Abstract
Free DNA in solution exhibits an untwisting of the double helix with increasing temperature. We have shown previously that when DNA is reconstituted with histones to form nucleosome core particles, both the core DNA and the adjacent linker DNA are constrained from thermal untwisting. The origin of this constraint is unknown. Here we examine the effect of two modifications of nucleosome structure on the constraint against thermal untwisting, and also on DNA topology. In one experiment, we removed the highly positively charged histone amino and carboxy termini by trypsinization. Alternatively, we added histone H5, a histone H1 variant from chick erythrocytes. Neither of these modifications had any major effect on DNA topology or twist in the nucleosome.
Full text
PDF




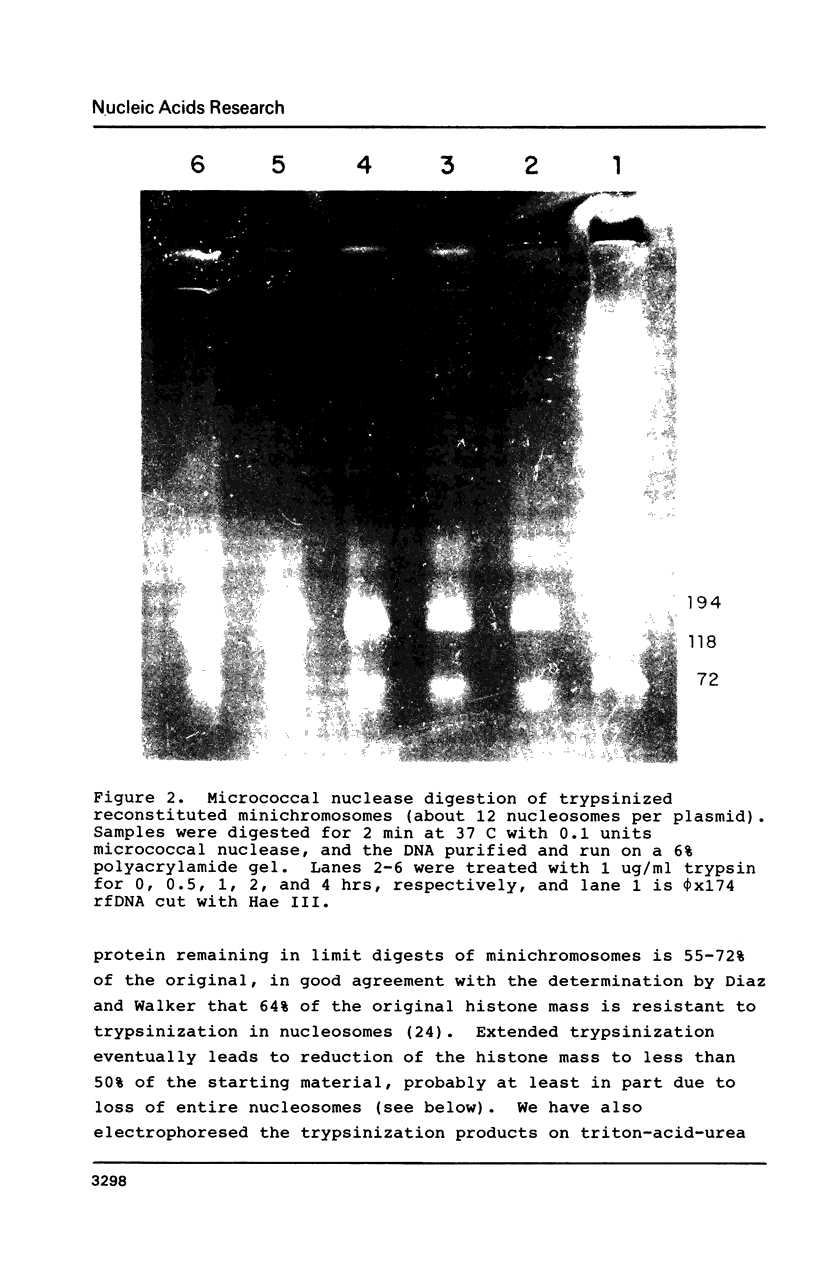

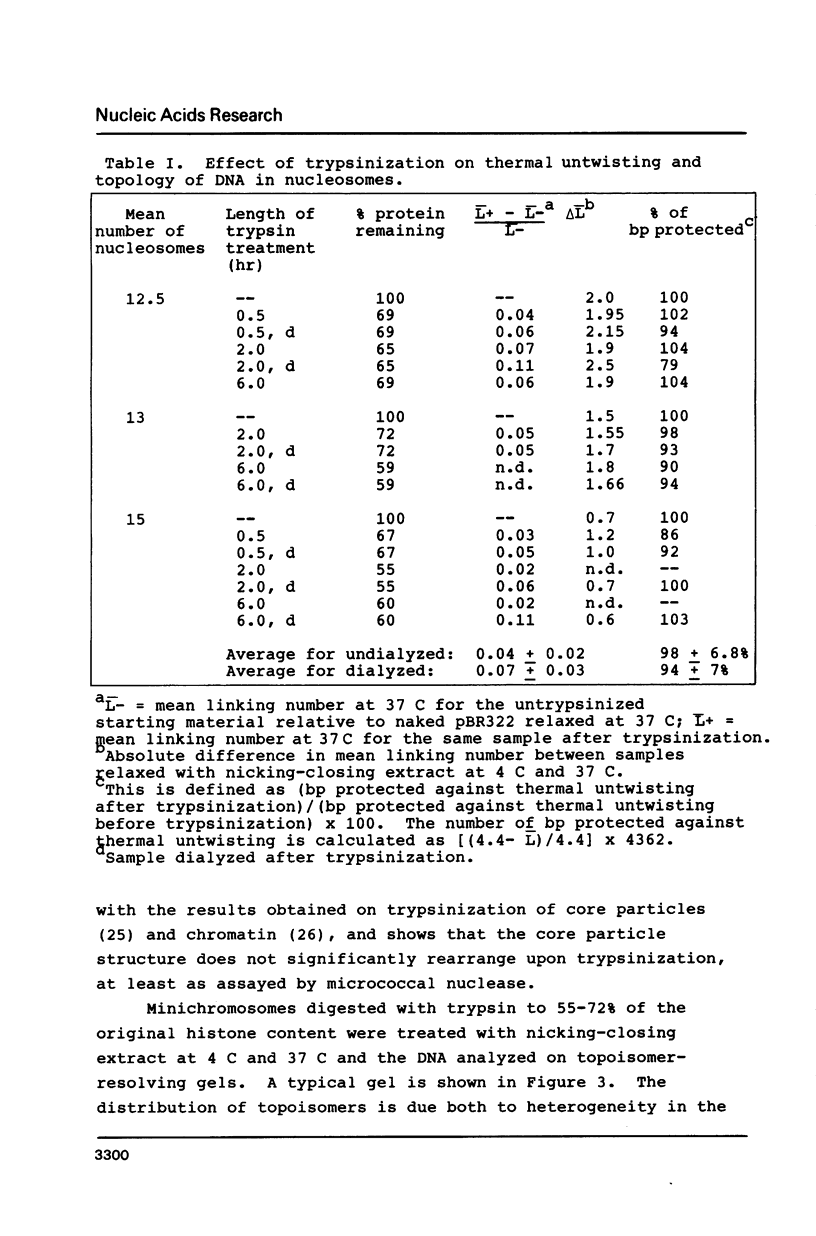



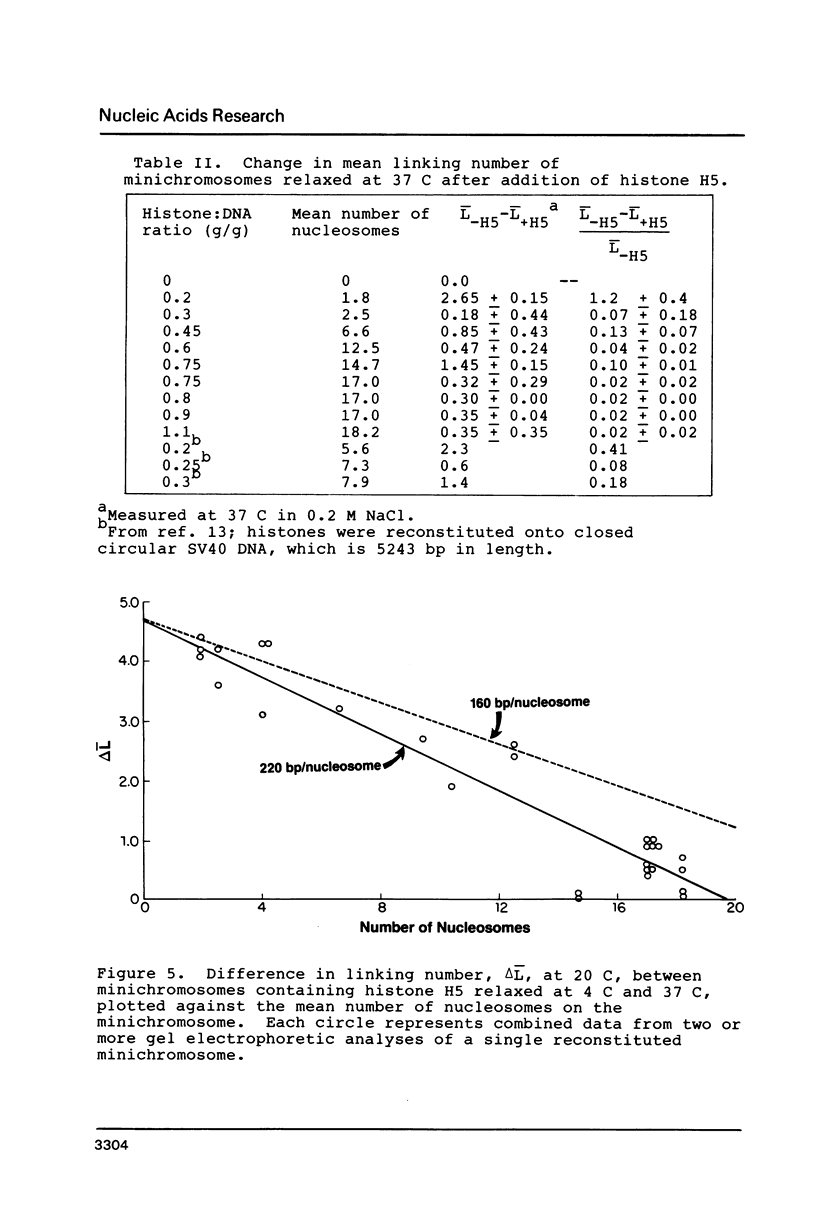






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J., Feldman J., Nasmyth K. A., Strathern J. N., Klar A. J., Broach J. R., Hicks J. B. Sites required for position-effect regulation of mating-type information in yeast. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):989–998. doi: 10.1101/sqb.1983.047.01.113. [DOI] [PubMed] [Google Scholar]
- Allan J., Harborne N., Rau D. C., Gould H. Participation of core histone "tails" in the stabilization of the chromatin solenoid. J Cell Biol. 1982 May;93(2):285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baase W. A., Johnson W. C., Jr Circular dichroism and DNA secondary structure. Nucleic Acids Res. 1979 Feb;6(2):797–814. doi: 10.1093/nar/6.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina-Stein M., Vogel T., Singer D. S., Singer M. F. H5 Histone and DNA-relaxing enzyme of chicken erythrocytes. Interaction with superhelical DNA. J Biol Chem. 1976 Dec 10;251(23):7363–7366. [PubMed] [Google Scholar]
- Böhm L., Crane-Robinson C. Proteases as structural probes for chromatin: the domain structure of histones. Biosci Rep. 1984 May;4(5):365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- Chan A., Kilkuskie R., Hanlon S. Correlations between the duplex winding angle and the circular dichroism spectrum of calf thymus DNA. Biochemistry. 1979 Jan 9;18(1):84–91. doi: 10.1021/bi00568a013. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz B. M., Walker I. O. Trypsin digestion of core chromatin. Biosci Rep. 1983 Mar;3(3):283–292. doi: 10.1007/BF01122461. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon R., Bateman E., Allan J., Harborne N., Gould H. Control of RNA polymerase binding to chromatin by variations in linker histone composition. J Mol Biol. 1984 Nov 25;180(1):131–149. doi: 10.1016/0022-2836(84)90434-0. [DOI] [PubMed] [Google Scholar]
- Harland R. M., Weintraub H., McKnight S. L. Transcription of DNA injected into Xenopus oocytes is influenced by template topology. Nature. 1983 Mar 3;302(5903):38–43. doi: 10.1038/302038a0. [DOI] [PubMed] [Google Scholar]
- Karpov V. L., Bavykin S. G., Preobrazhenskaya O. V., Belyavsky A. V., Mirzabekov A. D. Alignment of nucleosomes along DNA and organization of spacer DNA in Drosophila chromatin. Nucleic Acids Res. 1982 Jul 24;10(14):4321–4337. doi: 10.1093/nar/10.14.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautiainen T. L., Jones P. A. Effects of DNA binding proteins on DNA methylation in vitro. Biochemistry. 1985 Feb 26;24(5):1193–1196. doi: 10.1021/bi00326a021. [DOI] [PubMed] [Google Scholar]
- Keller W., Müller U., Eicken I., Wendel I., Zentgraf H. Biochemical and ultrastructural analysis of SV40 chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):227–244. doi: 10.1101/sqb.1978.042.01.025. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Tatchell K. Chromatin core particle unfolding induced by tryptic cleavage of histones. Nucleic Acids Res. 1977 Jun;4(6):2039–2055. doi: 10.1093/nar/4.6.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Nucleosome core particles suppress the thermal untwisting of core DNA and adjacent linker DNA. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4653–4657. doi: 10.1073/pnas.82.14.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. F., Wallace R. B., Bonner J. Altered nucleosome spacing in newly replicated chromatin from Friend leukemia cells. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5903–5907. doi: 10.1073/pnas.75.12.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D., Sommer K. R., Panyim S., Spiker S., Chalkley R. A modified procedure for fractionating histones. Biochem J. 1972 Sep;129(2):349–353. doi: 10.1042/bj1290349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina M. A model chromatin assembly system. Factors affecting nucleosome spacing. J Mol Biol. 1984 Sep 15;178(2):341–363. doi: 10.1016/0022-2836(84)90148-7. [DOI] [PubMed] [Google Scholar]
- Stein A. DNA wrapping in nucleosomes. The linking number problem re-examined. Nucleic Acids Res. 1980 Oct 24;8(20):4803–4820. doi: 10.1093/nar/8.20.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Künzler P. Histone H5 can correctly align randomly arranged nucleosomes in a defined in vitro system. Nature. 1983 Apr 7;302(5908):548–550. doi: 10.1038/302548a0. [DOI] [PubMed] [Google Scholar]
- Walker I. O. Differential dissociation of histone tails from core chromatin. Biochemistry. 1984 Nov 6;23(23):5622–5628. doi: 10.1021/bi00318a037. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Van Lente F. Dissection of chromosome structure with trypsin and nucleases. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4249–4253. doi: 10.1073/pnas.71.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Localization of the sites along nucleosome DNA which interact with NH2-terminal histone regions. J Biol Chem. 1977 Sep 25;252(18):6516–6520. [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Stein A. Folding of DNA by histones which lack their NH2-terminal regions. J Biol Chem. 1978 Jun 10;253(11):3857–3861. [PubMed] [Google Scholar]
- Zweidler A. Resolution of histones by polyacrylamide gel electrophoresis in presence of nonionic detergents. Methods Cell Biol. 1978;17:223–233. [PubMed] [Google Scholar]
- van den Broek H. W., Noodén L. D., Sevall J. S., Bonner J. Isolation, purification, and fractionation of nonhistone chromosomal proteins. Biochemistry. 1973 Jan 16;12(2):229–236. doi: 10.1021/bi00726a009. [DOI] [PubMed] [Google Scholar]