Background: Transport of preproteins by the accessory Sec system requires a specific sequence (the AST domain) adjacent to the signal peptide.
Results: The AST domain interacts specifically with SecA2, and this interaction is modulated by the accessory Sec proteins Asp1–3.
Conclusion: The Asp-dependent AST-SecA2 interaction is essential for transport.
Significance: A better understanding of accessory Sec transport may aid the development of novel antimicrobials.
Keywords: ATPases, Bacterial Pathogenesis, Photoaffinity Labeling, Protein Cross-linking, Protein Secretion, Streptococcus, Trafficking, SecA2, SecY2
Abstract
The accessory Sec systems of streptococci and staphylococci mediate the transport of a family of large, serine-rich glycoproteins to the bacterial cell surface. These systems are comprised of SecA2, SecY2, and three core accessory Sec proteins (Asp1–3). In Streptococcus gordonii, transport of the serine-rich glycoprotein GspB requires both a unique 90-residue N-terminal signal peptide and an adjacent 24-residue segment (the AST domain). We used in vivo site-specific photo-cross-linking to identify proteins that interact with the AST domain during transport. To facilitate this analysis, the entire accessory Sec system of S. gordonii was expressed in Escherichia coli. The determinants of GspB trafficking to the accessory Sec system in E. coli matched those in S. gordonii, establishing the validity of this approach. When the photo-cross-linker was placed within the AST domain, the preprotein was found to cross-link to SecA2. Importantly, no cross-linking to SecA was detected. Cross-linking of the N-terminal end of the AST domain to SecA2 occurred regardless of whether Asp1–3 were present. However, cross-linking to the C-terminal end was dependent on the Asps. The combined results indicate that full engagement of the AST domain by SecA2 is modulated by one or more of the Asps, and suggest that this process is important for initiating transport.
Introduction
The serine-rich repeat (SRR)2 glycoproteins of Gram-positive bacteria are a unique family of bacterial adhesins that have a significant impact on biofilm formation and disease (1–10). The SRR glycoproteins all have a similar domain organization, with an unusually long N-terminal signal peptide followed by a short SRR domain, a ligand binding domain, a long SRR domain, and a C-terminal LPXTG cell wall anchoring motif. Despite the similar domain organization, the ligand binding domains are surprisingly diverse in terms of amino acid sequences, predicted structures, and binding properties (11–15).
The SRR glycoproteins are transported to the bacterial cell surface by a specialized transporter known as the accessory Sec (aSec) system (3, 16–18). These specialized transporters are invariably comprised of five proteins: SecA2, SecY2, Asp1, Asp2, and Asp3. In some streptococcal species such as Streptococcus gordonii, the aSec system may include one or two additional proteins (Asp4 and Asp5). Studies of the aSec system in S. gordonii have indicated that this transport system has some striking similarities to, and some important differences from, the general Sec system. SecA2 resembles SecA both in primary amino acid sequence and in the presence of conserved residues and domains, except it lacks the C-terminal SecB binding region (17). SecA2 can hydrolyze ATP and is likely to function as the transport motor (19). SecY2 resembles SecY and is likely to form the transmembrane channel (20). Asp4 or Asp5 might interact with SecY2 and function similarly to SecE or SecG, but this has not yet been verified (21).
The most notable difference between the Sec and aSec systems is that the latter has three proteins (Asp1–3) that are essential for transport (22, 23), but have no obvious counterpart in the general Sec system. Although recent studies in both S. gordonii and Streptococcus parasanguinis have begun to uncover interactions that occur among the Asps (termed Gap1–3 in S. parasanguinis) (23, 24), the precise role of these proteins in transport remains obscure. Asp2 and Asp3 can bind the serine-rich regions of GspB (the SRR glycoprotein of S. gordonii), suggesting that one or both Asps might function as chaperones to facilitate targeting (25). However, why the aSec system should require two specialized chaperones is unknown.
There are also distinct differences in the preproteins that are transported by the aSec and general Sec systems. We have previously used a truncated version of GspB (GspB736FLAG; Fig. 1A) as a model substrate to analyze both the means by which the aSec system recognizes the preprotein substrate, and the features of the preprotein that facilitate trafficking to the aSec versus Sec system. Although the carbohydrate moieties on GspB strongly impede Sec transport, they are not necessary for recognition by the aSec system, since nonglycosylated GspB736FLAG is efficiently transported by the aSec system. Instead, an atypical 90 amino acid signal sequence at the N terminus of GspB is required for transport (26). Trafficking is largely determined by glycine residues in the hydrophobic core of the signal peptide (the H region; Fig. 2), which interfere with export via the Sec pathway and concomitantly facilitate aSec transport (27). Replacing one or more of the glycines with amino acids that increase the H region hydrophobicity or α-helicity (properties known to be important for the interaction of signal peptides with SecA and SecY), can re-route the protein, so that it is transported more readily by the general Sec system. Conversely, replacing one of the glycine residues with proline (G75P), which decreases the helical propensity, abrogates Sec transport but only partially interferes with aSec transport. Whether the H-region glycine residues affect trafficking by facilitating a direct interaction with an aSec component, or simply render the preprotein unrecognizable by the general Sec system (a “Sec avoidance” mechanism), is not yet known.
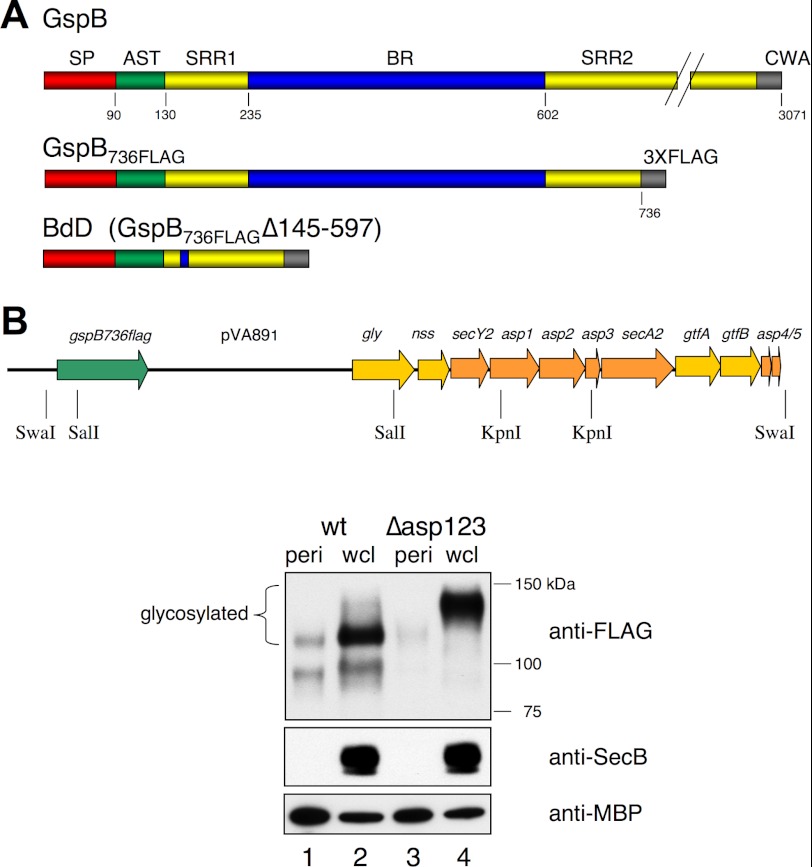
FIGURE 1.
The S. gordonii accessory Sec system and preprotein substrates. A, GspB is the native, cell wall-anchored (CWA) substrate. SP, signal peptide; AST, accessory Sec transport domain, SRR1 and SRR2, serine-rich repeat regions 1 and 2, respectively; BR, the ligand-binding region. GspB736FLAG is a model aSec substrate that is freely secreted into the S. gordonii culture medium. BdD is a derivative of GspB736FLAG that lacks residues 145–597 (26). B, map of the accessory Sec operon in S. gordonii PS2163, and expression of the aSec system in E. coli. The SwaI restriction site upstream of gspB736flag was engineered in to facilitate rescue of the entire operon. The transport components are indicated by orange arrows, the substrate is indicated in green, and glycosyl transferases are indicated in yellow. The integrated plasmid pVA891 has a chloramphenicol resistance gene and pACYC origin for selection and replication, respectively, in E. coli (35). A 23 kb plasmid that corresponds to the circularly-ligated SwaI chromosomal fragment (pSwaR), or a Δasp1–3 derivative plasmid (pSwaRΔKpn), was used to transform E. coli strain C43/DE3. Periplasmic proteins (peri) or whole-cell lysates (wcl) were analyzed by Western blotting with anti-FLAG, anti-SecB or anti-MBP (maltose-binding protein) as indicated. SecB (a cytoplasmic protein) and MBP are negative and positive controls for periplasmic proteins, respectively. Glycosylated forms of GspB736FLAG were detected by lectin blot analysis, using succinylated wheat germ agglutinin (not shown).
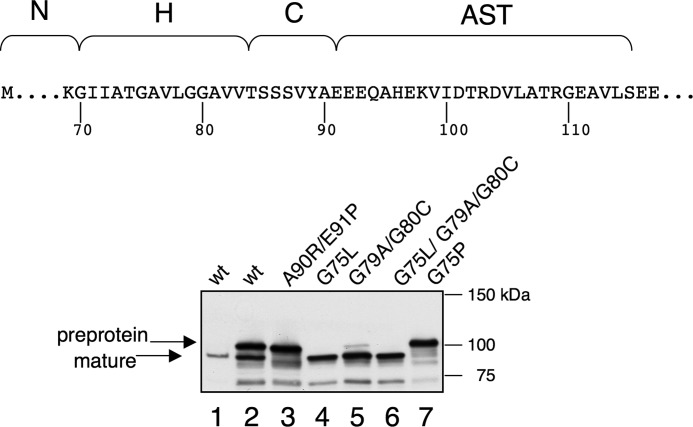
FIGURE 2.
Accessory Sec-independent transport of GspB736FLAG and selected variants in E. coli. The amino acid sequence of the H and C regions of the signal peptide, along with the AST domain of GspB is indicated. The GspB736FLAG variants were expressed constitutively from pVA891 (26, 27), and were detected by Western blotting with anti-FLAG monoclonal antibodies. Lane 1, periplasmic proteins; lanes 2–8, whole-cell lysates.
Perhaps the most unique aspect of aSec transport is that it requires a specific 24-residue segment known as the accessory Sec transport (AST) domain adjacent to the signal peptide (20). A leucine residue at the C-terminal end of the AST domain (L114) is especially critical for aSec transport. Replacement of L114 with the helix-breaking residues glycine or proline nearly abolishes transport by the aSec system, whereas replacement with hydrophobic residues such as isoleucine, valine, or phenylalanine has no adverse effect. Extensive mutational analysis of the AST domain indicates that it might form an amphipathic helix. Experimental evidence also suggests that the AST domain affects both the targeting of GspB to the SecY2 translocon and gating of the transmembrane channel, but precisely how the AST domain might facilitate these processes is undetermined.
Although the AST domain is essential for GspB transport, no component of the aSec system has been found to bind specifically to this region of the preprotein (25). We therefore chose to use site-specific photo-cross-linking, to capture proteins that may interact only transiently with the AST domain during transport. This powerful technique has been increasingly employed to monitor protein-protein interactions in vivo (28–30). The key to this method is the incorporation of a photo-activatable amino acid at a single, selected site in a protein of interest, through a process of “amber suppression” (31, 32). In brief, this method involves the placement of a TAG stop codon at a specific site within a gene of interest, and the co-expression of a nonsense suppressor tRNA along with a unique amino-acyl tRNA synthetase. The latter is engineered to transfer an “unnatural” amino acid such as benzoylphenylalanine (BzF), which is provided in the culture medium. Once the BzF-labeled protein is expressed, it can be photoactivated and cross-linked to any closely-associated proteins by exposing the culture to 350–365 nm light. The amber codon supression technique has been utilized in eukaryotes, E. coli and mycobacteria but, to our knowledge, it has not yet been used in any Gram-positive bacterial species. Because of the documented efficiency, and lack of toxicity, of amber suppression in E. coli, we opted to express the S. gordonii aSec system along with the amber suppression components in E. coli. The combined results indicate that the aSec system is functional in E. coli, and that trafficking of GspB736FLAG and variant preproteins recapitulates that in S. gordonii. In addition, results in both E. coli and S. gordonii indicate that the AST domain interacts specifically with SecA2 to facilitate transport.
EXPERIMENTAL PROCEDURES
Cloning and Expression of the S. gordonii aSec Operon in E. coli
The entire aSec operon was cloned from the S. gordonii chromosome by using a marker rescue strategy. A unique SwaI restriction site exists just downstream of asp5 (Fig. 1) and a second SwaI site was incorporated just upstream of gspB736flag by using forward primer: 5′-AAAAATGCATTTAAATTAAGTAGAGGGGATTAC-3′ (SwaI site is underlined) and reverse primer 96XR (20) to amplify the 5′-end of gspB. The resulting fragment was digested with NsiI and XhoI and used to replace the NsiI-XhoI fragment of pB736flag/96X (20). The plasmid (pB736flag/96X/NsiSwa) was used to transform the gspB deletion strain PS1740 (23) resulting in strain PS2163 (Fig. 1B). Chromosomal DNA was extracted from PS2163, digested with SwaI, treated with T4 DNA ligase, and then used to transform E. coli TOP10 cells. The 23 kb plasmid carrying gspB736flag and the entire accessory Sec operon (pSwaR) was stably maintained as long as the host strain was grown at or below 30 °C. Attempts to manipulate the plasmid, such as selective deletions of one or more genes, generally led to re-arrangements. However, a stable Δasp1–3 derivative plasmid (pSwaRΔKpn) was obtained by deletion of a KpnI fragment. Plasmids pSwaR and pSwaRΔKpn were used to transform E. coli strain C43/DE3. To assess the expression, glycosylation and transport of GspB736FLAG, strains were grown at 30 °C for 18 h in LB containing chloramphenicol (15 μg/ml), diluted 5-fold into fresh medium, and then incubated 90 min at 30 °C. Periplasmic proteins were obtained by osmotic shock (33). The glycosylation of GspB736FLAG was assessed by lectin blot analysis, using biotinylated succinylated wheat germ agglutinin as described (34).
Subcloning of the aSec Operon
To facilitate the separate expression of GspB736FLAG and the aSec system, a SalI fragment spanning from the middle of gly to codon 220 of gspB (i.e. a SalI-SwaI-SalI fragment; Fig. 3) was subcloned into pCOLADuet (Novagen), generating the plasmid pCOLAaSec. The subcloned fragment in pCOLAaSec was found to have a 1 kb insertion of an IS10 mobile genetic element in gtfA. The insertion apparently did not have a polar effect on the expression of the downstream genes, since the glycosylation of GspB736FLAG (which requires the expression of both GtfA and GtfB) could be restored by supplying gtfA on a separate plasmid (data not shown).
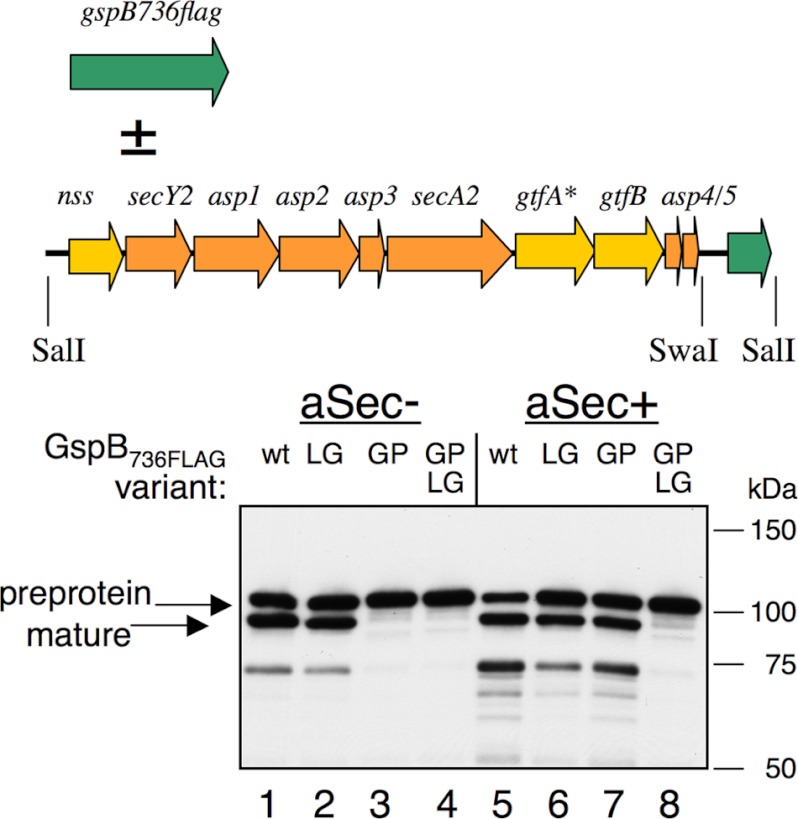
FIGURE 3.
Transport of GspB736FLAG and selected variants in the absence or presence of the aSec system in E. coli (aSec−, or aSec+, respectively). GspB736FLAG (wt) and variants with the substitutions L114G (LG), G75P (GP), and L114G/G75P (LG GP) were expressed constitutively from pVA891 (20, 27). The aSec components were expressed from pCOLAaSec/gtfA::IS10. Note that gtfA* is disrupted by an insertion sequence, so GspB736FLAG does not undergo glycosylation. Anti-FLAG monoclonal antibodies were used for the detection of GspB736FLAG by Western blotting of whole cell lysates.
To generate an otherwise isogenic plasmid that lacked asp1–3, attempts were made to similarly subclone the SalI fragment from pSwaRΔKpn into pCOLADuet, or to delete the asp1–3 KpnI fragment from pCOLAaSec/gtfA::IS10. For unknown reasons it was not possible to recover the aSecΔasp1–3 fragment in pCOLADuet using either of these methods. However, it was possible to subclone the SalI fragment from pCOLAaSec/gtfA::IS10 into pVA891 (35), and then delete the asp1–3 KpnI fragment from this construct, resulting in plasmids pVA891aSec/gtfA::IS10 and pVA891aSecΔKpnI/gtfA::IS10, respectively.
Construction of pCOLA-pBpF
A BamHI-XhoI fragment containing the amber suppression components was removed from pEVOL-pBpF and ligated to pCOLADuet that had been digested with BamHI and SalI. This plasmid was used instead of pEVOL-pBpF in experiments where the aSec components were expressed from pVA891aSec/gtfA::IS10 or pVA891aSecΔKpnI/gtfA::IS10, since pVA891 and pEVOL-pBpF have the same origin of replication and selective marker.
Construction of Amber Codon Variants of gspB736flag and BdD
The gspB736flag or BdD coding regions were amplified from plasmid pBflagR or pBflagRΔ145–597 (26), respectively, using forward primer 5′-GGAATTCCATATG-TTTTTTAAACGTCAAAAGGGTC-3′ and reverse primer 5′-TAATTTTAATTAATTTTACTTGTCATCGTCATCCTTGTAG-3′. The PCR products were digest with NdeI and PacI, and then cloned in pCDFDuet (Novagen) that had been digested similarly. Amber codons were incorporated into the gspB736flag or BdD coding sequences using a two-stage PCR strategy. The presence of only the intended change in any segments generated by PCR was confirmed by DNA sequence analysis (Sequetech).
Amber Codon Suppression and Photocross-Linking
E. coli C43/DE3 was transformed by electroporation, using simultaneous combinations of two or three plasmids as indicated. Transformants were selected by plating on medium containing chloramphenicol (15 μg/ml), spectinomycin (50 μg/ml), and/or kanamycin (50 μg/ml) as appropriate. Strains were grown at 37 °C for 18 h, diluted 5-fold into fresh medium, and then incubated at 37 °C for 90 min. BzF (Bachem) and IPTG (AllStar Scientific) were each added to 100 μm. In cases where the amber suppression components were expressed from pEVOL-pBpF, arabinose was added to 200 μm. The cultures were then transferred to 30 °C and incubated for 1.5 to 2 h. Culture samples were exposed to 365 nm light for 20 min, by placement under a Blak-Ray 100 Watt UV lamp (UVP). Parallel culture samples were left under ambient lighting for 20 min at room temperature. Cells were lysed by boiling in 2× SDS-PAGE sample buffer, and the equivalent of 30 μl cell culture was typically used for Western blot analysis. After separation of proteins by SDS-PAGE, samples were transferred to BioTraceNT (Pall) and cross-linked complexes were detected by using anti-FLAG antibody (Sigma).
Identification of Cross-linked Products
To obtain sufficient material for identification by mass spectrometry, the expression of BzF-labeled BdD and cross-linked products was scaled up as follows. Strains were grown at 37 °C for 18 h, 40 ml was added to 160 ml fresh medium, and then incubated at 37 °C for 90 min. BzF and IPTG were added to 100 μm, and arabinose was added to 200 μm. The cultures were transferred to 30 °C and incubated for 1.5 to 2 h. Cross-linking was then induced in 100 ml of the culture by transferring two 50 ml samples to a large Petri dish (144 mm2) and exposing to UV light with occasional mixing for 20 min. The other half of the culture was not exposed to UV but was otherwise processed similarly. After centrifugation for 20 min at 3200 × g, cells were lysed by suspension in CelLytic reagent (Sigma) followed by sonication. GspB736FLAG or BdD cross-linked products were purified using anti-FLAG-agarose (Sigma). The eluted proteins were separated by SDS-PAGE, stained with Coomassie, and then submitted for analysis by tandem mass spectrometry (Stanford PAN Lab). In some cases, cell lysates or purified cross-linked complexes were treated with 2 units (100 ng) of thrombin for 3 h at room temperature, and the thrombin-treated complexes were analyzed by Western blotting using either anti-FLAG or anti-SecA2 polyclonal antibodies.
Construction of the GspB736FLAG/spAST Chimera
Chromosomal DNA was extracted from S. pneumoniae strain TIGR4. The putative AST domain of psrP was amplified by PCR, using the foward primer 5′-AGTACTCGAGAAAACTGTAGAGAAAACGGATGCTTTGGC-3′ and the reverse primer 5′- CCGGATCCCGCTGAATTACTTGTAGATATCGTACC-3′. The PCR product was digested with XhoI and BamHI and then used to replace the XhoI-BamHI fragment of pB736flag/96X118B (20), to generate plasmid pB736flag/spAST. The NsiI-SpeI fragment spanning codons 1 to 296 of gspB736flag/spAST was then used to replace the NsiI-SpeI fragment containing the wild-type sequence in pS326B602 (36), to generate pS326B602/spAST, which was used to transform the S. gordonii strain PS1226 (19). A double crossover between sequences in pS326B602/spAST and the chromosome of PS1226 to generate PS2812 (M99 5′gspB::spec gspB736flag/spAST::pB736flagC ΔsecA2) was verified by sequence analysis of the chromosomal DNA. A spec resistance marker just upstream of gspB does not affect the expression or transport of GspB (36).
Construction of SecA2 Chimeras
The secA2 chimera CH1 was generated and cloned in pMSP3535 using the same strategy previously described for construction of secA-secA2 chimeras (19). In brief, primer pairs spA2F (5′-AAGGATCCCAAGGAGATCAGAAATGTTTAGACGT-3′) and IRA2R were used to amplify the pneumococcal secA2 NBD1, PPXD, and NBD2 regions, and primers IRA2F and 3535MCS were used to amplify the C-domain of S. gordonii secA2. The two first-stage PCR reactions were combined and amplified using primers spA2F and 3′A2, and the second-stage PCR product was then cloned in pMSP3535 (37). For CH2, primers spA2F and spPPXDR (5′-CATACACGTGTCTCAGGAGATAATTTGACATG-3′) were used to amplify the pneumococcal secA2 NBD1 and PPXD regions. The PCR product was digested with BamHI and PmlI and then used to replace the corresponding fragment in pMSPsecA2 (a PmlI site is present at the 3′-flank of the PPXD region). To facilitate the construction of CH3, a silent mutation was first introduced at codon 222 of secA2 by two-stage PCR, using forward primer gpAscF (5′-ATCKGGCGCGCCKCGTGTYCAGTC-3′) and reverse primer 5′-GACTGRACACGMGGCGCGCCMG-3′. The silent mutation generates a unique AscI restriction site (underlined) in the 5′-flank of the PPXD (pM99secA2/asc). The pneumococcal secA2 PPXD region was then amplified using primers gpAscF and the reverse primer spPPXDR. The PCR product was digested with AscI and PmlI, and used to replace the corresponding region of pM99secA2/asc. The resulting plasmids were used to transform the ΔsecA2 strains PS1226 and PS2812 (described above).
Analysis of GspB736FLAG Secretion from S. gordonii
Strains were grown at 37 °C for 18 h in Todd-Hewitt broth containing 60 μg/ml erythromycin. Proteins from S. gordonii culture supernatants or protoplasted cells were prepared and analyzed by Western blotting as described previously (20).
RESULTS
Functional Expression of the aSec System in E. coli
To assess the validity of studying the S. gordonii aSec system in E. coli, we first examined the transport of GspB736FLAG when co-expressed with the entire aSec system. It should be noted that attempts to assemble the aSec system by combining the individual genes into single or multiple plasmids were largely unsuccessful. However, it was possible to retrieve the entire operon with the native gene organization by using a marker rescue strategy (see “Experimental Procedures”). With the entire aSec operon present, GspB736FLAG became glycosylated and was transported to the E. coli periplasm (Fig. 1B, lane 1). Upon deletion of asp1–3, GspB736FLAG migrated with a higher apparent mass, which is indicative of more extensive glycosylation, and remained mainly in the cytosol (lanes 3 and 4). These results indicate that the S. gordonii aSec system can indeed function in E. coli.
The expression of BzF-labeled GspB736FLAG along with the entire aSec system was subsequently examined. Although UV-induced mass shifts were evident (data not shown), characterization of the cross-linked products tended to be complicated by the mass heterogeneity that results from carbohydrate modification (glycosylated GspB736FLAG ranges from 120 to 150 kDa). It was therefore more feasible to assess cross-linking to the AST domain in the absence of glycosylation, but it was then necessary to determine whether the preprotein features that affect trafficking of the non-glycosylated preprotein in S. gordonii were recapitulated in E. coli. As mentioned above, transport of GspB736FLAG via the general Sec system in S. gordonii is inhibited not only by the carbohydrate moieties associated with the mature region, but also by glycine residues in the H region of the signal peptide. In the absence of the S. gordonii aSec system and glycosyl transferases, some transport of non-glycosylated GspB736FLAG was evident, as determined by both cleavage of the signal peptide and the appearance of the mature protein in the periplasm (Fig. 2, lanes 1 and 2). The signal peptide appeared to be cleaved at the normal cleavage site (between residues 90 and 91), since the periplasmic form was ∼10 kDa smaller than the unprocessed preprotein. Moreover, a variant of GspB736FLAG that has an alteration predicted to interfere with signal peptidase cleavage (A90R/E91P; reference 26), did not show the same mature protein, but instead appeared to be cleaved somewhat heterogeneously (lane 3). Substitution of one or more of the H-region glycine residues with residues predicted to increase the H region hydrophobicity or helical propensity resulted in more efficient transport (lanes 4–6), whereas substitution with proline nearly abolished transport (lane 7). These results indicate that, as in S. gordonii, the native hydrophobic core of the signal peptide is not optimal for transport by the general Sec system of E. coli, and that Sec transport can be blocked by placement of a proline residue in the H region.
The effects of signal peptide and AST mutations on transport of non-glycosylated GspB736FLAG in the absence or presence of the aSec system was also assessed (note that the expression of non-glycosylated GspB736FLAG in the presence of the aSec system was facilitated by a fortuitous non-polar insertion in gtfA, as described under “Experimental Procedures”). With the aSec system present, most of the wild-type GspB736FLAG underwent transport and processing of the signal peptide (Fig. 3, lane 5). Transport of the L114G variant appeared reduced by comparison (lane 6), but was not abolished. However, since GspB736FLAG/L114G was transported (albeit inefficiently) by the Sec system (lane 2), it was not possible to determine the extent to which the L114G substitution impacted aSec transport. We therefore assessed the effect of the AST domain mutation in combination with the signal peptide mutation G75P, which selectively blocks Sec transport. As expected, the G75P variant was transported only when the aSec system was present (lane 7 versus lane 3). However, the G75P/L114G double mutant was not transported by either pathway (lanes 4 and 8). This indicated that the L114G mutation interferes specifically with aSec transport. The combined results demonstrate that trafficking of non-glycosylated GspB736FLAG in E. coli is similar to trafficking in S. gordonii and, importantly, that aSec transport is dependent on the preprotein AST domain.
Cross-linking to the AST Domain of GspB in E. coli
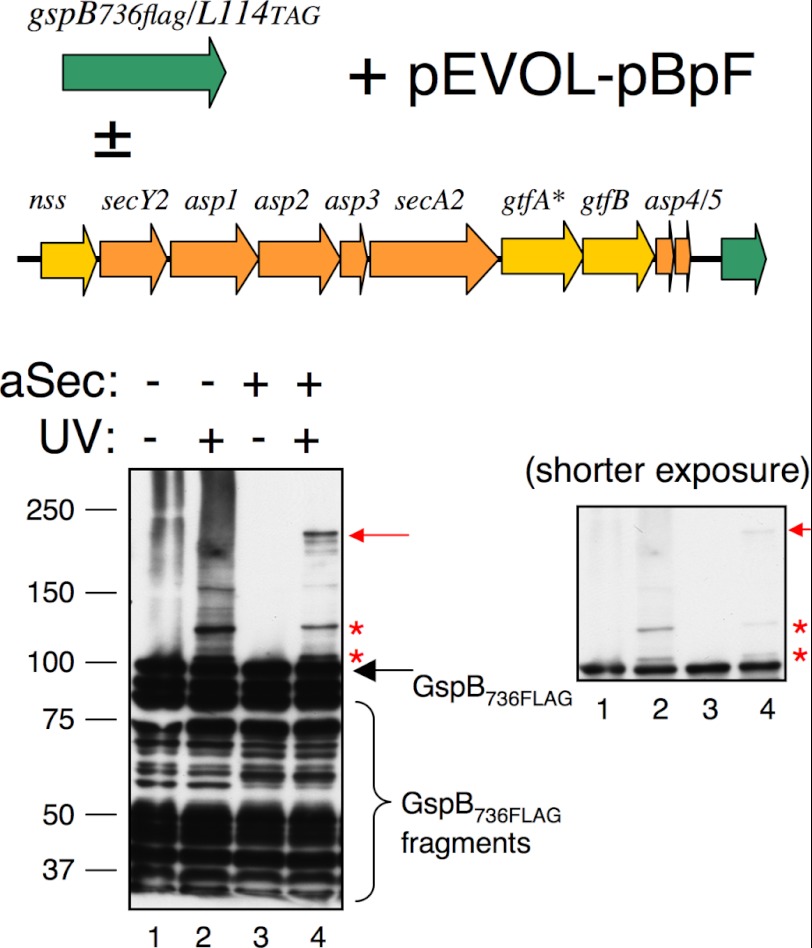
We next examined cross-linking to the AST domain of non-glycosylated GspB736FLAG in the absence or presence of the aSec system. The photoactivatable amino acid BzF was initially incorporated at position 114, since studies in S. gordonii had indicated that bulky hydrophobic residues at this position did not negatively impact transport (20). In the absence of the aSec system, two major cross-linked products were evident, migrating at 105 and 120 kDa (Fig. 4, lane 2), corresponding to GspB736FLAG cross-linked to E. coli proteins with apparent masses of ∼5 and 20 kDa, respectively. A minor cross-linked product migrating at 150 kDa, corresponding to GspB736FLAG and a 50 kDa E. coli protein, was also evident. When the aSec system was present, one additional cross-linked product migrating at 200 kDa was seen (lane 4), corresponding to a mass shift of ∼100 kDa.
FIGURE 4.
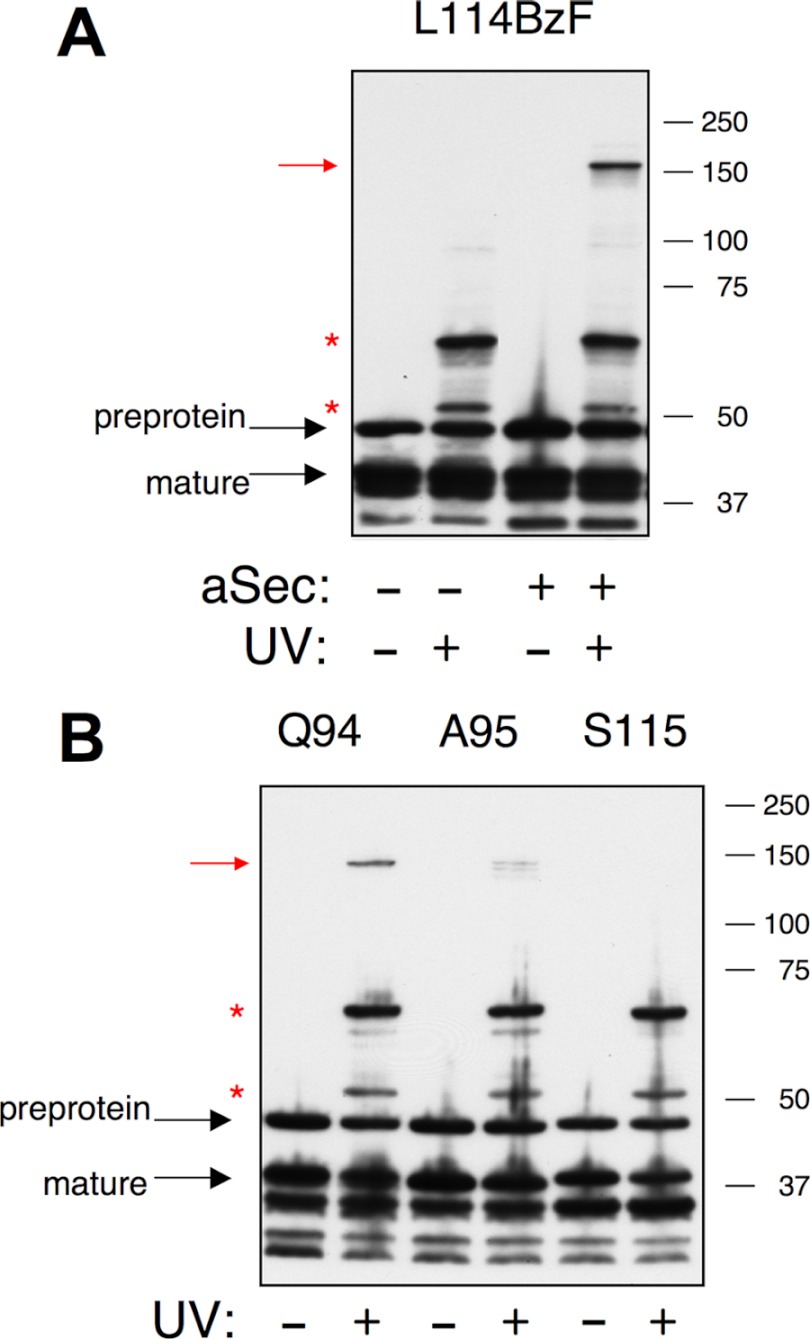
Cross-linking to position 114 in the AST domain of GspB736FLAG. GspB736FLAG/L114TAG was expressed from pCDF, the BzF-specific amber suppression components (amino-acyl tRNA synthase and tRNACUA) were expressed from pEVOL-pBpF, and the aSec operon was expressed from pCOLAaSec/gtfA::IS10. GspB736FLAG was detected by Western blotting with anti-FLAG antibodies, and the full-length preprotein is indicated by a black arrow. Red asterisks indicate cross-linked complexes that occur in both the presence and absence of the aSec system. Red arrow indicates an aSec-specific cross-linked complex. The panel on the right shows a shorter exposure of the Western blot, in which the 105 kDa cross-linked complex is more readily visible.
To optimize the yield of cross-linked products for purification and identification by mass spectrometry, we then examined cross-linking under a variety of conditions. Although the same three cross-linked products were consistently obtained, extensive fragmentation of GspB736FLAG within the ligand-binding domain occurred prior to and during purification (Fig. 4 and data not shown) making it difficult to track and recover the cross-linked protein by anti-FLAG affinity chromatography. The ligand-binding domain of GspB is highly susceptible to proteolytic degradation and is not essential for aSec transport (Ref. 26).3 Additional studies were therefore carried out using a derivative of GspB736FLAG that lacks most of this region (GspB derivative D, or BdD; Fig. 1). The photo-activatable BzF was again incorporated at position 114 in the AST domain. UV-dependent cross-linked products of apparent masses of 55 and 70 kDa were obtained in the absence of the aSec system, and an additional cross-linked product migrating at 150 kDa was obtained when the aSec system was present (Fig. 5A). The observed masses of the three cross-linked proteins represent UV-induced mass shifts of ∼5, 20, and 100 kDa (since BdD has an apparent mass of 50 kDa), which is the same as those in the major cross-linked complexes seen with BzF-labeled GspB736FLAG (Fig. 4).
FIGURE 5.
Cross-linking to the AST domain of BdD. The BdD amber codon variants were expressed from pCDF, the amber suppression components were expressed from pEVOL-pBpF, and the aSec operon was expressed from pCOLAaSec/gtfA::IS10. BdD was detected by Western blotting with anti-FLAG antibodies. Red asterisks indicate cross-linked complexes that occur in both the presence and absence of the aSec system. Red arrow indicates an aSec-specific cross-linked complex. A, crosslinking to position 114 in the absence and presence of the aSec system. B, cross-linking to positions 94, 95, and 115 in the presence of the aSec system.
Cross-linking to BdD at three additional positions (94, 95, and 115) was also examined. BdD labeled at positions 94 and 95 showed UV-dependent crosslinking similar to that of BdD labeled at position 114, with products of apparent masses of 55 and 70 kDa generated in the absence of the aSec system (data not shown) and an additional 150 kDa product generated in the presence of the aSec system (Fig. 5B). BdD labeled at position 115 yielded cross-linked products of 55 and 70 kDa in both the absence and presence of the aSec system (data not shown and Fig. 5B, respectively), but no 150 kDa cross-linked complex was detected. Thus, labeling at positions 94, 95, or 114 in the mature region results in identical cross-linking patterns, whereas labeling at position 115 showed no aSec-specific cross-linked product.
Characterization of the Cross-linked Proteins
Since cross-linking to the 100 kDa protein was clearly dependent on the aSec system, the protein cross-linked to BdD/Q94BzF was purified and was analyzed by mass spectrometry. The proteins identified in the complex were BdD and SecA2. The 150 kDa complex generated by using BdD/L114BzF was also purified, and Western blot analysis indicated it also contained SecA2.
Because cross-linking to the 5 and 20 kDa proteins occurred in both the presence and absence of the aSec system (Fig. 5 and data not shown), and BdD can be transported by either the Sec or aSec route, it was unclear whether cross-linking to these proteins was dependent on transport by either pathway. We therefore re-examined cross-linking to the AST domain of BdD in combination with the G75P substitution to inhibit general Sec transport. When using the BdD/G75P variant labeled at position 94, cross-linked complexes were again seen at 55, 70, and 150 kDa only when the aSec system was present (Fig. 6, lane 4 versus lane 2). This suggested that cross-linking of the AST domain to the 5 and 20 kDa proteins requires transport to the periplasm, and occurs upon transport by either pathway. If so, the cross-linked complexes may actually contain mature BdD, and the masses of the two E. coli proteins are likely to be 15 and 30 kDa.
FIGURE 6.
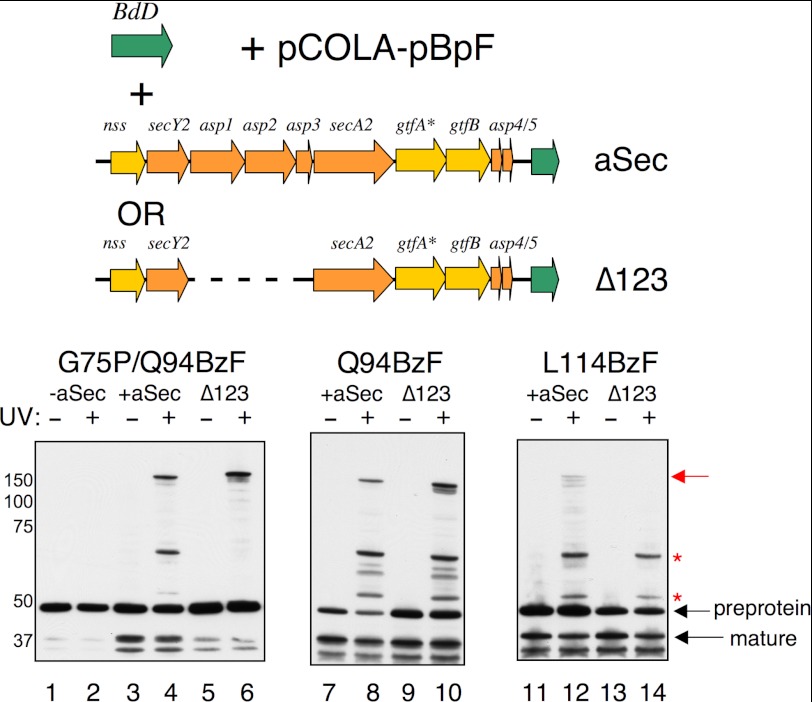
Asp-dependent cross-linking to the AST domain of BdD or BdD/G75P. The BdD variants were expressed from pCDF, and were detected by Western blotting with anti-FLAG antibodies. The amber suppression components were expressed from pCOLA-pBpF, and the aSec operon or operonΔasp1–3 was expressed from pVA891aSec/gtfA::IS10 or pVA891aSecΔKpnI/gtfA::IS10, respectively. Left panel (lanes 1–6), crosslinking to position 94 of BdD/G75P; middle panel (lanes 7–10) cross-linking to position 94 of BdD; right panel (lanes 11–14), cross-linking to position 114 of BdD. Samples in the even-numbered lanes were exposed to UV light to induce cross-linking. Red arrow indicates BdD cross-linked to SecA2. Red asterisks indicate transport-dependent, pathway-independent cross-linked products.
Dependence of the AST Interactions on Asp1–3
Our recent studies have indicated that two of the accessory Sec system components, Asp2 and Asp3, can bind GspB directly (25). We have proposed that these Asps might function as chaperones and, along with Asp1, facilitate the targeting of GspB to the SecA2/Y2 translocon. We therefore examined whether cross-linking of the AST domain to SecA2 and the two E. coli proteins occurred in the absence of Asp1–3. With the BdD/G75P variant labeled at position 94, the 55 and 70 kDa complexes were not detected in the absence of Asp1–3 (Fig. 6, lane 6). Moreover, there was little evidence of signal peptide cleavage, which confirms that the Asps are essential for aSec transport in E. coli. Thus, the results further support the possibility that formation of the 55 and 70 kDa complexes is dependent on transport to the periplasm. Surprisingly cross-linking to SecA2 was readily apparent. This indicated that the Asps were not necessary for preprotein targeting to SecA2.
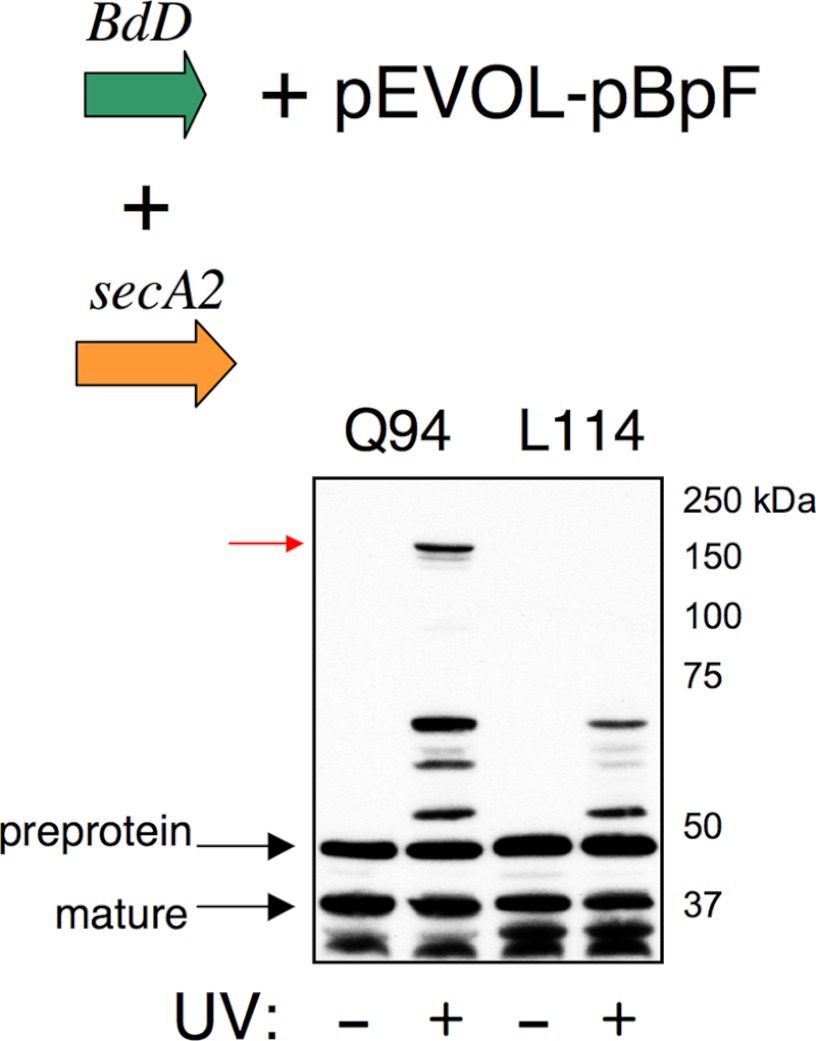
Since G75P partially interferes with aSec transport (Fig. 3, lanes 5 and 7 and Ref. 27), we also examined cross-linking to the AST domain of BdD that has the wild-type H region. With BdD labeled at position 94, cross-linking to SecA2 was again readily detected in the absence of Asp1–3 (Fig. 6, lane 10). Cross-linking to SecA2 was likely the result of targeting to, but not transport by, the aSec system (as in lane 6), whereas cross-linking to the 15 and 30 kDa proteins was due to transport by the general Sec system (as in Fig. 5). In contrast, with BdD labeled at position 114, cross-linking to the two smaller proteins, but not SecA2, was detected in the absence of the Asps (lane 14). To re-examine the ability of BdD labeled at position 94 or 114 to cross-link to SecA2, we co-expressed the labeled preproteins with SecA2 alone. Again, cross-linking of SecA2 to BdD labeled at position 94 was readily detected, but cross-linking of SecA2 to position 114 was not (Fig. 7). Therefore, although SecA2 can bind the preprotein at position 94 independently of the Asps, cross-linking of SecA2 to position 114 of the AST domain (i.e. full engagement of the AST domain) requires the entire aSec system.
FIGURE 7.
Cross-linking of BzF-labeled BdD to SecA2 alone. The BdD variants were expressed from pCDF, and were detected by Western blotting with anti-FLAG antibodies. The amber suppression components were expressed from pEVOL-pBpF, and SecA2 was expressed from pET28c (19). Red arrow indicates BdD cross-linked to SecA2.
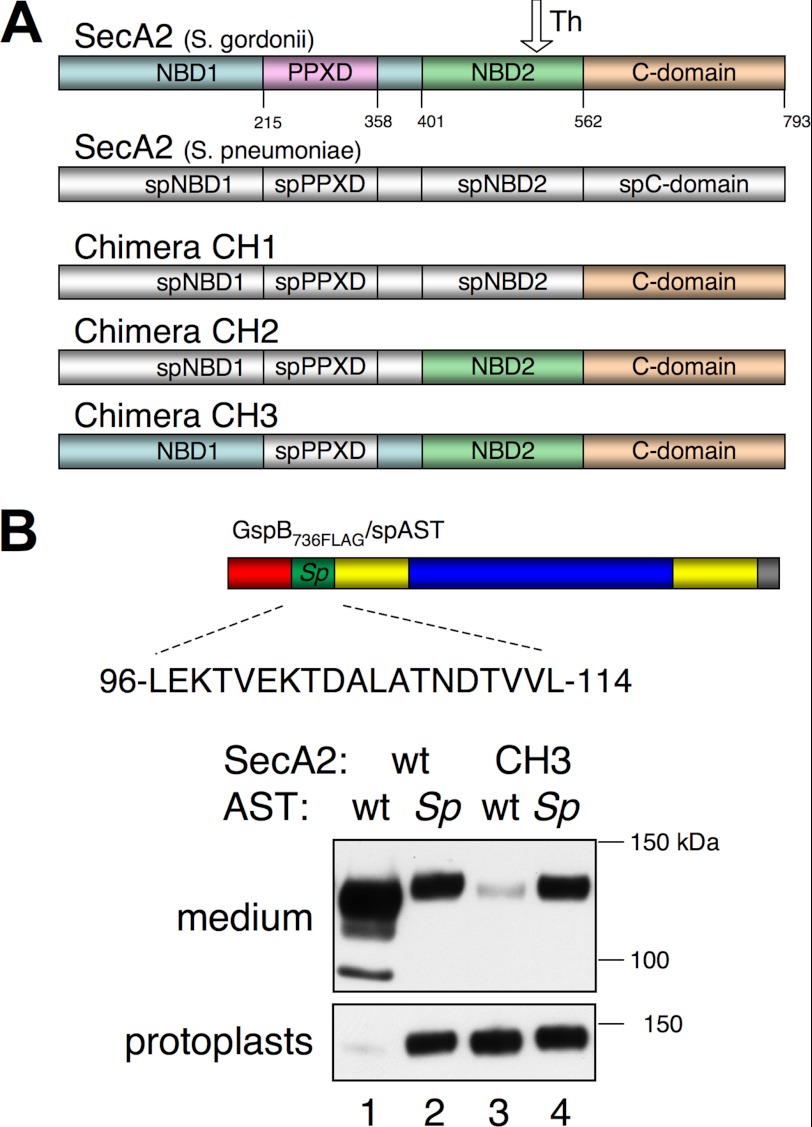
Interaction of the AST Domain with the PPXD Domain of SecA2
To identify the region of SecA2 that cross-linked to the AST domain at position 94 or 114, the 150 kDa complexes were treated with thrombin, which cleaves SecA2 at residue 490 (Fig. 8A) but does not cleave BdD. Western blot analysis of the resulting proteolytic fragments indicated that BdD was linked in both cases to the N-terminal two-thirds of SecA2 (data not shown). This region includes a domain similar to the preprotein binding domain (PPXD) of SecA, which has been implicated in the binding of both the signal peptide and mature region of preproteins (38–40). We reasoned that, if a highly specific interaction between the AST region and SecA2 was necessary for transport, it might be possible to detect such an export requirement in S. gordonii by simultaneously substituting the native AST domain of GspB and the AST-binding domain of SecA2 with a heterologous pair of regions.
FIGURE 8.
Transport of a GspB736FLAG chimera by SecA2 chimeras in S. gordonii. A, diagram of SecA2 and the SecA2 chimeras. The putative functional domains are demarcated as described previously (19). NBD, nucleotide binding domain; PPXD, preprotein binding domain. A thrombin cleavage site in the S. gordonii SecA2 is indicated (Th). B, diagram of the GspB736FLAG/spAST preprotein chimera, and secretion of the GspB736FLAG and GspB736FLAG/spAST from S. gordonii. GspB736FLAG and GspB736FLAG/spAST were detected by Western blotting with anti-FLAG antibodies. Lane 1, PS1226 with pM99secA2/asc; lane 2, PS2812 with pM99secA2/asc; lane 3, PS1226 with pM99secA2CH3; lane 4, PS2812 with pM99secA2CH3.
When selecting candidate homologues for domain exchange, we noticed that the sequences of SecA2, as well as the putative AST domains of the SRR proteins, diverge fairly rapidly across species. For example, the SecA2 proteins from Streptococcus sanguinis SK36, Streptococcus pneumoniae TIGR4, and Streptococcus agalactiae COH1 show 91, 79, and 70% similarity (81, 61, and 49% identity), respectively, to that of S. gordonii M99. Although the S. sanguinis SecA2 can partially complement a ΔsecA2 mutant of S. gordonii, the S. pneumoniae SecA2 is unable to transport GspB (data not shown).
The putative AST domains vary comparably. A chimeric form of GspB736FLAG in which the AST domain was replaced with the corresponding region of the S. sanguinis homologue SrpA (GspB736FLAG/ssAST) was readily transported by S. gordonii (data not shown), whereas a preprotein chimera containing the putative AST domain of the pneumococcal homologue PsrP (GspB736FLAG/spAST) showed reduced transport (Fig. 8B, lane 2 versus lane 1). For the compensatory exchange experiments, we therefore chose to incorporate regions from the pneumococcal SecA2 into the S. gordonii SecA2, and then examined the ability of these SecA2 chimeras to transport either GspB736FLAG or GspB736FLAG/spAST. Two of the SecA2 chimeras (CH1 and CH2, Fig. 8A) were unable to transport either wild-type GspB736FLAG or the chimeric preprotein (data not shown). However, a third SecA2 chimera (CH3) that contains only the pneumococcal PPXD showed markedly reduced transport of the wild-type preprotein (lane 3 versus lane 1), but was better able to transport GspB736FLAG/spAST (lane 4 versus lane 3). These data support the possibility that the AST domain interacts with the PPXD of SecA2 to facilitate transport. However, since transport of the preprotein chimera by the SecA2 chimera was reduced, as compared with transport of the wild-type preprotein by wild-type SecA2 (lane 4 versus lane 1), the results also suggest that the heterologous PPXD may be defective for interactions with the preprotein signal peptide or with a component of the aSec system.
DISCUSSION
Protein transport by the accessory Sec system of S. gordonii is distinctly different from transport via the general Sec system in that it requires a specific segment in the mature region of the preprotein, adjacent to the signal peptide, known as the AST domain. Our previous genetic analyses indicated that the AST domain of the streptococcal glycoprotein GspB might affect both targeting of the preprotein to the SecA2/Y2 translocon and gating of the SecY2 transmembrane channel. The studies presented here were undertaken to determine which aSec components might interact with the AST domain to facilitate these processes. With the preprotein labeled at position 94, 95, or 114 of the AST domain, cross-linking to SecA2 was detected. No cross-linking to SecA2 occurred when the preprotein was labeled at position 115, which is consistent with genetic data suggesting that position 114 is the C-terminal end of the AST domain.
In addition to SecA2, cross-linking to two E. coli proteins was detected. Cross-linking to these proteins only occurs if the labeled preprotein is transported (Figs. 4B and 5C), but the interactions are not aSec-specific. Cell fractionation experiments have indicated that these complexes are membrane-associated (data not shown), but sufficient quantities for identification of the constituent proteins have not yet been obtained. However, the combined results are consistent with the possibility that cross-linking to the two proteins occurs after the AST domain reaches the periplasm, and that the apparent masses of these proteins are therefore 15 and 30 kDa.
A notable outcome of these experiments is that no interaction with SecA was detected, even in cases where the labeled preprotein was transported by the general Sec system (i.e. in the absence of the aSec components, and when the preprotein carried the wild-type signal peptide H region). This is consistent with the fact that the AST domain is not required for, and does not interfere with, general Sec transport. This also implies that a more intimate or prolonged interaction between the AST domain and the translocon motor protein occurs during, or prior to, transport by the aSec versus Sec pathway, and that cross-linking between the AST domain and SecA2 does not merely reflect incidental contact.
One surprising result is that the interaction of SecA2 with the preprotein does not require Asp1–3, but is instead modulated by one or more of the Asps. That is, the interaction of SecA2 with the N terminus of the AST domain (position 94) did not require Asp1–3, whereas interaction with the C terminus (position 114) did. These results indicate that full engagement of the AST domain by SecA2 is dependent on one or more of the Asps, and help to explain why the Asps are essential components of the aSec system. The finding that SecA2 contacts position 94 (and presumably the adjacent signal peptide), but not position 114, in the absence of Asp1–3 suggests two possible roles for the Asps. Rather than targeting the preprotein to SecA2, the Asps might maintain the preprotein in a conformation in which the entire AST domain is accessible to SecA2, or the Asps may directly alter SecA2 such that the AST binding site is more exposed (Fig. 9). This latter possibility would be analogous to a proposed role for SecB in post-translational transport. In E. coli, the C-terminal SecB binding domain of SecA is thought to occupy or perhaps occlude a signal peptide binding site that resides within the preprotein binding domain of SecA (39). This “auto-inhibition” can be relieved by SecB interaction with the C-terminal domain as it delivers a preprotein to SecA. The Asps might similarly alter SecA2 conformation to unmask the AST binding site.
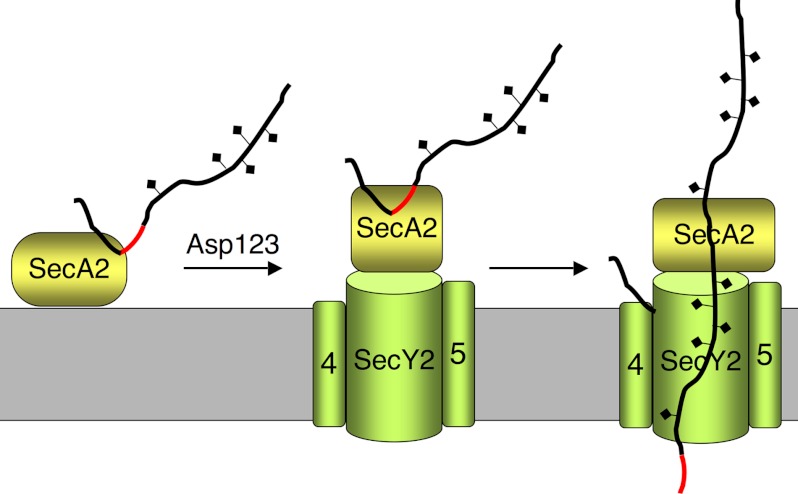
FIGURE 9.
Model for Asp-dependent engagement of the AST domain and transport of GspB. The AST domain of GspB is indicated in red. Preprotein binding to SecA2 is likely mediated by the signal peptide, and can occur in the absence of Asp1–3, although the Asps could maintain the preprotein in an export-competent state. One or more of the Asps directly or indirectly alter the conformation of SecA2 to facilitate binding of the entire AST domain. This leads to insertion of the preprotein into the SecY2 channel.
Although it was suspected that the AST domain might interact with SecY2 to facilitate gating (i.e. opening) of the transmembrane channel, cross-linking was detected only to SecA2 but no other aSec protein. This suggests that the previously-described “gating” and “targeting” functions of the AST domain may actually reflect two different interactions between the preprotein and SecA2. The two purported functions were based on the finding that some AST domain mutations could be compensated for by using a pre-gated, or partially opened, form of SecY2 (SecY2I382N), whereas other mutations could not (20). The presumed gating mutations were generally alterations in the N-terminal end of the AST domain, whereas the presumed targeting mutations were generally located near the C-terminal end of the AST domain. Since cross-linking of SecA2 to position 94 of the AST domain occurs independently of aSec transport, whereas cross-linking of SecA2 to position 114 is correlated with aSec transport (Figs. 6 and 7), it is possible that the full engagement of the AST domain by SecA2 is analogous to the “trapping” step of general Sec transport initiation described by Gouridis et al. (41). The trapping event is a prolonged, high affinity interaction between mature regions of preproteins and SecYEG-bound SecA, and corresponds to an early translocation intermediate. In the general Sec system, this role of the mature domain is not sequence-specific, whereas aSec transport is strictly dependent on the AST domain.
Results of domain-swapping experiments in S. gordonii support the possibility that the AST domain interacts specifically with the PPXD domain of SecA2 to mediate transport. However, replacement of the S. gordonii SecA2 PPXD with the pneumococcal SecA2 PPXD did not fully restore transport of a chimeric preprotein that contained the putative pneumococcal AST domain. The most likely explanation for this is that, in addition to being precisely matched to the AST domain, the PPXD must be matched to the signal peptide. Alternatively, the SecA2 chimera CH3 might not optimally interact with another component of the aSec system such as SecY2. Indeed, the PPXD of SecA makes several contacts with SecY during transport (28, 30, 42). An additional possibility is that the heterologous AST domain does not interact properly with the GspB signal peptide. We are currently undertaking experiments to examine these possibilities directly.
Acknowledgments
We thank Peter Schultz and The Scripps Research Institute for the generous gift of pEVOL-pBpF, Jon Beckwith for providing anti-SecB antiserum, and Gary Cecchini for comments on the manuscript.
This work was supported, in whole or in part, by the National Institutes of Health Grant RO1AI41513 (to P. S.), the Dept. of Veterans Affairs, and the VA Merit Review program, the Northern California Institute for Research and Education, and by a Postdoctoral Fellowship Award from the American Heart Association (to Y. Y.).
P. M. Sullam, unpublished data.
- SRR
- serine-rich repeat
- aSec
- accessory Sec
- AST
- accessory Sec transport
- BzF
- benzoylphenylalanine
- MBP
- maltose-binding protein.
REFERENCES
- 1. Froeliger E. H., Fives-Taylor P. (2001) Streptococcus parasanguis fimbria-associated adhesin Fap1 is required for biofilm formation. Infect. Immun. 69, 2512–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Handley P. S., Correia F. F., Russell K., Rosan B., DiRienzo J. M. (2005) Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral. Microbiol. Immunol. 20, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mistou M. Y., Dramsi S., Brega S., Poyart C., Trieu-Cuot P. (2009) Molecular dissection of the secA2 locus of group B streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J. Bacteriol 191, 4195–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Obert C., Sublett J., Kaushal D., Hinojosa E., Barton T., Tuomanen E. I., Orihuela C. J. (2006) Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74, 4766–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rose L., Shivshankar P., Hinojosa E., Rodriguez A., Sanchez C. J., Orihuela C. J. (2008) Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J. Infect. Dis. 198, 375–383 [DOI] [PubMed] [Google Scholar]
- 6. Seifert K. N., Adderson E. E., Whiting A. A., Bohnsack J. F., Crowley P. J., Brady L. J. (2006) A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 7. Siboo I. R., Chambers H. F., Sullam P. M. (2005) Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 73, 2273–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takahashi Y., Takashima E., Shimazu K., Yagishita H., Aoba T., Konishi K. (2006) Contribution of sialic acid-binding adhesin to pathogenesis of experimental endocarditis caused by Streptococcus gordonii DL1. Infect. Immun. 74, 740–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Sorge N. M., Quach D., Gurney M. A., Sullam P. M., Nizet V., Doran K. S. (2009) The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J. Infect. Dis. 199, 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiong Y. Q., Bensing B. A., Bayer A. S., Chambers H. F., Sullam P. M. (2008) Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb. Pathog. 45, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takamatsu D., Bensing B. A., Cheng H., Jarvis G. A., Siboo I. R., López J. A., Griffiss J. M., Sullam P. M. (2005) Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibα. Mol. Microbiol. 58, 380–392 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi Y., Sandberg A. L., Ruhl S., Muller J., Cisar J. O. (1997) A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to α2–3-linked sialic acid-containing receptors. Infect. Immun. 65, 5042–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shivshankar P., Sanchez C., Rose L. F., Orihuela C. J. (2009) The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol. Microbiol. 73, 663–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Samen U., Eikmanns B. J., Reinscheid D. J., Borges F. (2007) The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect. Immun. 75, 5405–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Plummer C., Wu H., Kerrigan S. W., Meade G., Cox D., Ian Douglas C. W. (2005) A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br. J. Haematol. 129, 101–109 [DOI] [PubMed] [Google Scholar]
- 16. Siboo I. R., Chaffin D. O., Rubens C. E., Sullam P. M. (2008) Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol 190, 6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bensing B. A., Sullam P. M. (2002) An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44, 1081–1094 [DOI] [PubMed] [Google Scholar]
- 18. Chen Q., Wu H., Fives-Taylor P. M. (2004) Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol. Microbiol. 53, 843–856 [DOI] [PubMed] [Google Scholar]
- 19. Bensing B. A., Sullam P. M. (2009) Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J. Bacteriol. 191, 3482–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bensing B. A., Sullam P. M. (2010) Transport of preproteins by the accessory Sec system requires a specific domain adjacent to the signal peptide. J. Bacteriol. 192, 4223–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takamatsu D., Bensing B. A., Sullam P. M. (2005) Two additional components of the accessory Sec system mediating export of the Streptococcus gordonii platelet-binding protein GspB. J. Bacteriol. 187, 3878–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takamatsu D., Bensing B. A., Sullam P. M. (2004) Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 52, 189–203 [DOI] [PubMed] [Google Scholar]
- 23. Seepersaud R., Bensing B. A., Yen Y. T., Sullam P. M. (2010) Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii. Mol. Microbiol. 78, 490–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou M., Zhang H., Zhu F., Wu H. (2011) Canonical SecA associates with an accessory secretory protein complex involved in biogenesis of a streptococcal serine-rich repeat glycoprotein. J. Bacteriol. 193, 6560–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yen Y. T., Seepersaud R., Bensing B. A., Sullam P. M. (2011) Asp2 and Asp3 interact directly with GspB, the export substrate of the Streptococcus gordonii accessory Sec System. J. Bacteriol. 193, 3165–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bensing B. A., Takamatsu D., Sullam P. M. (2005) Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol. Microbiol. 58, 1468–1481 [DOI] [PubMed] [Google Scholar]
- 27. Bensing B. A., Siboo I. R., Sullam P. M. (2007) Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J. Bacteriol 189, 3846–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mori H., Ito K. (2006) Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc. Natl. Acad. Sci. U.S.A. 103, 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ieva R., Bernstein H. D. (2009) Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Das S., Oliver D. B. (2011) Mapping of the SecA, SecY and SecA. SecG interfaces by site-directed in vivo photocross-linking. J. Biol. Chem. 286, 12371–12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Young T. S., Schultz P. G. (2010) Beyond the canonical 20 amino acids: expanding the genetic lexicon. J. Biol. Chem. 285, 11039–11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu C. C., Schultz P. G. (2010) Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 [DOI] [PubMed] [Google Scholar]
- 33. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1997) Current Protocols in Molecular Biology, John Wiley & Sons, Inc [Google Scholar]
- 34. Takamatsu D., Bensing B. A., Sullam P. M. (2004) Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J. Bacteriol. 186, 7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macrina F. L., Evans R. P., Tobian J. A., Hartley D. L., Clewell D. B., Jones K. R. (1983) Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25, 145–150 [DOI] [PubMed] [Google Scholar]
- 36. Pyburn T. M., Bensing B. A., Xiong Y. Q., Melancon B. J., Tomasiak T. M., Ward N. J., Yankovskaya V., Oliver K. M., Cecchini G., Sulikowski G. A., Tyska M. J., Sullam P. M., Iverson T. M. (2011) A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog. 7, e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bryan E. M., Bae T., Kleerebezem M., Dunny G. M. (2000) Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44, 183–190 [DOI] [PubMed] [Google Scholar]
- 38. Auclair S. M., Moses J. P., Musial-Siwek M., Kendall D. A., Oliver D. B., Mukerji I. (2010) Mapping of the signal peptide-binding domain of Escherichia coli SecA using Forster resonance energy transfer. Biochemistry 49, 782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gelis I., Bonvin A. M., Keramisanou D., Koukaki M., Gouridis G., Karamanou S., Economou A., Kalodimos C. G. (2007) Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131, 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zimmer J., Rapoport T. A. (2009) Conformational flexibility and peptide interaction of the translocation ATPase SecA. J. Mol. Biol. 394, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gouridis G., Karamanou S., Gelis I., Kalodimos C. G., Economou A. (2009) Signal peptides are allosteric activators of the protein translocase. Nature 462, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zimmer J., Nam Y., Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]