Abstract
The packaging of double-stranded DNA into bacteriophages leads to the arrangement of the genetic material into highly-packed and ordered structures. Although modern experimental techniques reveal the most probable location of DNA inside viral capsids, the individual conformations of DNA are yet to be determined. In the current study we present the results of molecular dynamics simulations of the DNA packaging into several bacteriophages performed within the framework of a coarse-grained model. The final DNA conformations depend on the size and shape of the capsid, as well as the size of the protein portal, if any. In particular, isometric capsids with small or absent portals tend to form concentric spools, whereas the presence of a large portal favors coaxial spooling; slightly and highly elongated capsids result in folded and twisted toroidal conformations, respectively. The results of the simulations also suggest that the predominant factor in defining the global DNA arrangement inside bacteriophages is the minimization of the bending stress upon packaging.
Keywords: Bacteriophages, Genome packaging, Coarse-grained models, DNA conformations
1. Introduction
Double-stranded DNA bacteriophage genomes vary in length from tens of thousands to hundreds of thousands of base-pairs, and they are packed inside polyhedral protein capsids. Capsids come in a variety of shapes, ranging from icosahedral to highly elongated, and with linear dimensions ranging from hundreds to thousands of Ångstroms (Ackermann and DuBow, 1987; Granoff and Webster, 1999). Packaging of the DNA genome into the pre-formed protein capsid is aided by an ATP-driven motor that is part of a complex of portal proteins (Guo et al., 1987). Some bacteriophages also contain a cylindrical protein core connected to the portal and extending into the capsid's interior. DNA packaging requires the DNA to be highly compacted and must be done against substantial forces (Smith et al., 2001) to overcome the repulsion between the DNA strands, its elastic bending and entropy associated with the confinement. Here we describe the results of modeling studies showing how the capsid geometry determines the DNA conformation.
Early structural studies, using X-ray diffraction and electron microscopy, provided information on the spacing between the DNA strands and low-resolution hints about the overall conformation. These led to the coaxial spool model (Richards et al., 1973), which remains the most widely-accepted model for DNA in icosahedral capsids. Other models include the ball of string (Earnshaw and Harrison, 1977), the folded chain (Earnshaw and Harrison, 1977), the interwound toroid (Earnshaw et al., 1978), toroidal winding (Kosturko et al., 1979), the kinked chain (Serwer, 1986), and the folded toroid(Hud, 1995).
The development of cryo-electron microscopy (cryo-EM) (Frank, 2002) offered the opportunity to view bacteriophage DNA in its native conformation, without possible artifacts from negative staining and/or fixing. Cryo-EM provides images of individual phage, and three-dimensional density reconstructions from thousands of individual images can lead to resolutions around 10 Å. 2D studies on the icosahedral phage T7 with a large protein core revealed that DNA forms a set of rings around the protein core and supported the coaxial spooling model (Cerritelli et al., 1997). Similar conformations were revealed by higher-resolution 3D reconstructions on phages ε15 (Jiang et al., 2006) and P22 (Chang et al., 2006; Lander et al., 2006), which are both isomorphous to T7 (Agirrezabala et al., 2005). 3D density reconstruction of prolate ϕ29 showed a set of concentric ellipsoidal shells of DNA density (Tao et al., 1998; Xiang et al., 2006) but failed to resolve individual DNA strands. The results of these experimental advances have been recently summarized in the review (Johnson and Chiu, 2007). Unfortunately, individual DNA conformations still remain unknown, because cryo-EM images of individual phage lack sufficient resolution, and because 3D reconstructions require averaging over hundreds or thousands of particles and, therefore, represent most probable locations of DNA inside capsids.
Significant progress has also been achieved in theoretical and computer-aided modeling studies. Some of these have used a statistical-thermodynamic approach to describe DNA packaging (Kindt et al., 2001; Purohit et al., 2005). Such models are capable of reproducing and predicting some thermodynamic properties such as packaging force, internal pressure, and internal energy. However, they provide no insights into DNA structure, because they require a priori assumption of the DNA conformation. Another group of studies overcomes this difficulty by simulating the packaging process, generating DNA conformations using coarsegrained DNA models and refining these by various molecular mechanics algorithms. These include molecular dynamics (MD) (Arsuaga et al., 2002; LaMarque et al., 2004; Locker, 2006; Petrov and Harvey, 2007), Brownian dynamics (Kindt et al., 2001; Spakowitz and Wang, 2005) and Langevin dynamics (Forrey, 2006). Most of these studies have been limited to isometric (spherical or icosahedral) capsids and simple elastic DNA models, which precludes accurate prediction of thermodynamic properties. Ali et al. have shown that a simple elastic model for DNA and the reduced size for bacteriophage's capsids reveal the effect of the cavity shape on DNA conformations (Ali et al., 2006). We have recently provided methods for treating capsids and cores of arbitrary shape, along with a proper potential of mean force for long-range DNA-DNA interactions (Petrov and Harvey, 2007), leading to the match of the experimental density distribution (Xiang et al., 2006) and force versus distance curves (Smith et al., 2001) for the packaging of ϕ29 as well as providing a set of the individual conformations. Here we apply our approach to capsids of different sizes and shapes (shown in Figure 1) to determine how these influence the conformation of DNA inside bacteriophages.
Figure 1.

Models for polyhedral capsids used in the present study. (A) ε15, (B) ϕ29, (C) and (D) highly elongated icosahedrons. Each triangular face of the polyhedra was filled with pseudoatoms of radius 8 Å. Protein portals and short DNA fragments are also shown.
2. Methods
The details of model construction, parameterization and optimization are presented elsewhere (Locker, 2006; Petrov and Harvey, 2007; Tan, 2006). Here we give only a brief summary. DNA is represented as a collection of spherical pseudoatoms (“beads”) connected together by suitably parameterized springs. In this discretized version of a continuum elastic model, each pseudoatom represents six basepairs. The Hamiltonian includes stretching and bending terms, DNA-DNA interactions and a DNA-capsid volume exclusion term (modeled as a semiharmonic repulsion between soft spheres).
All force field constants were derived to match the thermodynamic properties of DNA molecules (but independently of any virus related data). The same set of constants was applied to all models described in the Results section. The DNA elastic parameters are chosen to match the known elastic moduli for stretching and bending; the latter is derived from the persistence length. Specifically, the numerical values of constants are kb=3.5 kcal/(mol·Å2), b0=19.9 Å, kθ=22.4 kcal/(mol·rad2), θ0=π rad, kDNA-DNA=3.5 kcal/(mol·Å2), d0,DNA-DNA=25.0 Å, kDNA-Capsid =8.8 kcal/(mol·Å2), d0,DNA-Capsid =20.5 Å. A cutoff distance of 50 Å was used for calculating the volume exclusion terms.
The DNA-DNA interaction was parameterized to match data from osmotic pressure measurements for a solution containing 100 mM NaCl + 10 mM MgCl2 (Petrov and Harvey, 2007), which closely corresponds to the experimental conditions under which the packaging forces were measured (Smith et al., 2001). Under these conditions, the DNA-DNA interaction is repulsive, and we have shown (Petrov and Harvey, 2007) that it can be approximated by a modified Debye-Hückel function with qeff= -12.6 e per pseudoatom, κeff = 0.31 Å−1.
The spherical capsid was modeled using a semiharmonic restraining force that adds an energy penalty for any DNA pseudoatom that moves outside the specified capsid radius (Arsuaga et al., 2002; LaMarque et al., 2004; Locker, 2006). All other capsids and cores were represented by sets of pseudoatoms anchored at appropriate points in space and interacting with the DNA only through a semiharmonic nonbonded repulsive term (Petrov and Harvey, 2007). These capsid geometries are shown in Figure 1.
Packaging of DNA into the capsid was aided by five additional pseudoatoms (studs) fixed inside the cylindrical protein core. Packaging was driven by ratcheting DNA pseudoatoms attached to the studs into the capsid in a series of 10 Å steps, each followed by extensive MD equilibration of the injected portion of genome. This mechanism is not meant to mimic any physical characteristics of the actual packaging, where at each cycle a motor consumes a molecule of ATP and translocates DNA by ∼1.8 bp. The only purpose of this translocation mechanism is to guarantee a smooth insertion of a new portion of DNA represented by the coarse-grained model without significant perturbation of the already packaged fraction. Each simulation began with a rate of 6 ns per step; this was linearly increased by 4-8 ps per monomer as packaging progressed, to achieve equilibrium at each step. Total trajectory time depended on the size of the model genome and ranged from ∼70 to ∼230 μs. The packaging performed according to the described protocol is highly accelerated compared to DNA packaging in vivo or in vitro, which takes place on timescales from seconds to minutes and goes far beyond the capabilities of modern computers. Therefore, this protocol does not reproduce any kinetic parameters of packaging process but it simply guaranties that at each point along the packaging pathway DNA conformations are at or very close to thermodynamic equilibrium (Locker, 2006; Petrov and Harvey, 2007). All simulations were performed using YUP, a molecular simulation package designed primarily as a tool for coarse-grain modeling (Tan, 2006).
3. Results and Discussion
3.1. Isometric capsids with no core
Some bacteriophages are isometric icosahedra. Simulations on such viruses have often approximated the capsid by a sphere (Arsuaga et al., 2002; Kindt et al., 2001; LaMarque et al., 2004; Locker, 2006; Spakowitz and Wang, 2005), although a recent study did treat the icosahedral shape explicitly (Forrey, 2006). We have previously shown that, in the absence of long-range repulsive interactions, the minimum energy conformation for an elastic model DNA in a sphere is a concentric spool, rather than a coaxial spool (LaMarque et al., 2004). Energy minimization effectively takes the system to a very low temperature. Similar conformations, but more disordered, are found at room temperature for both spherical (Locker, 2006) and icosahedral (Forrey, 2006) capsids.
The model in this section has a 19,272 basepair DNA (3212 pseudoatoms) and a spherical capsid of radius 225 Å. It also includes our modified Debye-Hückel term for long-range DNA-DNA interactions. We note that the capsid diameter is less than the DNA persistence length (510Å), so elastic forces should be important.
Figure 2a shows a typical packaging trajectory for this system. The DNA is organized in concentrically spooled layers. This is similar to the minimum energy structure for a simple elastic model (LaMarque et al., 2004), so the long-range potential has little effect on the global structure of the packaged DNA. Other studies with long-range electrostatic repulsions have also reported little effect on the final conformation (Forrey, 2006; Spakowitz and Wang, 2005). During the initial stage of packaging, the DNA stiffness pushes the DNA against the spherical surface of the capsid. Once the outer layer of the packaged structure is formed, the DNA begins to form a second layer inside it. However, since the first layer does not completely cover the interior surface of the capsid, DNA in the second layer is packed into a somewhat elongated cavity. It arranges itself along the longest possible dimension to minimize the bending stress, which favors the concentric arrangement over purely coaxial spooling. As the internal pressure builds up, strands are forced closer together, and strands from later layers occasionally push through gaps in earlier layers. This leads to considerable variation between different packaging trajectories, all of which deviate substantially from the idealized concentric spool. Although this description provides a general idea about the formation of the coaxial spools, we point out that in the final structure each concentric layer is composed of several coaxially oriented sub-layers. The final pattern will probably depend on the size of the capsid because the larger the size, the more sub-layers it can accommodate before the inner surface becomes significantly elongated.
Figure 2.
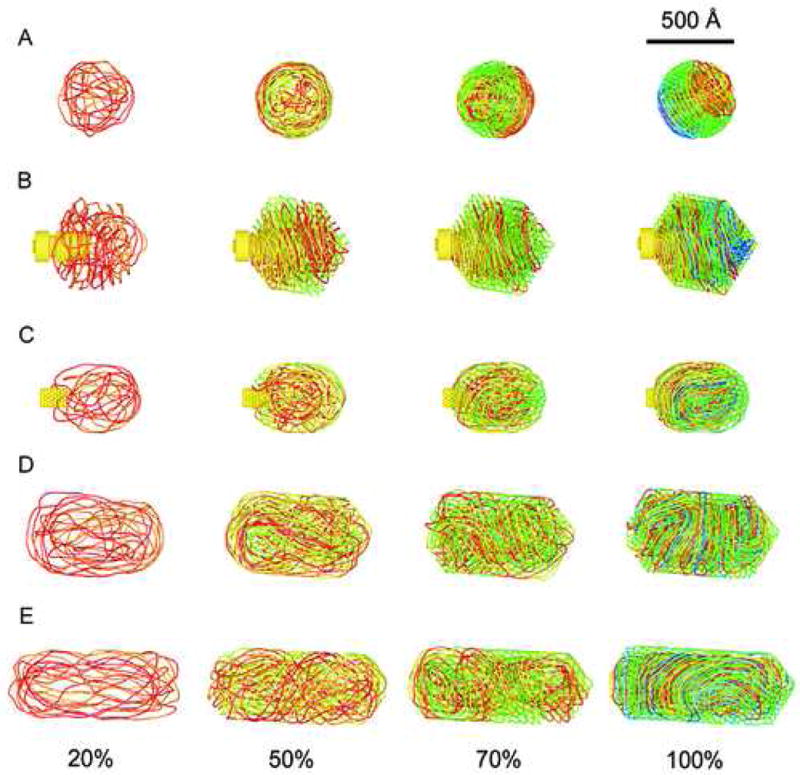
DNA conformations at 20%, 50%,70% and 100% of packed genomes inside the capsids used in the present study. (A) Concentric spooling, (B) Coaxial spooling, (C) Folded totoid, (D) and (E) twisted toroids.
3.2. Isometric capsids with a core: simulation of ε15
Recently, there have been several high-quality cryo-EM reconstructions of the structure of icosahedral bacteriophages e.g. ε15 (Jiang et al., 2006), T7 (Agirrezabala et al., 2005) and P22 (Chang et al., 2006; Lander et al., 2006), that contain a large protein portal complex located at one of the capsid's vertices. Inspired by these results we have simulated the packaging of dsDNA into a detailed model for ε15 (Petrov et al., 2007). We modeled the 39,671 bp genome as a chain of 6601 beads, and the capsid by a regular icosahedron; we also treated the shape of the portal and core proteins in detail (Figure 1a). A typical packaging trajectory is shown in Figure 2b. The final structure has a coaxially-spooled motif, which agrees with the results of the cryo-EM reconstructions. The structures derived from packaging with electrostatic interactions, reported here, are similar to the coaxially-spooled structures reported in previous molecular mechanics studies on purely elastic DNA models packed into capsids with cylindrical cores (Forrey, 2006; Locker, 2006). As in the spherical case discussed above, the elastic properties of DNA are the dominant factor in determining the overall organization of the final structure, with long-range forces playing a secondary role.
We argued in the previous section that concentrically spooled structures have a lower elastic bending energy than coaxially-spooled structures. The presence of the protein core in e15 precludes concentric spooling, consistent with suggestions that cylindrical cores extending well into the capsid help to organize the DNA into coaxial spools (Cerritelli et al., 1997). Therefore, for such structures the minimum of the bending energy would be achieved if DNA forms loops of largest possible radii around the cylindrical portals and avoids sharp bending. At the same time, DNA near the center of the capsid is highly disordered even though the global organization is coaxially-spooled. This is consistent with the DNA structures observed by cryo-EM in ε15 (Jiang et al., 2006) and P22 (Chang et al., 2006; Lander et al., 2006). Besides this, neither of the two packaging motifs considered so far (Figures 2a and 2b) exhibit geometrically idealized conformations such those of Arsuaga et al. (Arsuaga et al., 2002). Instead, we observed that the outer layers of DNA strands exist in a locally disordered near-crystalline or “glassy” state. It has been proposed that this disorder appears due to the stochastic character of DNA packaging (Forrey, 2006).
3.3. Slightly elongated capsids: Simulation of ϕ29
Bacteriophage ϕ29 has served as a model system and has been intensively studied both experimentally (Tao et al., 1998; Xiang et al., 2006) and theoretically (Locker, 2006; Petrov and Harvey, 2007). Its capsid has a prolate ellipsoidal shape with a length of 560Å and a width of 420Å. The packaging portal complex of ϕ29 is much smaller than that in e15 and does not penetrate much inside the capsid (Figure 1b). Originally, the structure of the DNA inside ϕ29 was reconstructed from cryo-EM images at relatively low resolution (∼36 Å), which showed that the outer layers of DNA are organized into elliptical shells, whereas the inner part is highly disordered (Tao et al., 1998). In neither the original study nor a more recent reconstruction at 16 Å (Xiang et al., 2006) did the DNA density show distinct rings similar to those seen in ε15 and P22.
We recently performed a detailed computational study of DNA packaging inside ϕ29 (Petrov and Harvey, 2007). The capsid was modeled as an elongated pentakisdodecahedron, and the 20 kilobase-pair genome was represented by 3212 beads. The DNA is organized into a folded toroidal structure, which allows a substantial fraction of the genome to be arranged along the longest principal axis of the capsid. This reduces the overall elastic bending energy below that which would be required for coaxial spooling. A representative DNA structure is shown in Figure 2c. A similar pattern has also been seen in the simulation study on an elongated virus with the same axial ratio (Ali et al., 2006), in which the dimensions of the capsid were significantly reduced. Arrangement of the DNA along the long axis was not considered in earlier theoretical work (Purohit et al., 2005), presumably because it was unclear what global structure would permit this. The folded toroid, first proposed by Nicholas Hud (Hud, 1995) solves this problem.
Movies of the entire packaging process (http://rumour.biology.gatech.edu/publications/showcase) show that the DNA arranges itself into the folded toroidal conformation from the early stages of packaging, during which the bending term is the major contributor to the total internal energy. By the time about half of the genome is packaged and the electrostatic energy begins to be significant, the characteristic folded toroid has already formed. Folded toroidal structures are also formed when the DNA is modeled as a simple elastic chain and long-range DNA-DNA electrostatic repulsions are ignored (Petrov and Harvey, 2007). Once again, the global structure of the packed genome is determined by the elastic properties of DNA (persistence length) and the capsid's shape and dimensions.
3.4. Highly elongated capsids
So far we have considered bacteriophages that have isometric or slightly elongated shapes. However, many bacteriophages are elongated, with axial ratios of two or more (Ackermann and DuBow, 1987). Most commonly, these have large genomes (up to several hundreds of thousands base pairs) and very long capsids (thousands of Ångstroms). For example, the giant T4 (Doermann et al., 1973) is ∼5000 Å×1000 Å. Long before the development of cryoscopic methods of sample preparation, electron microscopy studies on partially disrupted giant T4 phage suggested that the DNA is organized in a twisted toroidal structure (Earnshaw et al., 1978).
Genomes as large as giant T4 are too large for our current packaging protocols, for which the upper limit is ∼40-50 kbp. In order to study the effects of capsid elongation, we built models with genome sizes and capsid volumes close to those of ε15, maintaining the fractional packing volume (VDNA/Vcapsid) equal to 0.45. The capsids were modeled as elongated icosahedrons (five faces at each end, with ten additional triangular faces for elongation). The first model had a capsid with a length of 945 Å and an inradius of 225 Å, which contained a 37.8 kbp genome (6312 beads); these parameters were 1137 Å, 200 Å, and 39.1 kbp (6522 beads), respectively, for the second model (see Figure 1c and 1d). DNA was treated with the same set of parameters as the models described above. The key issues in constructing these models were that the capsid's longer dimension be significantly longer than the persistence length of DNA (∼500 Å), and that the shorter capsid dimension be less than the persistence length.
Figures 2d and 2e show the conformations of DNA obtained inside the elongated bacteriophage models. Both reveal a pattern of twisted toroids similar to that observed in the experimental studies of the giant T4 mutant (Earnshaw et al., 1978). As in the case of the slightly elongated capsid just discussed, the preferred DNA arrangement would be expected to lie along the longest axis of the capsid. The capsids are longer than the persistence length, so flexibility and entropic considerations favor some DNA bending. As the packaging progresses, the bends of numerous DNA strands become cooperative to avoid strand interpenetration and minimize the interactions between the strands. This leads to the appearance of a superstructure that resembles a twisted toroid. As in the previous simulations, the superstructure appears during the packaging of the first half of the genome, again confirming the importance of elastic forces in the determination of the global structure.
The structure in Figure 2d has a counterclockwise twist, while that in Figure 2e is twisted clockwise. Our DNA model does not currently treat torsional stiffness. It has recently been shown that DNA twisting does not contribute significantly to the energetic cost of packaging, but it can affect the final conformation, particularly when one compares structures obtained by packaging when the motor rotates relative to the capsid versus when it does not (Spakowitz and Wang, 2005). It is possible that the torsional stiffness of DNA would favor one chirality (clockwise or counterclockwise) over the other, in which case the handedness of one of the structures in Figures 2d and 2e might be an artifact of the simulations. While the preference of one chiral conformation over the other may appear upon the inclusion of torsional stiffness, we would still expect the global structure to be a twisted toroid. This minimizes the bending energy and is the experimentally observed conformation. We are currently working on the implementation of the torsional potential in our DNA models.
3.5. Structure development during packaging
Since the results presented above were obtained by using the same force field and packaging protocol, the differences between the final DNA structures are due to the size and shape of the capsid and the core. Nevertheless, each of these structures undergoes a similar series of structural and energetic stages upon packaging. During the initial stage when less than ∼20% of genome has been packaged, the DNA moves through all available space inside the capsid and behaves much like a free worm-like chain. Nevertheless, because it slightly pushes against the capsid walls, DNA density at the outer region is somewhat higher then that in the middle part of the capsid. The conformational energy change is not significant, and the structure is highly dynamic and disordered, without any distinct patterns.
As packaging continues up to ∼50%, confinement effects become significant, and well-defined structural patterns are formed. Although electrostatic repulsions work to keep the strands widely separated, there is still enough space that the electrostatic energy is smaller than the cost of the elastic deformations. Thus, the polymer begins to adopt a conformation that has minimal bending stress. If the longest dimension of the capsid is smaller than or comparable to the DNA persistence length, the molecule adopts whichever path has the smallest curvature (largest radius or linear dimension); this minimizes the enthalpic penalty of bending. If any capsid dimension is larger than the persistence length, dynamic conformational fluctuations become important, and entropic considerations preclude the DNA from strictly following the path of lowest curvature. During this stage, the principal effect of electrostatic repulsions is to distribute the viral genome fairly evenly over the entire capsid volume. Purely elastic models produce the same global organization as do models with long-range electrostatic forces, but the former favor a higher average density in the outer shell(s) at this stage than do models that include electrostatic repulsions. Despite the formation of global conformations, at the local scale DNA remains in a disordered (isotropic) phase.
As the packaging progresses further (up to ∼80%), the average distance between the DNA strands decreases, leading to a sharp increase in the electrostatic energy, which becomes comparable to the elastic deformation energy and then surpasses it (Petrov and Harvey, 2007). The significant volume confinement at this stage leads to a remarkable decrease in mobility, which is seen most easily in movies of the packaging (http://rumour.biology.gatech.edu/publications/showcase). The DNA appears to undergo a gradual phase transition to a near-crystalline (nematic) phase. The point at which this phase transition occurs depends on the amount of genome packed and on the size and morphology of the capsid.
Packaging of the last ∼20% of the DNA is characterized by minor changes in both local and global organization. There is almost no DNA motion except for a slithering motion that moves the DNA along a fixed track, allowing the most recently packaged segment to occupy the recently vacated position of its immediate predecessor. The slithering is occasionally interrupted by the forcing of one segment between two others, or by the extrusion of a new loop, presumably in a region of high bending stress. The increasing density requires a gradual decrease in the distance between the DNA strands, down to ∼25-28 Å, as observed experimentally (Cerritelli et al., 1997). This is accompanied by rapid growth in the DNA-DNA interaction energy, including both electrostatic repulsions and hydration forces (Parsegian et al., 1995). Orthogonal views of the final conformations of all structures are shown in Supplementary Data (Supplementary Figures 1-5).
4. Concluding Remarks
Our simulations reveal a variety of DNA conformations inside capsids of different morphologies. The results of the study are summarized in Table 1. The major determinants of the packed DNA structure inside bacteriophages are the size and shape of the capsid and the core, if any. Interestingly, the overall structural pattern is formed at those phases in packaging where elastic forces dominate. As a consequence, a simple elastic model can be used to qualitatively describe the packaging process. On the other hand, accurate treatment of the long-range DNA-DNA interaction potential is required to accurately reproduce the experimentally observed packaging forces and energy changes (Petrov and Harvey, 2007).
Table 1.
Effect of the capsid shape on DNA conformation inside bacteriophages.
| Bacteriophage | Capsid shape | Protein core | DNA conformation | Idealized conformation |
|---|---|---|---|---|
| Isometric model | Sphere | None | Concentric spool |

|
| ε15 | Icosahedron | Large | Coaxial spool |

|
| ϕ29 | Slightly elongated capsid | Small | Folded toroid |

|
| Highly elongated models | Highly elongated icosahedron | Small | Twisted toroid |

|
In models that include the long-range electrostatic potential, the DNA is fairly evenly distributed throughout the entire capsid volume during the entire packaging process, due to the strong repulsions. By contrast, packaging in purely elastic models pushes the DNA outward against the capsid wall to minimize the bending stress. Elastic models have higher DNA density in the outer layers compared to the capsid's interior at all stages of the packaging process. In both models (those that include electrostatics and those that don't), DNA ordering is more pronounced at the outer layers than in the interior, where the bending stresses are higher and there is more conformational variability. Although the DNA is significantly ordered overall, the conformations are far from the ideal close-packed conformations that one might infer from the concentric rings of density seen in cryo-EM studies, both two-dimensional projections (Cerritelli et al., 1997) and three-dimensional reconstructions (Jiang et al., 2006; Lander et al., 2006 Chang, 2006 #4; Xiang et al., 2006).
How are the reconstructed density maps related to the individual DNA conformations? In general, the three-dimensional reconstructions, which use images from hundreds or thousands of individual particles, represent the density probability distribution of DNA inside the capsid; they cannot reveal the details of individual structures. There is a common feature seen in all reconstructed density maps of different bacteriophages (Johnson and Chiu, 2007): on average DNA density is organized in concentric shells. However, the individual conformations may not necessarily have the same concentric spooling motif, e.g. we have shown that individual conformations inside resemble folded toroids (Petrov and Harvey, 2007). When images from individual ϕ29 models were averaged, they revealed the concentric shells, faithfully reproducing the experimental reconstructions due to the variability of individual structures. In some cases, like P22 and e15, the outer shells near portal structure are resolved into concentric rings because of the coaxial organization of DNA. Whereas the individual conformations may exhibit the same coaxial organization, it does not mean that single rings of the density seen in the reconstructions are comprised of single DNA strands (Petrov et al., 2007).
Our current model neglects the torsional stiffness of DNA, which is not expected to make a significant contribution to the total energy but may have some effect on the structure (Spakowitz and Wang, 2005). The model is also incapable of representing the base-pair disruptions that might occur under high bending stresses, leading to DNA kinking. As a consequence, we are not able to determine the probability of kinking, which is an essential component of one model for DNA packaging (Serwer, 1986). Finally, our electrostatic treatment does not include the possible presence of polycations, which might play a significant role in the structural and energetic aspects of packaging. All of these issues will be the subject of future studies.
Supplementary Material
Acknowledgments
This research was supported by NIH grant GM70785 to S.C.H.
Abbreviations footnote
- MD
molecular dynamics
- cryo-EM
cryo-electron microscopy
- dsDNA
double-stranded DNA
Footnotes
Appendix A. Supplementary Data: Supplementary data associated with this article can be found, in the online version, at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann HW, DuBow MS. Viruses of prokaryotes. CRC Press; Boca Raton, Fla: 1987. [Google Scholar]
- Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta M, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24(21):3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I, Marenduzzo D, Yeomans JM. Polymer packaging and ejection in viral capsids: Shape matters. Phys Rev Lett. 2006;96(20) doi: 10.1103/PhysRevLett.96.208102. [DOI] [PubMed] [Google Scholar]
- Arsuaga J, Tan RKZ, Vazquez M, Sumners DW, Harvey SC. Investigation of viral DNA packaging using molecular mechanics models. Biophys Chem. 2002;101:475–484. doi: 10.1016/s0301-4622(02)00197-7. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Cheng NQ, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91(2):271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14(6):1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Doermann AH, Eiserlin FA, Boehner L. Genetic-Control Of Capsid Length In Bacteriophage-T4. 1. Isolation And Preliminary Description Of 4 New Mutants. J Virol. 1973;12(2):374–385. doi: 10.1128/jvi.12.2.374-385.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature. 1977;268(5621):598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, King J, Harrison SC, Eiserling FA. Structural organization of DNA packaged within heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978;14(3):559–568. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct. 2002;31:303–319. doi: 10.1146/annurev.biophys.31.082901.134202. [DOI] [PubMed] [Google Scholar]
- Granoff A, Webster RG. Encyclopedia of virology. Academic Press; San Diego, Ca: 1999. [Google Scholar]
- Guo PX, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage-Phi-29. J Mol Biol. 1987;197(2):229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- Hud NV. Double-stranded DNA organization in bacteriophage heads: An alternative toroid-based model. Biophys J. 1995;69(4):1355–1362. doi: 10.1016/S0006-3495(95)80002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439(7076):612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol. 2007;17(2):237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proc Natl Acad Sci U S A. 2001;98(24):13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosturko LD, Hogan M, Dattagupta N. Structure of DNA within 3 isometric bacteriophages. Cell. 1979;16(3):515–522. doi: 10.1016/0092-8674(79)90026-6. [DOI] [PubMed] [Google Scholar]
- LaMarque JC, Le TVL, Harvey SC. Packaging double-helical DNA into viral capsids. Biopolymers. 2004;73(3):348–355. doi: 10.1002/bip.10529. [DOI] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Locker CR, Harvey SC. A model for viral geome packing. Multiscale Model Simul. 2006;5:1264–1279. [Google Scholar]
- Parsegian VA, Rand RP, Rau DC. Macromolecules and water: Probing with osmotic stress. Energetics Of Biological Macromolecules; 1995. pp. 43–94. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Harvey SC. Structural and thermodynamic principles of viral packaging. Structure. 2007;15(1):21–27. doi: 10.1016/j.str.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Lim-Hing K, Harvey SC. Packaging of DNA by Bacteriophage Epsilon15: Structure, Forces and Thermodynamics. Structure. 2007;15(7):807–812. doi: 10.1016/j.str.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Forces during Bacteriophage DNA packaging and ejection. Biophys J. 2005;88(2):851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KE, Williams RC, Calendar R. Mode of DNA packing within bacteriophage heads. J Mol Biol. 1973;78(2):255–259. doi: 10.1016/0022-2836(73)90114-9. [DOI] [PubMed] [Google Scholar]
- Serwer P. Arrangement of double-stranded DNA packaged in bacteriophage capsids: An alternative model. J Mol Biol. 1986;190(3):509–512. doi: 10.1016/0022-2836(86)90019-7. [DOI] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001;413(6857):748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Spakowitz AJ, Wang ZG. DNA packaging in bacteriophage: Is twist important? Biophys J. 2005;88(6):3912–3923. doi: 10.1529/biophysj.104.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RKZ, Petrov AS, Harvey SC. YUP: A molecular simulation program for coarse-grained and multiscaled models. J Chem Theory Comput. 2006;2:529–540. doi: 10.1021/ct050323r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao YZ, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95(3):431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage phi 29 upon DNA packaging and release. EMBO J. 2006;25(21):5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


