Abstract
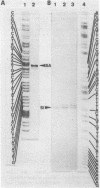
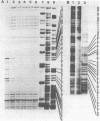
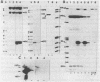
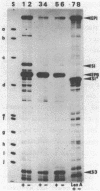
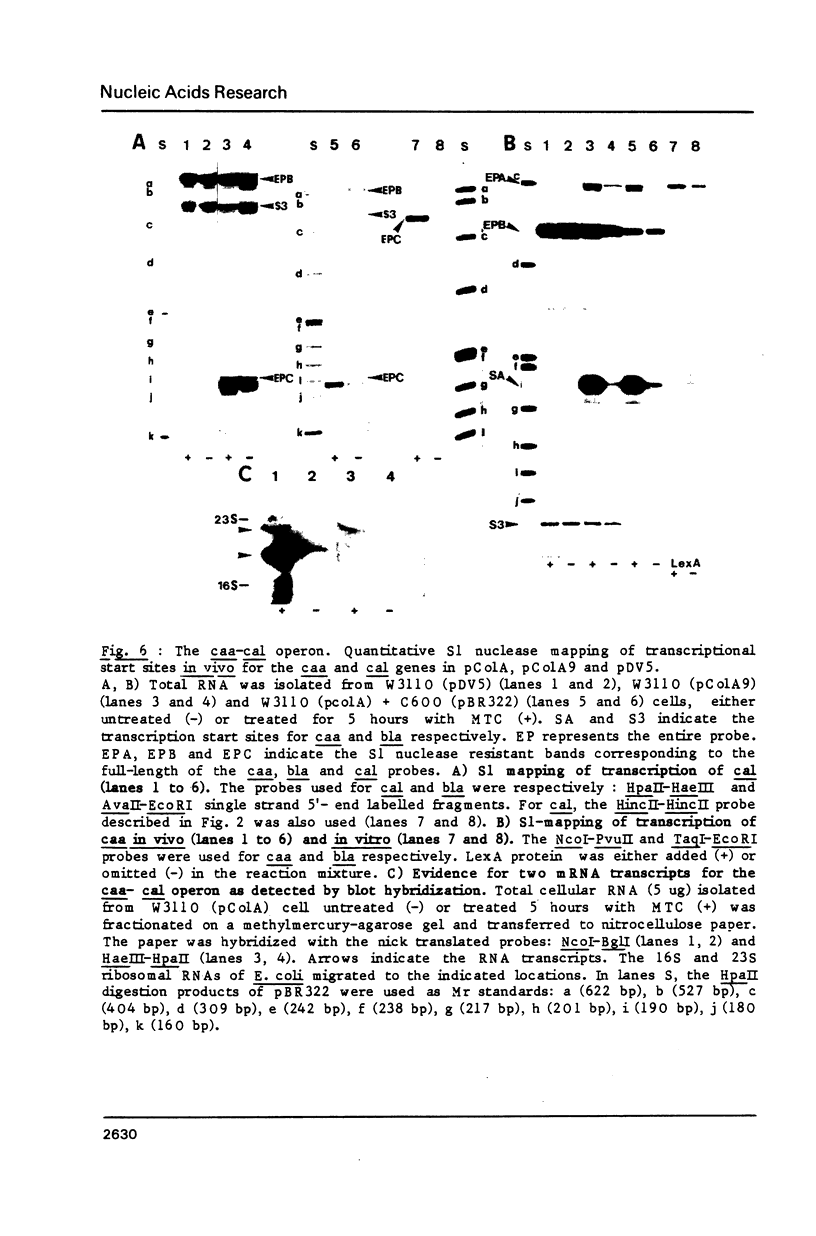
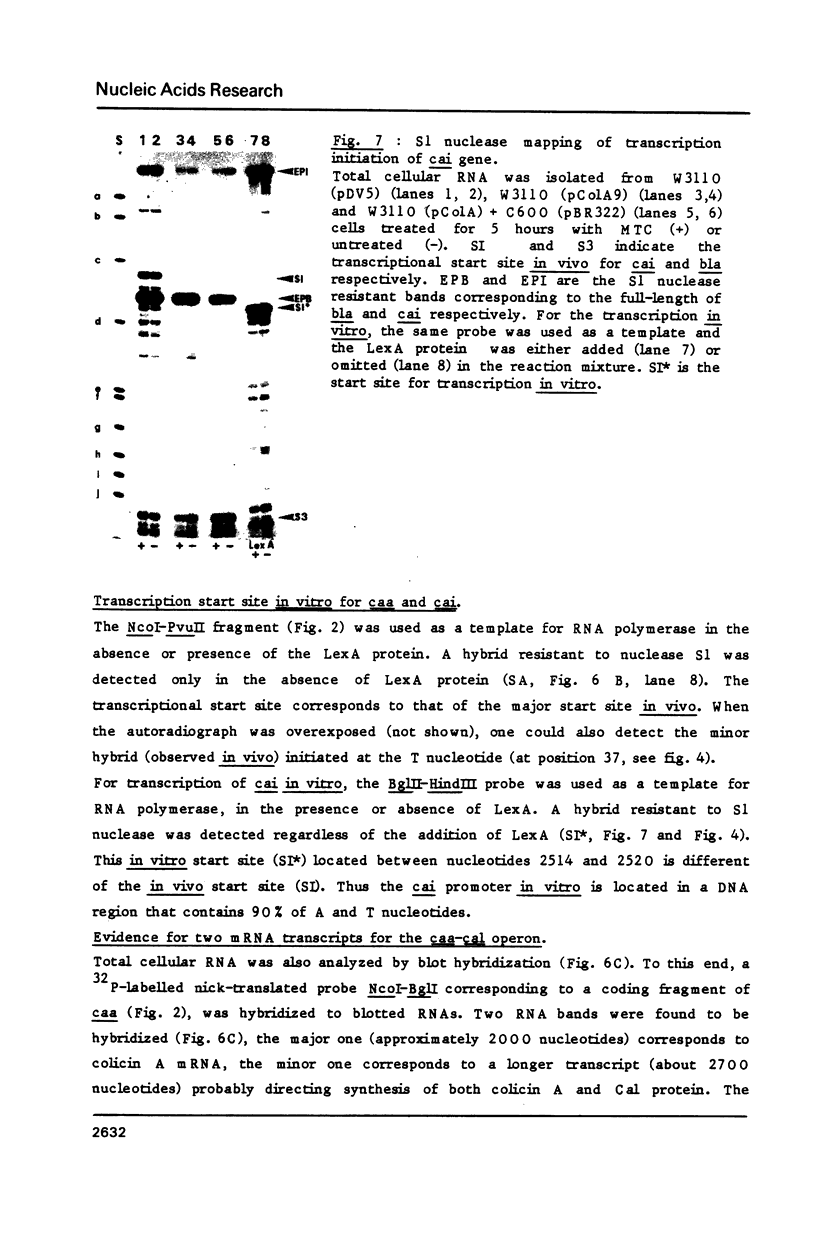
The initiation sites of transcription in vivo for the three genes caa, cai and cal encoding respectively colicin A (Caa), the immunity protein (Cai) and the pColA lysis protein (Cal) have been analysed by nuclease S1 mapping. This analysis demonstrates that caa and cal form an operon. cai is located between these two genes and transcribed in the opposite direction from its own promoter. The start sites for caa and cai have also been determined in vitro. For caa, the same start site was found in vivo and in vitro. In contrast, for cai the most efficient start site in vitro was not used in vivo. LexA protein strongly repressed the in vivo and in vitro transcription of the caa-cal operon. As determined by DNase 1 protection experiments, LexA protein binds with a high affinity to an approximately 40 bp long sequence just downstream of the Pribnow box. The sequence of the binding site is composed of two overlapped "SOS boxes". Two transcripts of the caa-cal operon were detected by blot hybridization. The longer mRNA can direct the synthesis of both Caa and Cal while the shorter one is terminated at the end of caa. When the transcription of the caa-cal operon is induced, there is a strong interference with cai transcription.
Full text
PDF



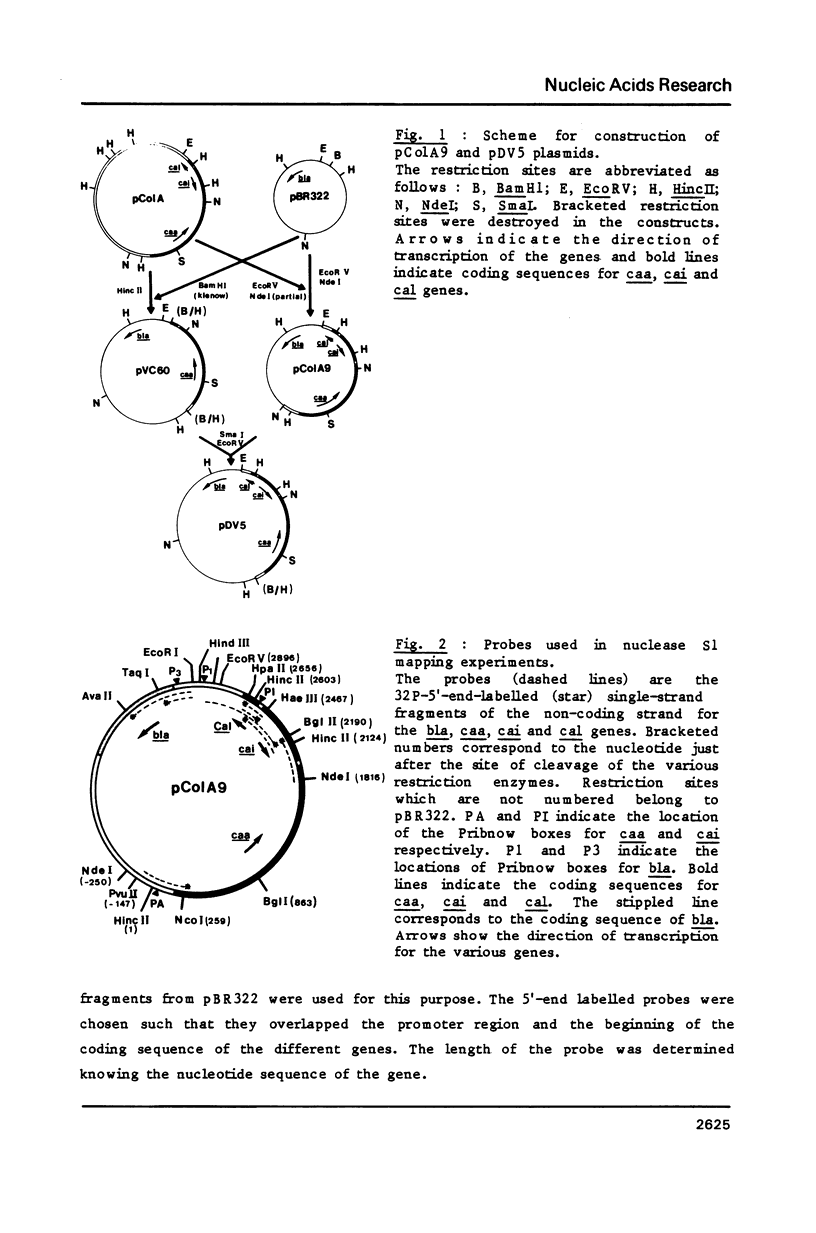
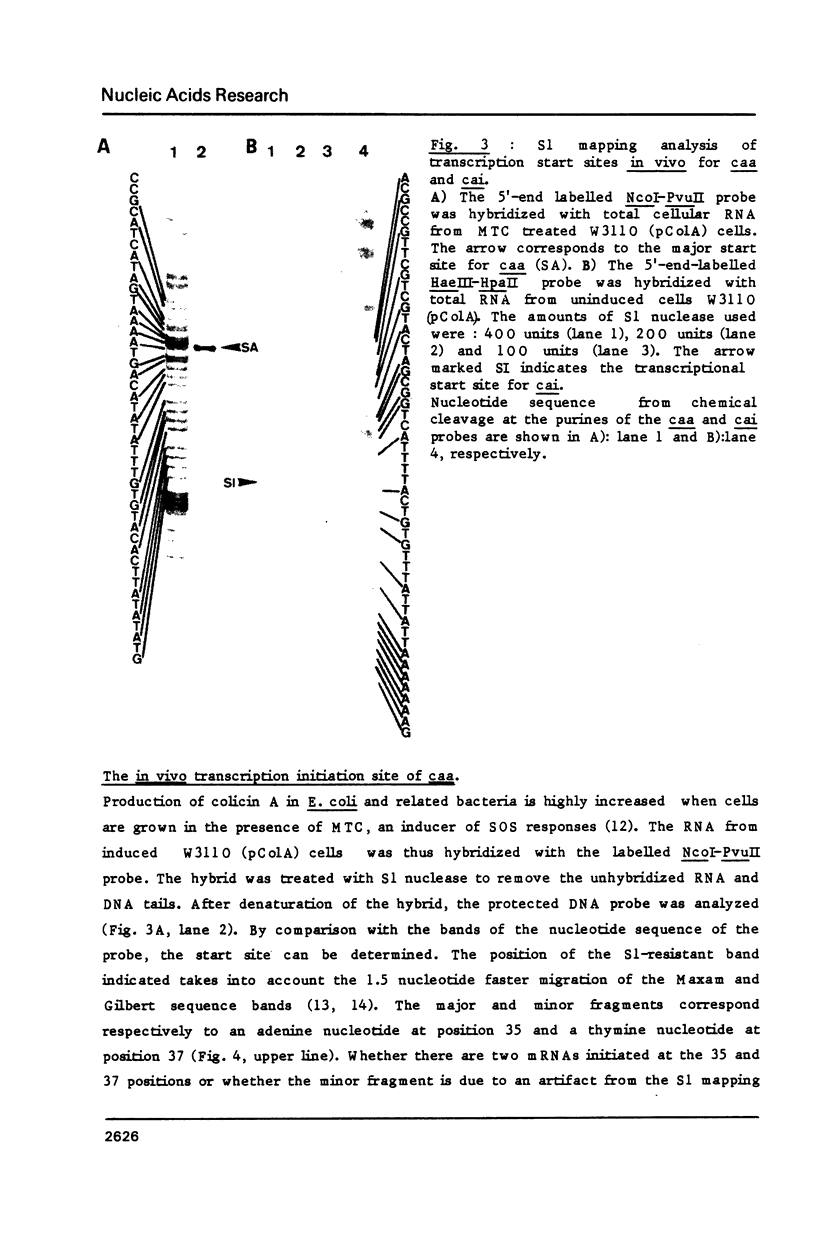
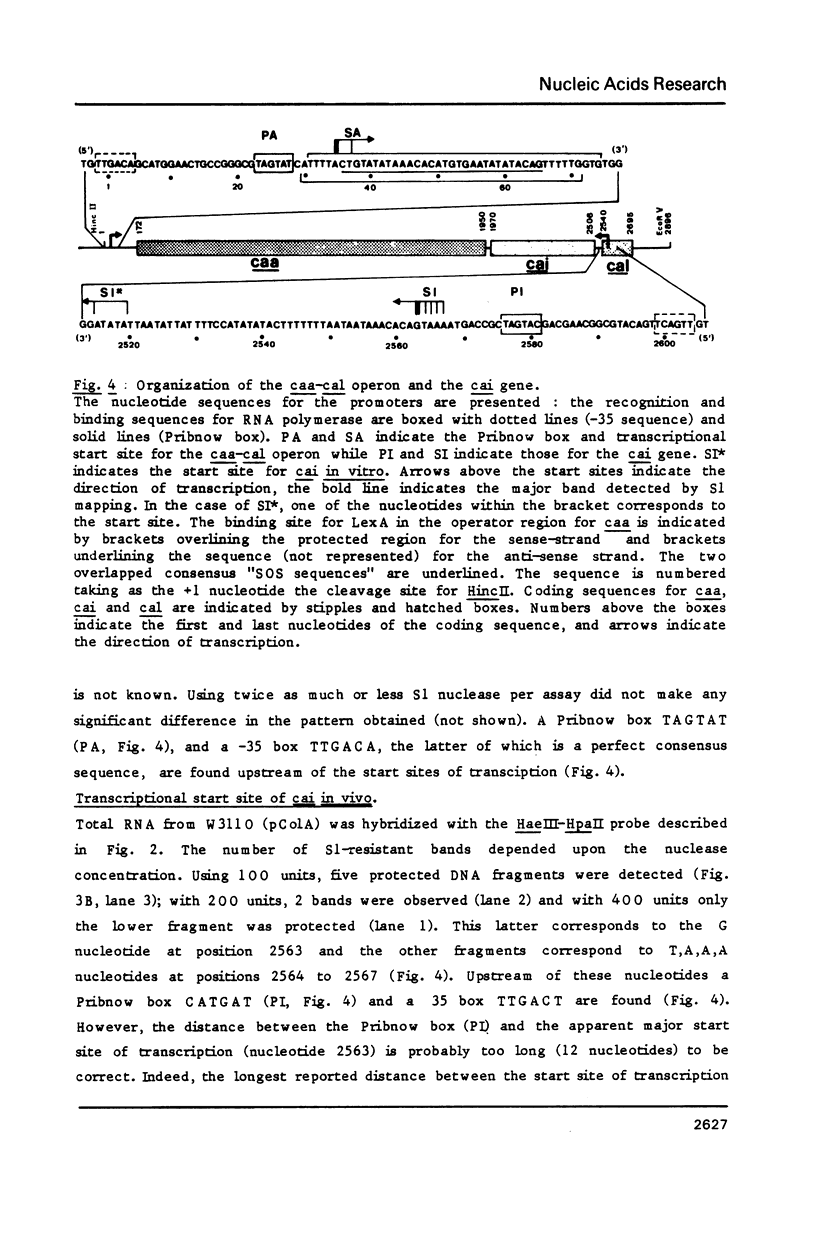
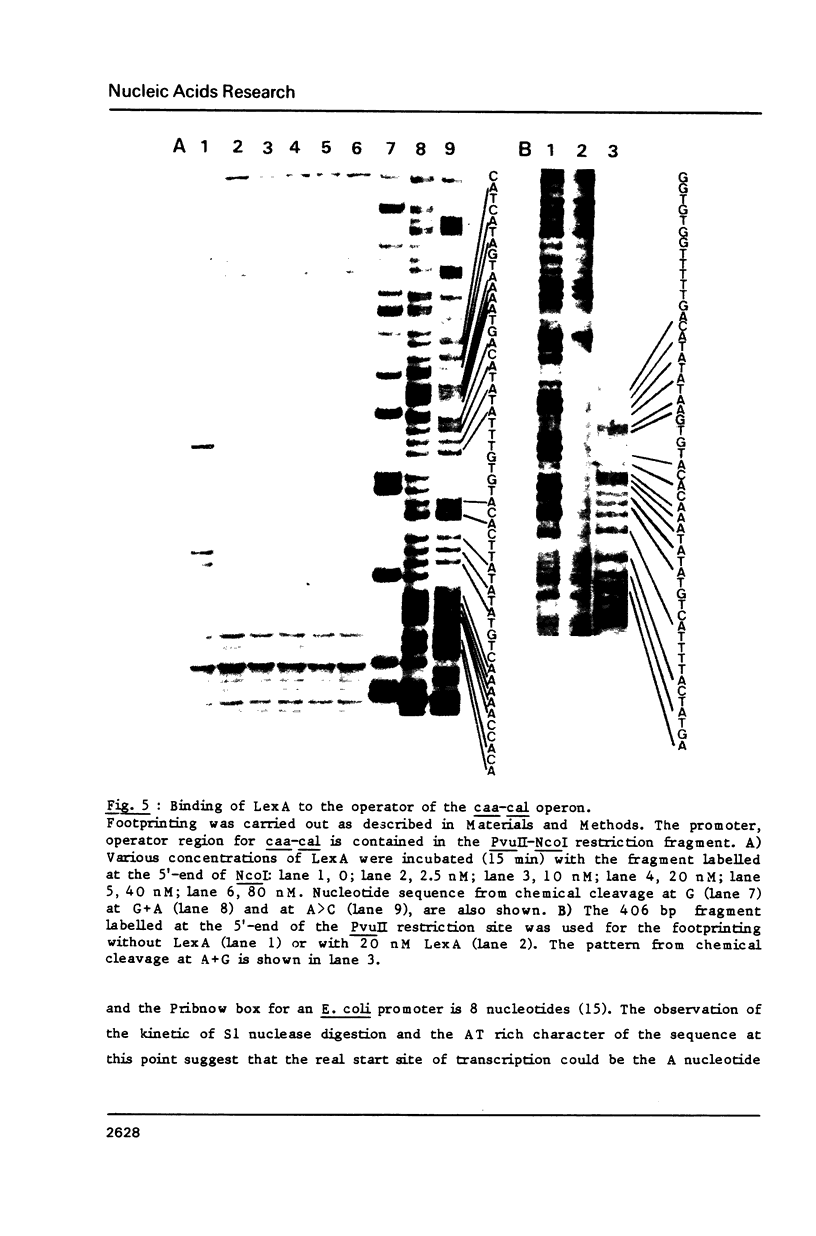






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Cavard D., Lloubès R., Morlon J., Chartier M., Lazdunski C. Lysis protein encoded by plasmid ColA-CA31. Gene sequence and export. Mol Gen Genet. 1985;199(1):95–100. doi: 10.1007/BF00327516. [DOI] [PubMed] [Google Scholar]
- Crozel V., Lazdunski C., Cavard D. Localization of genes responsible for replication and immunity to colicin A on plasmid ColA-CA31. Mol Gen Genet. 1983;192(3):500–505. doi: 10.1007/BF00392196. [DOI] [PubMed] [Google Scholar]
- Ebina Y., Nakazawa A. Cyclic AMP-dependent initiation and rho-dependent termination of colicin E1 gene transcription. J Biol Chem. 1983 Jun 10;258(11):7072–7078. [PubMed] [Google Scholar]
- Ebina Y., Takahara Y., Kishi F., Nakazawa A., Brent R. LexA protein is a repressor of the colicin E1 gene. J Biol Chem. 1983 Nov 10;258(21):13258–13261. [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloubes R. P., Chartier M. J., Journet A. M., Varenne S. G., Lazdunski C. J. Nucleotide sequence of the gene for the immunity protein to colicin A. Analysis of codon usage of immunity proteins as compared to colicins. Eur J Biochem. 1984 Oct 1;144(1):73–78. doi: 10.1111/j.1432-1033.1984.tb08432.x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morlon J., Cavard D., Lazdunski C. Physical map of pColA-CA31, an amplifiable plasmid, and location of colicin A structural gene. Gene. 1982 Mar;17(3):317–321. doi: 10.1016/0378-1119(82)90148-2. [DOI] [PubMed] [Google Scholar]
- Morlon J., Lloubes R., Chartier M., Bonicel J., Lazdunski C. Nucleotide sequence of promoter, operator and amino-terminal region of caa, the structural gene of colicin A. EMBO J. 1983;2(5):787–789. doi: 10.1002/j.1460-2075.1983.tb01501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon J., Lloubès R., Varenne S., Chartier M., Lazdunski C. Complete nucleotide sequence of the structural gene for colicin A, a gene translated at non-uniform rate. J Mol Biol. 1983 Oct 25;170(2):271–285. doi: 10.1016/s0022-2836(83)80148-x. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Shirabe K., Ebina Y., Miki T., Nakazawa T., Nakazawa A. Positive regulation of the colicin E1 gene by cyclic AMP and cyclic AMP receptor protein. Nucleic Acids Res. 1985 Jul 11;13(13):4687–4698. doi: 10.1093/nar/13.13.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Tapper D. P., Clayton D. A. Altered mobility of polydeoxyribonucleotides in high resolution polyacrylamide gels due to removal of terminal phosphates. Nucleic Acids Res. 1981 Dec 21;9(24):6787–6794. doi: 10.1093/nar/9.24.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S., Knibiehler M., Cavard D., Morlon J., Lazdunski C. Variable rate of polypeptide chain elongation for colicins A, E2 and E3. J Mol Biol. 1982 Jul 25;159(1):57–70. doi: 10.1016/0022-2836(82)90031-6. [DOI] [PubMed] [Google Scholar]
- Ward D. F., Murray N. E. Convergent transcription in bacteriophage lambda: interference with gene expression. J Mol Biol. 1979 Sep 15;133(2):249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Matthes H., Wintzerith M., Chambon P. Transcription from the SV40 early-early and late-early overlapping promoters in the absence of DNA replication. EMBO J. 1983;2(9):1605–1611. doi: 10.1002/j.1460-2075.1983.tb01631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman K. F., Mount D. W. Nucleotide sequence binding specificity of the LexA repressor of Escherichia coli K-12. J Bacteriol. 1985 Jul;163(1):376–384. doi: 10.1128/jb.163.1.376-384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]