Abstract
In this review, we elucidate the mechanisms of Aβ oligomer toxicity which may contribute to Alzheimer’s disease (AD). In particular, we discuss on the interaction of Aβ oligomers with the membrane through the process of adsorption and insertion. Such interaction gives rises to phase transitions in the sub-structures of the Aβ peptide from α-helical to β-sheet structure. By means of a coarse-grained model, we exhibit the tendency of β-sheet structures to aggregate, thus providing further insights to the process of membrane induced aggregation. We show that the aggregated oligomer causes membrane invagination, which is a precursor to the formation of pore structures and ion channels. Other pathological progressions to AD due to Aβ oligomers are also covered, such as their interaction with the membrane receptors, and their direct versus indirect effects on oxidative stress and intraneuronal accumulation. We further illustrate that the molecule curcumin is a potential Aβ toxicity inhibitor as a β-sheet breaker by having a high propensity to interact with certain Aβ residues without binding to them. The comprehensive understanding gained from these current researches on the various toxicity mechanisms show promises in the provision of better therapeutics and treatment strategies in the near future.
Keywords: molecular dynamics simulation, Alzheimer’s disease, amyloid β peptide, amyloid β oligomer toxicity mechanism, curcumin
1. Introduction
The pathogenesis of Alzheimer’s disease (AD) is characterized by the aggregation of amyloid β peptides leading to extracellular senile plaque, and the formation of intracellular neurofibrillary tangle by the hyperphosphorylated tau protein. These structures have the detrimental effects of causing a significant loss of neurons and synapses, which gives rise to the state of Alzheimer’s disease. Amyloid β peptide (Aβ) is cleaved from the amyloid precursor protein (APP) and it usually possesses 36–43 amino acids. Aβ peptides are known to self-assemble into dimer, trimer and higher-order oligomers, which are believed to be the main source of toxicity by causing the death of neurons [1,2]. The mechanism of the toxicity has been studied extensively from both the experimental and theoretical perspectives which are summarized here: (1) activation of inflammatory effects by interacting directly with the membrane [3]; (2) induction of oxidative stress [4] through the formation of metal-Aβ complex [5,6]; (3) disruption of membrane receptors’ function by intimate binding [7]; (4) formation of membrane pore [8–11] and alteration of ionic homeostasis [12,13] across the membrane; and (5) modification of the structure of certain DNA by the process of attachment [14].
In this review, we shall focus on the oligomers-membrane interaction and discuss the underlying mechanisms of the toxicity and their consequences. In this respect, we shall first exhibit the structure of small oligomers in aqueous environment, the process of Aβ adsorption, insertion, aggregation and ion channel/pore formation, as well as a brief review on the toxicity of intra-neuronal Aβ. We then bring out the relationship between Aβ and the membrane receptors, and discuss on the contribution of Aβ to oxidative stress. The secondary structure evolution of Aβ is then highlighted by means of the α-helix to β-sheet phase transition from the point of view of statistical physics of coarse-grained models. Finally, we examine into the potential inhibitory influence of the curcumin molecules on Aβ oligomers formation from the viewpoint of molecular dynamic (MD) simulations, with the inclusion of relevant experimental evidences and validations.
2. Aβ Oligomers in Aqueous Environment
Presently, there is intense interest in elucidating the structures of Aβ oligomers. Unlike amyloid fibril, whose structural understanding has been developed over the past decades, less is known on the structures of Aβ oligomers in aqueous environment. A mature Aβ1–42 amyloid fibril is known to consist of a β-strand-turn-β-strand motif, which is adopted by its residues 18–42 with the β-sheet being in a parallel, in-register organization, while its residues 1–17 are mainly in the form of a disordered structure [15]. It is formed by the nucleation-dependent self-assembly of Aβ [16] via a series of cascade neuropathogenic process [17]. The β-sheet-rich mature amyloid fibril and the monomeric Aβ are found to be far less toxic than the soluble Aβ oligomers.
Although there is limited experimental information on the structures of Aβ oligomers due to its propensity to form soluble aggregates in comparison to the amyloid fibrils, improved understanding has been achieved through computer simulations based on REMD, MD and Monte Carlo approaches [18–24]. These computations investigate into the formation and conformational properties of small Aβ oligomers from dimers to hexamers (in particular, dimers). They serve to explore into the nature of the toxicity of these oligomers and to complement the limitation of experiments in capturing the transient oligomeric states. These studies have revealed that Aβ1–42 oligomers are quasi-spherical in shape with a hydrophobic core and a hydrophilic surface, with enhanced exposure to the solvent at the region D1 to D7 of its N terminal. This indicates that oligomerization is driven by hydrophobic effects since it is more favorable energetically for the hydrophobic residues to hide away from the solvent and to form inter-molecular contacts with each other, although electrostatic forces are also known to play an important role. The identical interaction governing the oligomerization and fibrillization may explain the similar cross-β structure that is found in the Aβ oligomers (especially the toxic Aβ1–42 fibrillar oligomers) and the mature amyloid fibril. However, in contrast to the amyloid fibril, antiparallel-β sheets are observed in the oligomers instead of the fibrill-like parallel, in-register β-sheet structure [24]. During dimerization, it is also observed that there is a consistent reduction in the α-helical content with an increase in β-sheet structure. The residues ILE-41 and ALA-42 and the formation of salt-bridge between D23-K28 are found to be important in the dimer formation [23]. Notably, the free energy landscape of Aβ42 dimers is found to be complex and broad, indicating its greater tendency to form hydrophobic contacts and β-sheet structures relative to the dimers of Aβ40 [22,23]. More importantly, it is uncovered that a small difference in the Aβ primary structures can lead to significant differences in the resulting oligomer structures. This implies that to anticipate and fathom the associated toxicity properties of the eventual oligomer is not a straightforward task.
3. Aβ Adsorption and Insertion Mechanism
The influence of membrane surface on the adsorption and aggregation of Aβ peptides have been investigated on solid surfaces, monolayer bilayers, self-assembled monolayers (SAMs), implicit membrane models, and models that mimic membrane structures [25–30]. The general observation is that solid surfaces promote the self-assembly of Aβ peptides. The main driving forces for the association have been attributed to dehydration effects and electrostatic interactions. A detailed review on Aβ interaction with solid surface can be found in Reference [25]. On the other hand, the adsorption process of Aβ, ranging from dimer to hexamer, towards a self-assembled monolayers surface which is capped by the COOH, CH3 and OH groups, is also explored [26,27]. These researches have determined that the strength and extent of the adsorption on the SAMs surface are related to the binding sites. Furthermore, the Aβ is observed to change its conformation and reorient itself in order to adopt a more energetically favorable association during the adsorption process. However, the conformational change of the oligomers is found to be slowed down by the SAMs relative to oligomers that are placed solely in water. Extensive MD simulation studies have found that trimers and tetramers have well-preserved β-sheet structures that act as seed for future oligomerization and fibrillization. The hydrophobic effects, electrostatic interactions and water-mediated dewetting transition have been highlighted as the main driving forces behind these aggregation process [16,31–33].
Implicit membrane model with short Aβ fragments has been used to investigate the interaction of Aβ with the membrane and the folding process of Aβ in the membrane environment [28]. An interesting observation in this study is that the C-terminus of Aβ (25–35) first forms an α-helical structure in the hydrophilic region of the membrane before the Aβ attempts to knock into the hydrophobic membrane core through its hydrophobic residues. The study of Aβ peptides adsorption on different membrane monolayers has revealed that the adsorbed Aβ exhibits β-sheet structure with its orientation aligned parallel to the air-water interface and the lower pressure lipid surface [29]. The association of Aβ with the monolayer has the effect of increasing the surface pressure of the layer. On the other hand, the interaction of Aβ with the membrane bilayer affects both the order and fluidity of the bilayer. This is observed when the two fragments of Aβ1–28 and Aβ25–40 are incorporated into the bilayer. While the hydrophobic group of Aβ25–40 is observed to locate inside the hydrophobic core region, Aβ1–28 shows a greater propensity to interact with the hydrophilic region of the membrane [30]. In comparison to Aβ1–28, Aβ25–40 is found to induce larger membrane perturbation and alteration.
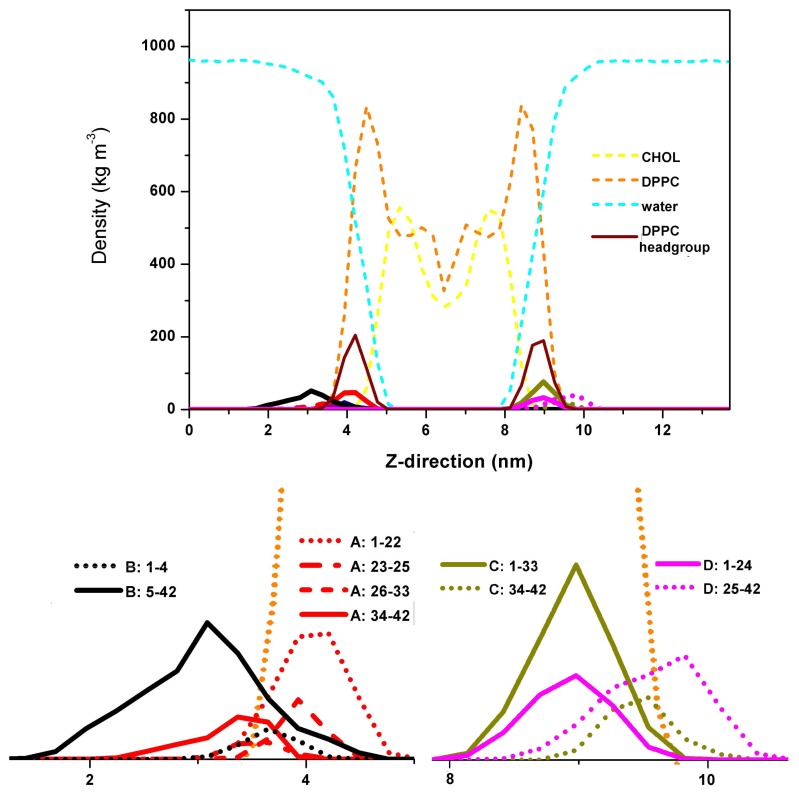
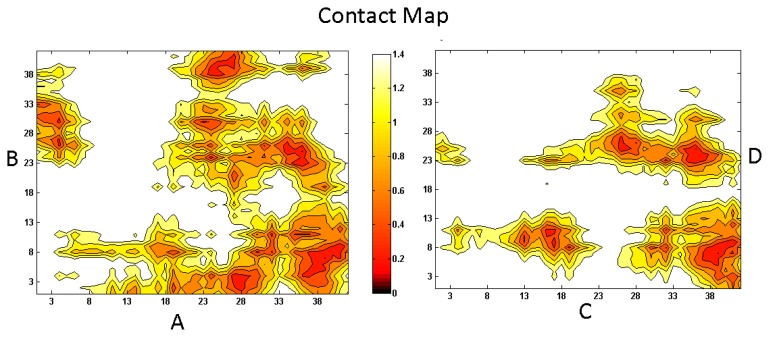
In our investigations, however, we have uncovered that while the C-terminus of the Aβ peptide remains outside the membrane, the N-terminus tends to bury inside the DPPC headgroup as a result of the strong protein-lipid interactions. We have considered two pre-formed Aβ dimers in our simulations, which consist of 4 Aβ peptide chains: A, B, C and D. They are represented by the GROMOS96 43a1 force field. These peptides are rich in β-sheet contents and they are placed at each side of the pre-equilibrated dipalmitoylphosphatidylcholine (DPPC) and cholesterol (CHOL) mixed bilayer model, with an initial 2 nm distance from the membrane surface. The membrane model is represented by the 43A1-S3 parameter set [34–38]. We have used 31β 777 water molecules with the addition of 103 Na+ and 95 Cl− ions to neutralize the system and reach the ions concentration of 0.1 mol/L. The temperature was kept at 323 K using the Nosé–Hoover coupling scheme [39,40]. The linear constraint solver (LINCS) algorithm [41] with a 2 fs integration time step was employed to constrain all the bonds in the simulation. Our results have indicated that the dimers mainly adsorb and reside on the membrane surface during the 1.2 μs simulations. The partial density functions of the dimers, water molecules, and the constituents of the mixed bilayer during the last 100 ns are given in Figure 1. The result here shows that the N-terminus of chain A, from residues 1–22 and 26–33, is fully inserted into the DPPC headgroup, while the C-terminus remains outside the membrane. On the other hand, chain B is observed to interact with chain A and has correspondingly less interactions with the membrane. Similar to chain A, the N-termini of chains C and D from 1–33 and 1–24 respectively are totally buried within the DPPC headgroup. Their C-termini, however, are found to stay outside the membrane. From Figure 2, we see that the dimer which is made up of chains A and B is more stable than the dimer formed by chains C and D. Furthermore, it is observed that Aβ peptides adsorption onto membrane surface do not always have stronger protein-protein interaction within the dimer. The perturbation of the bilayer due to the association of Aβ dimers is also found to be insignificant.
Figure 1.
Partial density function along the Z-direction.
Figure 2.
The residue contact map of each dimer. The coloring scheme is based on the inter-residue distance.
4. Membrane-Mediated Aβ Aggregation Mechanism
The toxicity of Aβ oligomers originates from the cleavage of APP with subsequent deposition of excessive Aβ peptides on the membrane surface, and to the cleaved peptides that fail to exit and release from the membrane into the extracellular space which form intracellular neurofibrillary tangle. Thus, a good understanding on the toxicity mechanisms requires knowledge on the possible structures of the transmembrane parts of the peptide and the detailed cleavage scenario of APP [42]. In particular, in order to find the possible aggregation behavior inside the membrane [18], it is necessary to examine the interaction of Aβ with the membrane, and cumulative evidence has suggested that elevated cholesterol level in the membrane plays an important role in increasing the risk of Alzheimer’s disease [43]. Indeed, research has found that Aβ is produced in the cholesterol-rich areas, which is also known as the lipid rafts. Such raft-like heterogeneous membrane environment which consists of ganglioside and cholesterol [44–46] has been extensively used to study the interaction of Aβ with the membrane environment. Study has proposed that lipid rafts accelerate the aggregation of Aβ [47], with the controversial results of cholesterol inhibiting the interaction of Aβ with the gangliolipids [48] while promoting the interaction of Aβ with 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayer[49]. Our research has revealed that the interaction between Aβ and lipids has facilitated the aggregation of the Aβ peptides. However, the interaction between Aβ and the cholesterol is inversely correlated with the extent of the peptide-peptide interactions [18]. The depletion of cholesterol or gangliosides has been shown to significantly reduce the amount of Aβ and its accumulation [46,50]. In fact, the aggregation of Aβ to fibrills is mediated by the gangliosides on the lipid rafts, where a transition from the alpha-helix-rich conformation to the β-sheet-rich conformation is observed. Thus, the constituents of the raft-like membrane strictly control the amyloid formation [51].
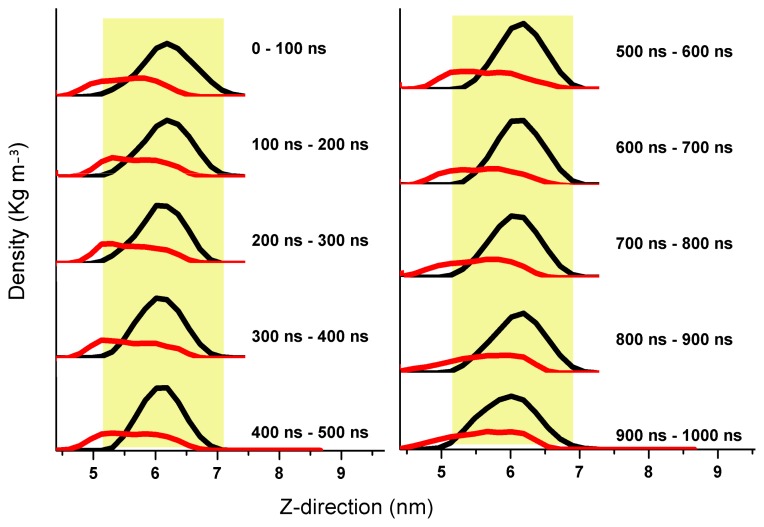
To gain a better understanding on the underlying interaction mechanism between Aβ, cholesterol and lipids, and the aggregation process of Aβ in a raft-like environment, we have arranged three full-length Aβ peptides aligning in a parallel configuration in the vicinity of a DPPC and cholesterol mixed bilayer, with residues 1–27 on the surface of the membrane and 28–42 inside the membrane. The detailed simulation information can be accessed from Ref. [18]. Our 1 μs long simulation shows that while residues 1–27 predispose to interact with the aqueous-membrane interface region, residues 28–42 incline to remain inside the hydrophobic core region (see Figure 3) [18]. The oligomer is found to attach to the sunken raft-like membrane surface forming a conglomerate of defects and disordered cholesterol molecules. Cholesterol further enhances the pre-existing hydrogen-bond network between Aβ and DPPC and promotes the incorporation of Aβ into the membrane [52]. However, the interaction between cholesterol and Aβ competes with the Aβ peptide-peptide interactions such that cholesterol hardly facilitates the aggregation of Aβ once Aβ has been immersed into the membrane [18]. Nonetheless, cholesterol is observed to facilitate the formation of pore/channel in the membrane by binding directly to the Aβ during the adsorption process [53].
Figure 3.
The density function of Aβ in the mixed bilayer environment along the Z-direction. Residues 1–27 are indicated by black line, and residues 28–42 by red line. The light yellow background shows the DPPC headgroup region. For detailed system information see Reference [18].
5. Intraneuronal Aβ Accumulation Mechanism
The intraneuronal accumulation of Aβ may precede the generation of Aβ plaque and neurofibrillary tangles formation [54], which may or may not [55] result in a series of pathological alterations like cognitive impairment, selective neuron loss, and axonopathy [56]. Since the intraneuronal Aβ17–42 secreted by α- and γ-secretases from APP has been suggested to be a normal product of neuronal metabolism, it is controversial as to whether its presence represents a sign of neuronal pathology. In fact, the manner in which Aβ comes to exist inside the neuron is still not well understood. One theory presupposes that the Aβs get inside the neuron directly after cleavage. These Aβs then lead to neuronal death. After that they are released into the extracellular space and form amyloid plaques subsequently [57]. Another theory suggests that Aβ is first being released outside the neuron before its re-uptake into the neuron through endocytosis or via membrane receptors. Several receptors have been reported to be able to mediate and internalize the Aβ located outside the neurons. These receptors are: alpha 7 nicotinic acetylcholine receptor [58,59], the low-density lipoprotein receptor-related protein 1 [60], and scavenger receptor for advanced glycation end products (RAGE) [61]. A detailed review on different uptake and internalization processes can be found in References [62,63].
Next, let us look at several aspects of intraneuroal Aβ toxicity. The accumulation of intraneuronal Aβ has been speculated to affect intracellular trafficking [64] which disrupts fast axonal transport [65]. Furthermore, results from APP/PS1KI mouse AD models indicate that the intracellular Aβ can cause early synaptic deficits, cholinergic neuron loss, and hippocampus atrophy [66–69]. There are also emerging evidences showing that the disruption of neuronal functions and survival is indirectly attributed to the intraneuronal Aβ [70].
6. Ion Channels/Pore Formation by the Incorporation of Aβ into the Cell Membrane
While Aβ oligomers exert toxicity from disruption of membrane integrity through altering its dielectric property to binding and activation of membrane receptors, they can also give rise to pores that leak Ca2+ ions which causes an elevation in cytosolic calcium. The pores formed by Aβ oligomers are much less efficient in transporting ions than other membrane pores, such as the nuclear pore, the ion channels, and the bacterial pores, which have all undergone the process of optimization during evolution [71]. Here we shall mainly focus on pores formed by Aβ oligomers on synthetic and cellular membrane based on experimental observations, as well as those generated from computational models.
Arispe et al. first proposed the possibility of Aβ forming an ion channel in 1992 [72] which serves to increase the level of intracellular Ca2+ ions. Later, several models have emerged which includes the helix-turn-helix [12] and β-sheet-twist-antiparallel-β-sheet [10,11] morphologies for the membrane-bound Aβ pore structures. The secondary structure of the membrane-bound Aβ is affected by many factors [44,73], such as the pH, the constituents and property of the membrane, peptide concentration, and others. By means of basin-hopping global optimization, the strand-turn-strand motif has been identified to be the most stable membrane-spanning structure for monomers, with tetrameric and hexameric β-sheet subunits constituting the pore structure [74]. Other proposal includes a hexamer-of-hexamer ion channel model with stable 36-stranded β-barrel in the membrane. This model is consistent with experimental observations and has been further used to explain the consequent channel selectivity [12]. Images from solid state NMR and atomic force microscopy (AFM) has revealed the presence of hexagonal annular channels in Aβ containing membrane. These images strongly suggest a pore-like assembly with 6 subunits [75] (in the case of rectangular assembly, 4 subunits in the membrane were observed). Zn2+ ions, as well as other small molecules like MRS2481 and its enatiomeric species, have been proposed to inhibit the toxicity by blocking the calcium-permeable channels formed by the Aβ oligomer [76–78].
7. The Relationship of Aβ with Different Receptors
We have discussed the role of adsorption, insertion, aggregation and ion channel pore formation as key determinants of Aβ toxicity via interaction of Aβ with the membrane. In this section, we shall extend our review to the area when Aβ oligomers serve as pathogenic ligands by binding to different receptors and inducing deterioration and loss of synapses through a redistribution of receptors, which further leads to alteration of neuronal plasticity accompanied with oxidative stress [79,80]. Indeed, it is well known that Aβ oligomers can bind to several receptors [81–86] and initiate numerous signaling cascades and surface expression regulations.
Recent studies have indicated that Aβ oligomers can induce impairment to the transduction of signal in neuronal insulin receptors [81], and suppress the activation of insulin receptor substrate [87]. Further studies have shown that the insulin receptor impairment and synaptic deterioration can be mitigated by insulin via down-regulation of the Aβ oligomers binding sites [79]. Omega-3 fatty acids and curcumin [87] are also reported to be able to prevent synaptic dysfunction and neuronal loss by suppressing the inactivation of insulin receptors due to the Aβ oligomers.
Several mechanisms have been proposed on the role of N-methyl-D-aspartate (NMDA) receptors in Aβ toxicity [88,89]. This includes the possibility that Aβ activates NMDA receptors directly [90] or indirectly by regulating the downstream NMDA receptors. Experimental observations have shown that both synthetic and naturally-secreted Aβ possesses the ability to reduce surface NMDA receptors, and reducing Aβ would restore the surface expression of NMDA receptors [82]. On the other hand, the activation of NMDA receptors affects synaptic Aβ generation [91,92] which further induces the elevation of intracellular Ca2+ and apoptosis [86]. In order to prevent the Aβ oligomer toxicity, various NMDA receptor antagonists have been suggested [90,93,94].
It is recently proposed that the natively folded cellular prion protein (PrPC), which is involved in the development of nervous system through mediation of synaptic and neuroprotective roles [95–100] and promoting neurite outgrowth [101], is an Aβ oligomer receptor and is related to AD by mediating synaptic dysfunction induced via the Aβ oligomers [102]. However, it is currently controversial as to whether the presence of PrPC is necessary for Aβ to induce synaptic dysfunction because there are experimental evidences which indicate that PrPC is not essential and has no effects on the impairment of synaptic plasticity induced by the Aβ oligomers [103,104].
Aβ has been reported to show a very high binding affinity towards the alpha 7 nicotinic acetylcholine receptors (nAChRs) [105], and it directly modulates [106] and blocks the response of these nAChRs [107]. Meanwhile, nAChRs is known to promote intraneuronal Aβ aggregation [58] and exacerbates cognitive deficits and synaptic pathology [108]. However, this view is in conflict with another report which states that the absence of nAChRs can enhance Aβ accumulation [109] and hence worsen the cognitive deficit. Nonetheless, the neuroprotective role of nAChRs by counterbalancing the toxicity of Aβ oligomers has been proposed based on experimental observation [109]. For example, drugs like 2-(2-(4-bromophenyl)-2-oxoethyl)-1-methyl pyridinium (S 24, 795), which have been assessed to be able to reduce the interaction between Aβ and nAChRs, have been shown to enhance long-term potentiation [110–112]. Interestingly, one research group has found that Aβ does not bind with nAChRs and has no direct relationship with the nAChRs expression and activity. Instead, the Aβ may affect the nAChRs indirectly by attaching to the membrane and altering the property of the membrane, which then influences the membrane receptors inadvertently [113]. Finally, note that there are other receptors like P75 neurotrophin receptor (P75NTR) [85,114,115], serpin-enzyme complex receptor (SEC-R) [116,117], receptor for advanced glycosylation end-products (RAGE) [118,119], and scavenger receptor CD36 [120–122], which bind with the Aβ oligomers.
8. Oxidative Stress
Patients with AD are found to show an elevated level of oxidative stress, which is mainly characterized by protein, DNA and RNA oxidation, and lipid peroxidation [123,124]. Oxidative stress may induce the overproduction of Aβ peptides through the activation of β-secretase [125]. The excessive Aβ peptides may aggregate into toxic oligomers which in turn initiate the free radical process. This results in new oxidative stress, accompanied with increased macroautophagy and lysosomal ensuing apoptosis [126], and any additional overexpression of oxide synthase can bring about extra neuronal damage [124]. Oxidative stress can also change the protein structure and affect its function, leading to physiological alteration and pathological induction [124,127,128]. In fact, a series of oxidatively modified proteins have been identified in the brain of AD patients [124,129–131]. Met-35 of Aβ is believed to be the key residue involved in oxidative stress and the mutation of Met-35 has been shown to reduce the effect of toxicity in Aβ [123,132,133]. There are many food and compounds, such as the walnut and turmeric extract, which are anti-oxidant in nature. These products have been reported to be able to prevent the oxidative stress induced by Aβ and its associated apoptosis [134].
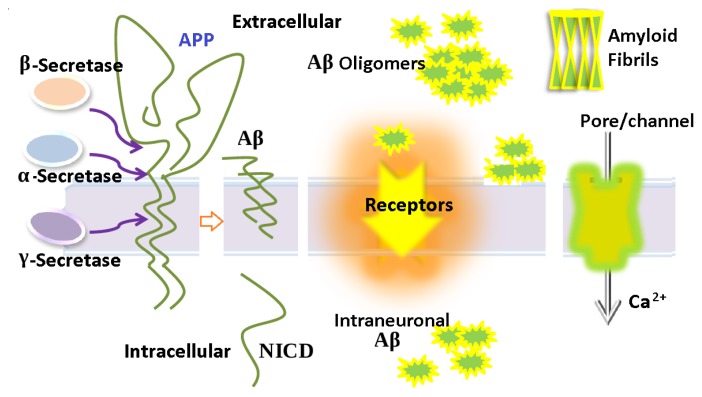
In summary, we have discussed the various mechanisms which contribute to Aβ oligomer toxicity: adsorption, insertion, aggregation and pore formation in the membrane, as well as the interaction of Aβ with the membrane receptors and oxidative stress. The pleiotropic effects of Aβ peptides can be seen in Figure 4. Currently, there are lots of therapeutic strategies being proposed to suppress the Aβ induced toxicity, such as the β- and γ-secretase inhibitors whose function is to reduce the production of Aβ peptides. There are also other strategies to overcome Aβ oligomer toxicity, such as the aggregation inhibitors, the pore/channel blockers etc.; a detailed review on these therapeutic approaches is available in Ref. [135].
Figure 4.
The cleavage of APP by α-, β- and γ-secretase and the production of Aβ peptides are shown on the left side of the figure. The following toxic mechanisms are illustrated in the figure: formation of Aβ oligomers and its further conversion to fibrils; disruption of membrane receptors; adsorption on membrane surface which alters the property of the membrane; formation of pore which causes the leakage of Ca2+; and the accumulation of intraneuronal Aβ.
9. Secondary Structure Phase Transition
One of the important underlying mechanisms that leads to the formation of Aβ oligomer is the occurrence of secondary structure phase transition within the Aβ peptide. In particular, the transition typically involves a phase transition from an α-helix or a random coil to a β-sheet configuration [136–138]. Such structural changes or misfolding can result in a loss of normal biological functions and is well known to be a source of diseases such as the Creutzfeldt–Jakob disease, in addition to the Alzheimer’s disease [139,140]. The phenomenon of protein misfolding has led to intensive studies on the subject of protein secondary structure phase transition. A fruitful approach in these studies involved coarse-grained models which include those developed by Zimm–Bragg, Lifson–Roig, Yakubovich et al., Ding et al., Hong–Lei, Yasar–Demir, Gibbs–DiMarzio etc. [141–149].
In our investigation, we have explored a coarse-grained protein model based on a polypeptide consisting of backbone atoms and hard-sphere side chains. The hard-sphere side chain serves to simulate the amino acid residue. In our approach, we have defined the base unit as crank instead of residue [150,151]. Each crank has a pair of dihedral angles φ and ψ, which are the only degrees of freedom in the polypeptide. In addition, bond lengths and bond angles are being held rigid in our model. We have studied phase transition from α-helix to β-sheet, and from β-sheet to random coil, using this model both numerically and theoretically. While our numerical study employs the model of Conditioned Self-Avoiding Walk (CSAW), our theoretical approach is based on the Hamiltonian formulation. Note that in both of these cases, we have assumed that the non-covalent forces arise solely from the hydrogen bonds. In our theoretical analysis, we have restricted the pair of dihedral angles of a crank to only five distinct states, such that the combination of crank states can adopt only the α-helix, β-sheet and random coil conformations. Such a protein model has shown to be reliable based on our computation of the normal modes of the α-helix and β-sheet [152].
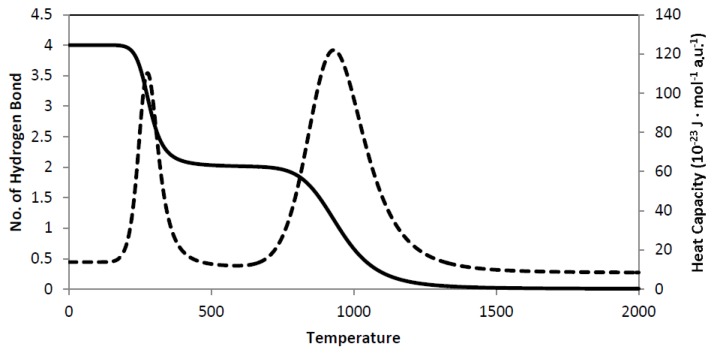
By taking polyalanine as a prototypical example, we have plotted an α-β -coil phase transition from the ensemble average of the number of hydrogen bonds against temperature (see Figure 5 and note that 1 a.u. is to be associated with a temperature of 1 K). Such a plot allows us to predict the transition temperature of the phase transition. We have also plotted an ensemble average of the heat capacity against temperature, where we observed two significant peaks for the α-β and the β-coil transition. These results have demonstrated that the transitions are associated with a first-order phase transition. In our model, we have assumed polyalanine to be in the gas phase, which implies that lower transition temperatures are to be expected in an aqueous environment [153].
Figure 5.
Plots of α-β-coil secondary structure phase transition for a seven crank polyalanine. Solid line represents the number of hydrogen bonds; dotted line represents the corresponding heat capacity. The transition temperatures are Tα–β = 300 a.u, and Tβ–coil = 950 a.u., respectively.
A key finding in both our theoretical and numerical studies relates to a biased in terms of stability for the hydrogen bond formed within the β-sheet and the α-helix. Our results show that hydrogen bonds are more stable in β-sheet due to a stronger hydrogen bond-to-hydrogen bond co-operative interaction [153]. This has important implication as we believe it is such a strong hydrogen bond-to-hydrogen bond interaction that leads to the aggregation behavior of the Aβ oligomer.
10. Curcumin
There are various small molecules, such as resveratrol [154], cyclodextrin [155,156], mitoxantrone and pixantrone [157], derivatives of Congo Red [158], curcumin and other compounds, which have been used to investigate the inhibitory effects on Aβ aggregation [159]. In fact, these molecules can serve as drugs or potential labelling agents for the diagnosis and monitoring of AD [160]. In particular, the molecule curcumin and its derivatives, with their antioxidant, anti-inflammatory, and anticancer properties, have made them very promising candidates for the treatment of Alzheimer’s disease.
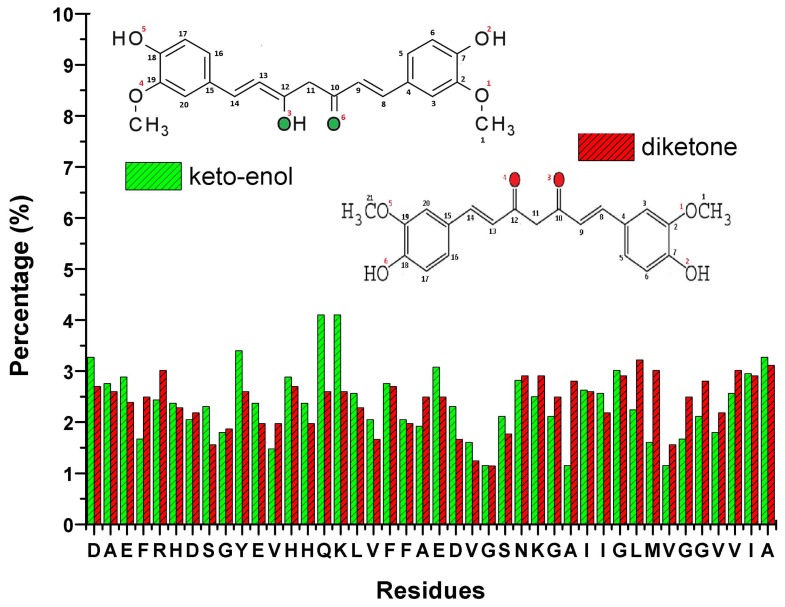
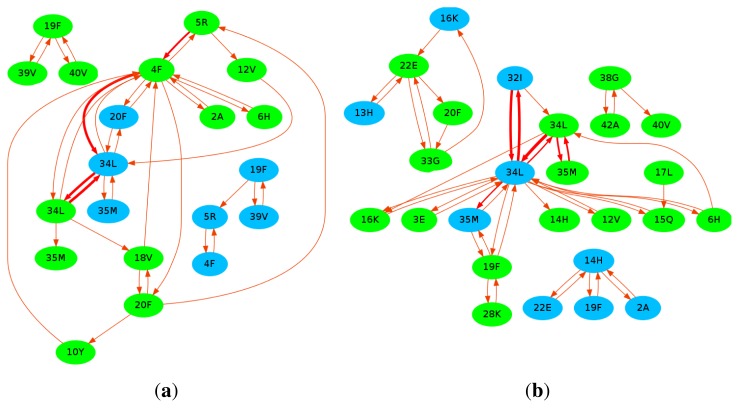
Curcumin can suppress the Tau phosphorylation and insulin receptor inactivation induced by Aβ. It can also improve insulin signaling and overcome cognitive deficits [87]. It is able to disrupt the existing Aβ plaques in vivo and partially restore the dystrophic dendrites at the same time [161]. Meanwhile, research has proposed that curcumin can reduce neurotoxicity by promoting the conversion of oligomers into fibril [162], with the inhibition of oxidative damage and tau hyperphosphorylation [163]. Solid state NMR has been employed to uncover the binding sites of curcumin on the full-length Aβ fibrils [164]. It was found that residues 12, 17–21, and the β-sheet structure of Aβ interact with the methoxy and/or hydroxy groups of curcumin. In order to investigate the potential binding sites of curcumin on Aβ, we had also performed 20 all-atom MD simulations for a system consisting of pre-formed Aβ dimer and curcumin molecules of both the diketone and keto-enol form. The detailed simulation information can be obtained from Reference [19]. After a total simulation time of 10 μs, we plotted the binding propensity of both the diketone and keto-enol form of curcumin in Figure 6. We observed that no specific residues strongly bind to curcumin although some residues have a higher chance of forming hydrogen bonds with curcumin. Furthermore, we observed that the –OH group of curcumin has more than twice the chance of forming a hydrogen bond than the methoxy group (diketone: –OH ~ 62%, –O–CH3 ~ 17%; keto-enol: –OH~ 50%, –O–CH3 ~ 20%). Interestingly, we discover that curcumin travels along certain common pathways as it moves around Aβ. These pathways are defined to consist of at least 4 steps and are common among at least two trajectories, where each step is being made from one residue of Aβ to another. The common pathways are displayed in Figure 7. Our results show that the most popular pathways are: 34LB → 34LA → 32IA → 34LA and 34LB → 34LA → 34LB → 34LA (see Figure 7). As curcumin traverses about Aβ, we observe that it serves the role of a β-sheet breaker. The frequent π–π stacking interactions between its aromatic ring and the aromatic side chain of HIS, TYR and PHE are important. Although these interactions are transient, they contribute indirectly to a reduction in the β-sheet content in the Aβ dimer [19].
Figure 6.
The binding propensity of curcumin towards the Aβ dimer.
Figure 7.
The common pathways of curcumin in the diketone (a) and the keto-enol form (b) of Aβ dimer. The blue colored domains represent the residues from chain A, while the green colored domains represent the residues from chain B. The thickness of the arrow indicates the popularity of the pathway. This figure was generated by Graphviz [165].
11. Conclusions
At present, there are still many unanswered and challenging questions for the experimentalists and theorists regarding the detailed mechanism of Aβ oligomers toxicity. Nevertheless, there are many excellent works that give a deeper insight into this area which unfortunately we are unable to cover and cite in this short review. To summarize, we list the current outstanding problems of Aβ oligomers toxicity as follow: (1) deciphering the unknown transmembrane structure of APP; (2) uncovering the configurational and structural diversity of Aβ oligomers; (3) elucidating the pathogenesis of intraneuronal Aβ accumulation; and (4) curing Alzheimer’s disease by directing the best antibodies at Aβ peptides. While there are pan-Aβ antibodies available at this point in time, these antibodies have the problem of discriminating the Aβ peptides from the much more abundant normal full-length APP, as well as the Aβ that is embedded in the APP fragments cleaved by the β-secretase. In conclusion, our review has demonstrated that the toxicity of Aβ oligomers arises from many factors, and in order for potential therapeutics and treatment strategy to be effective against them, extensive future research which aim to gain a more comprehensive account of the various toxicity mechanisms discussed here is both necessary and important.
Acknowledgements
The authors would like to thank Hwee Jin Soh from the High Performance Computing Center for his kind help in the provision of computational support and See-Wing Chiu for his suggestions. The authors would like to thank the unknown reviewers, whose comments have substantially improved the quality of this paper. This work is supported by the IDA Cloud Computing Call 4 and NTU Tier 1 grant RG 23/11 and the MOE AcRF Tier 1 grant RG52/08.
References
- 1.Kirkitadze M.D., Bitan G., Teplow D.B. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J. Neurosci. Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 2.Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C.M., Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 3.Verdier Y., Penke B. Binding sites of amyloid beta-peptide in cell plasma membrane and implications for Alzheimer’s disease. Curr. Protein Pept. Sci. 2004;5:19–31. doi: 10.2174/1389203043486937. [DOI] [PubMed] [Google Scholar]
- 4.Mutisya E.M., Bowling A.C., Beal M.F. Cortical cytochrome oxidase activity is reduced in Alzheimer’s disease. J. Neurochem. 1994;63:2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 5.Huang H.C., Lin C.J., Liu W.J., Jiang R.R., Jiang Z.F. Dual effects of curcumin on neuronal oxidative stress in the presence of Cu(II) Food Chem. Toxicol. 2011;49:1578–1583. doi: 10.1016/j.fct.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Atamna H., Boyle K. Amyloid-β peptide binds with heme to form a peroxidase: Relationship to the cytopathologies of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2006;103:3381–3386. doi: 10.1073/pnas.0600134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Walsh D.M., Rowan M.J., Selkoe D.J., Anwyl R. Block of long-term potentiation by naturally secreted and synthetic amyloid beta-peptide in hippocampal slices is mediated via activation of the kinases c-Jun N-terminal kinase, cyclin-dependent kinase 5, and p38 mitogen-activated protein kinase as well as metabotropic glutamate receptor type 5. J. Neurosci. 2004;24:3370–3378. doi: 10.1523/JNEUROSCI.1633-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang H., Zheng J., Lal R., Nussinov R. New structures help the modeling of toxic amyloidβ ion channels. Trends Biochem. Sci. 2008;33:91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Jang H., Arce F.T., Ramachandran S., Capone R., Lal R., Nussinov R. Structural convergence among diverse, toxic beta-sheet ion channels. J. Phys. Chem. B. 2010;114:9445–9451. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang H., Arce F.T., Ramachandran S., Capone R., Azimova R., Kagan B.L., Nussinov R., Lal R. Truncated β-amyloid peptide channels provide an alternative mechanism for Alzheimer’s disease and Down syndrome. Proc. Natl. Acad. Sci. USA. 2010;107:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang H., Arce F.T., Ramachandran S., Capone R., Lal R., Nussinov R. β-Barrel Topology of Alzheimer’s β-Amyloid Ion Channels. J. Mol. Biol. 2010;404:917–934. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafrir Y., Durell S., Arispe N., Guy H.R. Models of membrane-bound Alzheimer’s Abeta peptide assemblies. Proteins Struct. Funct. Bioinforma. 2010;78:3473–3487. doi: 10.1002/prot.22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sepulveda F.J., Parodi J., Peoples R.W., Opazo C., Aguayo L.G. Synaptotoxicity of Alzheimer beta amyloid can be explained by its membrane perforating property. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng J., Zhao C., Ren J., Qu X. Alzheimer’s disease Amyloid beta converting left-handed Z-DNAs back to right-handed B-form. Chem. Commun. 2010;46:7187–7189. doi: 10.1039/c0cc02049d. [DOI] [PubMed] [Google Scholar]
- 15.Lührs T., Ritter C., Adrian M., RiekLoher D., Bohrmann B., Dbeli H., Schubert D., Riek R. 3D structure of Alzheimer’s amyloid-β (1–42) fibrils. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi T., Mihara H. Peptide and protein mimetics inhibiting amyloid beta-peptide aggregation. Acc. Chem. Res. 2008;41:1309–1318. doi: 10.1021/ar8000475. [DOI] [PubMed] [Google Scholar]
- 17.Kirkitadze M.D., Kowalska A. Molecular mechanisms initiating amyloid beta-fibril formation in Alzheimer’s disease. Acta Biochim. Pol. 2005;52:417–423. [PubMed] [Google Scholar]
- 18.Zhao L.N., Chiu S.W., Benoit J., Chew L.Y., Mu Y. Amyloid β peptide aggregation in a mixed membrane bilayer: A molecule dynamic study. J. Phys. Chem. B. 2011;115:12247–12256. doi: 10.1021/jp2065985. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L.N., Chiu S.W., Benoit J., Chew L.Y., Mu Y. The effect of curcumin on the stability of Aβ dimers. J. Phys. Chem. B. 2012 doi: 10.1021/jp3034209. (in Press) [DOI] [PubMed] [Google Scholar]
- 20.Jang S., Shin S. Amyloid β-peptide oligomerization in silico: Dimer and trimer. J. Phys. Chem. B. 2006;110:1955–1958. doi: 10.1021/jp055568e. [DOI] [PubMed] [Google Scholar]
- 21.Urbanc B., Cruz L., Ding F., Sammond D., Khare S., Buldyrev S.V., Stanley H.E., Dokholyan N.V. Molecular dynamics simulation of amyloid β dimer formation. Biophys. J. 2004;87:2310–2321. doi: 10.1529/biophysj.104.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barz B., Urbanc B. Dimer formation enhances structural differences between amyloid β-protein (1–40) and (1–42): An explicit-solvent molecular dynamics study. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Côté S., Laghaei R., Derreumaux P., Mousseau N. Distinct dimerization for various alloforms of the amyloid-beta protein: Aβ1–40, Aβ1–42, and Aβ1–40(D23N) J. Phys. Chem. B. 2012;116:4043–4055. doi: 10.1021/jp2126366. [DOI] [PubMed] [Google Scholar]
- 24.Mitternacht S., Staneva I., Härd T., Irbäck A. Monte carlo study of the formation and conformational properties of dimers of Aβ 42 variants. J. Mol. Biol. 2011;410:357–367. doi: 10.1016/j.jmb.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Williams T.L., Serpell L.C. Membrane and surface interactions of Alzheimer’s Aβ peptide-insights into the mechanism of cytotoxicity. FEBS J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J., Wang Q.M., Liang G.Z., Zheng J. Molecular dynamics simulations of low-ordered Alzheimer beta-amyloid oligomers from dimer to hexamer on self-assembled monolayers. Langmuir. 2011;27:14876–14887. doi: 10.1021/la2027913. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q., Zhao J., Yu X., Zhao C., Li L., Zheng J. Alzheimer Abeta(1–42) monomer adsorbed on the self-assembled monolayers. Langmuir. 2010;26:12722–12732. doi: 10.1021/la1017906. [DOI] [PubMed] [Google Scholar]
- 28.Tsai H.H., Lee J.B., Tseng S.S., Pan X.A., Shih Y.C. Folding and membrane insertion of amyloid-beta (25–35) peptide and its mutants: Implications for aggregation and neurotoxicity. Protein. 2010;78:1909–1925. doi: 10.1002/prot.22705. [DOI] [PubMed] [Google Scholar]
- 29.Maltseva E., Brezesinski G. Adsorption of amyloid beta (1–40) peptide to phosphatidyl-ethanolamine monolayers. ChemPhysChem. 2004;5:1185–1190. doi: 10.1002/cphc.200400045. [DOI] [PubMed] [Google Scholar]
- 30.Ionov M., Klajnert B., Gardikis K., Hatziantoniou S., Palecz B., Salakhutdinov B., Cladera J., Zamaraeva M., Demetzos C., Bryszewska M. Effect of amyloid beta peptides A β1–28 and Aβ25–40 on model lipid membranes. J. Therm. Anal. Calorim. 2010;99:741–747. [Google Scholar]
- 31.Sgourakis N.G., Yan Y., McCallum S.A., Wang C., Garcia A.E. The Alzheimer’s peptides Aβ 40 and 42 adopt distinct conformations in water: A combined MD/NMR study. J. Mol. Biol. 2007;368:1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krone M.G., Hua L., Soto P., Zhou R., Berne B.J., Shea J.E. Role of water in mediating the assembly of Alzheimer amyloid Aβ 16–22 protofilaments. J. Am. Chem. Soc. 2008;130:11066–11072. doi: 10.1021/ja8017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glabe C.C. Amyloid accumulation and pathogensis of Alzheimer’s Disease: Significance of monomeric, oligomeric and fibrillar Aβ. Subcell. Biochem. 2005;28:167–177. doi: 10.1007/0-387-23226-5_8. [DOI] [PubMed] [Google Scholar]
- 34.Chiu S., Pandti S., Scott H.L., Jakobsson E. An improved united atom force field for simulation of mixed lipid bilayers. J. Phys. Chem. B. 2009;113:2748–2763. doi: 10.1021/jp807056c. [DOI] [PubMed] [Google Scholar]
- 35.Pandit S.A., Chiu S.W., Jakobsson E., Grama A., Scott H.L. Cholesterol packing around lipids with saturated and unsaturated chains: A simulation study. Langmuir. 2008;24:6858–6865. doi: 10.1021/la8004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandit S.A., Chiu S.W., Jakobsson E., Grama A., Scott H.L. Cholesterol Surrogates: A comparison of cholesterol and 16:0 ceramide in POPC Bilayers. Biophys. J. 2007;92:920–927. doi: 10.1529/biophysj.106.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandit S.A., Vasudevan S., Chiu S.W., Jakobsson E., Scott H.L. Sphingomyelin-cholesterol domains in phospholipid membranes: Atomistic simulation. Biophys. J. 2004;87:1092–1100. doi: 10.1529/biophysj.104.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu S.W., Vasudevan S., Jakobsson E., Mashl R.J., Scott H.L. Structure of sphingomyelin bilayers: A simulation study. Biophys. J. 2003;85:3624–3635. doi: 10.1016/S0006-3495(03)74780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;55:255–268. [Google Scholar]
- 40.Hoover W.G. Canonical dynamics: Equilibrium phase space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 41.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;113:1463–1472. [Google Scholar]
- 42.Miyashita N., Straub J.E., Thirumalai D., Sugita Y. Transmembrane structures of amyloid precursor protein dimer predicted by replica-exchange molecular dynamics simulations. J. Am. Chem. Soc. 2009;131:3438–3439. doi: 10.1021/ja809227c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leoni V., Solomon A., Kivipelto M. Links between ApoE brain cholesterol metabolism, tau and amyloid β-peptide in patients with cognitive impairment. Biochem. Soc. Trans. 2010;38:1021–1025. doi: 10.1042/BST0381021. [DOI] [PubMed] [Google Scholar]
- 44.Okada T., Wakabayashi M., Ikeda K., Matsuzaki K. Formation of toxic fibrils of Alzheimer’s amyloid beta-protein-(1–40) by monosialoganglioside GM1, a neuronal membrane component. J. Mol.Biol. 2007;371:481–489. doi: 10.1016/j.jmb.2007.05.069. [DOI] [PubMed] [Google Scholar]
- 45.Molander-Melin M., Blennow K., Bogdanovic N., Dellheden B., Månsson J.E., Fredman P. Structural membrane alterations in Alzheimer brains found to be associated with regional disease development: Increased density of gangliosides GM1 and GM2 and loss of cholesterol in detergent-resistant membrane domains. J. Neurochem. 2005;92:171–182. doi: 10.1111/j.1471-4159.2004.02849.x. [DOI] [PubMed] [Google Scholar]
- 46.Wakabayashi M., Matsuzaki K. Ganglioside-induced amyloid formation by human islet amyloid polypeptide in lipid rafts. FEBS Lett. 2009;583:2854–2858. doi: 10.1016/j.febslet.2009.07.044. [DOI] [PubMed] [Google Scholar]
- 47.Kakio A., Nishimoto S.I., Kozotsumi Y., Matsuzaki K. Formation of a membrane-active form of amyloid β-protein in raft-like model membranes. Biochem. Biophys. Res. Commun. 2003;303:514–518. doi: 10.1016/s0006-291x(03)00386-3. [DOI] [PubMed] [Google Scholar]
- 48.Yahi N., Aulas A., Fantini J. How cholesterol constrains glycolipid conformation for optimal recognition of Alzheimer’s β amyloid peptide (Aβ1-40) PLoS One. 2010;5 doi: 10.1371/journal.pone.0009079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X., Zheng J. Cholesterol promotes the interaction of Alzheimer β-amyloid monomer with lipid bilayer. J. Mol. Biol. 2011 doi: 10.1016/j.jmb.2011.11.006. (in Press) [DOI] [PubMed] [Google Scholar]
- 50.Wakabayashi M., Okada T., Kozutsumi Y., Matsuzaki K. GM1 ganglioside-mediated accumulation of amyloid beta-protein on cell membranes. Biochem. Biophys. Res. Commun. 2005;328:1019–1023. doi: 10.1016/j.bbrc.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 51.Kakio A., ichi Nishimoto S., Yanagisawa K., Kozutsumi Y., Matsuzaki K. Interactions of amyloid β-protein with various gangliosides in raft-like membranes: Importance of GM1 ganglioside-bound form as an endogenous seed for Alzheimer amyloid? Biochemistry. 2002;41:7385–7390. doi: 10.1021/bi0255874. [DOI] [PubMed] [Google Scholar]
- 52.Abramov A.Y., Ionov M., Pavlov E., Duchen M.R. Membrane cholesterol content plays a key role in the neurotoxicity of β-amyloid: Implications for Alzheimer’s disease. Aging Cell. 2011;10:595–603. doi: 10.1111/j.1474-9726.2011.00685.x. [DOI] [PubMed] [Google Scholar]
- 53.Fantini J., Yahi N. Molecular insights into amyloid regulation by membrane cholesterol and sphingolipids: Common mechanisms in neurodegenerative diseases. Expert Rev. Mol. Med. 2010;12 doi: 10.1017/S1462399410001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouras G.K., Tsai J., Naslund J., Vincent B., Edgar M., Checler F., Greenfield J.P., Haroutunian V., Buxbaum J.D., Xu H., et al. Intraneuronal Aβ42 accumulation in human brain. Am. J. Pathol. 2000;156:15–20. doi: 10.1016/s0002-9440(10)64700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wegiel J., Kuchna I., Nowicki K., Frackowiak J., Mazur-Kolecka B., Imaki H., Wegiel J., Mehta P.D., Silverman W.P., Reisberg B., et al. Intraneuronal Aβ immunoreactivity is not a predictor of brain amyloidosis-β or neurofibrillary degeneration. Acta Neuropathol. 2007;113:389–420. doi: 10.1007/s00401-006-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bayer T.A., Wirths O. Intracellular accumulation of amyloid-beta—A predictor for synaptic dysfunction and neuron loss in Alzheimer’s disease. Front. Aging Neurosci. 2010;2:1–9. doi: 10.3389/fnagi.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuello A.C. Intracellular and extracellular Abeta, a tale of two neuropathologies. Brain Pathol. 2005;15:66–71. doi: 10.1111/j.1750-3639.2005.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagele R.G., DÁndrea M.R., Anderson W.J., Wang H.Y. Intracellular accumulation of beta-amyloid (1-42) in neurons is facilitated by the alpha 7 nicotinic acetylcholine receptor in Alzheimer’s disease. Neuroscience. 2002;110:199–211. doi: 10.1016/s0306-4522(01)00460-2. [DOI] [PubMed] [Google Scholar]
- 59.DÁndrea M.R., Nagele R.G. Targeting the alpha 7 nicotinic acetylcholine receptor to reduce amyloid accumulation in Alzheimer’s disease pyramidal neurons. Curr. Pharm. Des. 2006;12:677–684. doi: 10.2174/138161206775474224. [DOI] [PubMed] [Google Scholar]
- 60.Fuentealba R.A., Liu Q., Zhang J., Kanekiyo T., Hu X., Lee J.M., LaDu M., Bu G. Low-density lipoprotein receptor-related protein 1 (LRP1) mediates neuronal Aβ42 uptake and lysosomal trafficking. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takuma K., Fang F., Zhang W., Yan S., Fukuzaki E., Du H., Sosunov A., McKhann G., Funatsu Y., Nakamichi N., et al. RAGE-mediated signaling contributes to intraneuronal transport of amyloid-β and neuronal dysfunction. Proc. Natl. Acad. Sci. USA. 2009;106:20021–20026. doi: 10.1073/pnas.0905686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.LaFerla1 F.M., Green1 K.N., Oddo S. Intracellular amyloid-β in Alzheimer’s disease. Nat. Rev. Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 63.Lai A.Y., McLaurin J. Mechanisms of amyloid-beta peptide uptake by neurons: The role of lipid rafts and lipid raft-associated proteins. Int. J. Alzheimer’s Dis. 2011;2011 doi: 10.4061/2011/548380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christensen D.Z., Schneider-Axmann T., Lucassen P.J., Bayer T.A., Wirths O. Accumulation of intraneuronal Aβ correlates with ApoE4 genotype. Acta Neuropathol. 2010;119:555–566. doi: 10.1007/s00401-010-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pigino G., Morfini G., Atagi Y., Deshpande A., Yu C., Jungbauer L., LaDu M., Busciglio J., Brady S. Disruption of fast axonal transport is a pathogenic mechanism for intraneuronal amyloid beta. Proc. Natl. Acad. Sci. USA. 2009;106:5907–5912. doi: 10.1073/pnas.0901229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christensen D.Z., Bayer T.A., Wirths O. Intracellular Aβ triggers neuron loss in the cholinergic system of the APP/PS1KI mouse model of Alzheimer’s disease. Neurobiol. Aging. 2010;31:1153–1163. doi: 10.1016/j.neurobiolaging.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 67.Christensen D.Z., Kraus S.L., Flohr A., Cotel M.C., Wirths O., Bayer T.A. Transient intraneuronal Aβ rather than extracellular plaque pathology correlates with neuron loss in the frontal cortex of APP/PS1KI mice. Acta Neuropathol. 2008;116:647–655. doi: 10.1007/s00401-008-0451-6. [DOI] [PubMed] [Google Scholar]
- 68.Bayer T.A., Wirths O. Review on the APP/PS1KI mouse model: Intraneuronal Aβ accumulation triggers axonopathy, neuron loss and working memory impairment. Genes Brain Behav. 2008;7:6–11. doi: 10.1111/j.1601-183X.2007.00372.x. [DOI] [PubMed] [Google Scholar]
- 69.Breyhan H., Wirths O., Duan K., Marcello A., Rettig J., Bayer T.A. APP/PS1KI bigenic mice develop early synaptic deficits and hippocampus atrophy. Acta Neuropathol. 2009;117:677–685. doi: 10.1007/s00401-009-0539-7. [DOI] [PubMed] [Google Scholar]
- 70.Gouras G.K., Tampellini D., Takahashi R.H., Capetillo-Zarate E. Intraneuronal β-amyloid accumulation and synapse pathology in Alzheimer’s disease. Acta Neuropathol. 2010;119:523–541. doi: 10.1007/s00401-010-0679-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lashuel H.A., Hartley D., Petre B.M., Walz T., Lansbury P.T. Neurodegenerative disease: Amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 72.Arispe N., Rojas E., Pollard H.B. Alzheimer disease amyloid β protein forms calcium channels in bilayer membranes: Blockade by tromethamine and aluminum. Proc. Natl. Acad. Sci. USA. 1993;90:567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong P.T., Schauerte J.A., Wisser K.C., Ding H., Lee E.L., Steel D.G., Gafni A. Amyloid-beta membrane binding and permeabilization are distinct processes influenced separately by membrane charge and fluidity. J. Mol. Biol. 2009;386:81–96. doi: 10.1016/j.jmb.2008.11.060. [DOI] [PubMed] [Google Scholar]
- 74.Strodel B., Lee J.W.L., Whittleston C.S., Wales D.J. Transmembrane structures for Alzheimer’s Aβ1–42 oligomers. J. Am. Chem. Soc. 2010;132:13300–13312. doi: 10.1021/ja103725c. [DOI] [PubMed] [Google Scholar]
- 75.Lin H., Bhatia R., Lal R. Amyloid beta protein forms ion channels: Implications for Alzheimer’s disease pathophysiology. FASEB J. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 76.Lin H., Zhu Y.J., Lal R. Amyloid β protein (1-40) forms calcium-permeable, Zn2+-sensitive channel in reconstituted lipid vesicles. Biochemistry. 1999;38:11189–11196. doi: 10.1021/bi982997c. [DOI] [PubMed] [Google Scholar]
- 77.Kagan B.L., Hirakura Y., Azimov R., Azimova R., Lin M.C. The channel hypothesis of Alzheimer’s disease: Current status. Peptides. 2002;23:1311–1315. doi: 10.1016/s0196-9781(02)00067-0. [DOI] [PubMed] [Google Scholar]
- 78.Diaz J.C., Simakova O., Jacobson K.A., Arispe N., Pollard H.B. Small molecule blockers of the Alzheimer Aβ calcium channel potently protect neurons from Aβ cytotoxicity. Proc. Natl. Acad. Sci USA. 2009;106:3348–3353. doi: 10.1073/pnas.0813355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Felice F.G.D., Vieira M.N.N., Bomfim T.R., Decker H., Velasco P.T., Lambert M.P., Viola K.L., Zhao W.Q., Ferreira S.T., Klein W.L. Protection of synapses against Alzheimer’s-linked toxins: Insulin signaling prevents the pathogenic binding of Aβ oligomers. Neuroscience. 2009;106:1971–1976. doi: 10.1073/pnas.0809158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lacor P.N., Buniel M.C., Furlow P.W., Clemente A.S., Velasco P.T., Wood M., Viola K.L., Kleinaalina W.L. Aβ oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer’s disease. J. Neurosci. 2007;27:796–807. doi: 10.1523/JNEUROSCI.3501-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao W.Q., Felice F.G.D., Fernandez S., Chen H., Lambert M.P., Quon M.J., Krafft G.A., Klein W.L. Amyloid beta oligomers induce impairment of neuronal insulin receptors. FASEB J. 2008;22:246–260. doi: 10.1096/fj.06-7703com. [DOI] [PubMed] [Google Scholar]
- 82.Snyder E.M., Nong Y., Almeida C.G., Paul S., Moran T., Choi E.Y., Nairn A.C., Salter M.W., Lombroso P.J., Greengard G.K.G.P. Regulation of NMDA receptor trafficking by amyloid-β. Nat. Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 83.Yan S.D., Chen X., Fu J., Chen M., Zhu H., Roher A., Slattery T., Zhao L., Nagashima M., Morser J., et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 84.Parri R.H., Dineley T.K. Nicotinic acetylcholine receptor interaction with beta-amyloid: Molecular, cellular, and physiological consequences. Curr. Alzheimer Res. 2010;7:27–39. doi: 10.2174/156720510790274464. [DOI] [PubMed] [Google Scholar]
- 85.Diarra A., Geetha T., Potter P., Babu J.R. Signaling of the neurotrophin receptor p75 in relation to Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2009;390:352–356. doi: 10.1016/j.bbrc.2009.09.116. [DOI] [PubMed] [Google Scholar]
- 86.Alberdi E., Sánchez-Gómez M.V., Cavaliere F., Pérez-Samartín A., Zugaza J.L., Trullas R., Domercq M., Matute C. Amyloid β oligomers induce Ca2+ dysregulation and neuronal death through activation of ionotropic glutamate receptors. Cell Calcium. 2010;47:264–272. doi: 10.1016/j.ceca.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 87.Ma Q., Yang F., Rosario E.R., Ubeda O.J., Beech W., Gant D.J., Chen P.P., Hudspeth B., Chen C., Zhao Y., et al. β-Amyloid oligomers induce phosphorylation of Tau and inactivation of insulin receptor substrate via c-Jun N-Terminal kinase signaling: Suppression by omega-3 fatty acids and curcumin. J. Neurosci. 2009;29:9078–9089. doi: 10.1523/JNEUROSCI.1071-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malinow R. New developments on the role of NMDA receptors in Alzheimer’s disease. Curr. Opi. Neurobiol. 2011 doi: 10.1016/j.conb.2011.09.001. (in Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S., Jin M., Koeglsperger T., Shepardson N.E., Shankar G.M., Selkoe D.J. Soluble Aβ oligomers inhibit long-term potentiation through a mechanism involving excessive activation of extrasynaptic NR2B-containing NMDA receptors. J. Neurosci. 2011;31:6627–6638. doi: 10.1523/JNEUROSCI.0203-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Texido L., Martin-Satue M., Alberdi E., Solsona C., Matute C. Amyloid β peptide oligomers directly activate NMDA receptors. Cell Calcium. 2011;49:184–190. doi: 10.1016/j.ceca.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 91.Verges D.K., Restivo J.L., Goebel W.D., Holtzman D.M., Cirrito J.R. Opposing synaptic regulation of amyloid-β metabolism by NMDA receptors in vivo. J. Neurosci. 2011;31:11328–11337. doi: 10.1523/JNEUROSCI.0607-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bordji K., Becerril-Ortega J., Nicole O., Buisson A. Activation of extrasynaptic, but not synaptic, NMDA receptors modifies amyloid precursor protein expression pattern and increases amyloid-β production. J. Neurosci. 2010;30:15927–15942. doi: 10.1523/JNEUROSCI.3021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rönicke R., Mikhaylova M., Rönicke S., Meinhardt J., Schröder U.H., Fändrich M., Reiser G., Kreutz M.R., Reymann K.G. Early neuronal dysfunction by amyloid β oligomers depends on activation of NR2B-containing NMDA receptors. Neurobiol. Aging. 2011;32:2219–2228. doi: 10.1016/j.neurobiolaging.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 94.Bicca M.A., Figueiredo C.P., Piermartiri T.C., Meotti F.C., Bouzon Z.L., Tasca C.I., Medeiros R., Calixto J.B. The selective and competitive N-methyl-D-aspartate receptor antagonist, (−)-6-phosphonomethyl-deca-hydroisoquinoline-3-carboxylic acid, prevents synaptic toxicity induced by amyloid-β in mice. Neuroscience. 2011;192:631–641. doi: 10.1016/j.neuroscience.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 95.Maglio L.E., Martins V.R., Izquierdo I., Ramirez O.A. Role of cellular prion protein on LTP expression in aged mice. Brain Res. 2006;1097:11–18. doi: 10.1016/j.brainres.2006.04.056. [DOI] [PubMed] [Google Scholar]
- 96.Criado J.R., Sanchez-Alavez M., Conti B., Giacchino J.L., Wills D.N., Henriksen S.J., Race R., Manson J.C., Chesebro B., Oldstone M.B. Mice devoid of prion protein have cognitive deficits that are rescued by reconstitution of PrP in neurons. Neurobiol. Dis. 2005;19:255–265. doi: 10.1016/j.nbd.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 97.Khosravani H., Zhang Y., Tsutsui S., Hameed S., Altier C., Hamid J., Chen L., Villemaire M., Ali Z., Jirik F.R., et al. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J. Cell Biol. 2008;181:551–565. doi: 10.1083/jcb.200711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vassallo N., Herms J. Cellular prion protein function in copper homeostasis and redox signalling at the synapse. J. Neurochem. 2003;86:538–544. doi: 10.1046/j.1471-4159.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- 99.McLennan N.F., Brennan P.M., McNeill A., Davies I., Fotheringham A., Rennison K.A., Ritchie D., Brannan F., Head M.W., Ironside J.W., et al. Prion protein accumulation and neuroprotection in hypoxic brain damage. Am. J. Pathol. 2004;165:227–235. doi: 10.1016/S0002-9440(10)63291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shyu W.C., Lin S.Z., Chiang M.F., Ding D.C., Li K.W., Chen S.F., Yang H.I., Li H. Overexpression of PrPC by adenovirus-mediated gene targeting reduces ischemic injury in a stroke rat model. J. Neurosci. 2005;25:8967–8977. doi: 10.1523/JNEUROSCI.1115-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Santuccione A., Sytnyk V., Leshchyns’ka I., Schachner M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J. Cell Biol. 2005;169:341–354. doi: 10.1083/jcb.200409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Laurén J., Gimbel D.A., Nygaard H.B., Gilbert J.W., Strittmatter S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kessels H.W., Nguyen L.N., Nabavi S., Malinow R. The prion protein as a receptor for amyloid-β. Nature. 2010;457:1128–1132. doi: 10.1038/nature09217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Calella A.M., Farinelli M., Nuvolone M., Mirante O., Moos R., Falsig J., Mansuy I.M., Aguzzi A. Prion protein and Aβ-related synaptic toxicity impairment. EMBO Mol. Med. 2010;2:306–314. doi: 10.1002/emmm.201000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang H.Y., Lee D.H.S., D’Andrea M.R., Peterson P.A., Shank R.P., Reitz A.B. β-Amyloid(1-42) binds to α7 nicotinic acetylcholine receptor with high affinity: Implications for Alzheimer’s disease pathology. J. Biol. Chem. 2000;275:5626–5632. doi: 10.1074/jbc.275.8.5626. [DOI] [PubMed] [Google Scholar]
- 106.Pettit D.L., Shao Z., Yakel J.L. β-amyloid 1-42 peptide directly modulates nicotinic receptors in the rat hippocampal slice. J. Neurosci. 2001;21:RC120–RC125. doi: 10.1523/JNEUROSCI.21-01-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Q.S., Kawai H., Berg D.K. Beta-amyloid peptide blocks the response of alpha7-containing nicotinic receptors on hippocampal neurons. Proc. Natl. Acad. Sci. USA. 2001;48:4734–4739. doi: 10.1073/pnas.081553598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dougherty J.J., Wu J., Nichols R.A. β-Amyloid regulation of presynaptic nicotinic receptors in rat hippocampus and neocortex. J. Neurosci. 2003;23:6740–6747. doi: 10.1523/JNEUROSCI.23-17-06740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hernandez C.M., Kayed R., Zheng H., Sweatt J.D., Dineley K.T. Loss of α7 nicotinic receptors enhances β-amyloid oligomer accumulation, exacerbating early-stage cognitive decline and Septohippocampal pathology in a mouse model of Alzheimer’s disease. J. Neurosci. 2010;30:2442–2453. doi: 10.1523/JNEUROSCI.5038-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang H.Y., Bakshi K., Shen C., Frankfurt M., Trocmé-Thibíerge C., Morain P. S24795 limits β-amyloid-α7 nicotinic receptor interaction and reduces Alzheimer’s disease-like pathologies. Biol. Psychiatry. 2010;67:522–530. doi: 10.1016/j.biopsych.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 111.Lagostena L., Trocme-Thibierge C., Morain P., Cherubini E. The partial α7 nicotine acetylcholine receptor agonist S 24795 enhances long-term potentiation at CA3-CA1 synapses in the adult mouse hippocampus. Neuropharmacology. 2008;54:676–685. doi: 10.1016/j.neuropharm.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 112.Beracochea D., Boucard A., Trocme-Thibierge C., Morain P. Improvement of contextual memory by S24795 in aged mice: Comparison with memantine. Psychopharmacology. 2008;196:555–564. doi: 10.1007/s00213-007-0987-5. [DOI] [PubMed] [Google Scholar]
- 113.Small D.H., Maksel D., Kerr M.L., Ng J., Hou X., Chu C., Mehrani H., Unabia S., Azari M.F., Loiacono R., et al. The beta-amyloid protein of Alzheimer’s disease binds to membrane lipids but does not bind to the alpha7 nicotinic acetylcholine receptor. J. Neurochem. 2007;101:1527–1538. doi: 10.1111/j.1471-4159.2006.04444.x. [DOI] [PubMed] [Google Scholar]
- 114.Knowles J.K., Rajadas J., Nguyen T.V.V., Yang T., LeMieux M.C., Griend L.V., Ishikawa C., Massa S.M., Wyss-Coray T., Longo F.M. The p75 neurotrophin receptor promotes amyloid-β (1-42)-induced neuritic dystrophy in vitro and in vivo. J. Neurosci. 2009;29:10627–10637. doi: 10.1523/JNEUROSCI.0620-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bengoechea T.G., Chen Z., O’Learya D.A., Masliahb E., Lee K.F. p75 reduces β-amyloid-induced sympathetic innervation deficits in an Alzheimer’s disease mouse model. Proc. Natl. Acad. Sci. USA. 2009;106:7870–7675. doi: 10.1073/pnas.0901533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boland K., Manias K., Perlmutter D.H. Specificity in recognition of amyloid-beta peptide by the serpin-enzyme complex receptor in hepatoma cells and neuronal cells. J. Biol. Chem. 1995;270:28022–28028. doi: 10.1074/jbc.270.47.28022. [DOI] [PubMed] [Google Scholar]
- 117.Joslin G., Griffin G.L., August A.M., Adams S., Fallon R.J., Senior R.M., Perlmutter D.H. The serpin-enzyme complex (SEC) receptor mediates the neutrophil chemotactic effect of alpha-1 antitrypsin-elastase complexes and amyloid-beta peptide. J. Clin. Invest. 1992;90:1150–1154. doi: 10.1172/JCI115934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schmidt A.M., Sahagan B., Nelson R.B., Selmer J., Rothlein R., Bell J.M. The role of RAGE in amyloid-beta peptide-mediated pathology in Alzheimer’s disease. Curr. Opin. Investig. Drugs. 2009;10:672–680. [PubMed] [Google Scholar]
- 119.Baiguera S., Fioravanzo L., Grandi C., Liddo R.D., Parnigotto P.P., Folin M. Involvement of the receptor for advanced glycation-end products (RAGE) in beta-amyloid-induced toxic effects in rat cerebromicrovascular endothelial cells cultured in vitro. Int. J. Mol. Med. 2009;24:9–15. doi: 10.3892/ijmm_00000199. [DOI] [PubMed] [Google Scholar]
- 120.Prk L., Wng G., Zhou P., Zhou J., Pitstick R., Previtic M.L., Younkind L., Younkind S.G., Nostrndc W.E.V., Choe S., et al. Scavenger receptor CD36 is essential for the cerebrovascular oxidative stress and neurovascular dysfunction induced by amyloid-β. Proc. Natl. Acad. Sci. USA. 2011;108:5063–5068. doi: 10.1073/pnas.1015413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Silverstein R.L., Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009;2:1–8. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coraci I.S., Husemann J., Berman J.W., Hulette C., Dufour J.H., Campanella G.K., Luster A.D., Silverstein S.C., El-Khoury J.B. CD36, a class B scavenger receptor, is expressed on microglia in Alzheimer’s disease brains and can mediate production of reactive oxygen species in response to beta-amyloid fibrils. Am. J. Pathol. 2002;160:101–112. doi: 10.1016/s0002-9440(10)64354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Varadarajan S., Yatin S., Aksenova M., Butterfield D.A. Review: Alzheimer’s amyloid β-peptide-associated free radical oxidative stress and neurotoxicity. J. Struct. Biol. 2000;130:184–208. doi: 10.1006/jsbi.2000.4274. [DOI] [PubMed] [Google Scholar]
- 124.Butterfield D.A., Reed T., Newman S.F., Sultana R. Roles of amyloid β-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer’s disease and mild cognitive impairment. Free Radic. Biol. Med. 2007;43:658–677. doi: 10.1016/j.freeradbiomed.2007.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jo D.G., Arumugam T.V., Woo H.N., Park J.S., Tang S.C., Mughal M., Hyun D.H., Park J.H., Choi Y.H., Gwon A.R., et al. Evidence that γ-secretase mediates oxidative stress-induced β-secretase expression in Alzheimer’s disease. Neurobiol. Aging. 2010;31:917–925. doi: 10.1016/j.neurobiolaging.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zheng L., Kågedal K., Dehvari N., Benedikz E., Cowburn R., Marcusson J., Terman A. Oxidative stress induces macroautophagy of amyloid β-protein and ensuing apoptosis. Free Radic. Biol. Med. 2009;46:422–429. doi: 10.1016/j.freeradbiomed.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 127.Abdul H.M., Butterfield D.A. Involvement of PI3K/PKG/ERK1/2 signaling pathways in cortical neurons to trigger protection by cotreatment of acetyl-l-carnitine and α-lipoic acid against HNE-mediated oxidative stress and neurotoxicity: Implications for Alzheimer’s disease. Free Radic. Biol. Med. 2007;42:371–384. doi: 10.1016/j.freeradbiomed.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Naoi M., Maruyama W., Shamoto-Nagai M., Yi H., Akao Y., Tanaka M. Oxidative stress in mitochondria: Decision to survive and death of neurons in neurodegenerative disorder. Mol. Neurobiol. 2005;31:81–93. doi: 10.1385/MN:31:1-3:081. [DOI] [PubMed] [Google Scholar]
- 129.Castegna A., Aksenov M., Aksenova M., Thongboonkerd V., Klein J.B., Pierce W.M., Booze R., Markesbery W.R., Butterfield D.A. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part I. Creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic. Biol. Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 130.Castegna A., Aksenov M., Thongboonkerd V., Klein J.B., Pierce W.M., Booze R., Markesbery W.R., Butterfield D.A. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part II. Dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J. Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 131.Sultana R., Perluigi M., Butterfield D.A. Redox proteomics identification of oxidatively modified proteins in Alzheimer’s disease brain and in vivo and in vitro models of AD centered around Abeta(1-42) J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2006;833:3–11. doi: 10.1016/j.jchromb.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 132.Butterfield D.A., Boyd-Kimball D. The critical role of methionine 35 in Alzheimer’s amyloid β- peptide (1-42)-induced oxidative stress and neurotoxicity. Biochimi. Biophys. Acta (BBA)-Protein Proteomics. 2005;1703:149–156. doi: 10.1016/j.bbapap.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 133.Butterfield D.A., Galvan V., Lange M.B., Tang H., Sowell R.A., Spilman P., Fombonne J., Gorostiza O., Zhang J., Sultana R., et al. In vivo oxidative stress in brain of Alzheimer disease transgenic mice: Requirement for methionine 35 in amyloid β-peptide of APP. Free Radic. Biol. Med. 2010;48:136–144. doi: 10.1016/j.freeradbiomed.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Muthaiyah B., Essa M.M., Chauhan V., Chauhan A. Protective effects of walnut extract against amyloid beta peptide-induced cell death and oxidative stress in PC12 cells. Biomed. Life Sci. 2011;36:2096–2103. doi: 10.1007/s11064-011-0533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barten D.M., Albright C.F. Therapeutic strategies for Alzheimer’s disease. Mol. Neurobiol. 2008;37:171–186. doi: 10.1007/s12035-008-8031-2. [DOI] [PubMed] [Google Scholar]
- 136.Tomaselli S., Esposito V., Vangone P., van Nuland N.A.J., Bonvin A.M.J.J., Guerrini R., Tancredi T., Temussi P.A., Picone D. The α-to-β conformational transition of Alzheimer’s Aβ-(1-42) peptide in aqueous media is reversible: A step by step conformational analysis suggests the location of β conformation seeding. ChemBioChem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 137.Hayat S., Helms V. Towards understanding the early events in the conformational transition of amyloid beta peptides. Comput. Biophys. Syst. Biol. (CBSB08) 2008;40:227–230. [Google Scholar]
- 138.Xu Y., Shen J., Luo X., Zhu W., Chen K., Ma J., Jiang H. Conformational transition of amyloid β-peptide. Proc. Natl. Acad. Sci. USA. 2005;102:5403–5407. doi: 10.1073/pnas.0501218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Creighton T.E. Protein Folding. W.H. Freeman & Company; New York, NY, USA: 1992. [Google Scholar]
- 140.Prusiner S.B. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zimm H., Bragg J.K. Theory of the phase transition between helix and random coil in polypeptide chains. J. Chem. Phys. 1959;31:526–535. [Google Scholar]
- 142.Lifson S., Roig A. On the theory of helix-coil transition in polypeptides. J. Chem. Phys. 1961;34:1963. [Google Scholar]
- 143.Kao Y.M., Jiang T.F. Transition temperatures of the trapped ideal spinor Bose gas. Eur. Phys. J. D. 2006;40:363–269. [Google Scholar]
- 144.Yakubovich A.V., Solov’yov I.A., Solov’yov A.V., Greiner W. Ab initio theory of helix-coil phase transition. Eur. Phys. J. D. 2008;46 doi: 10.1140/epjd/e2007-00328-9. [DOI] [Google Scholar]
- 145.Solovyov I.A., Yakubovich A.V., Solovyov A.V., Greiner W. α-helix ↔ random coil phase transition: Analysis of ab initio theory predictions. Eur. Phys. J. D. 2008;46:227–240. [Google Scholar]
- 146.Ding F., Borreguero J.M., Buldyrey S.V., Stanley H.E., Dokholyan N.V. Mechanism for the alpha-helix to beta-hairpin transition. Proteins. 2003;53:220–228. doi: 10.1002/prot.10468. [DOI] [PubMed] [Google Scholar]
- 147.Hong L., Lei J. Statistical mechanical model for helix-sheet-coil transitions in homopolypeptides. Phys. Rev. E. 2008;78:051904. doi: 10.1103/PhysRevE.78.051904. [DOI] [PubMed] [Google Scholar]
- 148.Yasar F., Demir K. The study of helix-coil transition of polyalanine with single histogram method. Comput. Phys. Commun. 2006;175:604–611. [Google Scholar]
- 149.Gibbs H., Dimarzio E.A. Statistical mechanics of helix-coil transitions in biological macromolecules. J. Chem. Phys. 1959;30:271–282. [Google Scholar]
- 150.Huang K. Protein folding as a physical stochastic process. Biophys. Rev. Lett. 2008;3:1–2. [Google Scholar]
- 151.Huang K. Conditioned self-avoiding walk (csaw): Stochastic approach to protein folding. Biophys. Rev. Lett. 2007:139–154. [Google Scholar]
- 152.Leong H.W., Chew L.Y., Huang K. Normal modes and phase transition of the protein chain based on the Hamiltonian formalism. Phys. Rev. E. 2010;82 doi: 10.1103/PhysRevE.82.011915. [DOI] [PubMed] [Google Scholar]
- 153.Goh B.C., Leong H.W., Qu X., Chew L.Y. The mechanism of antiparallel β-sheet formation based on conditioned self-avoiding walk. Eur. Phys. J. E. 2012;35 doi: 10.1140/epje/i2012-12027-8. [DOI] [PubMed] [Google Scholar]
- 154.Jiang P., Li W.F., Shea J.E., Mu Y. Resveratrol inhibits the formation of multiple-layered β-sheet oligomers of the human islet amyloid polypeptide segment 22–27. Biophys. J. 2011;100:1550–1558. doi: 10.1016/j.bpj.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Qin X.R., Abe H., Nakanishi H. NMR and CD studies on the interaction of Alzheimer β-amyloid peptide (12-28) with β-cyclodextrin. Biochem. Biophys. Res. Commun. 2002;297:1011–1015. doi: 10.1016/s0006-291x(02)02337-9. [DOI] [PubMed] [Google Scholar]
- 156.Camilleri P., Haskins N.J., Hewlett D.R. β-Cyclodextrin interacts with the Alzheimer amyloid β-A4 peptide. FEBS Lett. 1994;341:256–258. doi: 10.1016/0014-5793(94)80467-2. [DOI] [PubMed] [Google Scholar]
- 157.Colombo R., Carotti A., Catto M., Racchi M., Lanni C., Verga L., Caccialanza G., Lorenzi E.D. CE can identify small molecules that selectively target soluble oligomers of amyloid β protein and display antifibrillogenic activity. Electrophoresis. 2009;30:1418–1429. doi: 10.1002/elps.200800377. [DOI] [PubMed] [Google Scholar]
- 158.Cohen A.D., Ikonomovic M.D., Abrahamson E.E., Paljug W.R., DeKosky S.T., Lefterov I.M., Koldamova R.P., Shao L., Debnath M.L., Mason N.S., et al. Anti-amyloid effects of small molecule Aβ-binding agents in PS1/APP mice. Lett. Drug. Des. Discov. 2009;6:437. doi: 10.2174/157018009789057526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang F., Lim G.P., Begum A.N., Ubeda O.J., Simmons M.R., Ambegaokar S.S., Chen P.P., Kayed R., Glabe C.G., Frautschy S.A., et al. Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 2005;280:5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- 160.Inbar P., Bautista M.R., Takayama S.A., Yang J. Assay to screen for molecules that associate with Alzheimer’s related beta-amyloid fibrils. Anal. Chem. 2008;80:3502–3506. doi: 10.1021/ac702592f. [DOI] [PubMed] [Google Scholar]
- 161.Garcia-Alloza M., Borrelli L.A., Rozkalne A., Hyman B.T., Bacskai B.J. Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. J. Neurochem. 2007;102:1095–1104. doi: 10.1111/j.1471-4159.2007.04613.x. [DOI] [PubMed] [Google Scholar]
- 162.Caesar I., Jonson M., Nilsson K.P.R., Thor S., Hammarström P. Curcumin promotes A-beta fibrillation and reduces neurotoxicity in transgenic drosophila. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Park S.Y., Kim H.S., Cho E.K., Kwon B.Y., Phark S., Hwang K.W., Sula D. Curcumin protected PC12 cells against beta-amyloid-induced toxicity through the inhibition of oxidative damage and tau hyperphosphorylation. Food Chem. Toxicol. 2008;46:2881–2887. doi: 10.1016/j.fct.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 164.Masuda Y., Fukuchi M., Yatagawa T., Tada M., Takeda K., Irie K., ichi Akagi K., Monobe Y., Imazawa T., Takegoshi K. Solid-state NMR analysis of interaction sites of curcumin and 42-residue amyloid β-protein fibrils. Bioorg. Med. Chem. 2011;19:5967–5974. doi: 10.1016/j.bmc.2011.08.052. [DOI] [PubMed] [Google Scholar]
- 165.Ellson J., Gansner E., Koutsofios L., North S., Woodhull G., Description S., Technologies L. Lecture Notes in Computer Science. Springer-Verlag; Heidelberger/Berlin, Germany: 2001. Graphviz-Open Source Graph Drawing Tools; pp. 483–484. [Google Scholar]