Abstract
Uncontrolled proliferation is one of the hallmarks of breast cancer. We have previously identified the human Ecd protein (human ortholog of Drosophila Ecdysoneless, hereafter called Ecd) as a novel promoter of mammalian cell cycle progression, a function related to its ability to remove the repressive effects of Rb-family tumor suppressors on E2F transcription factors. Given the frequent dysregulation of cell cycle regulatory components in human cancer, we used immunohistochemistry of paraffin-embedded tissues to examine Ecd expression in normal breast tissue versus tissues representing increasing breast cancer progression. Initial studies of a smaller cohort without outcomes information showed that Ecd expression was barely detectable in normal breast tissue and in hyperplasia of breast, but high levels of Ecd were detected in benign breast hyperplasia, ductal carcinoma in situ (DCIS) and infiltrating ductal carcinoma (IDCs) of the breast. In this cohort of 104 IDC patients, Ecd expression levels showed a positive correlation with higher grade (P = 0.04). Further analyses of Ecd expression using a larger, independent cohort (954) confirmed these results, with a strong positive correlation of elevated Ecd expression with higher histological grade (P = 0.013), mitotic index (P = 0.032), and Nottingham Prognostic Index score (P = 0.014). Ecd expression was positively associated with HER2/neu (P = 0.002) overexpression, a known marker of poor prognosis in breast cancer. Significantly, increased Ecd expression showed a strong positive association with shorter breast cancer specific survival (BCSS) (P = 0.008) and disease-free survival (DFS) (P = 0.003) in HER2/neu overexpressing patients. Taken together, our results reveal Ecd as a novel marker for breast cancer progression and show that levels of Ecd expression predict poorer survival in Her2/neu overexpressing breast cancer patients.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-011-1946-8) contains supplementary material, which is available to authorized users.
Keywords: Ecdysoneless, Cell cycle, Breast cancer, HER2/neu, DCIS, IDC
Introduction
Breast cancer still remains the most frequent cancer of women with nearly a million new cases worldwide each year with about 400,000 deaths annually [1]. Delineating the molecular pathways that contribute to aggressive behavior of human breast cancers to identify newer prognostic markers and therapeutic targets is therefore a critical research priority. The most important prognostic factor in breast cancer remains the lymph node status, which strongly correlates with disease-free and overall survival. Additional markers that have now become linked to molecular classification of breast cancer subtypes include the expression of hormone receptors (ER and PR) that predicts response to endocrine therapy and overexpression of HER2/neu status that predicts response to anti-HER2/neu therapy with Trastuzumab. Concurrently, ER +/PR + tumors have a more favorable prognosis while HER2/neu overexpression signifies a markedly worse prognosis. Absence of ER, PR and HER2/neu in the so called triple-negative breast cancers is also associated with a poor prognosis. Identification of newer molecular pathways important in oncogenesis is therefore expected to provide additional useful prognostic and predictive markers to help in the selection of appropriate targeted therapies and could provide new therapeutic targets.
Uncontrolled proliferation is a hallmark of cancer and it has now become clear that major drivers of breast cancer oncogenesis enhance the expression and/or activity of cell cycle progression-associated genes [2]. Extensive research over the past two decades has led to our current model of how quiescent cells enter cell cycle [3, 4]. E2F family transcription factors play a critical role to turn on the expression of a large set of genes required for cell cycle progression [5]. These transcription factors are held in a repressive complex by their association with hypophosphorylated form of the Retinoblastoma (Rb) protein family members (Rb/p105, p107 and Rb2/p130) [6]. Phosphorylation of Rb proteins by cell cycle-associated cyclin-dependent kinases (CDKs) [7–9] helps dissociate the Rb proteins from E2Fs, resulting in the transcription of E2F target genes [6, 10]. Consistent with this basic paradigm, genetic alterations of cell-cycle machinery components are frequent in cancer [11–15]. Thus, analyses of novel cell cycle regulatory components provide an opportunity to discover new prognostic markers in breast cancer.
Recently we identified a protein called Ecdysoneless as a novel cell cycle regulator. Ecd interacts with Rb and facilitates its separation from E2Fs [15]. Genetic deletion of Ecd in mice is embryonic lethal, while conditional deletion of Ecd in MEFs leads to retardation of the separation of Rb from E2F, delay/reduction in E2F-dependent gene expression and block in cell cycle progression. Therefore, we sought to investigate if the status of expression of Ecd might serve as a novel prognostic/predictive marker for breast cancer. To this end, we have analyzed a large set of breast cancer tissue specimens with known clinical parameters and our findings establish the overexpression of Ecd as a new prognostic marker for breast cancer.
Materials and method
The breast cancer tissue specimens used in the present study came from two independent patient cohorts as described below.
Cohort1 comprises of 269 breast cancer tissue specimens that were obtained from the Department of Pathology, Evanston Northwestern Healthcare (ENH; now renamed as Northshore University Health System Research Institute) or the Department of Pathology and Microbiology, University of Nebraska Medical Center (UNMC) or purchased as tissue arrays from US Biomax (US Biomax, Inc.Rockville, MD, USA). In this collection, 194 specimens were derived from patients with IDC (95 from ENH, 32 from UNMC and 67 from US Biomax tissue arrays). For comparison, the collection included 29 normal reduction mammoplasty specimens (19 from ENH and 10 from UNMC), 10 benign hyperplasia specimens (ENH) and 36 DCIS specimens (ENH). The tissue specimens at ENH and UNMC were obtained after Institutional Review Boards reviewed and approved the protocols.
Cohort 2 include Formalin fixed paraffin embedded tissue microarrays prepared from a series of cases of primary operable (stage I and II) breast carcinoma cases from patients age <70 presenting consecutively to the Nottingham Breast Unit with tumors of less than 5 cm diameter between 1988 and 1998 as previously reported [16]. This consecutive patient series is well characterized and contains clinical and pathological data including patients’ age, histological tumor types, primary tumor size, lymph node status, mitotic count and histological grades [17], Nottingham prognostic index (NPI) [18], vascular invasion (VI), therapy and follow-up data. Outcomes data include the survival status, survival time, cause of death, and disease free interval (DFI). The Breast Cancer Specific Survival (BCSS) is defined as time (in months) from the date of primary surgery to the date of breast cancer-related death. DFI is defined as the duration (in months) from the date of primary surgery to the appearance of loco-regional recurrence or distant metastasis. In addition, data on a large panel of biomarkers with strong relevance to breast cancer were available including ER, PR, HER2/neu, cytokeratins (CKs; basal CKs: CK5/6), p53 protein, and proliferation marker Ki67. The median age of patients was 55 years (range 18–70 years) with a median overall survival of 126 months (range 4–243 months) and median time of event-free survival of 114 months (range 2–243 months). Distant recurrence occurred in 501 cases (28%). 454 (25%) patients died from breast cancer, while 1,024 (58%) patients were alive at the end of follow-up.
Immunohistochemical analysis of Ecd expression
Formalin-fixed paraffin tissue sections (4 μm) or breast cancer tissue microarrays [19] were deparaffinized in xylene, rehydrated in descending alcohols and treated in a digital pressure cooker containing citrate buffer (pH 6.0; DakoCytomation, S1699). Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 10 min. The sections were rinsed in Tris-buffered saline with Tween and incubated for 15 min in Protein-Block buffer (DakoCytomation X0909) and sections were stained with primary monoclonal anti-bodies against Ecd (generated by us) overnight at 4°C. After rinsing in TBST, the sections were incubated for 15 min with anti-mouse antibody conjugated to a dextran-labeled polymer and HRP (DakoCytomation K4007) secondary, later on DAB solution for 7 min was added (DakoCytomation DAB + K4007, Solution 3a, b). The sections were then counter-stained in Mayer’s hematoxylin (Sigma MHS-80) and mounted under cover glasses using a xylene-based mounting medium.
Scoring of staining
For cohort 1, the Ecd staining intensity was evaluated under a light microscope by six independent observers including three of our pathology collaborators, and expressed on a scale of 0 to 3: 0, no staining; 1, weak; 2, moderate; 3, strong.
For cohort 2, the stained TMAs sections were manually scored from high-resolution digital images (NanoZoomer, Hamamatsu Photonics, Welwyn Garden City, UK) obtained at 20× magnification, using a web-based interface (Distiller, Slidepath Ltd., Dublin, Ireland) and percentage staining was determined. The cut-point for dichotomization was determined using X-tile bio-informatics software. The optimal cut-points were determined by locating the brightest pixels on the X-tile plot diagram of the training set. Statistical significance was tested by validating the obtained cut-point to the validation set [20]. All cases were scored without prior knowledge of the clinicopathological stages or outcomes data.
Generation of Ecd monoclonal antibody
Full length human Ecd cDNA was cloned into pProEx HT bacterial expression vector (that contain N-terminal 6× Histidine tag). The recombinant protein (hEcd) was purified from a large scale culture of BL21 E coli using Ni–Nta columns. The purified hEcd (with N terminal 6× histidine tag) was used as an antigen to produce monoclonal antibodies at the Monoclonal Antibody Core Facility, Lurie Cancer Center, Northwestern University, Chicago. The clones were screened by (i) western blotting with anti-flag or anti-Ecd antibodies, using flag-tagged Ecd overexpressing 293T cell lysates and also by (ii) immunoprecipitation of endogenous or exogenous Ecd from 293T cell lysates (data not shown). A few well reacting antibodies were selected among which the clone 4A8 was used for subsequent experiments. Clone 4A8 recognized a single band of estimated size in western blotting and immunoprecipitation as previously described [15].
Validation of Ecd antibody specificity for immunohistochemistry
For validation of Ecd antibody specificity for immunohistochemical analyses, 76 N.TERT cells (hTERT-immortalized normal human mammary epithelial cells) with or without hEcd overexpression or Ecd knockout (MEF Ecd−/−) and wild type mouse embryonic fibroblasts (MEF Ecd+/+) [15] were cultured on coverslips, fixed with 4% paraformaldehyde and stained with anti-Ecd antibody, as above. Western blotting was performed on whole cell lysates of 76 N.TERT cells or MEFs using a 1:2,000 dilution of anti-Ecd antibody, as previously described [15].
Statistical analysis
Statistical analysis was performed using the SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA). The Kruskal–Wallis and χ2 tests were used to determine relationships between various variables. Cut-off values for the different biomarkers included in this study were chosen before statistical analysis. Standard cut-offs were used for established prognostic factors and were the same as previously published for this patient series [21, 22]. Determination of the optimal Ecd cut-offs were performed using the X-tile bioinformatics software, version 3.6.1, 2003–2005, Yale University, USA. Analysis of categorical variables was performed with the appropriate statistical test. Survival curves were analyzed using the Kaplan–Meier method with significance determined by the Log Rank test. Multivariate analysis was performed using the Cox hazard analysis. A p value of 0.05 or lower was considered significant.
Results
Validation of the specificity of anti-Ecd monoclonal antibody for immunohistochemical staining
We have previously generated an anti-Ecd monoclonal antibody and established its specific recognition of both the human and mouse Ecd proteins in western blot [15, 23, 24]. Furthermore, we have shown that Ecd shuttles between the nucleus and the cytoplasm with a fast nuclear export resulting in a predominantly cytoplasmic localization in cells [23].
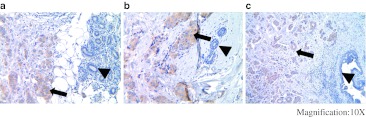
In order to assess the specificity of anti-Ecd monoclonal antibody in IHC staining, retroviral infection was used to overexpress Ecd in 76N. TERT human mammary epithelial cell line and compared with vector-infected cell line for IHC staining anti-Ecd mAb or nonspecific IgG as a staining negative control. As expected, essentially no staining was observed with the IgG control. Importantly, compared to a faint staining with anti-Ecd staining of vector-infected cells, a strong cytoplasmic signal was observed in Ecd-overexpressing (Supplementary Fig. 1a). Western blotting correlated with the IHC staining (Supplementary Fig. 1b). To further confirm the specificity of IHC staining of Ecd, we derived Ecd-null MEFs by adenovirus-Cre infection of Ecdfl/fl MEFs [15]. While specific Ecd staining was seen in MEFs without induced Ecd deletion, no staining was seen in MEFs with induced Ecd deletion (Supplementary Fig. 1c and 1d). Finally, we validated anti-Ecd staining by performing initial IHC staining of a small number of breast tumor specimens that contained adjacent normal tissue within the same section. While strong staining was observed in the tumor area, little staining was observed in the adjacent normal tissue (Fig. 1a–c). Altogether, these initial studies validated the use of our mAb for IHC staining, established its specificity, and suggested that Ecd protein levels were higher in breast tumor tissues compared to normal breast tissues.
Fig. 1.
IHC staining of IDC and adjacent normal duct. Three independent IDC (a–c) tumor specimens which contain adjacent normal ducts were used for IHC staining using anti-Ecd antibodies. Arrows indicate Ecd staining in tumor tissues and arrow heads indicate Ecd staining in the adjacent normal ducts at a magnification of ×10
Markedly increased Ecd expression in DCIS and IDC in comparison with normal breast tissues and hyperplasia
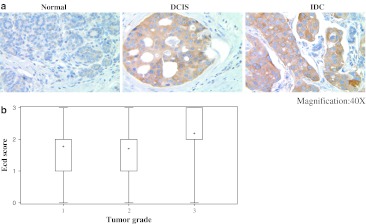
As Ecd functions to promote cell cycle progression, we assessed the relative Ecd expression by comparing the intensity of IHC staining with anti-Ecd mAb in 36 DCIS and 95 IDC tissue specimens in comparison with 19 normal breast tissues. In addition, we also performed similar analyses in 10 breast hyperplasia specimens. A clear cytoplasmic staining was observed in DCIS and IDC tumor tissues, whereas little or no staining was observed in normal breast tissues; interestingly, a similar low or no staining was observed in breast hyperplasia tissue specimens (Fig. 2a; Table 1). Expression of Ecd staining intensity according to the numeric scale indicated that none of the normal breast tissue specimen showed a high level of Ecd staining (score 2 or 3); while 20% of hyperplasia specimens showed a medium (score 2) level of Ecd staining, none scored at level 3 (Fig. 2b). In contrast, 17% of DCIS and 31% of IDC specimens showed a high level Ecd staining (score 3) (Table 1). These results suggested that the levels of Ecd expression may positively correlate with increasing grade of breast tumors.
Fig. 2.
a Analyses of Ecd expression in normal, DCIS and IDC breast tissues. Representative specimens (from Table 1) for Ecd staining are shown with magnification of ×40; b Correlation of Ecd expression in samples from cohort 1 with IDC tumor grade is shown. The distribution of IDC samples was: grade 1 = 18, grade 2 = 34, and grade 3 = 52, analyzed by Kruskal–Wallis test
Table 1.
Ecd expression in DCIS and IDC in comparison with normal and hyperplasia
| Tissue type | Normal | Hyperplasia | DCIS | IDC |
|---|---|---|---|---|
| Score | N (%) | N (%) | N (%) | N (%) |
| 0 | 17 (89) | 4 (40) | 8 (22) | 19 (20) |
| 1 | 2 (10) | 4 (40) | 8 (22) | 15 (16) |
| 2 | 0 (0) | 2 (20) | 14 (39) | 32 (34) |
| 3 | 0 (0) | 0 (0) | 6 (17) | 29 (31) |
Ecd expression correlates positively with increasing tumor grade in IDC specimens
Next, we examined an additional 104 IDC specimens within cohort 1 where tumor grade information was available. We observed a statistically significant positive correlation (P = 0.04) between the levels of Ecd expression and grade of IDC with high-grade cancers showing higher levels of Ecd protein (Supplementary Table 1, Fig. 2b).
Association of Ecd overexpression with clinicopathological parameters of poor prognosis
Cytoplasmic Ecd expression was dichotomised into negative-moderate (≤ 70% cytoplasmic staining) and high (>70% cytoplasmic staining) categories. Using these cut-points, a minority of patients (214 cases; 22.4%) showed negative-moderate levels of Ecd expression while a majority (740 cases; 77.6%) showed high levels of Ecd expression in cohort 2. High Ecd expression was positively associated with higher tumor grade (P = 0.013) (Table 2), further validating observations made with cohort 1. A positive correlation was also observed between high Ecd expression and known markers of poor prognosis, including increased mitotic index (P = 0.032) and poor NPI group (P = 0.014). No association between the levels of Ecd expression and other parameters observed (Table 2).
Table 2.
Relationship between Ecd expression and various clinicopathological parameters
| Parameter | Ecd expression | Significance | ||
|---|---|---|---|---|
| Low Ecd | High Ecd | Total | P value | |
| No. (%) | No. (%) | No. (%) | ||
| Patients age | ||||
| ≤50 years | 88 (41.1) | 265 (35.8) | 353 (37) | 0.156 |
| >50 years | 126 (58.9) | 475 (64.2) | 601 (63) | |
| Total | 214 | 740 | 954 | |
| Tumor size | ||||
| ≤1.5 cm | 72 (33.6) | 212 (28.8) | 284 (30) | 0.181 |
| >1.5 cm | 142 (66.4) | 522 (71.1) | 664 (70) | |
| Total | 214 | 734 | 948 | |
| Lymph node stage | ||||
| 1 | 132 (61.7) | 423 (57.6) | 555 (58.5) | 0.395 |
| 2 | 67 (31.3) | 240 (32.7) | 307 (32.4) | |
| 3 | 15 (7) | 71 (9.7) | 86 (9.1) | |
| Total | 214 | 734 | 984 | |
| Grade | ||||
| 1 | 41 (19.2) | 102 (13.9) | 143 (15.1) | 0.013 |
| 2 | 74 (34.6) | 211 (28.7) | 285 (30.1) | |
| 3 | 99 (46.3) | 421 (57.4) | 520 (54.9) | |
| Total | 214 | 734 | 984 | |
| Tubules | ||||
| 1 | 14 (6.7) | 33 (4.6) | 47 (5.1) | 0.273 |
| 2 | 70 (33.7) | 218 (30.5) | 288 (31.2) | |
| 3 | 124 (59.6) | 463 (64.8) | 587 (63.7) | |
| Total | 208 | 714 | 922 | |
| Pleomorphism | ||||
| 1 | 5 (2.4) | 6 (0.8) | 11 (1.2) | 0.151 |
| 2 | 73 (35.3) | 237 (33.3) | 310 (33.8) | |
| 3 | 129 (62.3) | 468(65.8) | 597(65) | |
| Total | 207 | 711 | 918 | |
| Mitosis | ||||
| 1 | 76 (36.5) | 216 (30.3) | 292 (31.7) | 0.032 |
| 2 | 46 (22.1) | 29 (18.1) | 175 (19) | |
| 3 | 86 (41.3) | 369 (51.7) | 455 (49.3) | |
| Total | 208 | 714 | 922 | |
| Nottingham prognostic index | ||||
| Good (3.4) | 69 (32.2) | 185 (25.3) | 254 (26.8) | 0.014 |
| Moderate (3.41–5.4) | 117 (54.7) | 393 (53.7) | 510 (53.9) | |
| Poor(4.41–5) | 28 (13.1) | 154 (21) | 182 (19.2) | |
| Total | 214 | 732 | 946 | |
| Vascular invasion | ||||
| Definite | 61 (29) | 253 (34.4) | 314 (33.2) | 0.145 |
| Negative | 149 (71) | 482 (65.6) | 631 (66.7) | |
| Total | 210 | 735 | 945 | |
Ecd expression shows a positive correlation with positive HER2/neu but not ER/PR status
Next we examined if the levels of Ecd expression correlate with molecular markers of breast cancer subtypes, ER/PR and HER2/neu. Notably, we observed a positive association between high Ecd expression and HER2/neu-positive status (P = 0.002) (Table 3). In contrast, we did not observe an association of Ecd expression with ER +, PR +, ER +/PR + or triple negative status (Table 3). The levels of Ecd expression also showed no association with p53 or Ki67 expression. Considering that a subtype of breast cancers is categorized by HER2/neu positive and expression of HER2/neu predicts poor outcome in patients, association of HER2/neu with Ecd is highly significant and could serve as a marker for poor outcome of patients.
Table 3.
Relationship between Ecd expression and other biomarkers
| Parameter | Ecd expression | Significance | ||
|---|---|---|---|---|
| Low Ecd | High Ecd | Total | P value | |
| No. (%) | No. (%) | No. (%) | ||
| ERα status | ||||
| Negative | 47 (22.3) | 206 (28.6) | 253 (27.1) | 0.070 |
| Positive | 164 (77.7) | 515 (71.4) | 679 (72.9) | |
| Total | 211 | 721 | 932 | |
| PgR status | ||||
| Negative | 78 (38) | 303 (42.9) | 381 (41.8) | 0.213 |
| Positive | 127 (62) | 403 (57.1) | 530 (58.2) | |
| Total | 205 | 706 | 911 | |
| HER-2 status | ||||
| Negative | 191 (92.3) | 612 (84) | 803 (85.8) | 0.002 |
| Positive | 16 (7.7) | 117 (16) | 133 (14.2) | |
| Total | 207 | 729 | 936 | |
| ER.PR. status | ||||
| ER-PR- | 44 (21.8) | 197 (28.5) | 241 (27) | 0.29 |
| ER-PR+ | 1 (0.5) | 2 (0.3) | 3 (0.3) | |
| ER + PR- | 31 (15.3) | 95 (13.7) | 126 (14.1) | |
| ER + PR+ | 126 (62.4) | 397 (57.5) | 523 (58.6) | |
| Total | 202 | 691 | 893 | |
| Triple negative status | ||||
| Non-TN | 172 (82.3) | 587 (81.4) | 759 (81.6) | 0.772 |
| TN | 37 (17.7) | 134 (18.6) | 171 (18.4) | |
| Total | 209 | 721 | 930 | |
| Basal type | ||||
| Negative | 153 (73.6) | 559 (80.3) | 712 (78.8) | 0.037 |
| Positive | 55 (26.4) | 137 (19.7) | 192 (21.2) | |
| Total | 208 | 696 | 904 | |
| p53 | ||||
| Negative | 154 (75.1) | 494 (71.1) | 648 (72) | 0.257 |
| Positive | 51 (24.9) | 201 (28.8) | 252 (28) | |
| Total | 205 | 695 | 900 | |
| Ki67 | ||||
| Moderate | 145 (79.2) | 455 (75.8) | 600 (76.6) | 0.341 |
| High | 38 (20.8) | 145 (24.2) | 183 (23.4) | |
| Total | 183 | 600 | 783 | |
The levels of Ecd expression correlate with disease outcome in breast cancer patients
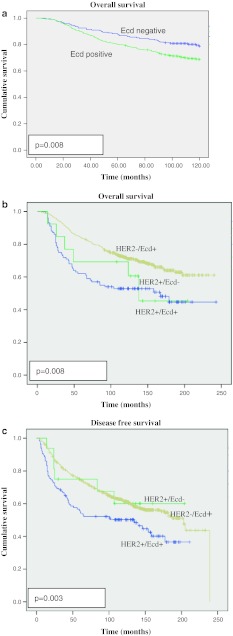
In view of a positive association of the levels of Ecd expression with several prognostic factors, we wished to assess if Ecd expression could predict the outcome of disease in breast cancer patients in cohort 2 where outcomes data was available. For this purpose, the estimated predictive power of Ecd expression was assessed using the Kaplan–Meier survival analysis. Patients with high Ecd expression exhibited a statistically significant reduction term for BCSS (120 months, P = 0.008) as compared to patients with no to moderate Ecd expression (Fig. 3a). These results suggest that Ecd expression can serve as a marker of overall survival outcome in breast cancer patients.
Fig. 3.
a Kaplan–Meier plot of Ecd expression in the whole series of breast cancer patients with respect to BCSS for 120 months (P = 0.008); b Kaplan–Meier plot of Ecd expression in Ecd alone, HER2 alone or HER2 + Ecd overexpressing breast cancer patients with respect to BCSS for 250 months (P = 0.04); c Kaplan–Meier plot of Ecd expression in Ecd alone, HER2 alone or HER2 + Ecd overexpressing breast cancer patients with respect to DFI for 250 months (P = 0.008)
Co-expression of Ecd predicts poorer outcome in HER2/neu + breast cancer patients
Next, we assessed whether levels of Ecd expression might predict different outcomes in HER2/neu + patients. For this purpose, we correlated Ecd expression levels in HER2/neu + patients with disease free survival as well as overall survival. This analysis revealed that the overall survival in HER2/neu + patients that expressed high levels of Ecd was significantly worse (P = 0.008) as compared to HER2/neu + patients with no to moderate Ecd expression. Furthermore, HER2/neu + patients with high levels of Ecd showed significantly reduced DFS (250 months, P = 0.003) as compared to HER2/neu + patients with no to moderate Ecd expression (Fig. 3b, c). However, Cox-multivariate regression analysis revealed that Ecd expression as a predictor of BCSS was not independent of tumor size, stage or grade (P = 0.126) (Table 4). Taken together, Ecd overexpression predicts poor prognosis and outcome in HER2/neu positive patients.
Table 4.
Multivariate COX regression model for predictors of BCSS in patient cohort
| Variable | P value | HR | 95% CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| Ecd | 0.126 | 2.34 | 0.539 | 1.079 |
| Tumor size | 0.00 | 13.86 | 0.313 | 0.697 |
| Tumor stage | 0.00 | 34.13 | 1.448 | 2.104 |
| Tumor grade | 0.00 | 40.02 | 1.799 | 3.046 |
Discussion
Breast cancer has emerged as group of diseases that are clinically and molecularly diverse and carry significantly different outcomes. Thus, newer prognostic markers that can help predict the course of disease, either by themselves or in association with established markers, can help devise better treatment strategies. Here, we have investigated the levels of expression of a novel cell cycle regulator Ecd in breast cancer patient specimens as a potential prognostic biomarker. Our studies show that expression of high levels of Ecd protein correlates with several known markers of poor prognosis in breast cancer, and predicts poorer breast cancer-specific survival, especially in the HER2/neu + breast cancer.
Ecd is an evolutionarily-conserved protein that we have demonstrated to be an essential component of mammalian cell cycle progression [15, 24]. Ecd plays an essential role in facilitating the removal of Rb-family of tumor suppressor proteins from E2F transcription factors such that lack of Ecd leads to prolonged Rb-E2F association and inhibition of cell cycle progression. In cell culture studies, we have observed that ectopic overexpression of Ecd in human mammary epithelial cells promotes proliferation (unpublished). To assess the expression of Ecd in breast cancer, we first established the ability of a monoclonal antibody generated in our laboratory to specifically detect in a quantitative manner the expression of human Ecd. By utilizing WT versus Ecd-null MEFs and human mammary epithelial cells with overexpression of Ecd versus vector controls, we established the ability of our mAb anti-Ecd to specifically detect Ecd and to provide an assessment of different levels of Ecd expression in a cell-based IHC. Importantly, this staining format was successfully adapted to paraffin-embedded human breast tissue specimens. Notably, these initial studies with breast tissue specimens from a smaller cohort that included normal reduction mammoplasty, benign hyperplasia, DCIS and IDC samples provided the first indication that Ecd expression increased in tissues with breast cancer progression: while essentially little Ecd signals were observed in normal breast tissue and hyperplasia, intense Ecd expression was observed in DCIS and IDC.
Analyses of patient cohort 1 showed a clear overexpression of Ecd in 56% of DCIS and 65% of IDC samples as compared to no or little expression of Ecd in normal and hyperplasia specimens. Two major conclusions arose from these analyses: (i) that Ecd is overexpressed starting at an early stage of breast cancer i.e. DCIS; (ii) that Ecd is overexpressed in IDC but not in normal and hyperplasia samples, suggesting that levels of Ecd expression could provide a potential marker of breast cancer progression. Indeed, further analyses using cohort 2 of IDC tissue specimens showed a positive association of Ecd expression with tumor grades. Furthermore, Ecd expression showed a significant positive correlation with the mitotic index, a result consistent with the role of Ecd in cell cycle progression [15].
Further analyses using a large cohort of breast cancer patient specimens with associated clinical outcomes data helped assess if the levels of Ecd overexpression correlate with patient outcome. Kaplan–Meier survival analysis showed that patients with high Ecd expression have a significantly reduced BCSS (120 months, P = 0.008). These results suggest that Ecd expression could serve as a marker of disease-specific survival outcome in breast cancer patients.
The expression of ER, PR and HER2/neu provides useful markers to help in the choice of post-operative adjuvant therapy with the strongest likelihood of better outcomes. The triple negative patients show a higher likelihood of disease recurrence and metastatic disease with poor overall survival, reflecting both the biology of these tumors and the current lack of suitable targeted therapies for this subset. It is notable that the maximum difference in the outcomes of TNBC versus other forms of breast cancer are seen at 3 years after diagnosis and the differences gradually diminish year by year for up to 10 years [25, 26]. Thus, we speculate that Ecd expression could provide a useful additional prognostic marker for use in conjunction with the established clinical-pathological prognostic markers.
Notably, our studies revealed that co-expression of high levels of Ecd in the HER2/neu + subset of patients predicted a significantly poorer outcome both in terms of overall and breast cancer specific survival. While the availability of HER2/neu-targeted therapies has significantly improved the outcomes of patients with HER2/neu + breast cancer, this subset of breast cancers continues to be associated with poorer outcomes compared to ER +/PR + tumors, and treatment-associated toxicities and high incidence of de novo and acquired resistance to Trastuzumab continue to be significant impediments in treatment. Thus, additional markers that might improve the prediction of outcomes in this subset of patients could help physicians and patients in deciding on options for conventional versus aggressive treatment strategies. While there are no known catalytic domains in Ecd to make it conventionally druggable, the increasing ability of researchers to design inhibitors that work by non-conventional mechanisms suggest that further studies in this direction are warranted.
It is notable that Ecd protein overexpression in breast cancer is not accompanied by increase in its mRNA levels (unpublished). This explains the lack of overexpression observed in microarray data and points to the need to further establish the mechanisms by which Ecd is overexpressed in breast cancer. Further studies will be needed to assess if translational and/or post-translational mechanisms contribute to Ecd overexpression.
In conclusion, studies presented here identify the overexpression of cell cycle regulatory protein Ecd as a potential prognostic marker in breast cancer. Ecd overexpression positively correlates with increasing tumor progression, predicts poorer overall and breast cancer-specific survival and defines a subset of HER2/neu + patients with a particularly worse outcome. As Ecd is expressed in a variety of normal tissues, further studies to assess Ecd overexpression in other tumors should help assess its broad role as a prognostic marker as well as help link its overexpression to oncogenesis of other organs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by the NIH grants CA96844 and CA144027 and Department of Defense grants W81XWH-07-1-0351 and W81XWH-11-1-0171 to V.B; the NIH grants CA87986, CA105489, CA99163, CA116552 and NCI 5U01CA151806-02 to H.B; and the NCI Core Support Grant to UNMC-Eppley Cancer Center. S.M and A.B are supported by a Susan G. Komen Postdoctoral Fellowship (KG111248) and a DOD predoctoral fellowship (W81XWH-11-1-0020), respectively. This work was initiated while the last author was on the faculty of Evanstan Northwestern Healthcare at Northwestern, Evanston, IL.
Conflict of interest
The authors disclose no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Xiangshan Zhao and Sameer Mirza contributed equally to this study.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez I, Dynlacht BD. New insights into cyclins, CDKs, and cell cycle control. Semin Cell Dev Biol. 2005;16(3):311–321. doi: 10.1016/j.semcdb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J Cell Biochem. 2007;102(6):1400–1404. doi: 10.1002/jcb.21609. [DOI] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30(11):630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70(6):993–1006. doi: 10.1016/0092-8674(92)90249-C. [DOI] [PubMed] [Google Scholar]
- 7.Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato J, Livingston DM. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73(3):487–497. doi: 10.1016/0092-8674(93)90136-E. [DOI] [PubMed] [Google Scholar]
- 8.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24(17):2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 9.Resnitzky D, Hengst L, Reed SI. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15(8):4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du W, Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25(38):5190–5200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8(9):671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24(17):2909–2915. doi: 10.1038/sj.onc.1208618. [DOI] [PubMed] [Google Scholar]
- 13.Mammas IN, Sourvinos G, Giannoudis A, Spandidos DA. Human papilloma virus (HPV) and host cellular interactions. Pathol Oncol Res POR. 2008;14(4):345–354. doi: 10.1007/s12253-008-9056-6. [DOI] [PubMed] [Google Scholar]
- 14.Band V. The role of retinoblastoma and p53 tumor suppressor pathways in human mammary epithelial cell immortalization. Int J Oncol. 1998;12(3):499–507. doi: 10.3892/ijo.12.3.499. [DOI] [PubMed] [Google Scholar]
- 15.Kim JH, Gurumurthy CB, Naramura M, Zhang Y, Dudley AT, Doglio L, Band H, Band V. Role of mammalian Ecdysoneless in cell cycle regulation. J Biol Chem. 2009;284(39):26402–26410. doi: 10.1074/jbc.M109.030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005;116(3):340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 17.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 18.Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham prognostic index in primary breast cancer. Breast Cancer Res Treat. 1992;22(3):207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- 19.McCarty KS, Jr, Miller LS, Cox EB, Konrath J, McCarty KS., Sr Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985;109(8):716–721. [PubMed] [Google Scholar]
- 20.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 21.Rakha EA, El-Sayed ME, Green AR, Paish EC, Powe DG, Gee J, Nicholson RI, Lee AH, Robertson JF, Ellis IO. Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype. J Clin Oncol. 2007;25(30):4772–4778. doi: 10.1200/JCO.2007.12.2747. [DOI] [PubMed] [Google Scholar]
- 22.Rakha EA, Putti TC, Abd El-Rehim DM, Paish C, Green AR, Powe DG, Lee AH, Robertson JF, Ellis IO. Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol. 2006;208(4):495–506. doi: 10.1002/path.1916. [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, Gurumurthy CB, Band H, Band V. Biochemical characterization of human Ecdysoneless reveals a role in transcriptional regulation. Biol Chem. 2010;391(1):9–19. doi: 10.1515/bc.2010.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Chen J, Gurumurthy CB, Kim J, Bhat I, Gao Q, Dimri G, Lee SW, Band H, Band V. The human orthologue of Drosophila ecdysoneless protein interacts with p53 and regulates its function. Cancer Res. 2006;66(14):7167–7175. doi: 10.1158/0008-5472.CAN-06-0722. [DOI] [PubMed] [Google Scholar]
- 25.Irvin WJ, Jr, Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44(18):2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 26.Dent R, Warner E. Screening for hereditary breast cancer. Semin Oncol. 2007;34(5):392–400. doi: 10.1053/j.seminoncol.2007.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.