of 150 mM glycine. Cleared supernatant from cell lysates was incubated with anti-HA monoclonal antibody (16B12 MMS-101P; Covance) overnight at 4°C. Next, rProtein A–Sepharose Fast Flow beads (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) were added and incubated for 2 h at 4°C. Immunoprecipitated material was analyzed by Western blotting using anti-HA (16B12 MMS-101P; Covance) or anti-Myc (9E10 MMS-150P; Covance) monoclonal antibodies.
Supplementary Material
Acknowledgments
This work was supported by Grants BIO2010-22146 (Ministerio de Ciencia e Innovacion, Spain), GR58/08 (Ref. 920640/Universidad Complutense de Madrid), S2010/BDM-2414, and 06-RNP-132 (European Science Foundation) to J.A. and a grant from the National Institutes of Health (GM49650) to C.L.P. A.B.S. is the recipient of a Formación del Profesorado Universitario Ph.D. Fellowship (Ministerio de Educación, Cultura y Deporte, Spain). Support for the Special Chair in Genomics and Proteomics (held by C.N.) is gratefully acknowledged. We thank P. Botías and R. Pérez-Díaz (Unidad de Genómica, Universidad Complutense de Madrid) for assistance with the DNA microarray experiments and qPCR, respectively; H. Martín and V. J. Cid for kindly providing plasmids; and J. Reese for α-Snf2 and α-Snf6 antibodies. We also thank G. Bennett and B. Manning for help with ChIP and SWI/SNF purification and M. Molina and H. Martín for critical reading of the manuscript. All members of the Research Unit 4 at the Department of Microbiology II are acknowledged for their support.
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- CR
Congo red
- CWI
cell wall integrity
- MAPK
mitogen-activated proten kinase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0278) on May 23, 2012.
REFERENCES
- Adkins MW, Williams SK, Linger J, Tyler JK. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol Cell Biol. 2007;27:6372–6382. doi: 10.1128/MCB.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004 doi: 10.1002/0471143030.cb1707s23. Chapter 17, Unit 17.7. [DOI] [PubMed] [Google Scholar]
- Arias P, Díez-Muñiz S, García R, Nombela C, Rodríguez-Peña JM, Arroyo J. Genome-wide survey of yeast mutations leading to activation of the yeast cell integrity MAPK pathway: novel insights into diverse MAPK outcomes. BMC Genomics. 2011;12:390–407. doi: 10.1186/1471-2164-12-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J, Hutzler J, Bermejo C, Ragni E, García-Cantalejo J, Botías P, Piberger H, Schott A, Sanz AB, Strahl S. Functional and genomic analyses of blocked protein O-mannosylation in baker's yeast. Mol Microbiol. 2011;79:1529–1546. doi: 10.1111/j.1365-2958.2011.07537.x. [DOI] [PubMed] [Google Scholar]
- Baetz K, Moffat J, Haynes J, Chang M, Andrews B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol Cell Biol. 2001;21:6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo C, Rodríguez E, García R, Rodríguez-Peña JM, Rodríguez de la Concepción ML, Rivas C, Arias P, Nombela C, Posas F, Arroyo J. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddick RK, Law GL, Young ET. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS One. 2008;3:e1436. doi: 10.1371/journal.pone.0001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns LG, Peterson CL. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Dodou E, Treisman R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkina TY, Tschetter PA, Erkine AM. Different requirements of the SWI/SNF complex for robust nucleosome displacement at promoters of heat shock factor and Msn2- and Msn4-regulated heat shock genes. Mol Cell Biol. 2008;28:1207–1217. doi: 10.1128/MCB.01069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fascher KD, Schmitz J, Horz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García R, Bermejo C, Grau C, Pérez R, Rodríguez-Peña JM, Francois J, Nombela C, Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J Biol Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- García R, Rodríguez-Peña JM, Bermejo C, Nombela C, Arroyo J. The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J Biol Chem. 2009;284:10901–10911. doi: 10.1074/jbc.M808693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J Biol Chem. 2002;277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- Jansen A, Verstrepen KJ. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2011;75:301–320. doi: 10.1128/MMBR.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- Jung US, Sobering AK, Romeo MJ, Levin DE. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol. 2002;46:781–789. doi: 10.1046/j.1365-2958.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- Kasten MM, Clapier CR, Cairns BR. SnapShot: chromatin remodeling: SWI/SNF. Cell. 2011;144:310. doi: 10.1016/j.cell.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Kim KY, Levin DE. Mpk1 MAPK association with the paf1 complex blocks sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Truman AW, Levin DE. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol Cell Biol. 2008;28:2579–2589. doi: 10.1128/MCB.01795-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagorce A, Hauser NC, Labourdette D, Rodríguez C, Martin-Yken H, Arroyo J, Hoheisel JD, Francois J. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J Biol Chem. 2003;278:20345–20357. doi: 10.1074/jbc.M211604200. [DOI] [PubMed] [Google Scholar]
- Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mas G, de Nadal E, Dechant R, Rodríguez de la Concepción ML, Logie C, Jimeno-González S, Chavez S, Ammerer G, Posas F. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J. 2009;28:326–336. doi: 10.1038/emboj.2008.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Peña JM, Díez-Muñiz S, Nombela C, Arroyo J. A yeast strain biosensor to detect cell wall-perturbing agents. J Biotechnol. 2008;133:311–317. doi: 10.1016/j.jbiotec.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Iyer VR. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol. 2008;28:2221–2234. doi: 10.1128/MCB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol. 2003;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Perlin JR, Leonelli L, Allis CD, Coonrod SA. Linking covalent histone modifications to epigenetics: the rigidity and plasticity of the marks. Cold Spring Harb Symp Quant Biol. 2004;69:161–169. doi: 10.1101/sqb.2004.69.161. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat Struct Mol Biol. 2007;14:540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.