Abstract
Macrophages are central players in immune response, manifesting divergent phenotypes to control inflammation and innate immunity through release of cytokines and other signaling factors. Recently, the focus on metabolism has been reemphasized as critical signaling and regulatory pathways of human pathophysiology, ranging from cancer to aging, often converge on metabolic responses. Here, we used genome-scale modeling and multi-omics (transcriptomics, proteomics, and metabolomics) analysis to assess metabolic features that are critical for macrophage activation. We constructed a genome-scale metabolic network for the RAW 264.7 cell line to determine metabolic modulators of activation. Metabolites well-known to be associated with immunoactivation (glucose and arginine) and immunosuppression (tryptophan and vitamin D3) were among the most critical effectors. Intracellular metabolic mechanisms were assessed, identifying a suppressive role for de-novo nucleotide synthesis. Finally, underlying metabolic mechanisms of macrophage activation are identified by analyzing multi-omic data obtained from LPS-stimulated RAW cells in the context of our flux-based predictions. Our study demonstrates metabolism’s role in regulating activation may be greater than previously anticipated and elucidates underlying connections between activation and metabolic effectors.
Keywords: constraint-based modeling, immunometabolism, metabolic network reconstruction, RAW 264.7
Introduction
Macrophages have a key role in coordinating immune response in mammalian systems through the production of regulatory cytokines, proteases, and coagulation factors. In addition, they are critical for digesting pathogens, necrotic cellular debris, and other foreign objects that are encountered within the host. Growing evidence indicates that macrophages manifest divergent metabolic phenotypes to mediate a variety of alternative functions, including immunosuppression and tissue repair (Mosser, 2003). While metabolic status can greatly vary depending on macrophage type or stimulated state, the murine leukemic monocyte macrophage cell line RAW 264.7 has been an amenable in-vitro model of macrophage and monocyte functions as it exhibits key characteristics representative of different macrophage types in vivo (Raschke et al, 1978; Chapekar et al, 1996; Scheel et al, 2009). For that reason, the RAW cell line has served as a host model to experimentally probe macrophage phenotypes under various exposure conditions (e.g., endotoxins and cytokines) as well as during direct parasitic infection (Cirillo et al, 1998; Gutierrez et al, 2004) and proliferative inflammatory response states (Moeslinger et al, 1999).
The phenotypic responses that macrophages display have been extensively studied and are generally categorized according to activation status (Mosser and Edwards, 2008). While macrophage activation functions at the front line of host defense, improper control of its activation has also been implicated to have major roles in disease progression. For example, infiltration of activated macrophages in tissues has been strongly associated with a number of pathological disorders including diabetes, obesity, and renal injury (Heilbronn and Campbell, 2008; Guo et al, 2011). The recruitment of specific, activated macrophage sub-populations in the tumor microenvironment is widely known to promote chemoresistance by enabling cancer cells to effectively evade host immune responses (De Palma and Lewis, 2011). Macrophage activation states are also modulated by parasitic organisms, which hijack host macrophage cells to promote their long-term, intracellular survival (Stempin et al, 2010). Macrophage activation is metabolically associated with the amino-acid arginine that diverges into classical (M1) and alternative (M2) pathways with the respective productions of: (1) nitric oxide (NO) for microbicidal purposes via NOS2 and (2) proline and polyamines for inducing local cell proliferation and collagen remodeling via arginase (Mosser, 2003; Odegaard and Chawla, 2011). These polarized functions are activated in response to bacterial and viral infections in the M1 phenotype, and to parasitic infection, tissue remodeling and angiogenesis in the M2 phenotype (Mosser, 2003; Odegaard and Chawla, 2011). Although there is a growing interest in understanding the interface between metabolism and immunity (Mathis and Shoelson, 2011), little systems-based approaches have been utilized in elucidating metabolic mechanisms that are linked to macrophage activation to date.
Molecular systems biology has arisen as a discipline to meet the challenges associated with the current era of high-throughput, data-rich biology. Genome-scale reconstructions provide mechanistic foundations for network-level modeling, biological discovery, and analyzing high-throughput data sets (Oberhardt et al, 2009). Metabolic networks bridge the gap between genomic and biochemical information and form a mechanistic context in which data sets can be incorporated to evaluate causal phenotypic relationships. This approach has been demonstrated in evaluating metabolic phenotypes for microbial and eukaryotic systems, ranging from industrial microbes to pathogens to human cells (Duarte et al, 2007; Feist et al, 2007; Jamshidi and Palsson, 2007). Recently, algorithmic approaches have leveraged genome-scale networks as a mechanistic scaffold for interpreting condition- and tissue-specific gene expression data (Bordbar et al, 2010; Chang et al, 2010; Jerby et al, 2010).
In this study, we present a genome-scale metabolic reconstruction and analysis for the RAW 264.7 cell line to evaluate metabolite effectors and mechanisms associated with macrophage activation. Highly effective metabolites identified by our analysis are richly supported in the published literature for their immunomodulatory properties, in which several predicted metabolites have been previously experimentally verified. Mechanisms for activation and inhibition by predicted metabolic immunomodulators were investigated through Monte Carlo sampling analysis. Finally, transcriptomic, proteomic, and metabolomic analysis of LPS-stimulated RAW cells demonstrates how model-based predictions enhance the mechanistic interpretation of high-throughput data to enable better understanding of macrophage metabolic activation phenotypes.
Results
RAW 264.7 metabolic network reproduces experimentally measured flux rates
A RAW 264.7 metabolic network was reconstructed based on the global human metabolic network Recon 1 (Duarte et al, 2007) by integrating gene expression and proteomic data with a Homologene-mapped metabolic network (see Materials and methods for workflow and details). The physiological capabilities of the network were evaluated using uptake rates derived from in-vitro data sets. Rates of biomass growth, ATP production, and NO synthesis were compared with experimental values. The maximum doubling time for the imposed in-vitro uptake rates was 16.99 h. Although the measured growth rate tends to vary for different experimental conditions, the calculated rate is consistent with the previously reported range of 11 h (Sakagami et al, 2009), 18–22 h (Zhuang and Wogan, 1997), and 24.7 h (Alldridge et al, 1999). For subsequent ATP and NO rate calculations, the lower bound on the biomass reaction was set to the lower experimental growth rate of 0.0281/h to mimic minimal maintenance of the macrophage. The maximum calculated ATP production rate was 0.796 mmol/h/g DW, similarly to the in-vitro rate of 0.712 mmol/h/g DW (Newsholme et al, 1999). The NO production rate (via NOS2) was 0.0399, mmol/h/g DW, similarly to an in-vitro measured rate of 0.0365, mmol/h/g DW (Griscavage et al, 1993). Overall, our results indicate that the metabolic network is predictive of physiologically relevant experimental rates when in-vitro uptake rates are imposed. To further evaluate the physiological accuracy of the cell-specific RAW 264.7 metabolic network compared with the larger global model from which it was derived, we calculated biomass growth, ATP production, and NO production rates for the Recon 1 network. We also performed sensitivity analyses presented in the later sections. For both analyses, we found Recon 1 predictions on the quantitative production rates to be considerably less accurate compared with the RAW 264.7-specific network (see Supplementary information).
Deletion analysis differentiates critically essential reactions of M1 and M2 activation phenotypes
To metabolically characterize macrophage phenotypes, we defined five metabolic objective functions associated with general macrophage function and activation: (1) energy (ATP) generation, (2) redox maintenance (NADH), (3) NO production for M1 activation, (4) synthesis of extracellular matrix precursors (proline), and (5) induction of local cell proliferation through polyamines (i.e., putrescine) to define M2 activation (Mosser, 2003). As a highly mobile and active cell, macrophages require high glycolytic activity to generate ATP and NADH for essential functional purposes (Newsholme et al, 1999). M1 and M2 activation phenotypes serve different functions in immune response, with the primary difference due to the differential upregulation of NOS2 and arginase (Mosser, 2003; Odegaard and Chawla, 2011). We first evaluated whether distinguishing M1 and M2 phenotypic features can be demonstrated through the optimization of the respective M1- and M2-associated objective functions without directly imposing regulatory constraints on NOS2 and arginase reactions.
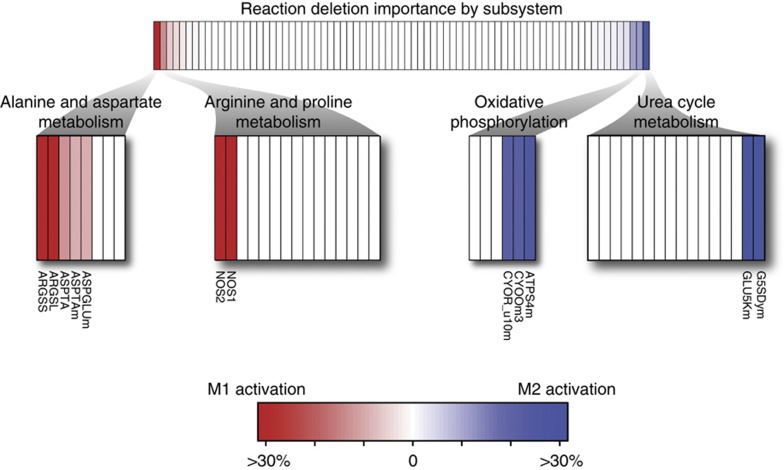
A single reaction deletion analysis was performed while maximizing for M1 (NO production) and M2 activation (proline and putrescine productions) to evaluate reactions that are differentially critical, or essential, for M1 and M2 phenotypes. In general, the essential reactions between M1 and M2 functions were similar, as has been previously demonstrated by the overall similarities between M1 and M2 metabolic flux distributions (Rodriguez-Prados et al, 2010). This result is primarily due to the close network proximity of NO, proline, and polyamine production. Of the 76 metabolic subsystems in the RAW 264.7 network, only 4 had a large difference in response to reaction deletions: alanine and aspartate metabolism, arginine and proline metabolism, oxidative phosphorylation, and urea cycle/amino group metabolism (Figure 1). In all, there were 37 reaction deletions that resulted in a large differential response (>10% difference from original objective function value) between M1 and M2 activation (Figure 1). As expected, reactions that were directly involved in the respective production of NO (NOS1 and NOS2), proline (G5SADrm), and putrescine (ORNDC) were critically essential as their deletion resulted in the inability to produce their respective products (i.e., value of 0).
Figure 1.
Reaction deletion analysis differentiates metabolic differences observed for M1 and M2 activation. The difference between reaction essentiality for M1 and M2 activation is shown. In the top portion, the reactions are grouped by subsystem and rank ordered in terms of importance for M1 activation. Only a few subsystems were differentially important. Largely differential subsystems are shown in reaction detail. The reaction importance differences seen for oxidative phosphorylation and the shuttling of NADH equivalent reflects known metabolic flux variations seen in M1 and M2 activation.
Outside of the reactions that were directly involved in the production of NO, proline, and putrescine, the majority of differential reactions were involved in oxidative phosphorylation and the malate-aspartate shuttle. The deletion of oxidative phosphorylation reactions (CYOR_u10, CYOOm3, and ATPS4m) reduced proline and putrescine production to 63–76% of its maximum capacity. The effect on NO production was far less detrimental at 88–96%, thus indicating oxidative phosphorylation to be more important for the M2 activation phenotype than for M1. Glycolysis-associated reaction deletions (GAPD, PGK, PGM, PYK and ENO) led to a reduced NO production of 73–74%, slightly lower than 75–78% for proline and putrescine. However, alanine and aspartate metabolism reactions (ASPTA, ASPGLUm) were significantly more important for NO production as shown for the reduction to 83–85%, compared with 93–96% for the M2-associated objective functions.
The reaction deletion analysis indicates that the M1-activation phenotype has a slightly higher dependency on the shuttling of glycolysis-associated NADH equivalents from the cytosol to the mitochondria. However, the removal of oxidative phosphorylation reactions is more detrimental in M2 activation. These results are consistent with a recent study showing that M1-activated macrophages have a higher glycolytic capacity whereas M2-activated macrophages are more dependent on oxidative phosphorylation (Rodriguez-Prados et al, 2010).
Sensitivity analysis identifies immunomodulatory metabolites of macrophage activation
To determine important metabolic immunomodulators of macrophage activation, we performed a sensitivity analysis on the metabolite exchanges in the RAW macrophage network for each of the five activation-based objective functions. The analysis quantifies metabolite exchanges that are strongly associated with the maximization of each objective function by prioritizing the slope values calculated from robustness analysis (Edwards and Palsson, 2000; Figure 2). For example, NO production is expected to be highly sensitive to L-arginine uptake as L-arginine is its precursor metabolite. In general, sensitivity scores for NO, proline, and putrescine did not vary significantly as their respective productions are coupled with arginine fate and their synthesis pathways are proximal to one another, as previously discussed. These results are supportive of the similarities in metabolic activation flux patterns of M1 and M2 phenotypes measured in murine peritoneal macrophages (Rodriguez-Prados et al, 2010).
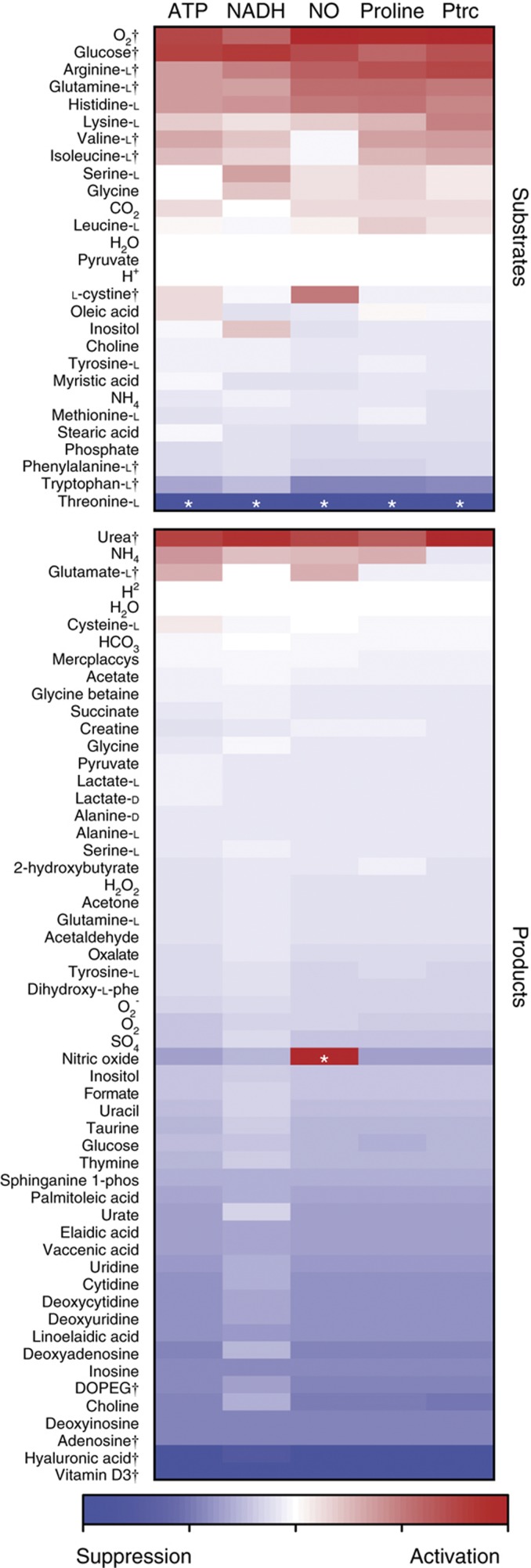
Figure 2.
Network sensitivity analysis recapitulates literature-supported immunomodulatory metabolites. Five objective functions were evaluated for activating and suppressing metabolites based on magnitude and directionality of slope. Support from previously published experimental studies was enriched toward metabolites that were predicted to be most effective. Metabolites with literature support and discussed in our analysis are denoted by (†). Metabolites denoted with (*) were excluded as those results are due to artifacts of the network.
Several computationally predicted activating metabolic substrates are well established to be critical for macrophage activation (Figure 2). Oxygen, glucose, and glutamine have all been previously shown to be of great importance in macrophage metabolism and activation as they are key for cellular respiration, energy production, and respiratory burst (Newsholme et al, 1996). High consumption of glutamine is particularly important for enhancing immunity (Newsholme et al, 1999), as it is required for arginine biosynthesis and nitrite/urea production (Murphy and Newsholme, 1998). The uptake of arginine and the branched chain amino acids (BCAAs) valine and isoleucine were also identified to be important for ATP, NADH, proline, and putrescine production; however, it was not found to be critical for NO production. Arginine is among the most critical amino acid as it is a direct precursor metabolite for NO (Baydoun et al, 1993) and its transport increases up to five-fold during activation and proliferation (Yeramian et al, 2006). Previous experiments verify these predictions as higher arginine and glucose concentrations of LPS- and LTA-activated macrophages increased NO production above levels seen without supplementation (Newsholme et al, 1999; de Souza et al, 2008). Though BCAAs are not as well studied as arginine in macrophages, supplementation of BCAAs has shown to help improve immune response in long distance runners (Bassit et al, 2002). During strenuous exercise, plasma glutamine levels drop having a large effect on peripheral blood mononuclear cells (PBMCs). Bassit et al, (2002) showed that BCAA supplementation before strenuous exercise allowed PBMC to retain their ability to proliferate. L-cystine was also found to be selectively activating for NO production. While L-cystine was not predicted to be an effective activator or suppressor of M2 activation, it has been previously shown that LPS-activated NO production induces L-cystine uptake in murine macrophages (Watanabe and Bannai, 1987).
Tryptophan and phenylalanine were highly ranked among substrates in which its catabolism had a suppressive effect on M1 and M2 metabolic activation phenotypes (Figure 2). Most notably, indoleamine 2,3-dioxygenase (IDO), a key enzyme in tryptophan catabolism, is a widely known suppressive mechanism by which IDO-expressing monocytes and macrophages inhibit T-cell proliferation in mammals (Mellor and Munn, 2004). Tryptophan catabolites have been widely implicated in mediating immune tolerance (Moffett and Namboodiri, 2003), and the tryptophan catabolite 3-hydroxyanthranilic acid has been directly shown to suppress NO synthase expression (Oh et al, 2004). A similar catabolic mechanism has been proposed for phenylalanine, by which the immunosuppressive enzyme IL4I1 has been shown to inhibit T-cell proliferation in vitro via L-phenylanine oxidase (Carbonnelle-Puscian et al, 2009). It is important to note that the uptake of threonine had the largest inhibitory effect on activation objectives, but was not under consideration during analysis. While constructing the model, it was noted that the RAW 264.7 cell line lacked threonine degradation pathways, confirmed through low threonine dehydrogenase transcript levels. As threonine uptake increased in the sensitivity analysis, the amino acid was being shuttled only to biomass production that resulted in the diversion of precursor metabolites away from M1 and M2 phenotypes, thus appearing to be an immunosuppressive metabolite.
Produced metabolites were also found to have profound effects on metabolic activation. Synthesis of most metabolites had a suppressive effect on M1- and M2-associated metabolic phenotypes (Figure 2), as there is a metabolic ‘cost’ by diverting precursor metabolites and energy resources away from the arginine-derived activation pathways. Vitamin D3 production was found to be the most effective at lowering flux across all activation functions, a metabolite which has been previously implicated as a critical negative feedback mechanism to control activated macrophages through paracrine signaling (Helming et al, 2005). A similar mechanism has been found to be employed by the immunosuppressant cyclosporine, which upregulates vitamin D3 synthesis via 1-α-hydroxylase (Overbergh et al, 2000). The production of several nucleotides and deoxynucleotides was also indicated to be negatively associated with macrophage activation as 8 of the top 13 inhibiting metabolites involved in nucleotide metabolism. In particular, adenosine is known to be released in tissues during injurious stimuli to mitigate pro-inflammatory responses across various immune cell types (Hasko and Cronstein, 2004; Palmer and Trevethick, 2008). Interestingly, the supply of purines and pyrimidines has previously been found to be vital for intracellular pathogens to proliferate within macrophages, as many pathogens have auxotrophies for nucleotides (Appelberg, 2006).
The complementary nature of a metabolite that causes macrophage inactivation due to its production and shows importance for pathogenic growth was also found for hyaluronan. Hyaluronan production was one of the most suppressive of macrophage activation and has been previously been shown to be a critical substrate that enhances Mycobacterium tuberculosis proliferation (Hirayama et al, 2009). In addition, our previous study indicated that hyaluronan synthase is active only in pulmonary M. tuberculosis infections versus latent infections (Bordbar et al, 2010). Finally, DOPEG production that occurs via monoamine oxidase (MAO)-mediated norepinephrine degradation was also suppressive of macrophage activation. Our network finding of the suppressive properties of DOPEG production is supported by a recent study that demonstrated metabolic inactivation of catecholamines occurs via upregulated MAO in LPS-stimulated macrophages to regulate intensity of inflammatory injury (Flierl et al, 2007).
In general, metabolite production is metabolically taxing to macrophage activation phenotypes (Figure 2); however, it is sometimes more efficient to secrete nitrogen by-products, such as for urea, ammonia, and glutamate, the only metabolites through which its production had a positive effect on activation. In particular, urea and glutamate production arises from the respective use of arginine and glutamine. It has been experimentally shown that urea supplementation in the media of RAW 264.7 cells inhibits NO production (Prabhakar et al, 1997). Prabhakar et al noted that the mechanism for NO production inhibition is not related to transcription, as inducible NO synthase mRNA levels are the same in control and urea spiked samples. However, it is possible that the addition of urea in the media perturbs homeostatic cycling of arginine, thereby inhibiting NO production. In addition, the inhibition of glutaminase, an enzyme that produces glutamate from glutamine, is known to mediate activated immune response in macrophages (Yawata et al, 2008). As demonstrated later, proteomic data from LPS-activated RAW 264.7 cells indicate increased glutaminase activity during macrophage activation.
Overall, our results demonstrate that metabolite effectors with the highest sensitivity scores across macrophage functions and activation phenotypes are widely known to have an immunomodulatory role. Supplementation of cell culture media with glucose and arginine has been previously shown to increase nitrite production. Predicted suppressive modulators have also been previously confirmed experimentally through signaling regulatory mechanisms. The high correlation between our findings and experimental literature is of particular interest as our network approach does not explicitly account for signaling or regulation of metabolism. However, it is highly possible that the converging changes in metabolism during macrophage activation and suppression, as mediated by regulatory and signaling factors, may in fact have an important role in directing macrophage phenotypes.
Sampling analysis identifies potential intracellular mechanisms linked to immunomodulatory metabolites
The results of the sensitivity analysis demonstrated that predicted metabolites were supported by previous experimental studies on metabolic effectors. To understand the underlying metabolic pathways that are associated with the metabolic effectors, Monte Carlo sampling analysis was used to identify intracellular reaction changes linked to metabolites (i.e., metabolite uptake and production) with suppressive properties. In particular, we studied the most effective inhibitor (tryptophan) and the most demanding group of produced metabolites (nucleotides and deoxynucleotides). In total, 90 different conditions were sampled to determine the perturbed metabolic pathways and reactions of 9 metabolic exchanges on the 5 activating objectives, during uptake or synthesis compared with those without such activities.
First, we evaluated the suppressive effects of tryptophan uptake. Our sampling analysis indicates that the macrophage enters a ketogenic-like state during increased tryptophan uptake (Figure 3). Increased tryptophan catabolism (i.e., IDO induction) results in an increase in the production of reduced glutathione and the degradation of two ketogenic amino acids, leucine and lysine, as shown by increased activity of hydroxymethylglutaryl-CoA lyase, as well as decreased pyruvate dehydrogenase and increased pyruvate carboxylase activity, a switch characteristic of a ketogenic state (Wang and De Vivo, 2011). Interestingly, rats that are fed a ketogenic diet result in elevated levels of reduced glutathione (Jarrett et al, 2008), and fasting individuals in ketosis are known to have decreased inflammation (Garai et al, 2005).
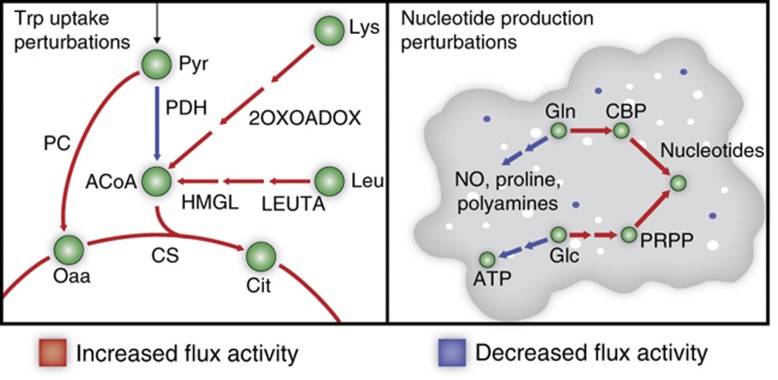
Figure 3.
Randomized sampling elucidates intracellular mechanisms for observed macrophage activation and suppression. Tryptophan induces a shift to a ketogenic-like state, increasing metabolic usage of leucine and lysine. To balance the redox potential shift, there is a significantly greater use of the malate-aspartate shuttle, diverting glutamate from activation pathways. In addition, increased nucleotide synthesis shifts metabolic resources toward nucleotide intermediates PRPP and CRP. PRPP and CRP are produced from glutamine and glucose, respectively, diverting metabolic resources from nitric oxide, proline, putrescine, and ATP generation.
Due to the change in redox potential, there is a cellular need to shuttle NAD+ and NADH across the mitochondrial membrane. Sampling analysis demonstrated a statistically significant increase in use of the malate-aspartate shuttle. The shuttle is dependent on glutamate, which in-silico analysis shows is derived from glutamine. Thus, glutamine is diverted from activation-associated metabolites (NO, proline, and putrescine) to balance the redox potential across the mitochondrial membrane. In the M1- and M2-reaction deletion analysis, we noticed a greater importance of the shuttle for M1 activation. With sampling, we saw the same result as the basal level of the malate-aspartate shuttle in M1 activation was greater than in M2 activation. However, forced tryptophan uptake significantly increased shuttle activity for both M1 and M2 activation.
Next, we evaluated eight nucleotides and deoxynucleotides whose synthesis was predicted to be especially taxing on activation properties. Intracellular changes induced by nucleotide production are drastically different than those for tryptophan uptake (Figure 3). As expected, de-novo purine and pyrimidine synthesis pathways are activated, including reactions pertaining to synthesis of IMP, UMP, and deoxynucleotides. Purine and pyrimidine synthesis requires 5-Phosphoribosyl diphosphate (PRPP) and carbamoyl phosphate (CBP), respectively. Sampling indicated that PRPP production via pentose phosphate pathway was increased resulting in suppression of glycolysis, pyruvate metabolism, and overall ATP production. There was also an increase in CBP synthesis from glutamine via CBP synthase; draining the availability of glutamine toward the arginine-derived synthesis of NO, proline, and putrescine.
We detected different inhibitory mechanisms for tryptophan uptake and nucleotide production on macrophage activation. However, the mechanisms often converged on glutamine and overall energy production, thus implicating intracellular metabolic shifts in glutamine utilization as a potential critical junction for immunomodulatory mechanisms. To further study and confirm the proposed mechanisms, we generated multiple levels of high-throughput data sets from LPS-activated RAW 264.7 cells.
Predicted metabolic activation phenotypes enhance mechanistic interpretation of experimental data for activated RAW 264.7 cells
Our network sensitivity and flux sampling analyses defined potential metabolic mechanisms associated with macrophage activation and inhibition. In particular, our results indicate that macrophage activation may be dependent on the flux divergence from glutamine/glutamate as a critical junction (Figure 4A). To experimentally confirm our predictions, we stimulated RAW 264.7 cells with LPS (derived from Salmonella typhimurium), a TLR4 agonist associated with M1 activation, to assess whether these mechanisms were associated with similar changes at the transcript, protein (2, 4, and 24 h after stimulation), and metabolite levels (24 h after stimulation) following macrophage activation.
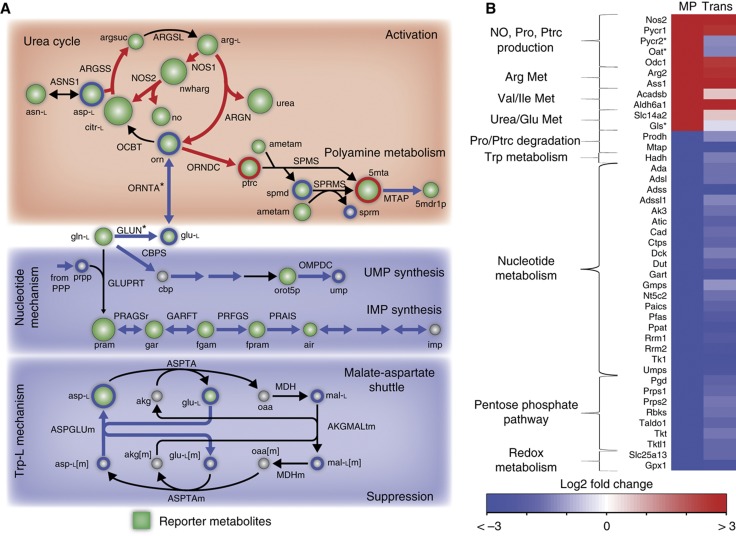
Figure 4.
High-throughput data support in-silico predictions. (A) Reporter metabolites provide a global analysis of the expression data. Major changes pertained to predicted pathways of activation and suppression. Green nodes are scaled by degree of enrichment. Circled metabolites in red and blue represent significantly changed metabolites detected by GC–MS. (B) Directionality of in-silico predictions was in high accordance with the transcriptional and proteomic response of LPS-stimulated cells. Pycr2, Oat, and Gls expression contradicted model predictions, but the proteomics data confirmed the predictions. Only 24 h transcriptomics data are shown due to sparsity of proteomic data. MP – Model Prediction, metabolite, and reaction abbreviations are provided in Supplementary information.
First, we performed a global, unbiased assessment of the primary metabolic changes during activation by calculating reporter metabolites based on the gene expression data in accordance with a published method (Patil and Nielsen, 2005). Reporter metabolites are nodes that are statistically enriched with transcriptional changes in the metabolic network and represent key regions that are significantly perturbed. A total of 105 statistically significant reporter metabolites (P<0.05) were identified for LPS-stimulated RAW cells. Reporter metabolites for LPS-stimulated macrophages at 24 h are shown (Figure 4A). Among the top 20 reporter metabolites, 12 are related to the predicted activating and suppressive mechanisms previously discussed: urea cycle, NO and putrescine production, and nucleotide biosynthesis. Thirteen extracellular reporter metabolites were also identified, eight of which were predicted to be important metabolic effectors through sensitivity analysis (including glutamine, arginine, glucose, and phenylalanine). Hence, the reporter metabolite results provide an unbiased confirmation that major transcriptional changes in metabolism during macrophage activation are highly correlated with the model-predicted regions of arginine and urea metabolism as well as nucleotide biosynthesis.
We then evaluated the directional changes of reactions involved with the most highly perturbed reporter metabolites (Figure 4B). We compared sampling predictions during activation and suppression with the associated genes and proteins in the transcriptomic and proteomic data. We found a highly significant (P<1e−5 through a permutation test) correlation between model predictions and high-throughput data (Figure 4B; see Supplementary information). As expected, LPS-stimulated cells showed significantly upregulated (P<0.05) genes and proteins involved with NO, proline, and putrescine production. In addition, metabolic pathways associated with proline and putrescine degradation were suppressed, suggesting accumulation of the characteristic M2-associated metabolites. Gene expression data also showed upregulated arginase (Arg2) and urea transporter (Slc14a2), as well as several genes (Acadsb, Aldh6a1) associated with BCAA degradation. In contrast with our predictions, we found that glutaminase (Gls) and ornithine aminotransferase (Oat) were significantly downregulated in the expression data. However, at the protein level, both Gls and Oat were upregulated and a previous study showed a respective 150% and 40% increase in Gls and Oat enzyme activity in LPS-activated versus resident macrophages (Newsholme et al, 1999). These experimental results provide further support that arginine and BCAA metabolism as well as urea and glutamate production are highly active during macrophage activation.
Conversely, metabolic genes and proteins involved in the predicted suppressive malate-aspartate shuttle and nucleotide synthesis were found to be significantly downregulated during LPS stimulation. The pentose phosphate pathway and nucleotide synthesis, including PRPP and CBP utilizing genes (Ppat and Cad), were significantly downregulated (Figure 4B). We also observed a gene involved with the malate-aspartate shuttle (Slc25a13) to be significantly downregulated.
Finally, we profiled ∼700 metabolites using GC–MS to determine altered metabolite levels during LPS activation. In all, 51 metabolites that were detected in both control and activated conditions had significant changes (P<0.05). In particular, we detected a significant increase in putrescine (ptrc), in accordance with transcription and proteomic data that suggested its accumulation (Figure 4A). Downstream metabolites spermine (sprm) and spermidine (spmd) were decreased, although an increase in 5′-deoxy-5′-(methylthio)adenosine (5mta), a by-product of spermine and spermidine synthesis, was indicated. Most notably, 11 metabolites associated with the model-predicted metabolic suppressors were significantly decreased: nucleotide metabolism (adn, amp, gmp, hxan, ins, ump), pentose phosphate pathway (r5p), malate-aspartate shuttle (asp-L, glu-L, mal-L), and tryptophan catabolism (kynate). Though metabolite level changes are not directly linked to predicted flux changes, these results are highly supportive of a systemic shift of metabolic resources away from nucleotide metabolism and other predicted suppressive pathways to activation-associated metabolic functions.
To further verify the observed divergence of nucleotide synthesis and activation functions, we re-evaluated time-course quantitative proteomic data of RAW 264.7 cells infected with Salmonella typhimurium from a previous study (Shi et al, 2009). Based on time-course proteomic data, seven detected proteins were associated with NO and proline production (Nos2 and Pycr2), nucleotide biosynthesis (Atic and Impdh2), and pentose phosphate pathway (G6pdx, Pgls, and Tkt). We found a similar trend between the LPS-stimulated and infection results (Supplementary Figure S3). In addition, the protein abundance changes followed a clear time-course trend, with the accumulation of activation proteins and a continual decrease in proteins associated with suppressive nucleotide pathways and pentose phosphate pathway during infection.
In summary, the intracellular metabolic signature predicted for macrophage activation from the tailored genome-scale metabolic network to the RAW cell line was significantly correlated with and was highly supported by three levels of high-throughput data sets. Our experimental results demonstrate that the regulation of glutamine/glutamate towards de-novo nucleotide synthesis and malate-aspartate shuttle may be an effective mechanism for modulating macrophage metabolic activation as the LPS-stimulated macrophage significantly downregulated those pathways during activation. In addition, we re-analyzed time-course proteomic data of infected RAW 264.7 cells, finding a similar trend as the LPS results. A comparison of our model-predicted changes with the full time-course transcriptomics and proteomics of LPS-stimulated macrophages as well as a list of significantly altered metabolites are available in Supplementary information.
Discussion
It is becoming clear that the two common denominators in many pathophysiological states are metabolism and inflammation (Mathis and Shoelson, 2011). Research areas, particularly in the cancer field which previously focused heavily on regulation and signaling controls, are now recognizing metabolic reprogramming as a predominant characteristic during disease progression. For example, well-established tumor suppressors that regulate cancer cell proliferation, such as p53, have been shown to converge on the control of central metabolism (Cairns et al, 2011). In this study, we present a case that metabolism similarly has a crucial role in the control of macrophage activation and, consequently, immune response. Notably, our systematic network analysis of macrophage metabolism highlights metabolic effectors that were previously established in the experimental literature and proposes potential mechanisms that are critically linked to activation.
In recent years, high-throughput technologies have emphasized the need to develop integrative data analysis tools with improved biological relevance (Palsson and Zengler, 2010). While most data-driven analysis approaches are purely statistical in nature, metabolic network-based approaches have demonstrated highly coherent dependencies at the gene expression level that are functionally correlated based on flux associations (Notebaart et al, 2008; Wessely et al, 2011). Our study further supports the notion that predicted flux-based associations provide a biologically accurate context for elucidating metabolic mechanisms from multiple layers of high-throughput data. By coupling model-based predictions with transcriptomic, proteomic, and metabolomic analyses on LPS-stimulated RAW 264.7 cells, we confirmed a consistent trend by which glutamine is a critical junction for activation and a possible contending link between nucleotide synthesis and macrophage activation via pentose phosphate pathway and CBP synthase exists. Our re-analysis of previously published proteomic data from Salmonella-infected RAW 264.7 cells demonstrated a similar divergent trend, indicating that metabolic network-based predictions can enhance the mechanistic interpretation of omics data than previously possible.
With a growing interest in metabolism as therapeutic targets (Vander Heiden, 2011), the results of our study indicate metabolic network-based approaches can be used to delineate metabolic mechanisms as immunotherapeutic targets. In particular, our study provides the framework for identifying metabolic signatures of macrophage activation that serve as potential therapeutic strategies during infectious states. Interestingly, our previous work on differential metabolism during M. tuberculosis infection in human alveolar macrophages showed that nucleotide synthesis, pentose phosphate pathway, hyaluronan synthase, and Vitamin D3 metabolism were solely active in macrophages with pulmonary and meningeal tuberculosis infections versus latent infections (Bordbar et al, 2010). Several macrophage-produced metabolites that suppressed activation phenotypes, particularly nucleotides and hyaluronan, have also been shown to be important substrates for intracellular pathogen replication and survival (Appelberg, 2006; Hirayama et al, 2009). Our current analysis suggests a complementary nature between pathogen auxotrophies and the mitigation of macrophage activation due to the synthesis of metabolites required for pathogen replication. These interactions are indicative of potential virulence mechanisms by intracellular pathogens that enable suppression of macrophage activation and serve as possible therapeutic strategies to treat persistent infectious states.
Regulation of immune cells has been traditionally explored from a signaling perspective, but recent studies have suggested that metabolic factors may influence the activities of immune cell responses (Newell et al, 2006; Kominsky et al, 2010; Mathis and Shoelson, 2011; Osborn and Olefsky, 2012). In particular, macrophage recruitment and activation has drawn considerable interest from the scientific community for its interfacing role between metabolism and immunity (Chawla et al, 2011; Odegaard and Chawla, 2011). While the relationship between metabolism and immunity has been historically explored from a nutritional perspective (Odegaard and Chawla, 2011), recent studies have demonstrated and characterized immune cell-secreted metabolites to be signaling modulators. For example, macrophage production of both Vitamin D3 and DOPEG have been shown to have potent immunosuppressive effects as a result of paracrine regulatory control to mitigate pro-inflammatory response (Helming et al, 2005; Flierl et al, 2007). While these regulatory mechanisms are well established, our analysis indicates alternative contributing factors that may be imposed by the complementing metabolic capabilities of effector immune cells. The ability to recapitulate most known metabolite effectors with our metabolic network analysis strongly suggests an underlying complementation in how metabolite signals are biochemically processed (i.e., degraded or synthesized) and its eventual immunomodulatory role (i.e., activating or suppressing). Hence, our study suggests metabolism as a potential remote sensor that mediates the activation status of macrophages through its metabolic capacities. Taken together, our results lend support to the notion that understanding reciprocity between metabolism and its regulatory signaling state may be critical to decoding mechanisms in immunity (Odegaard and Chawla, 2011) and other biological systems (McKnight, 2010), thus establishing potential application of metabolic systems biology toward the emerging field of immunometabolism.
Materials and methods
Tailoring the global human metabolic network to RAW 264.7
A full workflow for generating the reconstruction is provided in Supplementary information. The NCBI HomoloGene database was used to replace Recon 1 genes with murine Entrez Gene IDs. Expression data for untreated RAW 264.7 cells was obtained (Shell et al, 2005). Present and absent calls were made according to a probability distribution approximated for the log2-transformed, averaged values using a Gaussian mixture model (Chang et al, 2010). The processed data was integrated with reactions of the mouse metabolic network according to the gene-protein-reaction associations. The biomass function was adopted from Bordbar et al (2010) as the data used to construct it came mostly from mice. Two established algorithms that integrate high-throughput data with metabolic networks (Becker and Palsson, 2008; Shlomi et al, 2008) were used to construct the cell-specific networks. We also employed a new algorithm that improves upon the original GIMME algorithm through the integration of proteome-based objective functions called Gene Inactivity Moderated by Metabolism and Expression by Proteome (GIMMEp).
The GIMME algorithm (Becker and Palsson, 2008) was used to construct the RAW 264.7 cell line model through an integrated transcripto-proteomic approach. The original algorithm builds context-specific models by utilizing expression data to minimally activate a set of reactions that are necessary to fulfill a single user-defined reaction objective. This can be problematic given that there is no clear single objective in mammalian cells and tissues. To build the RAW 264.7-specific model, the GIMMEp algorithm integrated published proteomic data of untreated RAW 264.7 cells (Shi et al, 2009) as objective function cues that are evaluated separately with the original GIMME algorithm in an interative fashion. Hence, separate subnetworks are determined for each proteome-associated reaction objectives that are active on the basis of satisfying each objective function in turn. The subnetworks are then combined to create the finalized GIMMEp-based model. iMat (Shlomi et al, 2008) was also used to predict a flux distribution most consistent with the omics data. iMat was used as it is not biased by a particular objective. A macrophage-specific reaction subnetwork was derived from the iMat analysis.
The automated draft reconstruction process yielded three algorithmically tailored models (GIMME, iMAT, and GIMMEp) that were compared by their ability to perform the published 288 metabolic functions of Recon 1. Differences in metabolic functional capability were used as a classification system to target manual curation efforts in the literature. The final reconciled model was used in the study.
Functional characterization using flux balance analysis
A genome-scale reconstruction is converted into a mathematical format by representing the reactions and metabolites in a stoichiometric matrix (S). The rows of the matrix represent the metabolites in the network, while the columns represent the reactions. Flux balance analysis (FBA) is used to characterize the system. FBA is a linear programming-based mathematical operation to calculate the maximum flux through a particular reaction objective under mass balance (S·v=0) and thermodynamic/capacity constraints (lb,ub) (Equation 1) without the need for kinetic parameters. The objective vector (c) is a zero vector with a value of 1 corresponding to the reaction that is being maximized. An FBA primer is now available (Orth et al, 2010).
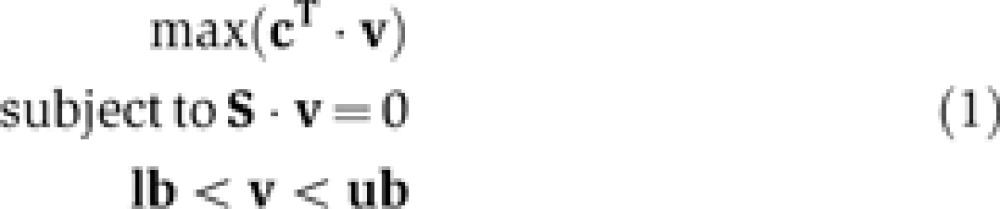 |
Uptake rates for major carbon sources (glucose, glutamine, pyruvate, and fatty acids) and oxygen were set from murine macrophage literature (Newsholme et al, 1986; Sato et al, 1987; Curi et al, 1988; Newsholme et al, 1999). Minimal supplementation of essential amino acids and choline of 0.1 mmol/cell gDW/h was required to generate biomass. In addition, sodium, chlorine, bicarbonate, and ammonia were allowed to enter the network freely.
Reaction deletion analysis
Deletion analysis was completed by iteratively removing a reaction from the network and determining the maximum value of the three objective functions associated with M1 and M2 activation. The two objective functions for M2 activation (proline and putrescine generation) were averaged and compared with M1 activation (NO production). Large differences (>10% difference from the original objective value) in reaction deletion for M1 and M2 were analyzed. Within the two M2 activation functions, some reactions had large differences (>10%) and were ignored in this analysis as they are not characteristic of M2 activation but rather specifically of proline or putrescine generation. There were only six such reactions.
Sensitivity and sampling analysis of metabolite exchanges
For each exchanged metabolite and objective function, sensitivity analysis was completed by iteratively fixing the exchange flux at 20 different flux values ranging from the minimum to maximum allowable flux and calculating the maximum objective value. To compare the effects on each of the objectives, we calculated the slope of the sensitivity curves. The sensitivity of metabolites to the objective functions was typically monotonic. However, 8 of the 355 tests were not monotonic (L-cysteine production on ATP, L-glutamate production on NADH, L-cystine uptake on NOS, NH4 uptake on PTRC, and NH4 production on ATP, NADH, NOS, PRO functions) where a change in direction of the function occurs at large non-physiological exchange values (>0.1 mmol/h/g cell DW). For these cases, the sensitivity slope was calculated for the monotonic region closer to an exchange of 0. Though threonine was predicted to be the most effective suppressant, the result is due to an artifact of the network analysis and those results were removed.
Randomized Monte Carlo sampling of the solution space constrained by metabolite exchanges and objective function value was used to identify reaction activity changes pertaining to the different phenotypes calculated from sensitivity analysis. Sampling was performed for both the first and last points by fixing the exchange flux and the objective, thus alternate solutions were inherently accounted for. The sampling procedure and differential reaction activity detection was done similarly to a previous study (Bordbar et al, 2010). Reactions that were differential in activity across a minimum of three objectives were only considered to filter out objective biased changes.
Quantitative analysis of stimulated RAW 264.7 macrophages
The RAW 264.7 (ATTC) cell line was stimulated for 0, 2, 4, and 24 h with LPS (from Salmonella enterica Serovar Typhimurium). Treated cells were washed twice with Dulbecco’s PBS and harvested for high-throughput analyses. Quantitative proteomics was done with accurate mass and time (AMT) tag. After digesting the proteins with trypsin, peptides were analyzed by liquid chromatography-tandem mass spectrometry on an LTQ XL (Thermo Scientific) (to build the mass tag (MT database) or an LTQ Orbitrap XL (Thermo Scientific) (for the quantitative measurements) mass spectrometer. Tandem mass spectra were searched with SEQUEST (Yates et al, 1995) against mouse IPI database and the reliable identifications were used to build the MT database, which was further matched to the orbitrap data using the VIPER tool (Monroe et al, 2007). Abundance values were processed and submitted to statistical test using DAnTE (Polpitiya et al, 2008).
Labeled cDNA was prepared as described (Jones et al, 2010). A mixture Cy3-labeled control cDNA and Cy5-labeled were hybridized to Agilent Mouse GE 4 × 44K v2 Microarray (Agilent Technologies) and processed. Image analysis and intra-chip normalization were performed with Feature Extraction 9.5.3.1 (Agilent). Data were analyzed with MeV (tm4.org) or with custom python scripts.
For metabolomics, cell suspensions were centrifuged. Ammonium bicarbonate was added to the cell pellet and metabolites were extracted with a chloroform/methanol mixture. Extracted metabolites were successively derivatized by two chemical reagents to enhance stability and volatility for gas chromatography-mass spectrometry (GC–MS) analysis (Kim et al, 2011). Samples were analyzed in biological triplicates and technical duplicates in a GC–MS system (Agilent GC 7890A coupled with a single quadrupole MSD 5975C) connected to HP-5 MS column (30 m × 0.25 mm × 0.25 μm; Agilent).
Omics data are available to the larger scientific community. The SysBEP.org project website provides links to each of the disseminated materials with the transcriptomics data disseminated via GEO (Barrett et al, 2011) under the accession number GSE35237 and proteomics data via ( http://omics.pnl.gov; Auberry et al, 2010). More detailed methods on experimental conditions and data generation are provided in Supplementary information.
Supplementary Material
Supplementary Figures S1–4, Supplementary Tables S1 and S2
All significantly changed metabolites
Raw 264.7 metablolic model in SBML and XLS format
Full heat map comparison of model predictions and transcriptions and proteomics data at all time points
Full data comparison of model predictions and transcriptomics and proteomics data at all time points with log2 fold changes and P-value
Differential metabolic functionality of three draft models and list of sources for curation for final reconciled model
Proteomic derived reactins used as GIMMEp objectives
Acknowledgments
We would like to thank Nathan E Lewis for helping us with the figures. This work was supported by the US National Institute of Allergy and Infectious Diseases agreement Y1-AI-8401-01 and the National Institutes of Health Grant GM068837. Proteomics analyses were performed in the Environmental Molecular Sciences Laboratory, a US DOE BER national scientific user facility at Pacific Northwest National Laboratory using instrumentation developed under support from the US Department of Energy (DOE) Office of Biological and the NIH National Center for Research Resources (RR018522).
Author contributions: AB, MLM, and DRH built the model, completed computational analysis, and wrote the manuscript. ESN and ACS-R grew and processed macrophage samples for omics analysis. MBJ, BCF, and SNP were responsible for transcriptomics analysis and interpretation. TOM, ESN, and Y-MK performed the metabolomics analysis and data interpretation. RDS provided access to unique mass spectrometry-capabilities. JNA conceived of and coordinated the omics generation and incorporation into the metabolic model. AB, MLM, DRH, and BOP conceived of the study. All authors read, edited, and approved the final manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alldridge LC, Harris HJ, Plevin R, Hannon R, Bryant CE (1999) The annexin protein lipocortin 1 regulates the MAPK/ERK pathway. J Biol Chem 274: 37620–37628 [DOI] [PubMed] [Google Scholar]
- Appelberg R (2006) Macrophage nutriprive antimicrobial mechanisms. J Leukoc Biol 79: 1117–1128 [DOI] [PubMed] [Google Scholar]
- Auberry KJ, Kiebel GR, Monroe ME, Adkins JN, Anderson GA, Smith RD (2010) Omics.pnl.gov: A Portal for the Distribution and Sharing of Multi-Disciplinary Pan-Omics Information. J Proteomics Bioinform 3: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A (2011) NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res 39(Database issue) D1005–D1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassit RA, Sawada LA, Bacurau RF, Navarro F, Martins E Jr., Santos RV, Caperuto EC, Rogeri P, Costa Rosa LF (2002) Branched-chain amino acid supplementation and the immune response of long-distance athletes. Nutrition 18: 376–379 [DOI] [PubMed] [Google Scholar]
- Baydoun AR, Bogle RG, Pearson JD, Mann GE (1993) Arginine uptake and metabolism in cultured murine macrophages. Agents Actions 38 Spec No: C127–C129 [DOI] [PubMed] [Google Scholar]
- Becker SA, Palsson BO (2008) Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol 4: e1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Lewis NE, Schellenberger J, Palsson BO, Jamshidi N (2010) Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol Syst Biol 6: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW (2011) Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95 [DOI] [PubMed] [Google Scholar]
- Carbonnelle-Puscian A, Copie-Bergman C, Baia M, Martin-Garcia N, Allory Y, Haioun C, Cremades A, Abd-Alsamad I, Farcet JP, Gaulard P, Castellano F, Molinier-Frenkel V (2009) The novel immunosuppressive enzyme IL4I1 is expressed by neoplastic cells of several B-cell lymphomas and by tumor-associated macrophages. Leukemia 23: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RL, Xie L, Bourne PE, Palsson BO (2010) Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput Biol 6: e1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapekar MS, Zaremba TG, Kuester RK, Hitchins VM (1996) Synergistic induction of tumor necrosis factor alpha by bacterial lipopolysaccharide and lipoteichoic acid in combination with polytetrafluoroethylene particles in a murine macrophage cell line RAW 264.7. J Biomed Mater Res 31: 251–256 [DOI] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, Goh YP (2011) Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol 11: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo DM, Valdivia RH, Monack DM, Falkow S (1998) Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30: 175–188 [DOI] [PubMed] [Google Scholar]
- Curi R, Newsholme P, Newsholme EA (1988) Metabolism of pyruvate by isolated rat mesenteric lymphocytes, lymphocyte mitochondria and isolated mouse macrophages. Biochem J 250: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma M, Lewis CE (2011) Cancer: Macrophages limit chemotherapy. Nature 472: 303–304 [DOI] [PubMed] [Google Scholar]
- de Souza LF, Jardim FR, Sauter IP, de Souza MM, Bernard EA (2008) High glucose increases RAW 264.7 macrophages activation by lipoteichoic acid from Staphylococcus aureus. Clin Chim Acta 398: 130–133 [DOI] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO (2000) Robustness analysis of the Escherichia coli metabolic network. Biotechnol Prog 16: 927–939 [DOI] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl MA, Rittirsch D, Nadeau BA, Chen AJ, Sarma JV, Zetoune FS, McGuire SR, List RP, Day DE, Hoesel LM, Gao H, Van Rooijen N, Huber-Lang MS, Neubig RR, Ward PA (2007) Phagocyte-derived catecholamines enhance acute inflammatory injury. Nature 449: 721–725 [DOI] [PubMed] [Google Scholar]
- Garai J, Lorand T, Molnar V (2005) Ketone bodies affect the enzymatic activity of macrophage migration inhibitory factor. Life Sci 77: 1375–1380 [DOI] [PubMed] [Google Scholar]
- Griscavage JM, Rogers NE, Sherman MP, Ignarro LJ (1993) Inducible nitric oxide synthase from a rat alveolar macrophage cell line is inhibited by nitric oxide. J Immunol 151: 6329–6337 [PubMed] [Google Scholar]
- Guo S, Wietecha TA, Hudkins KL, Kida Y, Spencer MW, Pichaiwong W, Kojima I, Duffield JS, Alpers CE (2011) Macrophages are essential contributors to kidney injury in murine cryoglobulinemic membranoproliferative glomerulonephritis. Kidney Int 80: 946–958 [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753–766 [DOI] [PubMed] [Google Scholar]
- Hasko G, Cronstein BN (2004) Adenosine: an endogenous regulator of innate immunity. Trends Immunol 25: 33–39 [DOI] [PubMed] [Google Scholar]
- Heilbronn LK, Campbell LV (2008) Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr Pharm Des 14: 1225–1230 [DOI] [PubMed] [Google Scholar]
- Helming L, Bose J, Ehrchen J, Schiebe S, Frahm T, Geffers R, Probst-Kepper M, Balling R, Lengeling A (2005) 1alpha,25-Dihydroxyvitamin D3 is a potent suppressor of interferon gamma-mediated macrophage activation. Blood 106: 4351–4358 [DOI] [PubMed] [Google Scholar]
- Hirayama Y, Yoshimura M, Ozeki Y, Sugawara I, Udagawa T, Mizuno S, Itano N, Kimata K, Tamaru A, Ogura H, Kobayashi K, Matsumoto S (2009) Mycobacteria exploit host hyaluronan for efficient extracellular replication. PLoS Pathog 5: e1000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO (2007) Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 1: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett SG, Milder JB, Liang LP, Patel M (2008) The ketogenic diet increases mitochondrial glutathione levels. J Neurochem 106: 1044–1051 [DOI] [PubMed] [Google Scholar]
- Jerby L, Shlomi T, Ruppin E (2010) Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol 6: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MB, Peterson SN, Benn R, Braisted JC, Jarrahi B, Shatzkes K, Ren D, Wood TK, Blaser MJ (2010) Role of luxS in Bacillus anthracis growth and virulence factor expression. Virulence 1: 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Metz TO, Hu Z, Wiedner SD, Kim JS, Smith RD, Morgan WF, Zhang Q (2011) Formation of dehydroalanine from mimosine and cysteine: artifacts in gas chromatography/mass spectrometry based metabolomics. Rapid Commun Mass Spectrom 25: 2561–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominsky DJ, Campbell EL, Colgan SP (2010) Metabolic shifts in immunity and inflammation. J Immunol 184: 4062–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D, Shoelson SE (2011) Immunometabolism: an emerging frontier. Nat Rev Immunol 11: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL (2010) On getting there from here. Science 330: 1338–1339 [DOI] [PubMed] [Google Scholar]
- Mellor AL, Munn DH (2004) IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol 4: 762–774 [DOI] [PubMed] [Google Scholar]
- Moeslinger T, Friedl R, Volf I, Brunner M, Baran H, Koller E, Spieckermann PG (1999) Urea induces macrophage proliferation by inhibition of inducible nitric oxide synthesis. Kidney Int 56: 581–588 [DOI] [PubMed] [Google Scholar]
- Moffett JR, Namboodiri MA (2003) Tryptophan and the immune response. Immunol Cell Biol 81: 247–265 [DOI] [PubMed] [Google Scholar]
- Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD (2007) VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23: 2021–2023 [DOI] [PubMed] [Google Scholar]
- Mosser DM (2003) The many faces of macrophage activation. J Leukoc Biol 73: 209–212 [DOI] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP (2008) Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8: 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C, Newsholme P (1998) Importance of glutamine metabolism in murine macrophages and human monocytes to L-arginine biosynthesis and rates of nitrite or urea production. Clin Sci (Lond) 95: 397–407 [PubMed] [Google Scholar]
- Newell MK, Villalobos-Menuey E, Schweitzer SC, Harper ME, Camley RE (2006) Cellular metabolism as a basis for immune privilege. J Immune Based Ther Vaccines 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme P, Costa Rosa LF, Newsholme EA, Curi R (1996) The importance of fuel metabolism to macrophage function. Cell Biochem Funct 14: 1–10 [DOI] [PubMed] [Google Scholar]
- Newsholme P, Curi R, Gordon S, Newsholme EA (1986) Metabolism of glucose, glutamine, long-chain fatty acids and ketone bodies by murine macrophages. Biochem J 239: 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme P, Curi R, Pithon Curi TC, Murphy CJ, Garcia C, Pires de Melo M (1999) Glutamine metabolism by lymphocytes, macrophages, and neutrophils: its importance in health and disease. J Nutr Biochem 10: 316–324 [DOI] [PubMed] [Google Scholar]
- Notebaart RA, Teusink B, Siezen RJ, Papp B (2008) Co-regulation of metabolic genes is better explained by flux coupling than by network distance. PLoS Comput Biol 4: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Palsson BO, Papin JA (2009) Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Chawla A (2011) Alternative macrophage activation and metabolism. Annu Rev Pathol 6: 275–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh GS, Pae HO, Choi BM, Chae SC, Lee HS, Ryu DG, Chung HT (2004) 3-Hydroxyanthranilic acid, one of metabolites of tryptophan via indoleamine 2,3-dioxygenase pathway, suppresses inducible nitric oxide synthase expression by enhancing heme oxygenase-1 expression. Biochem Biophys Res Commun 320: 1156–1162 [DOI] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BO (2010) What is flux balance analysis? Nat Biotechnol 28: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn O, Olefsky JM (2012) The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med 18: 363–374 [DOI] [PubMed] [Google Scholar]
- Overbergh L, Decallonne B, Valckx D, Verstuyf A, Depovere J, Laureys J, Rutgeerts O, Saint-Arnaud R, Bouillon R, Mathieu C (2000) Identification and immune regulation of 25-hydroxyvitamin D-1-alpha-hydroxylase in murine macrophages. Clin Exp Immunol 120: 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer TM, Trevethick MA (2008) Suppression of inflammatory and immune responses by the A(2A) adenosine receptor: an introduction. Br J Pharmacol 153(Suppl 1): S27–S34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson B, Zengler K (2010) The challenges of integrating multi-omic data sets. Nat Chem Biol 6: 787–789 [DOI] [PubMed] [Google Scholar]
- Patil KR, Nielsen J (2005) Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA 102: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG II, Anderson GA, Smith RD (2008) DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24: 1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar SS, Zeballos GA, Montoya-Zavala M, Leonard C (1997) Urea inhibits inducible nitric oxide synthase in macrophage cell line. Am J Physiol 273(6 Part 1): C1882–C1888 [DOI] [PubMed] [Google Scholar]
- Raschke WC, Baird S, Ralph P, Nakoinz I (1978) Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 15: 261–267 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, Cascante M, Bosca L (2010) Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 185: 605–614 [DOI] [PubMed] [Google Scholar]
- Sakagami H, Kishino K, Amano O, Kanda Y, Kunii S, Yokote Y, Oizumi H, Oizumi T (2009) Cell death induced by nutritional starvation in mouse macrophage-like RAW264.7 cells. Anticancer Res 29: 343–347 [PubMed] [Google Scholar]
- Sato H, Watanabe H, Ishii T, Bannai S (1987) Neutral amino acid transport in mouse peritoneal macrophages. J Biol Chem 262: 13015–13019 [PubMed] [Google Scholar]
- Scheel J, Weimans S, Thiemann A, Heisler E, Hermann M (2009) Exposure of the murine RAW 264.7 macrophage cell line to hydroxyapatite dispersions of various composition and morphology: assessment of cytotoxicity, activation and stress response. Toxicol In Vitro 23: 531–538 [DOI] [PubMed] [Google Scholar]
- Shell SA, Hesse C, Morris SM Jr, Milcarek C (2005) Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem 280: 39950–39961 [DOI] [PubMed] [Google Scholar]
- Shi L, Chowdhury SM, Smallwood HS, Yoon H, Mottaz-Brewer HM, Norbeck AD, McDermott JE, Clauss TR, Heffron F, Smith RD, Adkins JN (2009) Proteomic investigation of the time course responses of RAW 264.7 macrophages to infection with Salmonella enterica. Infect Immun 77: 3227–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E (2008) Network-based prediction of human tissue-specific metabolism. Nat Biotechnol 26: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Stempin CC, Dulgerian LR, Garrido VV, Cerban FM (2010) Arginase in parasitic infections: macrophage activation, immunosuppression, and intracellular signals. J Biomed Biotechnol 2010: 683485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG (2011) Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov 10: 671–684 [DOI] [PubMed] [Google Scholar]
- Wang D, De Vivo D (2011) Pyruvate carboxylase deficiency. In: GeneReviews at GeneTests Medical Genetics Information Resource (database online). Copyright, 1997–2012. University of Washington, Seattle. Available at http://www.genetests.org (accessed 6 June 2012)
- Watanabe H, Bannai S (1987) Induction of cystine transport activity in mouse peritoneal macrophages. J Exp Med 165: 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessely F, Bartl M, Guthke R, Li P, Schuster S, Kaleta C (2011) Optimal regulatory strategies for metabolic pathways in Escherichia coli depending on protein costs. Mol Syst Biol 7: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR III, Eng JK, McCormack AL, Schieltz D (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67: 1426–1436 [DOI] [PubMed] [Google Scholar]
- Yawata I, Takeuchi H, Doi Y, Liang J, Mizuno T, Suzumura A (2008) Macrophage-induced neurotoxicity is mediated by glutamate and attenuated by glutaminase inhibitors and gap junction inhibitors. Life Sci 82: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Yeramian A, Martin L, Arpa L, Bertran J, Soler C, McLeod C, Modolell M, Palacin M, Lloberas J, Celada A (2006) Macrophages require distinct arginine catabolism and transport systems for proliferation and for activation. Eur J Immunol 36: 1516–1526 [DOI] [PubMed] [Google Scholar]
- Zhuang JC, Wogan GN (1997) Growth and viability of macrophages continuously stimulated to produce nitric oxide. Proc Natl Acad Sci USA 94: 11875–11880 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–4, Supplementary Tables S1 and S2
All significantly changed metabolites
Raw 264.7 metablolic model in SBML and XLS format
Full heat map comparison of model predictions and transcriptions and proteomics data at all time points
Full data comparison of model predictions and transcriptomics and proteomics data at all time points with log2 fold changes and P-value
Differential metabolic functionality of three draft models and list of sources for curation for final reconciled model
Proteomic derived reactins used as GIMMEp objectives