Background: The generation of human-induced pluripotent stem cells (iPS) has raised expectations for disease modeling, drug discovery, and cell therapy.
Results: VP16-polycistronic vectors display enhanced reprogramming capacity.
Conclusion: Primary tubular renal cells are amenable for iPSC reprogramming in the absence of oncogenes.
Significance: Kidney-derived iPSCs provide a reliable cellular platform for the study of kidney pathology and drug discovery studies.
Keywords: Induced Pluripotent Stem (iPS) Cell, Induced Pluripotent Stem Cells, Kidney, Reprogramming, Stem Cells, Renal Tubular Cells, VP16
Abstract
The tubular epithelium of the kidney is susceptible to injury from a number of different causes, including inflammatory and immune disorders, oxidative stress, and nephrotoxins, among others. Primary renal epithelial cells remain one of the few tools for studying the biochemical and physiological characteristics of the renal tubular system. Nevertheless, differentiated primary cells are not suitable for recapitulation of disease properties that might arise during embryonic kidney formation and further maturation. Thus, cellular systems resembling kidney characteristics are in urgent need to model disease as well as to establish reliable drug-testing platforms. Induced pluripotent stem cells (iPSCs) bear the capacity to differentiate into every cell lineage comprising the adult organism. Thus, iPSCs bring the possibility for recapitulating embryonic development by directed differentiation into specific lineages. iPSC differentiation ultimately allows for both disease modeling in vitro and the production of cellular products with potential for regenerative medicine. Here, we describe the rapid, reproducible, and highly efficient generation of iPSCs derived from endogenous kidney tubular renal epithelial cells with only two transcriptional factors, OCT4 and SOX2. Kidney-derived iPSCs may provide a reliable cellular platform for the development of kidney differentiation protocols allowing drug discovery studies and the study of kidney pathology.
Introduction
Kidney-related diseases constitute one of the major health challenges in modern society. The incidence and poor prognosis of some kidney-associated diseases, together with the shortage in organ donors (kidney transplantation remains the best therapeutic option upon renal failure), highlight the need for the development of new therapeutic approaches. In addition to the generation of cellular products, the development of human disease models may help to gain a deeper understanding of the molecular and cellular alterations that lead to kidney pathologies.
The generation of induced pluripotent stem cells (iPSCs)4 may open the door for the potential translation of stem cell-related therapies into the clinic as well as for the generation of patient-derived pluripotent stem cells suitable for disease modeling in vitro (1). Most of the initial studies on iPSC generation have relied on the use of c-Myc and Klf4, two well known oncogenes (2). Although non-integrative approaches for iPSCs have been reported, the presence of such genes in the reprogramming mixture has been strongly linked to tumorigenesis due to oncogene re-activation and accumulation of point mutations (3, 4). Yet the non-integrative approaches have been recently reported to facilitate genetic stability when compared with that observed by traditional retroviral transduction (5). These findings suggest that the reprogramming process per se induces uncontrollable stochastic effects, which might interfere with drug discovery and disease modeling studies. To alleviate these concerns, a range of different methodologies is being developed to generate “safer and higher quality” iPSCs suitable for transplantation and disease modeling. Such approaches include a reduction in the number of factors used for reprogramming (6–8), the engineering of more potent transcription factors (9), the combination of chemical compounds alongside different reprogramming factors (10), as well as non-integrative approaches (11–18). However, and despite the progress achieved, current methodologies still suffer from low reproducibility as well as a large variability in the efficiency of the reprogramming process.
Furthermore, variables such as age and tissue of origin have a profound impact on the reprogramming efficiency, eventually requiring the expression of fewer factors and/or reducing the timing of the whole process (6–8). Of note is the fact that somatic cell reprogramming into iPSCs does not lead to full erasure of the epigenetic marks that define initial somatic cell identity (19–21). Thus, it is generally accepted that, upon differentiation, iPSCs will more efficiently give rise to the initial populations employed for reprogramming (19–21). Accordingly, and although there is little information regarding in vitro differentiation of iPSCs into kidney populations, the possibility to reprogram patient kidney samples might represent, once reliable differentiation protocols are established, a more reliable future alternative as compared with iPSCs derived from other somatic sources (21).
In summary, a number of different considerations have to be taken into account for iPSC production. First, the methodologies employed should avoid the use of oncogenes, such as c-Myc (22). Second, the number of integrations, and thus the number of independent viral particles encoding each reprogramming factor, has to be kept to a minimum while allowing for robust expression of the transgenes in the initial reprogramming phase. Importantly, and although a previous report highlighted the presence of genetic mutations in iPSCs generated by non-integrative approaches to levels comparable with those obtained by retroviral transduction (4), a more recent publication demonstrated that nonintegrative approaches contribute to genomic stability (5). Third, the reprogramming procedure has to be fast and efficient in terms of reproducibility and the number of iPSC colonies generated.
Here, we describe the rapid, reproducible, and highly efficient generation of iPSCs derived from renal proximal tubular epithelial cells by using the transcription factors Oct4 and Sox2, with the final goal of providing a cellular platform aimed toward the development of differentiation protocols for kidney populations suitable for disease modeling and eventually the treatment of human kidney pathologies.
EXPERIMENTAL PROCEDURES
Renal Proximal Tubular Cell Isolation
The isolation and culture of primary renal proximal tubular epithelial cells have been described previously (24–25). The ethics committee of “Hospital Clinic de Barcelona” approved the procedure, and signed consent forms are available upon request. Primary renal proximal epithelial cells from 24-, 60-, and 64-year-old men were used in this study. Briefly, tubular cells were prepared from renal tissue after nephrectomy from portions of kidney not involved in renal cell carcinoma. Prior to cell isolation, the fibrous capsule and the inner medulla were removed. 1-mm2 pieces of tissue were digested for 1 h with agitation at 37 °C in Iscove's modified Dulbecco's medium containing 1% collagenase IV (Invitrogen). The digested fragments of tissue were filtered in 300-, 100-, and 70-μm cell strainers (BD Biosciences), and cell suspensions were overlaid on a pre-cooled Percoll density gradient solution (starting density 1.07 g/ml, Amersham Biosciences) and centrifuged for 40 min at 4 °C at 16,000 rpm. This procedure established a gradient with densities between 1.019 and 1.139 g/ml. The fraction between 1.05 and 1.076 g/ml was collected and washed three times in three volumes of cold Hanks' buffered saline solution (Invitrogen). Finally, cells were plated on plastic plates with DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen), Glutamax (1 mm), penicillin/streptomycin, and nonessential amino acids (100 μm).
OCT3/4-VP16-SOX2 Plasmid Generation
A DNA fragment (3′-OctGlyVP16Bsp) containing the last amino acids of mouse Oct4 cDNA, followed by a glycine-rich spacer and a mammalian codon-optimized sequence encoding amino acids MLGDGDSPGPGFTPHDSAPYGALDMADFEFEQMFTDALGIDEYGG of the herpes simplex virus VP16 transcriptional activator, was created by annealing and ligating the three following primer pairs: 1F, 5′-TTCAAACACCAGCGGCCTGGGCGGCGGCAGCGGCGGCGGCGGCAGCGGCGGCGGCGGCAGCGGCATGCTGGGCGACGGCGACAGC-3′, and 1R, 5′-TCGCCGTCGCCCAGCATGCCGCTGCCGCCGCCGCCGCTGCCGCCGCCGCCGCTGCCGCCGCCCAGGCCGCTGGTGTTTGAATGCA-3′; 2F, 5′-CCCGGCCCCGGCTTCACCCCCCACGACAGCGCCCCCTACGGCGCCCTGGACATGGCCGACTTCGAGTTCG-3′, and 2R, 5′-CTCGAAGTCGGCCATGTCCAGGGCGCCGTAGGGGGCGCTGTCGTGGGGGGTGAAGCCGGGGCCGGGGCTG-3′; and 3F, 5′-AGCAGATGTTCACCGACGCCCTGGGCATCGACGAGTACGGCGGCGGATCCGGAG-3′, and 3R, 5′-TCGACTCCGGATCCGCCGCCGTACTCGTCGATGCCCAGGGCGTCGGTGAACATCTGCTCGAA-3′. 3′-OctGlyVP16 was then cloned into a unique NsiI site located at the 3′ end of the mouse Oct4 cDNA generating Oct4VP16Bsp. The polycistronic Oct4VP16-Sox2-mOrange cassette was generated as described elsewhere (26) and cloned in the pMXs retroviral vector.
Viral Transduction
Retroviral vectors containing Oct3/4, Sox2, Klf4, and c-Myc (Addgene: 20072, 20073, 20074, and 20075, respectively) or OCT3/4-VP16-SOX2 were transfected into Phoenix Ampho 293 cells (ATCC) with FuGENE 6 (Roche Applied Science). The next day, the media were changed, and virus-producing cells were cultured overnight at 32 °C in a 5% CO2 incubator. Viral supernatants were harvested on two consecutive days and collected every 12 h through a 0.45-μm PVDF filter (Millipore) to remove any cells. Finally, 1 μg/ml Polybrene (Chemicon) was added to viral supernatant preparations to increase infection efficiency. Infection efficiency was monitored separately with a pMXs GFP-expressing vector (Addgene). Pools of 80,000 tubular proximal cells were infected three times at 12-h intervals with retroviruses carrying tricistronic vectors or four factor combinations and centrifuged at 750 × g at 32 °C for 45 min.
Induced Pluripotent Stem Cell Generation and Subculture
One day after the last infection, cells were trypsinized and plated onto irradiated human fibroblasts (iHFs), supplemented with hES medium. hES medium is as follows: KO-DMEM, Glutamax (1 mm), penicillin/streptomycin, nonessential amino acids (100 μm), 2-mercaptoethanol (100 μm), bFGF (10 ng/ml). From day 15, the iPSC colonies were indistinguishable from ESC colonies (i.e. well defined borders, flat morphology, and big nuclei containing prominent nucleoli, scarce cytoplasm). On day 20, the iPSC colonies were picked manually and expanded in Matrigel (BD Biosciences) supplemented with human ES cell media conditioned on mouse embryonic fibroblasts feeder layers. From this stage the iPSC colonies were amplified either by trypsinization or manual dissection. Silencing of transgenes and expression of reactivation of endogenous pluripotent factors were analyzed by RT-PCR as described previously (8).
Immunofluorescence Analysis
Cells were grown on plastic cover slide chambers and fixed with 4% paraformaldehyde. The following antibodies were used: tumor rejection antigen 1 (TRA-1), TRA-1–60 (MAB4360, 1:200), TRA-1–81 (MAB4381, 1:200), sex-determining region Y-box 2 (SOX2; AB5603, 1:500; Chemicon); stage-specific embryonic antigens SSEA-4 (MC-813-70, 1:2) and SSEA-3 (MC-631, 1:2; Developmental Studies Hybridoma Bank); OCT-3/4 (sc-5279, 1:100; Santa Cruz Biotechnology); NANOG (EB06860, 1:100; Everest Biotech); neuron-specific class III β-tubulin (TUJ-1; 1:500; Covance); glial fibrillary acidic protein (GFAP; 1:1000; Dako); α1-fetoprotein (AFP; 1:400; Dako); Forkhead Box Protein A2 (FOXA-2, 1:50; R&D Systems); smooth muscle actin (ASMA; 1:400; Sigma); α-sarcomeric actin (ASA; 1:200; Sigma); occludin (OCCDN; 1:200) and cytokeratin (CYT; 1:200; Dako); CD13 (1:200; BD Biosciences); epithelial membrane antigen (EMA; 1:200; Dako); vimentin (VIM; 1:200; Dako); CD24 (1:200; BD Biosciences); CD133 (0.5 μl per million cells; Miltenyi Biotec, Bergisch Gladbach); E-cadherin-6 (CDH6; AP1415a, 1:200) from Abgent, cytokeratin 8 (CK8; MMS-162P-250, 1:1000) from Covance, and pan-cytokeratin (PAN-CK; C2562, 1:2000) from Sigma. Images were taken using a Leica SP5 confocal microscope. Transgene expression was evaluated as the expression of red fluorescent protein (Abcam) in the colonies generated with two factors and as the expression of FLAG (M2: Sigma) in the iPSC colonies generated with four factors.
RT-PCR Analysis
Total RNA from each cell line was isolated using All Prep RNA columns (Qiagen), following the manufacturer's guidelines. All RNA samples were treated with TURBO DNase inhibitor (Ambion) to remove any residual genomic DNA, and 1 μg of RNA was used to synthesize cDNA using the Invitrogen SuperScript III reverse transcriptase kit. 25 ng of cDNA was used to quantify typical pluripotency gene expression markers by using Platinum SYBR Green quantitative PCR super mix (Invitrogen) in an ABI Prism 7000 thermocycler (Applied Biosystems) and primers as described previously (26).
In Vitro Differentiation of the Generated iPSC Lines
After 3–4 days, embryoid bodies (EBs) were transferred to 0.1% gelatin-coated polystyrene chamber slides and cultured in differentiation medium (DMEM supplemented with 20% fetal bovine serum, 2 mm l-glutamine, 0.1 mm 2-mercaptoethanol, nonessential amino acids, and penicillin/streptomycin) for 2–3 weeks to allow spontaneous endoderm formation. The medium was changed every other day. For mesoderm differentiation, EBs were maintained on gelatin-coated plate in differentiation medium supplemented with 100 μm ascorbic acid (Sigma). For ectoderm differentiation, EBs were cultured on Matrigel-coated plates in 1% N2 and 0.5% B27 (Invitrogen) medium supplemented with 1 μm retinoic acid for 2–3 weeks.
In Vivo Teratoma Assays
Severe combined immune deficient-Beige male mice (n = 2 animal/iPS clone), ∼8 weeks old, were injected with iPSCs (1 million for each injection site, approximately) subcutaneously in the testicular parenchyma. All procedures involving animals were approved by the Institutional Animal Ethical Board, and the protocols were approved by the Conselleria De Salut of Cataluña. Mice were sacrificed 8 weeks after the injections or when a tumor was detected by palpation, whichever came first. Teratoma formation was assessed by hematoxylin/eosin staining and immunofluorescence techniques.
High Resolution, G-banded Karyotype
The analysis was performed on 85% confluent iPS cells growing on Matrigel. Cells were treated with colcemid at 20 ng/ml, followed by a 45-min incubation at 37 °C. Upon trypsinization, the cells were treated with Carnoy's fixative solution at −20 °C prior to analysis with the software Cytovision (Applied Imaging).
RESULTS
Isolation and Characterization of Renal Proximal Tubular Epithelial Cells from Human Kidney Samples
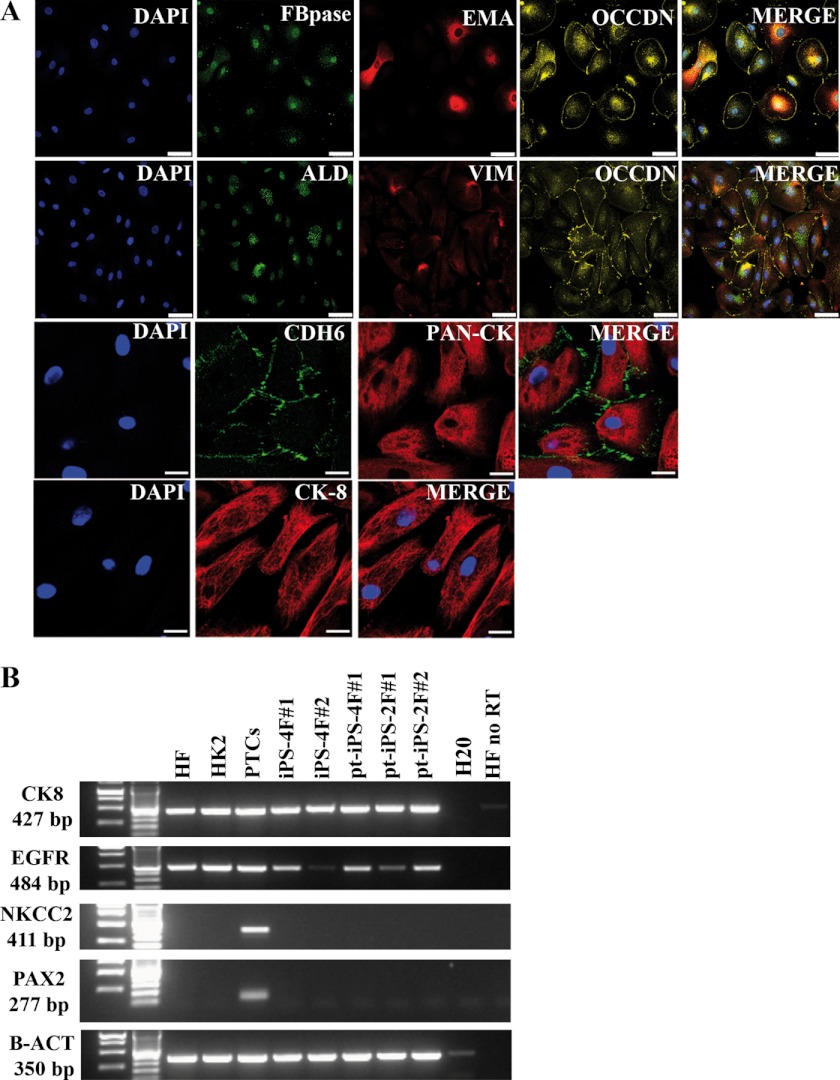
Upon approved consent, a total of three different patients were subjected to sample collection. Primary renal proximal tubular epithelial cells were cultured and further amplified prior to the reprogramming procedure with no foreseeable differences between the different patients. The cultures displayed a homogeneous population and typical epithelial morphology. To rule out the presence of contaminant cells, immunofluorescence characterization was performed. As shown in Fig. 1A, isolated cultured cells expressed typical renal tubular epithelial markers, including fructose-1,6-bisphosphatase, epithelial membrane antigen, occludin, aldolase, vimentin, cytokeratin, E-cadherin-6, pan-cytokeratin, and cytokeratin 8. Expression of the selected kidney markers was further validated at the RNA level (Fig. 1B), compared with nonrelated samples, fibroblasts, at the protein level (supplemental Fig. S1), as well as by staining of normal kidney sections (supplemental Fig. S2). Altogether, our results demonstrated that upon isolation and culture, pure populations of primary renal cells could be obtained and further subcultured.
FIGURE 1.
Characterization of proximal tubular cells. A, confocal immunofluorescence microscopy in proximal tubular cells. Fructose-1,6-bisphosphatase (FBPase) and aldolase (ALD) are enzymatic markers. Epithelial membrane antigen (EMA), occludin (OCCDN), and E-cadherin-6 (CDH6) are adherens junction markers. Vimentin (VIM) and cytokeratin (CYT), pan-cytokeratin (Pan-CK), and cytokeratin 8 (CK8) are markers of intermediate filaments; scale bars, 100 μm. B, RT-PCR for the indicated markers in the retroviral induced iPSCs and primary renal epithelial cells.
Generation of Induced Pluripotent Stem Cells from Isolated Proximal Tubular Cells
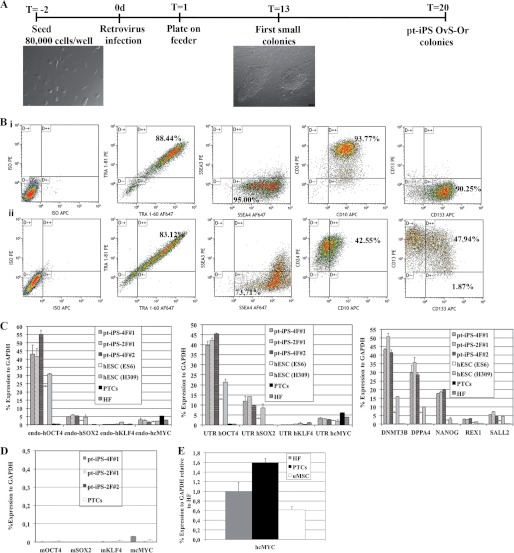
Because we were able to produce highly pure cultures of primary renal cells, as quantified by flow cytometry (supplemental Fig. S1), we next asked whether this population was suitable for reprogramming into iPSCs. To this end, we first attempted to generate iPSC lines using the four classical Yamanaka factors (Oct4, Sox2, Klf4, and c-Myc). Viral transduction of the four factors led to the rapid generation of embryonic stem (ES)-like colonies, which were further characterized upon manual picking and subculturing (supplemental Fig. S3). In light of the fact that our initial attempts demonstrated the amenability of proximal tubular epithelial cells to undergo reprogramming, we next aimed to eliminate the two best described oncogenes present in the reprogramming mixture, Klf4 and c-Myc (1). Accordingly, we further engineered a tricistronic vector encoding for Oct4 and Sox2 and separated by the VP16 transactivation domain, which has been described as an enhancer of reprogramming (supplemental Fig. S4) (9). Although this combination was not sufficient to reprogram human fibroblasts, reprogrammed proximal tubular epithelial cells, hereafter referred to as pt-iPSCs, were observed as early as 13 days after viral transduction. To shed new light into this observation, we wondered whether endogenous c-Myc levels were higher in proximal tubular renal cells and thus could potentially compensate for removal of the transgene in the reprogramming mixture as described previously (8). As expected, c-Myc levels were elevated in proximal tubular renal cells as compared with human fibroblasts (Fig. 2E).
FIGURE 2.
Generation of pt-iPS cell lines using OCT4 and SOX2 factors. A, time line of proximal tubular cells reprogramming onto iPSCs with OCT4VP16SOX2 retroviral particles. Three days post-infection, the proximal tubular cells were transferred onto human feeders. Compact and tight adherent colonies are observed already at day 13. Typical hES-like colonies are picked at day 20 after viral infection. B, flow cytometry analysis of pt-iPS-2F#1 and proximal tubular cells. Cells were analyzed for the embryonic stem cell markers, including CD24, CD133, SSEA-3, SSEA-4, TRA-1–60, and TRA-81. At the same time, cells were analyzed for the surface markers CD10 and CD13. C, RT-PCR for the indicated markers in OCT4VP16SOX2-pt-iPSCs. iPS lines generated with 4F from human fibroblasts (iPS-4F#1 and iPS-4F#2) and human ESC lines ES6 and H306 were used as stem cells control. Human fibroblasts (HF), HK-2 cells (HK2), and human proximal tubular cells were used as somatic controls. D, RT-PCR demonstrating transgene silencing in OCT4VP16SOX2-pt-iPSCs. C and E, RT-PCR in proximal tubular renal cells (PTCs) demonstrates higher levels of endogenous c-Myc expression as compared with unrelated fibroblasts samples (HF) and comparable with umbilical cord-derived mesenchymal stem cells (uMSCs).
Expression of Pluripotency Related Genes in iPSC Lines Generated from Proximal Tubular Epithelial Cells
Colonies with tight borders and large sizes were subsequently picked and expanded in less than 20 days after viral transduction (Fig. 2A). As expected, a down-regulation of the epithelial marker CD13 was observed upon reprogramming (Fig. 2B, panel i). Expression of surface markers associated with kidney development and/or progenitor cells such as CD10, CD24, and CD133 was sustained upon reprogramming (Fig. 2B, panel ii). To ascertain the stem cell status of the reprogrammed cells, we further complemented surface protein expression analysis with RNA analysis. Expression levels of different pluripotency markers in reprogrammed pt-iPSCs were comparable with those found in human ES cells (Fig. 2C). Specificity of the reprogramming process was exemplified by the absence of pluripotency-associated markers at the level of mRNA expression in both primary proximal tubular epithelial cells and fibroblasts, an unrelated somatic cell type (Fig. 2C). Furthermore, quantitative PCR analysis demonstrated robust expression of the endogenous pluripotency factors Oct4 and Sox2, with levels comparable with those found in both human ES cell lines ES6 and H306 (Fig. 2C), and expression of the transgenes was efficiently silenced (Fig. 2D). Altogether, our results demonstrated the pluripotent identity of the reprogrammed pt-iPSCs.
Characterization of iPSC Lines Generated from Proximal Tubular Epithelial Cells
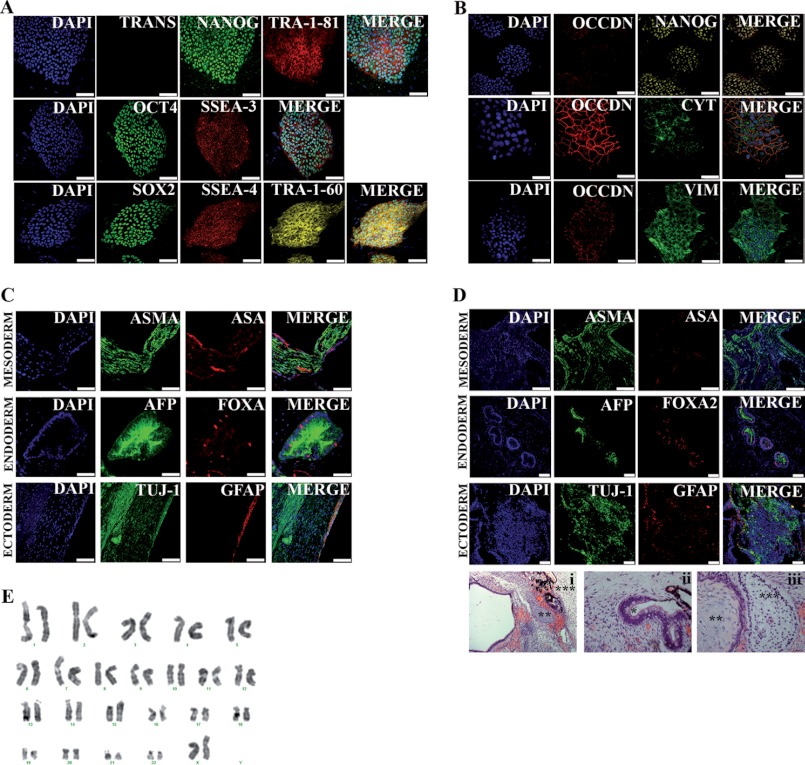
pt-iPSCs generated with Oct4 and Sox2 transcription factors demonstrated robust expression of the endogenous pluripotency-associated transcription nuclear factors OCT4, SOX2, and NANOG as well as the surface markers SSEA-3/4 and TRA-1–60/81 (Fig. 3A). As expected, pt-iPSCs maintained occludin and vimentin expression (Fig. 3B) and showed no evidence of oncogene re-activation (Fig. 3A).
FIGURE 3.
Characterization of pt-iPS cell lines. A, immunofluorescence analysis of pt-iPS-2F#1 cell line for pluripotency markers. Transgene expression was assessed by analysis of orange fluorescence expression. Colonies exhibit typical pluripotency markers, including SSEA-4, SSEA-3, TRA-1–60, TRA-1–81, and the transcription factors OCT4, SOX2, and NANOG. Underlying fibroblasts provide a negative control. Scale bars are 100 μm. B, immunofluorescence analysis of the pt-iPS-2F#1 cell line for epithelial and intermediate filament markers. Scale bar is 100 μm. C, representative images of in vitro differentiation of the pt-iPS-2F#1 line into the three primary germ cell layers (smooth-α-sarcomeric actin (ASMA) and α-sarcomeric actin (ASA), endoderm-α-fetoprotein (AFP), Forkhead box protein A2 (FOXA2), ectoderm-neuron-specific class III β-tubulin (TUJ-1), and glial fibrillary acidic protein (GFAP)). Scale bar is 50 μm for endoderm differentiation images. Scale bar is 100 μm for mesoderm and ectoderm differentiation images. D, teratoma formation assay by injection of pt-iPS into the testis of severe combined immune deficient-Beige male mice. Representative images of in vivo differentiation of pt-iPS-2F#1 line into the three primary germ cell layers by immunofluorescence (smooth-α-sarcomeric actin (ASMA) and α-sarcomeric actin (ASA), endoderm-α-fetoprotein (AFP), Forkhead box protein A2 (FOXA2), ectoderm-neuron-specific class III β-tubulin (TUJ-1), and glial fibrillary acidic protein (GFAP)). Scale bar is 100 μm for mesoderm and endoderm differentiation images. Scale bar is 50 μm for ectoderm differentiation images. After 12 weeks, teratomas were collected and analyzed by hematoxylin and eosin staining. pt-iPS-2F differentiated in vivo to endoderm (*), mesoderm (**), or ectoderm fates (***). E, high -resolution, G-banded karyotype indicating a normal diploid male chromosomal content in the pt-iPS-2F#1 line.
To fully characterize the generated pt-iPSCs, we next investigated their differentiation potential both in vitro and in vivo. Spontaneous in vitro differentiation was assessed by EB formation. EB differentiation led to the appearance of cell types from the three different germ layers of the embryo as demonstrated by specific immunostaining (Fig. 3C). Moreover, we did not detect any relevant genomic aberration by karyotype analysis (Fig. 3E). Finally, pt-iPSCs were injected into immunodeficient animals to assess teratoma-forming capacity, the most stringent test of pluripotency available for human cells. Injection of pt-iPSCs consistently led to teratoma formation in all animals analyzed. Teratomas were well defined and included tissues derived from the three germ layers as demonstrated by immunofluorescence of specific markers and H&E staining (Fig. 3D).
DISCUSSION
The derivation of ES cells from human embryos offered a new therapeutic scenario for disease modeling (27). However, the following two major obstacles impeded the further development of such technology for that specific purpose: immune rejection after cell transplantation and ethical concerns regarding the use of hES for research. Recently, Takahashi et al. (28) solved such inconveniences when they described, in 2007, that human fibroblasts could be reprogrammed to induce pluripotent stem cells by forced expression of only four transcription factors (Oct4, Sox2, Klf4, and c-Myc). By means of a simple methodology, the derivation of iPSCs from individuals with genetic diseases offered a unique scenario for the study of both pathology and drug discovery in a human model. In the last few years, intense research has proven that iPSC lines can be generated from almost any human somatic cell type (1–8). However, several crucial questions remain to be answered, such as what is the most amenable and efficient cell type to be reprogrammed. However, the development of different reprogramming methodologies has shown that it is possible to reprogram human somatic cells by means of non-integrative strategies (11–18), although such approaches are more labor intensive and less efficient than the integrative ones (e.g. viral transduction). Interestingly, different studies have shown that iPSCs retain an “epigenetic memory” of their cell type of origin, which can vary after cell expansion. Moreover, such works have demonstrated that iPSCs can differentiate back to their original cell type with higher efficiency than to other cell fates (19–21).
We believe that reprogramming of renal populations into iPSCs opens the possibility for patient-specific iPSC generation to study two unexplored issues in the kidney field as follows: human kidney development and the pathology of its related diseases. Furthermore, the generation of iPSCs from kidney tissue could allow for the study and development of novel human kidney therapies through disease modeling and drug discovery studies once differentiation protocols are widely available and are reproducible. For these purposes, we have described here, for the first time, the reprogramming of human renal proximal tubular cells to a pluripotent state. Moreover, it is important to notice that in our hands the pt-iPSCs were generated in a rapid (in only 13 days) and efficient manner and displayed all typical hallmarks of pluripotency, including differentiation into cell types comprising the three germ layers as well as in vivo teratoma formation. Recently, Zhou et al. (23) have shown the generation of iPSCs by a four-factor viral transduction, including Klf4 and c-Myc, upon isolation of different cell populations from urine. Thus, the presence of different cell populations, alongside the intrinsic stochastic nature of the reprogramming process, raises the possibility that the iPSCs originated from contaminant somatic cells, such as blood or squamous cells from the urethra and not necessarily from tubular renal cells. Our work extensively validates the renal nature of the cells prior to reprogramming, thus ruling out the possibility that the iPSCs originated from nonrenal contaminant cell populations. Most importantly, we have been able to generate pt-iPSCs with only two transcription factors, Oct4 and Sox2, by using a single tricistronic vector with no need of any additional chemical compounds. Therefore, pt-iPSCs can be efficiently generated in the absence of the oncogenes Klf4 and c-Myc (1). So far, only cord blood cells have been reprogrammed by means of Oct4 and Sox2 transcription factors (8). Our data showed that human renal proximal tubular cells, similar to cord blood cells, expressed higher endogenous levels of c-MYC when compared with other somatic cell types such as human fibroblasts, suggesting that higher endogenous levels were sufficient to compensate for omission of c-Myc in our reprogramming mixture.
Overall, we show here for the first time the generation of iPSCs from a new human cell source, renal proximal tubular cells. Our iPSC lines were generated in a short time period with only two transcription factors, omitting the use of two potent oncogenes, Klf4 and c-Myc. More importantly, and taking in consideration that pt-iPSCs might retain certain epigenetic memory as demonstrated for other cell sources (19–21), we are currently working on the identification of the transcription factors responsible for driving the differentiation to kidney progenitor-like cells from human iPSCs. Such information will be needed for iPSC-based modeling of kidney diseases, because, to our knowledge, no study has described reliable differentiation protocols allowing for the generation of kidney-like cells.
Supplementary Material
Acknowledgments
We are grateful to Meritxell Carrió for excellent assistance in cell culture; Lola Mulero Pérez, Cristina Morera, Cristina Pardo, and Mercé Martí for bioimaging assistance; and José Miguel Andrés Vaquero for assistance with flow cytometry.
This work was supported in part by grants from Ministerio de Economía y Competitividad, Fondo de Investigaciones Sanitarias Grants TERCEL and PI052847, Fundacion Cellex, Sanofi, The Leona M. and Harry B. Helmsley Charitable Trust, and the G. Harold and Leila Y. Mathers Charitable Foundation (to the J. C. I. B. laboratory).

This article contains supplemental Figs. S1–S4.
- iPSC
- induced pluripotent stem cell
- pt
- proximal tubular
- EB
- embryoid body
- PTC
- proximal tubular cells
- hES
- human ES.
REFERENCES
- 1. Yamanaka S. (2010) Patient-specific pluripotent stem cells become even more accessible. Cell Stem Cell. 7, 1–2 [DOI] [PubMed] [Google Scholar]
- 2. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 3. Sun N., Longaker M. T., Wu J. C. (2010) Human iPS cell-based therapy. Considerations before clinical applications. Cell Cycle 9, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gore A., Li Z., Fung H. L., Young J. E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M. A., Kiskinis E., Lee J. H., Loh Y. H., Manos P. D., Montserrat N., Panopoulos A. D., Ruiz S., Wilbert M. L., Yu J., Kirkness E. F., Izpisua Belmonte J. C., Rossi D. J., Thomson J. A., Eggan K., Daley G. Q., Goldstein L. S., Zhang K. (2011) Somatic coding mutations in human-induced pluripotent stem cells. Nature 471, 63–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng L., Hansen N. F., Zhao L., Du Y., Zou C., Donovan F. X., Chou B. K., Zhou G., Li S., Dowey S. N., Ye Z., NISC Comparative Sequencing Program, Chandrasekharappa S. C., Yang H., Mullikin J. C., Liu P. P. (2012) Low incidence of DNA sequence variation in human-induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 10, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim J. B., Greber B., Araúzo-Bravo M. J., Meyer J., Park K. I., Zaehres H., Schöler H. R. (2009) Direct reprogramming of human neural stem cells by OCT4. Nature 461, 649–653 [DOI] [PubMed] [Google Scholar]
- 7. Aasen T., Raya A., Barrero M. J., Garreta E., Consiglio A., Gonzalez F., Vassena R., Bilić J., Pekarik V., Tiscornia G., Edel M., Boué S., Izpisúa Belmonte J. C. (2008) Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276–1284 [DOI] [PubMed] [Google Scholar]
- 8. Giorgetti A., Montserrat N., Aasen T., Gonzalez F., Rodríguez-Pizà I., Vassena R., Raya A., Boué S., Barrero M. J., Corbella B. A., Torrabadella M., Veiga A., Izpisua Belmonte J. C. (2009) Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 5, 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., Chen J., Hu J. L., Wei X. X., Qin D., Gao J., Zhang L., Jiang J., Li J. S., Liu J., Lai K. Y., Kuang X., Zhang J., Pei D., Xu G. L. (2011) Reprogramming of mouse and human somatic cells by high performance engineered factors. EMBO Rep. 12, 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. (2010) Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 7, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou W., Freed C. R. (2009) Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 27, 2667–2674 [DOI] [PubMed] [Google Scholar]
- 12. Kim D., Kim C. H., Moon J. I., Chung Y. G., Chang M. Y., Han B. S., Ko S., Yang E., Cha K. Y., Lanza R., Kim K. S. (2009) Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 4, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Judson R. L., Babiarz J. E., Venere M., Blelloch R. (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 27, 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Warren L., Manos P. D., Ahfeldt T., Loh Y. H., Li H., Lau F., Ebina W., Mandal P. K., Smith Z. D., Meissner A., Daley G. Q., Brack A. S., Collins J. J., Cowan C., Schlaeger T. M., Rossi D. J. (2010) Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 7, 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W., Xue Y., Cai J., Guo X., Qin B., Zhang R., Wu J., Lai L., Teng M., Niu L., Zhang B., Esteban M. A., Pei D. (2011) MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 286, 17359–17364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Subramanyam D., Lamouille S., Judson R. L., Liu J. Y., Bucay N., Derynck R., Blelloch R. (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 29, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anokye-Danso F., Trivedi C. M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P. J., Epstein J. A., Morrisey E. E. (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 8, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K.-I., Shibata T., Kunisada T., Takahashi M., Takahashi J., Saji H., Yamanaka S.. (2011) A more efficient method to generate integration-free human iPS cells. Nat. Methods 5, 409–412 [DOI] [PubMed] [Google Scholar]
- 19. Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I., Yabuuchi A., Takeuchi A., Cunniff K. C., Hongguang H., McKinney-Freeman S., Naveiras O., Yoon T. J., Irizarry R. A., Jung N., Seita J., Hanna J., Murakami P., Jaenisch R., Weissleder R., Orkin S. H., Weissman I. L., Feinberg A. P., Daley G. Q. (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bar-Nur O., Russ H. A., Efrat S., Benvenisty N. (2011) Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 9, 17–23 [DOI] [PubMed] [Google Scholar]
- 21. Sullivan G. J., Bai Y., Fletcher J., Wilmut I. (2010) Induced pluripotent stem cells. Epigenetic memories and practical implications. Mol. Hum. Reprod. 16, 880–885 [DOI] [PubMed] [Google Scholar]
- 22. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 23. Zhou T., Benda C., Duzinger S., Huang Y., Li X., Li Y., Guo X., Cao G., Chen S., Hao L., Chan Y. C., Ng K. M., Ho J. C., Wieser M., Wu J., Redl H., Tse H. F., Grillari J., Grillari-Voglauer R., Pei D., Esteban M. A. (2011) Generation of induced pluripotent stem cells from urine. J. Am. Soc. Nephrol. 22, 1221–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baer P. C., Nockher W. A., Haase W., Scherberich J. E. (1997) Isolation of proximal and distal tubule cells from human kidney by immunomagnetic separation. Technical note. Kidney Int. 52, 1321–1331 [DOI] [PubMed] [Google Scholar]
- 25. Baer P. C., Geiger H. (2008) Human renal cells from the thick ascending limb and early distal tubule. Characterization of primary isolated and cultured cells by reverse transcription polymerase chain reaction. Nephrology 13, 316–321 [DOI] [PubMed] [Google Scholar]
- 26. Gonzalez F., Barragan Monasterio M., Tiscornia G., Montserrat Pulido N., Vassena R., Batlle Morera L., Rodriguez Piza I., Izpisua Belmonte J. C. (2009) Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc. Natl. Acad. Sci. U.S.A. 106, 8918–8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 28. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.