Abstract
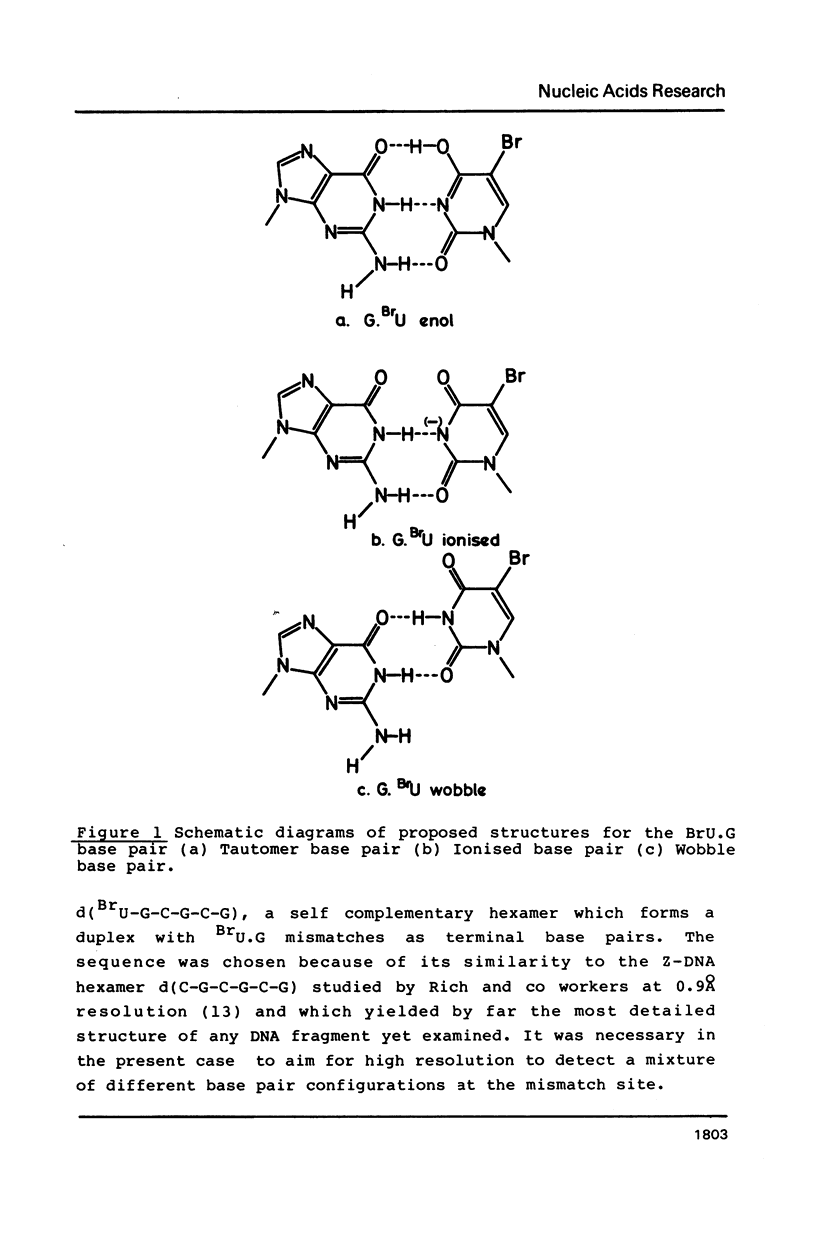
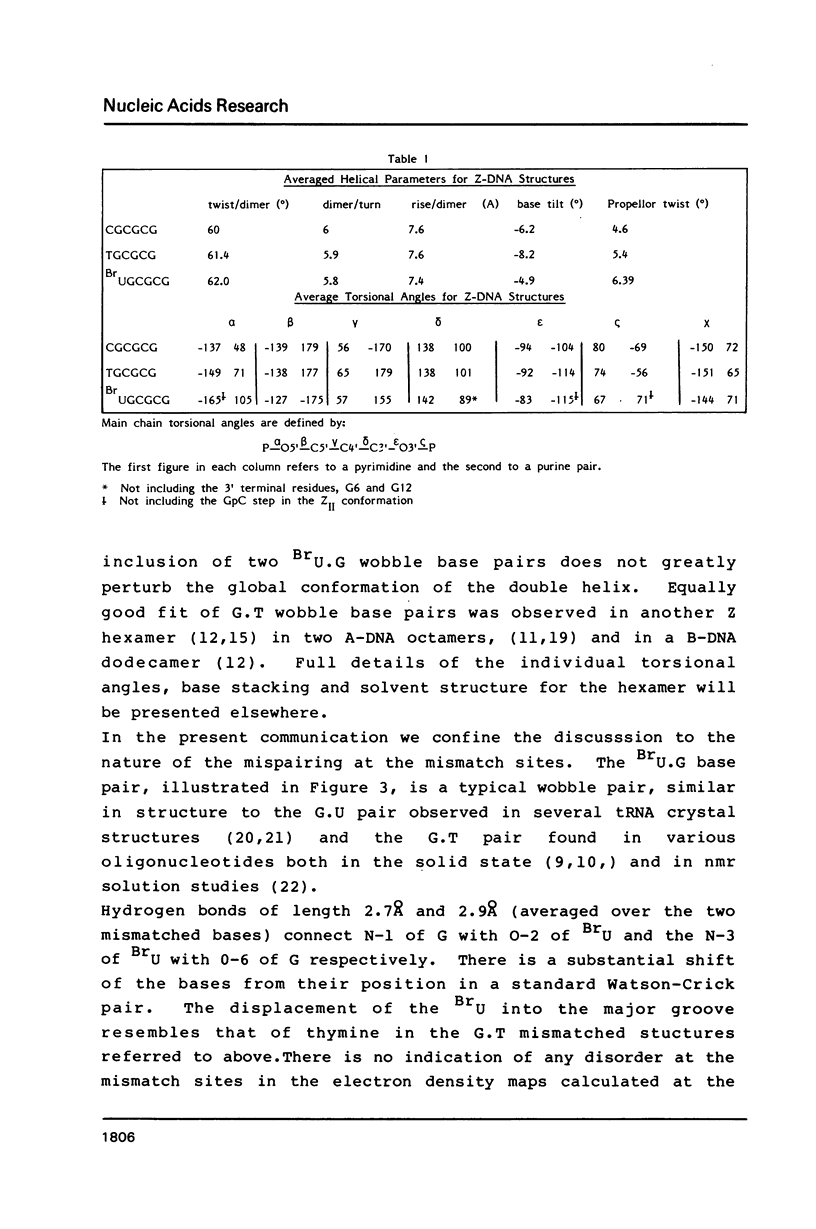
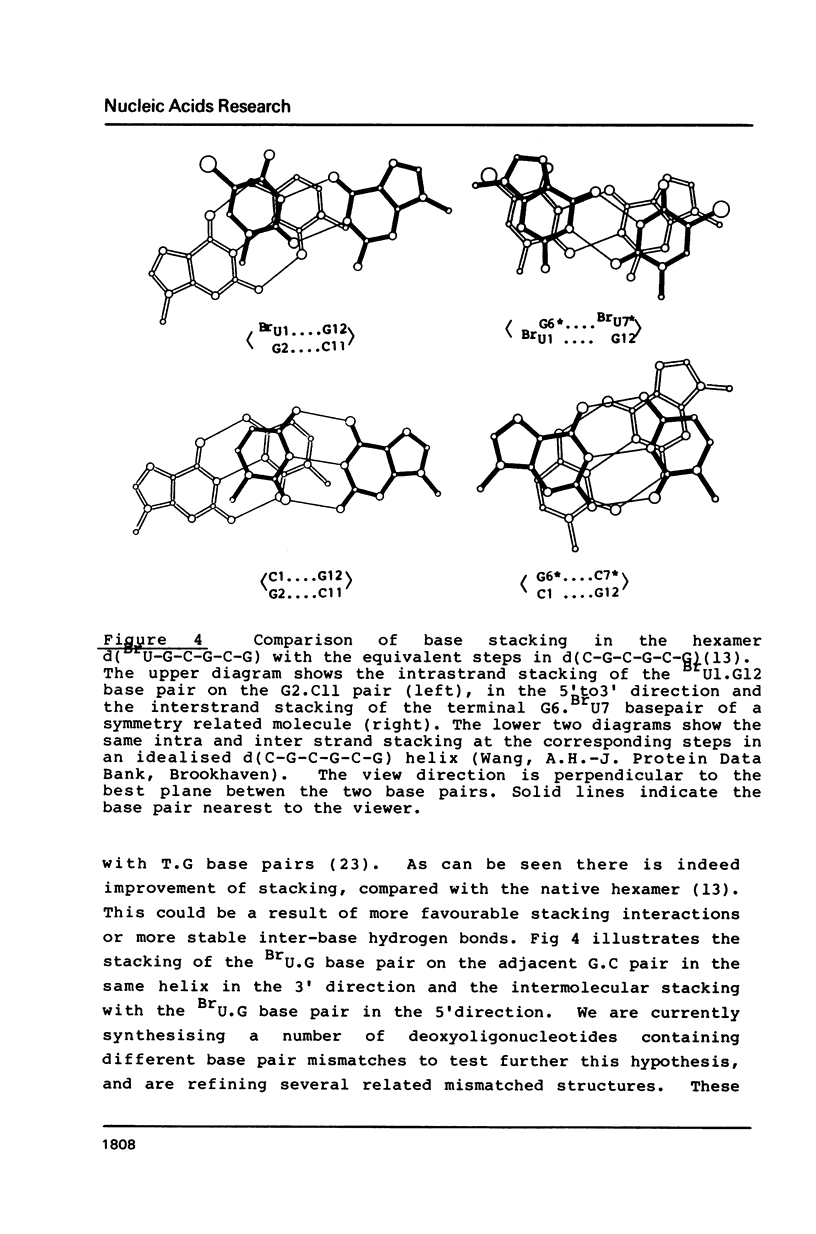
The deoxyoligonucleotide d(BrU-G-C-G-C-G) was crystallised at pH 8.2 and its structure analysed by X-ray diffraction. The unit cell, of dimensions a = 17.94, b = 30.85, c = 49.94A contains four DNA duplexes in space group P2(1)2(1)2(1). The duplexes are in the Z conformation, with four Watson-Crick G.C base pairs and two BrU.G base pairs. The structure was refined to an R factor of 0.16 at a resolution of 2.2A with 64 solvent molecules located. The BrU.G base pair mismatch is of the wobble type, with both bases in the major tautomer form and hydrogen bonds linking 0-2 of BrU with N-1 of G and N3 of BrU with 0-6 of G. There is no indication of the presence of ionised base pairs, in spite of the high pH of crystallisation. The results are discussed in terms of the mutagenic properties of 5- bromouracil.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown T., Kennard O., Kneale G., Rabinovich D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature. 1985 Jun 13;315(6020):604–606. doi: 10.1038/315604a0. [DOI] [PubMed] [Google Scholar]
- Gait M. J., Matthes H. W., Singh M., Sproat B. S., Titmas R. C. Rapid synthesis of oligodeoxyribonucleotides. VII. Solid phase synthesis of oligodeoxyribonucleotides by a continuous flow phosphotriester method on a kieselguhr-polyamide support. Nucleic Acids Res. 1982 Oct 25;10(20):6243–6254. doi: 10.1093/nar/10.20.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R. L., Goodman M. F. Deoxyribonucleotide pools, base pairing, and sequence configuration affecting bromodeoxyuridine- and 2-aminopurine-induced mutagenesis. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1801–1805. doi: 10.1073/pnas.77.4.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- LAWLEY P. D., BROOKES P. Ionization of DNA bases or base analogues as a possible explanation of mutagenesis, with special reference to 5-bromodeoxyuridine. J Mol Biol. 1962 Mar;4:216–219. doi: 10.1016/s0022-2836(62)80053-9. [DOI] [PubMed] [Google Scholar]
- Lasken R. S., Goodman M. F. A fidelity assay using "dideoxy" DNA sequencing: a measurement of sequence dependence and frequency of forming 5-bromouracil X guanine base mispairs. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1301–1305. doi: 10.1073/pnas.82.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken R. S., Goodman M. F. The biochemical basis of 5-bromouracil-induced mutagenesis. Heteroduplex base mispairs involving bromouracil in G x C----A x T and A x T----G x C mutational pathways. J Biol Chem. 1984 Sep 25;259(18):11491–11495. [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984 Aug;43(11):2663–2670. [PubMed] [Google Scholar]
- Skopek T. R., Hutchinson F. DNA base sequence changes induced by bromouracil mutagenesis of lambda phage. J Mol Biol. 1982 Jul 25;159(1):19–33. doi: 10.1016/0022-2836(82)90029-8. [DOI] [PubMed] [Google Scholar]
- Sternglanz H., Bugg C. E. Relationship between the mutagenic and base-stacking properties of halogenated uracil derivatives. The crystal structures of 5-chloro- and 5-bromouracil. Biochim Biophys Acta. 1975 Jan 6;378(1):1–11. doi: 10.1016/0005-2787(75)90130-6. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]


