Background: Methionine sulfoxide reductases have previously been studied mostly using low molecular weight substrates.
Results: Methionine sulfoxide reductases preferentially reduce unfolded oxidized proteins.
Conclusion: These enzymes serve a critical function in protein folding by repairing oxidized nascent polypeptides and unfolded proteins.
Significance: Understanding precise functions of methionine sulfoxide reductases will help define mechanisms of protein repair and identify their physiological substrates.
Keywords: Enzyme Catalysis, Oxidative Stress, Protein Folding, Stress, Yeast, Methionine Sulfoxide, Methionine Sulfoxide Reductase, Protein Unfolding, Oxidation
Abstract
Reduction of methionine sulfoxide (MetO) residues in proteins is catalyzed by methionine sulfoxide reductases A (MSRA) and B (MSRB), which act in a stereospecific manner. Catalytic properties of these enzymes were previously established mostly using low molecular weight MetO-containing compounds, whereas little is known about the catalysis of MetO reduction in proteins, the physiological substrates of MSRA and MSRB. In this work we exploited an NADPH-dependent thioredoxin system and determined the kinetic parameters of yeast MSRA and MSRB using three different MetO-containing proteins. Both enzymes showed Michaelis-Menten kinetics with the Km lower for protein than for small MetO-containing substrates. MSRA reduced both oxidized proteins and low molecular weight MetO-containing compounds with similar catalytic efficiencies, whereas MSRB was specialized for the reduction of MetO in proteins. Using oxidized glutathione S-transferase as a model substrate, we showed that both MSR types were more efficient in reducing MetO in unfolded than in folded proteins and that their activities increased with the unfolding state. Biochemical quantification and identification of MetO reduced in the substrates by mass spectrometry revealed that the increased activity was due to better access to oxidized MetO in unfolded proteins; it also showed that MSRA was intrinsically more active with unfolded proteins regardless of MetO availability. Moreover, MSRs most efficiently protected cells from oxidative stress that was accompanied by protein unfolding. Overall, this study indicates that MSRs serve a critical function in the folding process by repairing oxidatively damaged nascent polypeptides and unfolded proteins.
Introduction
Proteins may undergo various post-translational modifications altering their structure and function. Their sulfur-containing residue, methionine (Met), can be oxidized to R- and S-diastereoisomers of Met sulfoxide (MetO).2 This modification is reversible, as MetO can be reduced back to Met by methionine sulfoxide reductases (MSRs) A (MSRA) and B (MSRB), which are specific for the S- and R-forms of MetO, respectively. Theses enzymes are present in almost all living organisms and catalyze the reduction of their substrates at the expense of NADPH using thioredoxin (Trx) or glutaredoxin systems (1, 2).
Whereas the catalytic mechanisms of MSRs are well characterized (2, 3), their physiological functions remain elusive, mainly due to insufficient information on the identity of their cellular targets. The absence of clearly established substrates also limits studies on specificity of Met oxidation and MetO reduction in proteins. Met oxidation has been reported for proteins that could be classified into three groups (4): (i) enzymes activated by Met oxidation, such as the calcium/calmodulin-dependent protein kinase II (5), (ii) proteins not impaired by Met oxidation, which could fulfill together with MSRs a protective (antioxidant) function though cyclic oxidation and reduction of Met (6), and (iii) proteins damaged by Met oxidation, such as those involved in neurodegenerative diseases (7, 8). The consequence of Met oxidation is determined by the effects of this modification on structure and function of these proteins; however, these effects have been characterized only for a handful of proteins. For example, in human prion, oxidation of two Met residues converts a cellular α-helix-rich form to the infectious β-sheet-rich form by perturbing the network of stabilizing interactions (9). In addition, oxidation of two solvent-accessible Met residues in a human growth hormone increases its susceptibility to thermal denaturation (10), and oxidation of two Met residues in calmodulin prevents protein-protein interaction due to incompatibility of MetO for stable α-helixes (11).
MSRs, which repair oxidative modifications, play important roles in the protection of proteins from oxidative stress in various eukaryotes (12–14). Similar protective effects were also observed in prokaryotes, particularly in the case of the stress induced by hypochlorite, a strong antimicrobial agent found in household bleach that is also produced by mammalian neutrophils to kill invading microorganisms (15). For instance, a Helicobacter pylori strain deficient in the expression of MSRs could not survive in a neutrophil cell culture (16). Hypochlorite is known to have a dual effect in provoking oxidation and unfolding of proteins and trigger chaperone activation (17). These studies imply that MSR might directly participate in the protection from oxidative and unfolding stress through the reduction of MetO in proteins.
The MSR activity was quantified for several protein substrates, such as bacterial Ffh and several targets of plant MSRBs (13, 18, 19). However, little is known about enzyme kinetics of MSRs with oxidized protein substrates, their physiological targets. The yeast Saccharomyces cerevisiae possesses single MSRA and MSRB genes as well as a free Met-R-O reductase (fRMSR) that is specific for the reduction of the R-diastereoisomer of free MetO (20, 21). In this work we took advantage of the NADPH-coupled Trx system and used different forms of MetO-containing proteins as substrates to examine functions of yeast MSRA and MSRB.
EXPERIMENTAL PROCEDURES
Cloning and Site-directed Mutagenesis
Sequences coding for NADPH-dependent thioredoxin reductase 1, Trx1, and MSRB (from codon 30 to the stop codon), were amplified by PCR from the S. cerevisiae genomic DNA using Platinum® Pfx DNA Polymerase (Invitrogen) and specific pairs of primers shown in supplemental Table S1. Similarly, sequences coding for Met-rich protein 4 (MRP4) (from codon 23 to stop codon) and Met-rich protein 5 (MRP5) (from codon 21 to stop codon) were amplified from genomic DNA of Idiomarina loihiensis L2TR and Pseudomonas putida W619, respectively, using specific primers.3 Amplicons were purified and digested with NdeI and BamHI for cloning of thioredoxin reductase 1 and Trx1 in pET15b or with BamHI and XhoI or with NheI and NotI for cloning MSRB and MRP4 in pET21b (EMD Biosciences, Billerica, MA), respectively. Met-rich protein 1 (MRP1) (22) and MRP5 were cloned similarly in p425 yeast expression vector under glycerol-3-phosphate dehydrogenase (GPD) promoter using BamHI and SalI restriction sites.3 Site-directed mutagenesis of MSRA and MSRB was made by whole plasmid amplification with Phusion® High-Fidelity DNA Polymerase (Thermo Scientific, Billerica, MA) using primers containing mutated bases (supplemental Table S1). After amplification, the methylated template vector was digested by incubation with DpnI for 1 h at 37 °C. 5 μl of the digested PCR product was used to transform NEB 5-α competent Escherichia coli (High Efficiency) cells (New England Biolabs, Ipswich, MA), and clones were selected on Luria-Bertani plates containing ampicillin (50 μg·ml−1). All constructs were validated by DNA sequencing. The expression vectors pET28a-MSRA (20) and pGEX4T1 (GE Healthcare) were used to produce yeast MSRA and glutathione S-transferase (GST) from Schistosoma japonicum, respectively.
Expression and Purification of Recombinant Proteins
SoluBL21TM E. coli (Gelantis, San Diego, CA) cells were transformed with the expression vector and grown in Luria-Bertani containing ampicillin or kanamycin (50 μg·ml−1) at 37 °C. When the A600 reached ∼0.6, production of the recombinant protein was induced by the addition of 100 μm isopropyl β-d-1-thiogalactopyranoside. After overnight incubation at 25 °C, cells were harvested by centrifugation. For thioredoxin reductase 1, Trx1, MSRA, MSRB, and MRP4 containing His6 tag, pellets were resuspended in PBS containing 25 mm imidazole in the presence of Complete, EDTA-free, protease inhibitor mixture (Roche Applied Science), and for GST, the pellet was resuspended in PBS. Cells were disrupted by sonication, His6-tagged proteins were purified on nickel-containing His·Bind® Resin (Novagen, Billerica, MA), and GST was purified on glutathione-Sepharose 4 Fast Flow (GE Healthcare). Protein solutions were concentrated using 15-ml Amicon® Ultra concentrators with 30- or 10-kDa cutoffs (Millipore, Billerica, MA) and desalted in 30 mm Tris-HCl, pH 8, using 5-ml HiTrapTM Desalting columns (GE Healthcare). Protein concentrations were determined spectrophotometrically using the Pierce® BCA Protein assay kit (Thermo Scientific) and specific molar extinction coefficients at 280 nm. Protein purity was verified using SDS-PAGE gels stained with ImperialTM Protein Stain (Thermo Scientific).
Protein and N-Acetyl-MetO Preparation
MRP4, GST, bovine β-casein (Sigma), and Bacillus sp. α-amylase (Sigma) (all 1 mg·ml−1) were oxidized by incubation with 100 mm H2O2 in PBS overnight at room temperature, concentrated, and then desalted using HiTrapTM Desalting or Illustra NAP5TM columns (GE Healthcare) in 30 mm Tris-HCl, pH 8. For unfolding assays, oxidized proteins were incubated with urea at a final concentration ranging from 0.25 to 7 m for 1 h at room temperature before the assays. N-Acetyl-MetO was prepared as described (23).
MSR Absolute Stoichiometry and Activity with Dabsyl-MetO
Activities of recombinant MSRs (1 μm) were determined by monitoring the reduction of 0.5 mm dabsyl-MetO in the presence of 20 mm DTT. The absolute stoichiometry was determined similarly; 100 μm reduced and desalted MSRs was incubated with 1 mm dabsyl-MetO for 1 h at room temperature without the reducing agent. Dabsyl-Met and dabsyl-MetO were separated by HPLC using a C18 reverse phase column, SunFireTM 3.5 μm, 3.0 × 50 mm (Waters, Milford, MA) as described (2).
MSR Activity Assays
MSR activity was measured after NADPH oxidation at 340 nm in the presence of the Trx system (200–400 μm NADPH, 2 μm thioredoxin reductase 1, 25 μm Trx1) using 1–10 μm MSRA or MSRB in the presence of free MetO (0.25–20 mm), N-acetyl-MetO (62.5 μm to 5 mm), oxidized MRP4 (0.8–200 μm), oxidized β-casein (6.25–200 μm), oxidized GST (9.4–150 μm) or urea-treated oxidized GST (4.8–150 μm) as substrates. Reactions were carried out at 25 °C in a 500-μl reaction volume. MSR activities were calculated from the slope after subtracting the background (absence of the enzyme) considering that 1 mol of oxidized NADPH corresponds to 1 mol of MetO reduced. The apparent stoichiometry was determined similarly using subsaturating concentrations of substrates: 0.8–6.5 μm oxidized MRP4, 7–29 μm oxidized β-casein, 11–44 μm oxidized GST, and 5.6–22 μm 4 m urea-treated oxidized GST. The amount of oxidized NADPH was determined after the rate of oxidation reached the basal level. To test the activity using the urea-treated oxidized protein, 10–25 μm substrate was used, and urea was added in control assays at the same concentration (less than 50 mm). Kinetic and catalytic parameters were calculated from nonlinear regressions using GraphPad Prism 4.0 (GraphPad Software, Inc, La Jolla, CA). In the presence of urea-treated oxidized GST, the curves fit sigmoidal regressions described by Equation 1, with the Hill coefficient (h) > 1.
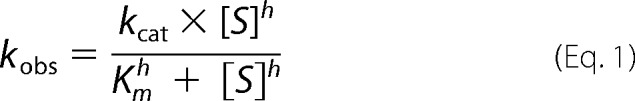 |
Fluorescence Analyses
Emission spectra of intrinsic fluorescence were recorded in 200 μl of 30 mm Tris-HCl, pH 8.0, containing 10 or 25 μm non-oxidized, oxidized, or urea-treated oxidized proteins with excitation at 280 nm. As a control, amounts of urea used in the urea-treated proteins were added to the oxidized protein samples (less than 50 mm). The same samples were used to determine 8-anilinonaphthalene-1-sulfonate (ANS) fluorescence. Emission at 466 nm (excitation at 377 nm) was recorded 15 min after the addition of 50 μm ANS. Fluorescence was recorded in 96-well microplates using a SpectraMax M5 fluorescence microplate reader (Molecular Devices, Sunnyvale, CA).
Mass Spectrometry Analysis
Oxidized MRP4, oxidized GST, and urea-treated oxidized GST (100 μm) were incubated with or without 5 μm MSRA or MSRB in the presence of 10 mm DTT in 30 mm Tris-HCl, pH 8, for 1 h at 25 °C. A solution containing 50 μg of substrate was incubated with 12.5 mm iodoacetamide for 30 min at 25 °C in 0.1 m ammonium sulfate then with 1 μg of sequencing grade modified trypsin (Promega, Madison, WI) and 1 mm CaCl2 overnight at 37 °C. An additional 1 μg of trypsin was added, and the solution was incubated for 2 h at 37 °C. The trypsin digests were frozen and stored at −80 °C until mass spectrometry analysis. LC-MS/MS analysis was performed on an LTQ-Orbitrap Discovery mass spectrometer (Thermo Fisher) coupled to an Agilent 1200 series HPLC system. Tryptic digest (30 μl) was pressure-loaded onto a 250-μm fused silica desalting column packed with 4 cm of Aqua C18 reverse phase resin (Phenomenex, Torrance, CA). The peptides were then eluted onto a C18 column (100-μm fused silica with a 5-μm tip, packed with 10 cm C18) using a gradient 5–100% buffer B in buffer A (buffer A: 95% water, 5% acetonitrile, 0.1% formic acid; buffer B: 20% water, 80% acetonitrile, 0.1% formic acid) and into the mass spectrometer. The flow rate through the column was set to ∼0.25 μl·min−1, and the spray voltage was set to 2.75 kV. One full MS scan (Fourier Transform Mass Spectrometry) (400–1800 Mr) was followed by 7 data-dependent scans (Ion Trap Mobility Spectrometry) of the nth-most intense ions with dynamic exclusion enabled.
Peptide Identification
The tandem MS data were searched using the SEQUEST algorithm (24) using a concatenated target/decoy variant of the human and mouse International Protein Index databases modified to include the sequences for the proteins used in this study. A static modification of +57.02146 on cysteine was specified to account for iodoacetamide alkylation, and a differential modification of +16 was specified on methionine to account for oxidation. SEQUEST output files were filtered using DTASelect 2.0 (25). Reported peptides were required to be fully tryptic, and discriminant analyses were performed to achieve a peptide false-positive rate below 5%. The percentage of oxidation per Met was calculated using the redundancy of the peptide containing the specific Met, oxidized or not. The percentage of oxidation values corresponds to the number of times a MetO was found divided by the total number of times the peptide was found (coverage), multiplied by 100. This method allows quantifying the oxidation of each Met found in peptide-containing several Met, which is not possible using area integration.
Yeast Spotting Assays
MSRA expressed under the glycerol-3-phosphate dehydrogenase promoter from the high copy number yeast expression vector p425 and MSR null strains was described previously (21). WT yeast cells were transformed with MRP1, MRP5, and/or yeast MSRA constructs. After colony formation, single colonies were picked up from the plates and grown overnight in the media lacking histidine or leucine. The following day, these cells were retransformed with MSRA and/or empty vectors and incubated on media lacking histidine and leucine. This procedure was also applied for empty vector transformation, which was used as a control. In the spotting assay, the indicated strains were grown overnight in appropriate media and diluted to an A600 of 0.3. Cells were washed with the prewarmed PBS buffer and incubated in 1 ml of PBS and the indicated concentrations of sodium hypochlorite (NaOCl). Every 5 min, 5 μl of each culture was spotted on the selective media. Yeast MSR mutant strains lacking MSRA, MSRB, or both genes were also tested as indicated above for NaOCl sensitivity. All plates were incubated for 3 days at 30 °C and photographed. Halo assays were performed to assess viability of MSRA-, MRP1-, and MRP5-overexpressing yeast cells under conditions of H2O2 stress. Cells were prepared as indicated above, and the A600 was adjusted to 0.5. Cells were washed with the prewarmed PBS, and 1 ml of each culture was spread onto agar plates missing the appropriate amino acids for selection. Plates were dried for 1 h at room temperature, and filter paper discs were placed in the middle of each plate. 4 μl of 30% hydrogen peroxide were applied onto each paper disc, and the plates were incubated for 3 days at 30 °C and photographed. The diameter of cleared zones in each plate was measured with a ruler. This experiment was repeated three times.
Modeling, Structural Analysis of Substrate Proteins, and Determination of MSR Hydrophobicity
The percentage of amino acids included in disordered regions was calculated using SPINE-D (26) and Multilayered Fusion-based Disorder predictor (27) web servers. The structural coordinates of MSRA and GST were obtained from the PDB repository (PDB codes 3PIL and 1DUG, respectively). To model MSRB, we used a -fold recognition algorithm, FAS03, to generate alternative profile-profile alignments. MSRB were modeled with Modeler (46) using the alignments generated with FAS03 and 2K8D as template. Detailed atomic exposure calculations were calculated with Surface Racer 227 (28). Structural profiles were generated as previously described (29). Briefly, all residues lying with one or more of their atoms found within 8 Å from the sulfur atom of the catalytic Cys residues of yeast MSRA and MSRB were considered and then separately analyzed for their hydrophobic content (implementing the standard Kyte-Doolittle scale, where each amino acid is described by numeric value ranging from negative, i.e. hydrophilic, to positive, i.e. hydrophobic (supplemental Fig. S4)) (30) and compositional features (e.g. content of basic, acidic, or aromatic residues) through in-house Python (v2.6) scripts.
RESULTS
Yeast MSRA Efficiently Reduces Oxidized Proteins and Free MetO, Whereas MSRB Is Specialized for the Reduction of Oxidized Proteins
To characterize the kinetics of yeast MSRA and MSRB, we utilized the NADPH-coupled Trx system, which is physiologically relevant and allows comparative analyses of MetO-containing substrates. Catalytic parameters of MSRs are usually determined using low molecular MetO-containing compounds, whereas little is known on the catalysis of MetO reduction in oxidized proteins. We examined MSRA and MSRB kinetics using three different oxidized protein substrates and used free MetO and N-acetyl-MetO for comparison. These proteins were chosen based on their high Met content and structural features; both Met-rich protein 4 (MRP4) and β-casein are predicted to be completely disordered and possess 31 (22%) and 7 (3.1%) Met residues, respectively. A third protein, GST, has Met content similar to that of β-casein (9 Met, 3.8%) but is a highly structured protein (supplemental Table S2). For all substrates, MSRA and MSRB catalysis followed the Michaelis-Menten kinetics (Table 1). MSRA displayed the kcat values from ∼1 to ∼13 s−1. The Km values were ∼0.5 mm for free MetO, N-acetyl-MetO, and GST and ∼10-fold lower for MRP4 and β-casein. The catalytic efficiency (kcat/Km) was ∼2.1 mm−1·s−1 for GST, and this value increased ∼5-, ∼13-, ∼15-, and ∼200-fold in the case of free MetO, β-casein, N-acetyl-MetO, and MRP4, respectively. For MSRB, the kcat values were ∼1 s−1 for all tested substrates, but a striking difference was observed in Km values, which were ∼6.5 mm for free MetO and ∼10 times lower for N-acetyl-MetO. The differences were more pronounced for proteins, i.e. the Km values were 45–650-fold lower for oxidized proteins than for free MetO (Table 1). Analysis of these data reinforces the idea that MSRB is far more efficient in the reduction of MetO in oxidized proteins (e.g. its catalytic efficiency was ∼1200-fold higher for MRP4 than for free MetO), whereas MSRA reduces both proteins and MetO-containing compounds with similar efficiency.
TABLE 1.
Kinetic parameters of MSRA and MSRB in the reduction of oxidized proteins
Assays were carried out under steady-state conditions following NADPH oxidation at 340 nm. Apparent stoichiometry (mol of NADPH oxidized per mol of substrate) was determined after full reduction of the substrate using substoichiometric substrate concentrations as described under “Experimental Procedures.” Data are represented as the means ± S.D.
| MetO-containing substrate | MSRA |
MSRB |
||||||
|---|---|---|---|---|---|---|---|---|
| Apparent stoichiometry | kcat | Km | kcat/Km | Apparent stoichiometry | kcat | Km | kcat/Km | |
| mol of NADPH ox. mol sub−1 | s−1 | μm | m−1.s−1 (×103) | mol NADPH ox. mol sub−1 | s−1 | μm | m−1.s−1 (×103) | |
| Free MetO | ||||||||
| Measured | 7.71 ± 0.03 | 1,120 ± 16 | 6.9 | 0.59 ± 0.02 | 13,014 ± 752 | 0.05 | ||
| Correcteda | 560 ± 8 | 13.8 | 6,507 ± 376 | 0.09 | ||||
| N-Acetyl-MetO | ||||||||
| Measured | 13.23 ± 1.83 | 896 ± 282 | 15.0 | 0.80 ± 0.06 | 1,348 ± 224 | 0.6 | ||
| Correcteda | 448 ± 141 | 30.0 | 674 ± 112 | 1.2 | ||||
| MRP4 | ||||||||
| Measured | 4.16 ± 0.16 | 13.01 ± 5.4 | 33 ± 18 | 386.3 | 7.18 ± 0.66 | 1.04 ± 0.16 | 10 ± 3 | 106.7 |
| Correctedb | 137 ± 76 | 95.3 | 70 ± 20 | 14.9 | ||||
| β-Casein | ||||||||
| Measured | 2.49 ± 0.15 | 1.13 ± 0.11 | 45 ± 13 | 25.1 | 3.19 ± 0.34 | 0.78 ± 0.05 | 54 ± 9 | 14.4 |
| Correctedb | 113 ± 32 | 10.1 | 172 ± 28 | 4.5 | ||||
| GST | ||||||||
| Measured | 1.21 ± 0.02 | 0.73 ± 0.15 | 356 ± 105 | 2.1 | 0.78 ± 0.02 | 0.41 ± 0.08 | 142 ± 48 | 2.9 |
| Correctedb | 428 ± 127 | 1.7 | 111 ± 37 | 3.7 | ||||
| Urea-treated GSTc | ||||||||
| Measured | 3.25 ± 0.05 | 0.97 ± 0.19 | 78 ± 24 | 12.4 | 4.66 ± 0.24 | 0.52 ± 0.03 | 55 ± 11 | 9.5 |
| Correctedb | 254 ± 78 | 3.8 | 256 ± 51 | 2.0 | ||||
a Considering that only the S- and R-diastereoisomers serve as substrates for MSRA and MSRB, respectively, and assuming that the other diastereoisomer does not act as inhibitor, the Km values were divided by 2.
b For comparison, the Km values were multiplied by the apparent stoichiometry, allowing the removal of variation due to the different numbers of MetO reduced in each substrate.
c The regression curves obtained for MSRA and MSRB with the urea-treated oxidized GST fit a sigmoidal described by the equation 1 (“Experimental Procedures”) with the h values equal to 1.4 ± 0.3 and 1.5 ± 0.1 for MSRA and MSRB, respectively.
Quantification of MetO Reduction in Protein Substrates
The absolute stoichiometry displayed by each MSR was 1 mol of MetO reduced/mol of enzyme (supplemental Fig. S1A). Two redox-active Cys were used by each enzyme, and mutation analyses verified the roles of Cys-25 and Cys-176 in MSRA and Cys-157 and Cys-97 in MSRB as catalytic and resolving Cys, respectively (supplemental Fig. S1B). We further estimated the number of MetO reduced by MSRA and MSRB in oxidized proteins as the apparent stoichiometry i.e. mol of NADPH oxidized/mol of substrate using subsaturating concentrations of substrates. Fig. 1 shows the data for MRP4. After the addition of the substrate, NADPH consumption was followed until the rate reached the background level, representing the state when all MetO residues reducible by MSR were reduced (Fig. 1A). The amount of oxidized NADPH was then plotted as a function of substrate concentration (Fig. 1B). The slope of the calculated linear regression corresponded to the apparent stoichiometry. Using MSRA and increasing concentrations of MRP4, the apparent stoichiometry of ∼4 mol of NADPH oxidized/mol of substrate was found (Table 1; Fig. 1B). With MSRB, this value was ∼7 mol of NADPH oxidized/mol of substrate (Table 1; Fig. 1B). As changes in oxidized NADPH were directly proportional to those in MetO (18, 31), the data indicate that MSRA and MSRB reduced 4 and 7 MetO equivalents, respectively, in the oxidized MRP4. Because this protein has 31 Met, 13 and 23% Met equivalents were reduced, respectively. To compare catalytic parameters for different substrates, the data were normalized by multiplying the Km values by the apparent stoichiometry, yielding values per MetO reduced and thus allowing removal of variation due to the different numbers of MetO reduced in each substrate (Table 1). For MSRA, this normalization gave a kcat/Km ratio similar to that for N-acetyl-MetO and free MetO. For MSRB, the corrected catalytic efficiency was 15- and 150-fold higher than that for N-acetyl-MetO and free MetO, respectively (Table 1). Similar analyses with β-casein showed that 2.5 and 3.2 MetO equivalents, which correspond to 36 and 46% of the 7 Met in the protein, were reduced by MSRA and MSRB, respectively. The corrected catalytic efficiency was similar to that obtained for MSRA with free MetO and 50-fold higher in the case of MSRB (Table 1).
FIGURE 1.
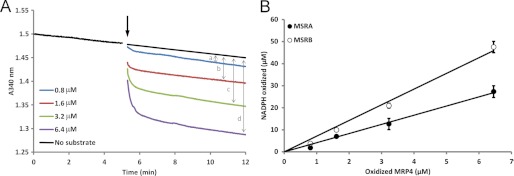
Apparent stoichiometry of MetO reduction in oxidized MRP4. A, NADPH consumption was followed with 0.8–6.4 μm oxidized MRP4 as the substrate. The initial reaction mixture contained 250 μm NADPH, 2 μm thioredoxin reductase 1, 25 μm Trx1, and 10 μm MSRA. After equilibration, the substrate was added (arrow), and NADPH consumption was followed until it reached the diaphorase level. The final absorbance was subtracted taking into account the diaphorase activity for each concentration of the substrate (two-sided arrows a, b, c, and d). B, oxidized NADPH was plotted as a function of substrate concentration for MSRA (●) and MSRB (○).
The analysis of apparent stoichiometries further indicated that 1.2 and 0.8 MetO equivalents were reduced per molecule of oxidized GST, corresponding to 13 and 9% of all Met in the case of MSRA and MSRB, respectively (Table 1). In the MSRA-catalyzed reaction, the corrected catalytic efficiency observed with GST as the substrate was 5–50-fold lower than with other tested substrates. In the MSRB-dependent reduction of oxidized GST, this correction gave a catalytic efficiency 45-fold higher than that obtained with free MetO but similar to those determined for the two other proteins. It is noteworthy that the oxidized GST was a better substrate for MSRB; its catalytic efficiency was 2-fold higher than the MSRA value. Altogether, these results indicated that MSRA efficiently reduced both free and protein-bound MetO, whereas MSRB showed a dramatic preference for oxidized proteins compared with low molecular weight MetO-containing molecules, such as free MetO and N-acetyl-MetO.
Differential Reduction of MetO Residues by MSRA and MSRB
Quantification of MetO reduced by MSRs in the tested proteins suggested that the various oxidized Met were not equivalent substrates. We subjected MRP4 and GST to tryptic digestion and LC-MS/MS analyses to determine the oxidation state of each Met before and after reduction by MSRs (Table 2; supplemental Table S3). The coverage of oxidized MRP4 was 64%, allowing us to determine precisely the oxidation state of 11 Met among the 31 Met present in the protein (supplemental Table S3). All detected Met were highly oxidized (more than 90% of Met were in the form of MetO), with the exception of Met-66 (48% oxidized). After the reduction by MSRA or MSRB, protein coverage was 89% in both cases, allowing us to estimate the oxidation status of 27 Met, which corresponded to 55 and 64% for the substrate reduced by MSRA and MSRB, respectively. Considering only the 11 Met, for which the oxidation state was determined in the oxidized MRP4 before MSR reduction, MSRA and MSRB reduced 28 and 21% of MetO, respectively. However, not all MetO were reduced with the same efficiency, and their reduction also depended on the MSR used. For instance, oxidized Met-31 was reduced only by MSRA and oxidized Met-66 only by MSRB. In addition, the last Met, Met-125, was completely oxidized but reduced by neither MSR (supplemental Table S3).
TABLE 2.
Surface accessibility of Met residues in GST and their differential oxidation as revealed by mass spectrometry analyses
Surface accessibility of each Met to the solvent in Å2 was determined using protein structure, and the percentage of oxidation of individual Met in the substrate was determined by mass spectrometry as described under “Experimental Procedures.” Met position, position of the Met in the primary sequence. Oxidized, the GST was oxidized with 100 mm H2O2 before tryptic digestion. Oxidized + MSRA, the oxidized GST was reduced by MSRA before tryptic digestion. Oxidized + MSRB, the oxidized GST was reduced by MSRB before tryptic digestion.
| Met position | Secondary structure | Surface accessibility |
Oxidized |
Oxidized + MSRA |
Oxidized + MSRB |
||
|---|---|---|---|---|---|---|---|
| Å2 | % Oxidation (coveragea) | % Oxidation (coveragea) | % Reductionb | % Oxidation (coveragea) | % Reductionb | ||
| Oxidized GST | |||||||
| 1 | coil | NDd | 93 (25) | 57 (65) | 39 | 79 (29) | 15 |
| 69 | α-helix | 8.025 | 67 (47) | 57 (110) | 14 | 66 (58) | 2 |
| 81 | coil | 2.071 | 97 (12) | 100 (22) | 0c | 31 (29) | 68 |
| 94 | α-helix | 0 | 53 (348) | 52 (383) | 1 | 50 (326) | 5 |
| 129 | α-helix | 0 | 80 (4) | 0 (11) | 100 | 50 (4) | 38 |
| 132 | α-helix | 9.977 | 100 (1) | NDd (0) | NDd | NDd (0) | NDd |
| 154 | coil | 0 | 16 (50) | 19 (79) | 0c | 6 (52) | 64 |
| 165 | coil | 0 | 26 (65) | 23 (96) | 10 | 26 (70) | 0 |
| 168 | coil | 37.720 | 99 (67) | 99 (100) | 0 | 99 (70) | 1 |
| Average | 7 ± 12 | 70 ± 33 (69 ± 108) | 51 ± 36 (96 ± 115) | 18 ± 37 | 51 ± 30 (71 ± 99) | 24 ± 29 | |
| 4 m urea-treated oxidized GST | |||||||
| 1 | coil | NDd | 100 (2) | 53 (15) | 47 | 43 (7) | 57 |
| 69 | α-helix | 8.025 | 60 (30) | 68 (31) | 0c | 59 (37) | 1 |
| 81 | coil | 2.071 | 100 (5) | 50 (4) | 50 | 0 (6) | 100 |
| 94 | α-helix | 0 | 57 (367) | 54 (393) | 5 | 53 (374) | 7 |
| 129 | α-helix | 0 | 100 (3) | 67 (3) | 33 | 50 (6) | 50 |
| 132 | α-helix | 9.977 | 100 (1) | 100 (1) | 0 | NDd (0) | NDd |
| 154 | coil | 0 | 38 (24) | 8 (64) | 79 | 9 (22) | 76 |
| 165 | coil | 0 | 63 (27) | 21 (87) | 67 | 15 (26) | 76 |
| 168 | coil | 37.720 | 100 (28) | 99 (94) | 1 | 88 (26) | 12 |
| Average | 7 ± 12 | 80 ± 25 (54 ± 118) | 58 ± 31 (77 ± 124) | 30 ± 33 | 40 ± 30 (56 ± 120) | 47 ± 37 | |
a Coverage represents the number of times the peptide in which Met, oxidized or not, was found.
b The percentage of reduction was calculated using the formula described in supplemental Table S3.
c Due to experimental approximation, calculation gave a slightly negative percentage of reduction when no reduction activity was observed.
d ND, not determined.
Met residues were on average 70% oxidized in the oxidized GST, with an oxidation status varying from 16% (Met-154) to 99% (Met-168). Surface accessibility of each Met, calculated using protein structure, was found to correlate with the oxidation status, with the 4 buried Met residues being less than 80% oxidized (Table 2). The average percentage of reduction by MSRA and MSRB was 18 and 24%, respectively, but the data varied significantly, indicating that, as in the case of MRP4, not all MetO served as substrates for MSRs. For instance, oxidized Met-69 and Met-81 were efficiently reduced by MSRA and MSRB, respectively, whereas oxidized Met-94, Met-165, and Met-168 were reduced by neither enzyme. These results indicate that MetO residues in the tested proteins were not equivalent MSR substrates and that sequence and structure properties influenced the capacity for their reduction. The data also suggest that the complete reduction of MetO in the protein substrate would require opening of the structure to give access to MSRs, as low reduction capacities were recorded for buried MetO.
MSRs Preferentially Reduce Unfolded Proteins
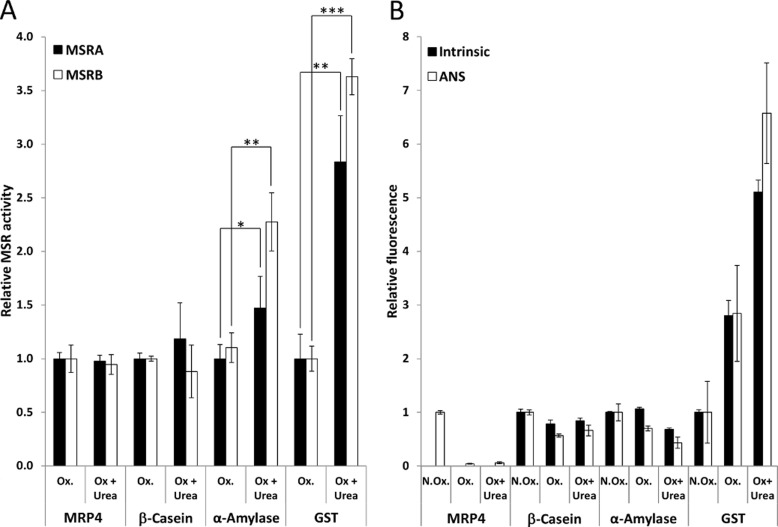
To examine the reduction of MetO present in the hydrophobic core of protein substrates, we assayed MSRA and MSRB activities with the three oxidized Met-rich proteins described above and Bacillus subtilis α-amylase that contains 12 Met (2.3%) and is highly structured (supplemental Table S2) after treatment with 4 m urea, a chaotropic agent that leads to protein unfolding (Fig. 2). These proteins were treated with urea in a small volume followed by dilution of the chaotropic agent when the treated proteins were transferred to the reaction mixture. As a control, the MSR activities were determined with untreated oxidized proteins added to the reaction mixture containing the same final amount of urea. No significant differences in MSRA or MSRB activities were observed in the case of urea-treated or untreated oxidized MRP4 and β-casein, both of which are unstructured proteins. In contrast, when oxidized α-amylase was used as a substrate, the MSRA and MSRB activities were significantly increased by 1.5- and 2.3-fold, respectively, with the urea-treated substrate compared with the non-treated protein. These activity increases were even more dramatic in the case of urea-treated oxidized GST, which showed a 2.8- and 3.6-fold increase in MSRA and MSRB activity, respectively, compared with the non-treated protein (Fig. 2A). Although the proteins unfolded by 4 m urea were rapidly assayed after the transfer to the MSR activity assay mixture, whereby diluting urea, we could not exclude a possibility of spontaneous partial refolding of proteins during the enzymatic reaction. To monitor the substrate folding state during activity measurements, we characterized their intrinsic fluorescence and used a ANS probe, which emits fluorescence upon binding to hydrophobic areas of proteins (Fig. 2B; supplemental Fig. S2). We assayed non-oxidized, oxidized, and urea-treated oxidized proteins. For all tested proteins, oxidation induced changes in intrinsic and ANS fluorescence (with the exception of MRP4, for which no intrinsic fluorescence could be detected), indicating modification of protein structure. However, when oxidized proteins were compared with the urea-treated oxidized proteins, changes were observed in intrinsic and ANS fluorescence in the case of α-amylase and GST. No changes were observed in the case of MRP4 and β-casein, indicating that urea treatment did not induce changes in the folding state as these proteins are not structured (supplemental Table S2). In contrast, after urea treatment, α-amylase and GST remained significantly changed under conditions used in the MSR activity assays (Fig. 2B; supplemental Fig. S2). Altogether, these results indicate that for the tested proteins, MSRA and MSRB activities were higher with unfolded oxidized than with folded oxidized proteins.
FIGURE 2.
MSR activities using oxidized and urea-treated oxidized proteins as substrates and characterization of their folding state by fluorometry. A, MSRA and MSRB activities were measured using the NAPDH-coupled Trx system with 25 μm oxidized protein (Ox.) and the oxidized protein treated with 4 m urea (Ox + Urea). As a control, for oxidized proteins, the same amount of urea present in the urea-treated oxidized proteins was added to the reaction mixture (less than 50 mm final concentration). *, **, and ***, significantly different with p < 0.05, p < 0.01, and p < 0.0001, respectively (t test). B, intrinsic and ANS fluorescence of 25 μm oxidized or urea-treated oxidized protein was determined under conditions similar to those used for MSR activity assays.
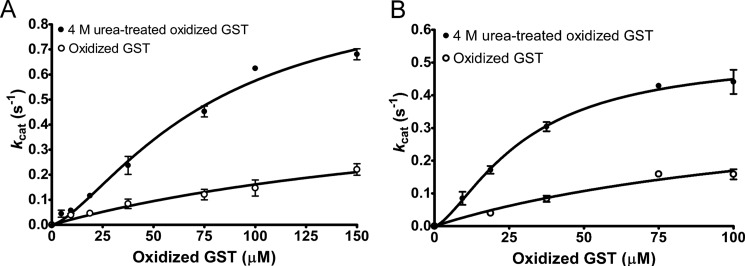
GST was further used to investigate MSRA and MSRB activities with unfolded proteins in detail. The kinetics determined using increasing concentrations of urea-treated oxidized GST could be described by a regression curve similar to an allosteric sigmoidal (Equation 1), with the h values equal to 1.4 ± 0.3 and 1.5 ± 0.1 for MSRA and MSRB, respectively (Fig. 3). However, these values are close to 1, and the known MSR mechanism with Trx as a reductant (32) suggest that modification of these kinetics could be due to an experimental factor, especially for the low values, and does not reflect an allosteric behavior. In the case of MSRA (Table 1; Fig. 3A), the catalytic efficiency (kcat/Km) was 6-fold higher with the urea-treated oxidized GST than the oxidized GST (Table 1; Fig. 3A). Similarly, for MSRB, the catalytic efficiency revealed a 3.2-fold increase in activity with the urea-treated oxidized GST compared with the oxidized protein (Table 1; Fig. 3B). The apparent stoichiometries determined for MSRA and MSRB revealed that 3.2 and 4.7 MetO equivalents, corresponding to 36 and 52% of the 9 Met, were reduced, respectively. Normalization of catalytic parameters to the stoichiometry revealed that both MSRA and MSRB were more efficient in reducing the urea-treated oxidized GST considering the whole protein as a substrate. Moreover, MSRA was more efficient even when the activity values were corrected to the number of MetO reduced in the protein substrate.
FIGURE 3.
Saturation curves of MSRA (A) and MSRB (B) activities using oxidized and urea-treated oxidized GST. MSRA and MSRB activities were measured using the NAPDH-coupled Trx system with 9.4–150 μm oxidized GST or 5–150 μm oxidized GST treated with 4 m urea. As a control, the amount of urea present in the sample of the urea-treated oxidized GST was added in the reaction mixture (less than 50 mm final concentration). The saturation curves obtained with the urea-treated oxidized GST best fit sigmoidal regression as described under “Experimental Procedures” with h = 1.4 ± 0.3 and h = 1.5 ± 0.1 for MSRA and MSRB, respectively.
The apparent stoichiometries indicated that a higher proportion of MetO was reduced in the urea-treated oxidized GST than in the oxidized protein. To determine the identity of MetO used as substrates after urea treatment, the unfolded protein was analyzed by mass spectrometry after incubation with MSRA or MSRB (Table 2). In both cases the average percentage of reduction was almost doubled for the urea-treated oxidized GST compared with the native protein. In particular, buried Met-154 and Met-165 were more efficiently reduced by both MSRA and MSRB, indicating that urea treatment allowed better access to buried MetO (Table 2).
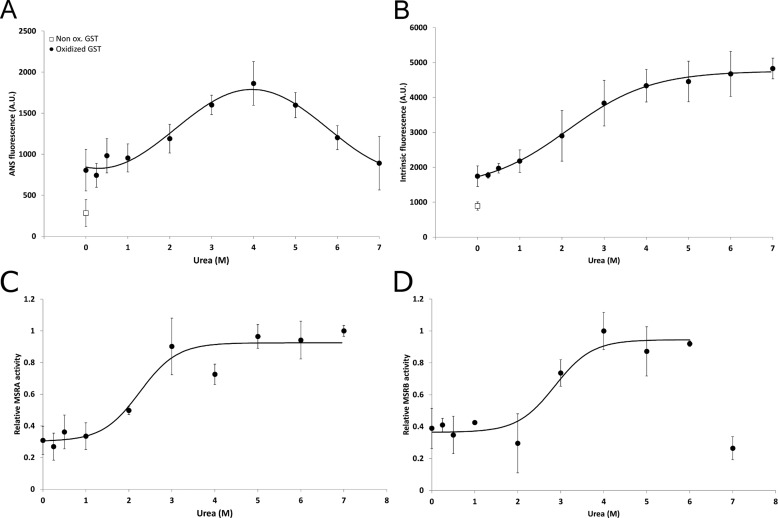
We further incubated oxidized GST with increasing concentrations of urea to examine MSRA and MSRB activities as a function of the unfolded state of protein substrate. To characterize the folding state, ANS and intrinsic fluorescence were measured for each urea concentration (Fig. 4). After oxidation, ANS fluorescence increased with the increase in urea concentration up to 4 m, presumably due to increased exposure of hydrophobic regions, and then decreased at higher concentrations either due to a decrease in exposed hydrophobic parts or the fact that ANS did not bind the protein under conditions of high urea (Fig. 4A). The intrinsic Trp fluorescence fitted a sigmoidal curve with a half-maximum of 2.2 ± 0.1 m of urea (Fig. 4B). Interestingly, MSRA and MSRB activities using the same urea-treated GST samples followed similar sigmoidal curves, with the half-maxima of 2.3 ± 0.2 and 2.9 ± 0.3 m urea, respectively (Fig. 4, C and D). These results suggest that MSRA and MSRB activities were proportional to the urea concentration and increased with the unfolding state of oxidized GST.
FIGURE 4.
Analysis of the folding state of the urea-treated oxidized GST and MSR activities. 10 μm non-oxidized or oxidized GST treated with 0–7 m urea were incubated in 30 mm Tris-HCl, pH 8.0, in the presence (A) or absence (B) of 50 μm ANS. Intrinsic and ANS fluorescence was then recorded. Relative MSRA (C) and MSRB (D) activities were recorded using 25 μm oxidized GST treated with 0–7 M urea. A.U., absorbance units.
Overall, these results showed that both MSRs were more efficient in the reduction of unfolded proteins due to better access to MetO in the hydrophobic core of the substrate. Moreover, regardless of MetO exposure, MSRA was intrinsically more efficient in the reduction of MetO in the unfolded oxidized GST.
MSRs Protect against Oxidative Unfolding Stress Induced by Hypochlorite
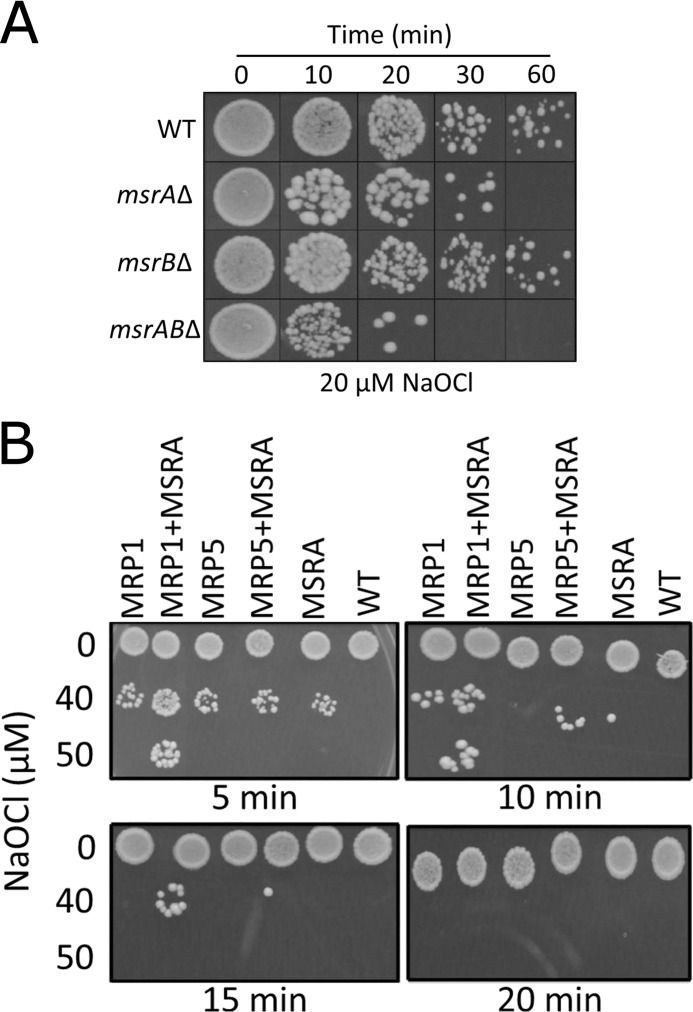
To test if the observed preferential reduction of unfolded proteins by MSRs is physiologically relevant, we utilized sodium hypochlorite (NaOCl), which leads to concomitant oxidation and unfolding of proteins (17). First, we examined if cells deficient in MSRs were more susceptible to NaOCl stress than WT cells. Growth of MSRA-null yeast cells was dramatically inhibited by treatment with 20 μm NaOCl (Fig. 5A). Although MSRB-deficient cells grew similarly to WT cells when treated with NaOCl, deletion of this gene in the context of MSRA deficiency made yeast cells more susceptible to NaOCl stress compared with the MSRA knock-out cells. Thus, both MSRA and MSRB contributed to the protection against NaOCl stress by reducing MetO formed in cellular proteins upon treatment with NaOCl. However, MSRA was more efficient in protecting yeast cells.
FIGURE 5.
Roles of MSRs and MRP in the protection of yeast cells from hypochlorite. A, viability of the indicated MSR mutants and WT cells under conditions of hypochlorite stress is shown. Yeast strains (5 μl each) were spotted onto agar plates and incubated at 30 °C. B, viability of cells overexpressing MRPs and/or MSRA is shown. Cells overexpressing an indicated MRP and/or MSRA were treated with different concentrations of NaOCl. Cells were plated, incubated at 30 °C, and photographed. Cells expressing empty vectors were used as controls.
We further took advantage of previously identified Met-rich proteins3 (22) to determine if these proteins could protect cells through cyclic oxidation of Met and reduction of MetO by MSRs. We overexpressed a structured MRP1 possessing 38 Met and a completely disordered MRP5, which has 31 Met3 residues (supplemental Table S2). Overexpression of MSRA, MRP1, and MRP5 independently protected yeast cells from oxidation. However, the most dramatic effect was observed when MSRA and an MRP were coexpressed (Fig. 5B). We conclude that MRPs alone or in combination with MSRs protect cellular proteins through cyclic oxidation of their Met residues during the oxidative and unfolding stress provoked by hypochlorite.
Finally, we tested the role of MSRA and MRP in the protection against H2O2, which induces oxidative stress but does not lead to obvious protein unfolding (17). Although the effects were less pronounced, overexpression of MSRA protected yeast cells from this oxidant; however, expression of MRPs did not offer additional protection (supplemental Fig. S3). Thus, whereas the Met oxidation/reduction cycle protects yeast during an oxidative and unfolding stress, the effect is less clear in the case of oxidative stress alone. Altogether, these results showed that both MSR types protected yeast against oxidative and unfolding stress and that the protection offered by MSRA was the most efficient.
DISCUSSION
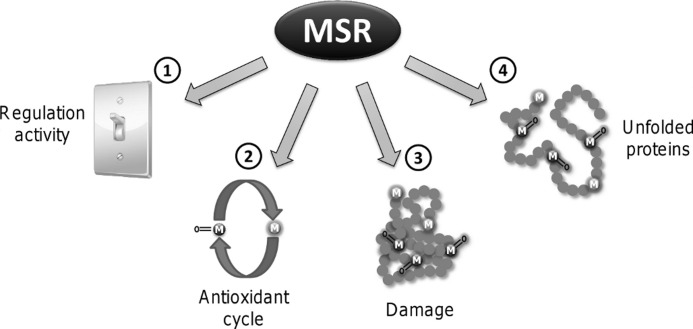
This study describes the first comparative kinetic analyses of MSRA and MSRB using MetO-containing proteins as substrates. MSRA reduced efficiently both free MetO and oxidized proteins, whereas MSRB was specialized for the reduction of oxidized proteins. Further experiments revealed that both MSR types were more efficient in the reduction of unfolded than folded oxidized proteins, and this effect was due to a better access to MetO present in the hydrophobic cores of substrate proteins. However, MSRA was intrinsically more efficient with unfolded proteins irrespective of MetO accessibility. The relevance of this finding was verified in vivo as yeast cells deficient in MSRs were highly sensitive to oxidative unfolding stress provoked by hypochlorite, whereas overexpression of MSRA and Met-rich proteins was highly protective. Our study suggests a new functional category of natural targets for MSRs, the unfolded proteins such as nascent polypeptides, proteins in route for subcellular compartments, and proteins unfolded because of oxidative stress (Fig. 6).
FIGURE 6.
Overview of MSR targets. Based on the effects of MetO oxidation on the structure and activity of proteins, three categories of MSR targets were proposed (4): 1, enzymes in which Met oxidation leads to an increased activity, such as calcium/calmodulin-dependent protein kinase II; 2, proteins that support cyclic Met oxidation and MetO reduction thereby providing antioxidant defense; 3, native proteins damaged by formation of MetO, such as those involved in neurodegenerative diseases. Based on our data, we propose a fourth type of MSR targets: 4, unfolded proteins and nascent polypeptides whose protein core Met are susceptible to oxidation thereby affecting their folding, structure and function.
Characterization of catalytic mechanisms of MSRA and MSRB was previously carried out using free MetO or its variants, such as N-acetyl-MetO or dabsyl-MetO (23, 32, 33). The catalytic parameters with a protein substrate were only known for bovine MSRA acting on oxidized calmodulin (35). We used three oxidized proteins as model substrates; an unstructured MRP4 that has an exceptionally high Met content (21%), another unstructured protein β-casein (3.1% Met) (36), and GST with 3.8% Met and a well defined structure (37) (supplemental Table S2). We showed that both MSRs displayed higher affinity for these proteins than for free MetO and N-acetyl-MetO (Table 1). Whereas MSRA was characterized by catalytic efficiencies similar for free MetO, N-acetyl-MetO, and oxidized proteins, MSRB displayed a striking preference for oxidized proteins (Table 1). The catalytic parameters for the reduction of free MetO and N-acetyl-MetO by MSRA and MSRB were in the range of those determined for prokaryotic and plant MSRs (23, 31, 33, 34), consistent with the idea that the preference for MetO reduction in proteins is a common feature of MSRBs, whereas MSRAs act on any MetO-containing substrates. This finding also agrees with in vivo analysis showing that, despite the presence of three MSRB isozymes, human cells could not reduce the R-diastereoisomer of free MetO (38).
The use of NADPH-coupled Trx system allowed us to determine the fraction of MetO actually used as a substrate in oxidized proteins. As the overall stoichiometry was 1 mol of NADPH oxidized per mol of substrate reduced, NADPH consumption directly reflected MetO reduction using low substrate concentrations (18, 31). This assay is independent of enzyme concentration, and the use of increasing substrate concentrations allows carrying out linear regression whose slope represents the apparent stoichiometry and, thus, the number of MetOs reduced in the protein (Fig. 1). MRP4 possesses 31 Met residues and the apparent stoichiometries were 4 and 7 mol of NADPH oxidized/mol of substrate, indicating that MSRA and MSRB reduced 13 and 23% of MetO, respectively (Table 1). Further analysis of the Met oxidation state in oxidized MRP4 by mass spectrometry showed that MSRA and MSRB reduced on average 33 and 24% of MetO, respectively (supplemental Table S3). In excellent agreement for MSRB, the value found for MSRA was 2.5-fold higher than that calculated by apparent stoichiometry. This could be due to the lower protein coverage observed in the oxidized sample, with 11 Met not covered, whereas only 4 were not covered in the sample repaired by MSRA or MSRB, indicating that Met oxidation affects efficiency of tryptic digestion as suggested previously (39). Similarly, the mass spectrometry analysis showed that MSRA and MSRB reduced 18 and 24% of MetO in GST. These values were higher than those determined by the calculation of apparent stoichiometry, i.e. 1.2 (13%) and 0.8 (9%) MetO reduced by MSRA and MSRB, respectively. This difference was likely due to enrichment of Met-containing peptides, as in almost all cases, the mass spectrometry signal for each peptide was higher for the MSR-reduced samples than for the oxidized proteins (Table 2). The amino acid immediately upstream of Met may influence both the susceptibility of Met to oxidation and its reduction by MSRs, particularly when a Pro flanks the Met (39). Our mass spectrometry analysis corroborated this observation as we found that the Met residues preceded by Pro residues in MRP4 and GST were completely oxidized by hydrogen peroxide but were not substrates for MSRs (Table 2; supplemental Table S3).
Treatment of the oxidized GST with urea dramatically affected the MSR catalytic parameters, increasing catalytic efficiency (Table 1; Fig. 4). Higher MSRA and MSRB activities were also recorded with the urea-treated oxidized α-amylase than with the corresponding untreated oxidized protein (Fig. 3). Further characterization revealed that protein unfolding increased with the increase in urea concentration, following a sigmoidal curve with the half-maximum at ∼2 m urea (Fig. 4); the maximum was reached when the oxidized protein was fully unfolded. In addition, a higher proportion of MetO was reduced in the 4 m urea-treated oxidized protein than in the untreated oxidized protein (Table 2). Moreover, unfolding allowed an almost complete reduction of MetO residues.
Correction of the Km for the apparent stoichiometry allowed estimation of the catalytic efficiency per each reduced MetO. MSRA was 3-fold more efficient in the reduction of MetO in the unfolded GST than in the folded oxidized protein. Apparently, MSRA is intrinsically more efficient in the reduction of MetO in unfolded proteins regardless of the accessibility to MetO. In the case of MSRB, the catalytic efficiency was higher with the unfolded proteins than with the folded one, but this could be explained by improved accessibility of MetO. These observations are consistent with the finding that MSRA preferentially reduced MetO in a disordered region of oxidized calmodulin and that tryptic digestion was required to open up the protein structure for access to all MetO (35).
An analysis of yeast MSR structures showed that hydrophobicity was particularly pronounced in the active sites, which were also enriched in aromatic residues (supplemental Fig. 4). These properties could make these enzymes better equipped to interact with unfolded proteins, which expose hydrophobic residues usually confined to the core regions of folded proteins (40, 41). Therefore, the high content of aromatic and aliphatic residues in MSRs could be an important factor promoting the ability of these enzymes to preferentially use unfolded protein substrates through hydrophobic or π-stacking interactions. This property should allow them to exhibit a higher affinity for hydrophobic regions of target proteins, such as protein cores and regions involved in protein-protein interactions through hydrophobic interactions, where Met is particularly enriched (40, 41). Contrary to the oxidation of surface-exposed Met in folded proteins, which may have little effect on protein function (42), Met oxidation in buried regions should dramatically affect folding and function of cellular proteins. Likewise, oxidation of Met in the regions involved in protein interactions is expected to affect protein structure and function as shown for calmodulin (35). MSRs may play a major role in the protein folding process by protecting Met from oxidation in nascent peptides.
Recently, the use of hypochlorite was found to induce Met oxidation in catalase, concomitant with inactivation and unfolding of this protein. This study showed that the reduction of MetO within the hydrophobic core of catalase was required for enzyme refolding by the chaperone GroEL (16). Hypochlorite treatment leads to both oxidative and unfolding stress as demonstrated by HSP33 activation (17). Indeed, whereas the concomitant treatment with H2O2 and thermal denaturation were required to activate HSP33, sodium hypochlorite alone led to its full activation. This could be due to these two oxidants oxidizing Met residues in different ways. For example, Met oxidation by NaOCl could result in dehydromethionine intermediates that are then converted to MetO (43).
An analysis of yeast cells grown in the presence of NaOCl showed an involvement of MSRs in protection against oxidative unfolding stress (Fig. 5). Indeed, the MSRA-null mutant and especially MSRA/MSRB-null cells were more sensitive than WT cells to hypochlorite treatment, and overexpression of MSRA protected cells from this stressor. Similar effects were observed in prokaryotes (15, 16). Interestingly, overexpression of two Met-rich proteins also conferred protection, very likely due to reversible Met oxidation/reduction. This observation shows that the higher MSR activity with unfolded proteins was relevant in vivo during the NaOCl stress that triggered unfolding of proteins. This property may be particularly important during the oxidative battle between neutrophils and pathogens wherein both opponents induce protein oxidation and use MSRs as a sword and shield strategy (44, 45). Overall, our findings suggest that a major protective function of MSRs in the cell is to rescue and repair oxidized nascent polypeptides and unfolded proteins.
Supplementary Material
Acknowledgment
We thank Dr. Pascal Rey (CEA-Cadarache) for the kind gift of dabsyl-MetO.
This work was supported, in whole or in part, by National Institutes of Health Grant AG021518.

This article contains supplemental Tables S1–S3 and Figs. S1–S4.
X. Liang, Y. Zhang, D. T. Le, D. Hua, and V. N. Gladyshev, submitted for publication.
- MetO
- methionine sulfoxide
- MSR
- MetO reductase
- ANS
- 8-anilinonaphthalene-1-sulfonate
- MRP
- methionine-rich protein
- Trx
- thioredoxin.
REFERENCES
- 1. Zhang X. H., Weissbach H. (2008) Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol. Rev. Camb. Philos. Soc. 83, 249–257 [DOI] [PubMed] [Google Scholar]
- 2. Tarrago L., Laugier E., Zaffagnini M., Marchand C., Le Maréchal P., Rouhier N., Lemaire S. D., Rey P. (2009) Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxide reductases B by glutaredoxins and thioredoxins. J. Biol. Chem. 284, 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boschi-Muller S., Gand A., Branlant G. (2008) The methionine sulfoxide reductases. Catalysis and substrate specificities. Arch. Biochem. Biophys. 474, 266–273 [DOI] [PubMed] [Google Scholar]
- 4. Oien D. B., Moskovitz J. (2008) Substrates of the methionine sulfoxide reductase system and their physiological relevance. Curr. Top. Dev. Biol. 80, 93–133 [DOI] [PubMed] [Google Scholar]
- 5. Erickson J. R., Joiner M. L., Guan X., Kutschke W., Yang J., Oddis C. V., Bartlett R. K., Lowe J. S., O'Donnell S. E., Aykin-Burns N., Zimmerman M. C., Zimmerman K., Ham A. J., Weiss R. M., Spitz D. R., Shea M. A., Colbran R. J., Mohler P. J., Anderson M. E. (2008) A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 133, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo S., Levine R. L. (2009) Methionine in proteins defends against oxidative stress. FASEB J. 23, 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barnham K. J., Ciccotosto G. D., Tickler A. K., Ali F. E., Smith D. G., Williamson N. A., Lam Y. H., Carrington D., Tew D., Kocak G., Volitakis I., Separovic F., Barrow C. J., Wade J. D., Masters C. L., Cherny R. A., Curtain C. C., Bush A. I., Cappai R. (2003) Neurotoxic, redox-competent Alzheimer's β-amyloid is released from lipid membrane by methionine oxidation. J. Biol. Chem. 278, 42959–42965 [DOI] [PubMed] [Google Scholar]
- 8. Breydo L., Bocharova O. V., Makarova N., Salnikov V. V., Anderson M., Baskakov I. V. (2005) Methionine oxidation interferes with conversion of the prion protein into the fibrillar proteinase K-resistant conformation. Biochemistry 44, 15534–15543 [DOI] [PubMed] [Google Scholar]
- 9. Colombo G., Meli M., Morra G., Gabizon R., Gasset M. (2009) Methionine sulfoxides on prion protein Helix-3 switch on the α-fold destabilization required for conversion. PLoS ONE 4, e4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mulinacci F., Bell S. E., Capelle M. A., Gurny R., Arvinte T. (2011) Oxidized recombinant human growth hormone that maintains conformational integrity. J. Pharm. Sci. 100, 110–122 [DOI] [PubMed] [Google Scholar]
- 11. Gao J., Yao Y., Squier T. C. (2001) Oxidatively modified calmodulin binds to the plasma membrane Ca-ATPase in a nonproductive and conformationally disordered complex. Biophys. J. 80, 1791–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minniti A. N., Cataldo R., Trigo C., Vasquez L., Mujica P., Leighton F., Inestrosa N. C., Aldunate R. (2009) Methionine sulfoxide reductase A expression is regulated by the DAF-16/FOXO pathway in Caenorhabditis elegans. Aging Cell 8, 690–705 [DOI] [PubMed] [Google Scholar]
- 13. Laugier E., Tarrago L., Vieira Dos Santos C., Eymery F., Havaux M., Rey P. (2010) Arabidopsis thaliana plastidial methionine sulfoxide reductases B, MSRBs, account for most leaf peptide MSR activity and are essential for growth under environmental constraints through a role in the preservation of photosystem antennae. Plant J. 61, 271–282 [DOI] [PubMed] [Google Scholar]
- 14. Shchedrina V. A., Kabil H., Vorbruggen G., Lee B. C., Turanov A. A., Hirosawa-Takamori M., Kim H. Y., Harshman L. G., Hatfield D. L., Gladyshev V. N. (2011) Analyses of fruit flies that do not express selenoproteins or express the mouse selenoprotein, methionine sulfoxide reductase B1, reveal a role of selenoproteins in stress resistance. J. Biol. Chem. 286, 29449–29461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee W. L., Gold B., Darby C., Brot N., Jiang X., de Carvalho L. P., Wellner D., St John G., Jacobs W. R., Jr., Nathan C. (2009) Mycobacterium tuberculosis expresses methionine sulphoxide reductases A and B that protect from killing by nitrite and hypochlorite. Mol. Microbiol. 71, 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahawar M., Tran V., Sharp J. S., Maier R. J. (2011) Synergistic roles of Helicobacter pylori methionine sulfoxide reductase and GroEL in repairing oxidant-damaged catalase. J. Biol. Chem. 286, 19159–19169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winter J., Ilbert M., Graf P. C., Ozcelik D., Jakob U. (2008) Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ezraty B., Grimaud R., El Hassouni M., Moinier D., Barras F. (2004) Methionine sulfoxide reductases protect Ffh from oxidative damages in Escherichia coli. EMBO J. 23, 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tarrago L., Kieffer-Jaquinod S., Lamant T., Marcellin M. N., Garin J. R., Rouhier N., Rey P. (2012) Affinity chromatography. A valuable strategy to isolate substrates of methionine sulfoxide reductases? Antioxid. Redox Signal. 16, 79–84 [DOI] [PubMed] [Google Scholar]
- 20. Koc A., Gasch A. P., Rutherford J. C., Kim H. Y., Gladyshev V. N. (2004) Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc. Natl. Acad. Sci. U.S.A. 101, 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le D. T., Lee B. C., Marino S. M., Zhang Y., Fomenko D. E., Kaya A., Hacioglu E., Kwak G. H., Koc A., Kim H. Y., Gladyshev V. N. (2009) Functional analysis of free methionine-R-sulfoxide reductase from Saccharomyces cerevisiae. J. Biol. Chem. 284, 4354–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le D. T., Liang X., Fomenko D. E., Raza A. S., Chong C. K., Carlson B. A., Hatfield D. L., Gladyshev V. N. (2008) Analysis of methionine/selenomethionine oxidation and methionine sulfoxide reductase function using methionine-rich proteins and antibodies against their oxidized forms. Biochemistry 47, 6685–6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vieira Dos Santos C., Cuiné S., Rouhier N., Rey P. (2005) The Arabidopsis plastidic methionine sulfoxide reductase B proteins. Sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiol. 138, 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 25. Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and Contrast. Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang T., Faraggi E., Xue B., Dunker A. K., Uversky V. N., Zhou Y. (2012) SPINE-D. Accurate prediction of short and long disordered regions by a single neural-network-based method. J. Biomol. Struct. Dyn. 29, 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizianty M. J., Stach W., Chen K., Kedarisetti K. D., Disfani F. M., Kurgan L. (2010) Improved sequence-based prediction of disordered regions with multilayer fusion of multiple information sources. Bioinformatics 26, i489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsodikov O. V., Record M. T., Jr., Sergeev Y. V. (2002) Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 23, 600–609 [DOI] [PubMed] [Google Scholar]
- 29. Marino S. M., Gladyshev V. N. (2009) A structure-based approach for detection of thiol oxidoreductases and their catalytic redox-active cysteine residues. PLoS Comput. Biol. 5, e1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 31. Grimaud R., Ezraty B., Mitchell J. K., Lafitte D., Briand C., Derrick P. J., Barras F. (2001) Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276, 48915–48920 [DOI] [PubMed] [Google Scholar]
- 32. Boschi-Muller S., Azza S., Branlant G. (2001) E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus or both retains the ability to reduce methionine sulfoxide. Protein Sci. 10, 2272–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boschi-Muller S., Azza S., Sanglier-Cianferani S., Talfournier F., Van Dorsselear A., Branlant G. (2000) A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J. Biol. Chem. 275, 35908–35913 [DOI] [PubMed] [Google Scholar]
- 34. Rouhier N., Kauffmann B., Tete-Favier F., Palladino P., Gans P., Branlant G., Jacquot J. P., Boschi-Muller S. (2007) Functional and structural aspects of poplar cytosolic and plastidial type a methionine sulfoxide reductases. J. Biol. Chem. 282, 3367–3378 [DOI] [PubMed] [Google Scholar]
- 35. Xiong Y., Chen B., Smallwood H. S., Urbauer R. J., Markille L. M., Galeva N., Williams T. D., Squier T. C. (2006) High affinity and cooperative binding of oxidized calmodulin by methionine sulfoxide reductase. Biochemistry 45, 14642–14654 [DOI] [PubMed] [Google Scholar]
- 36. Creamer L. K., Richardson T., Parry D. A. (1981) Secondary structure of bovine αs1- and β-casein in solution. Arch. Biochem. Biophys. 211, 689–696 [DOI] [PubMed] [Google Scholar]
- 37. Lim K., Ho J. X., Keeling K., Gilliland G. L., Ji X., Rüker F., Carter D. C. (1994) Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV. Protein Sci. 3, 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee B. C., Le D. T., Gladyshev V. N. (2008) Mammals reduce methionine-S-sulfoxide with MsrA and are unable to reduce methionine-R-sulfoxide, and this function can be restored with a yeast reductase. J. Biol. Chem. 283, 28361–28369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghesquière B., Jonckheere V., Colaert N., Van Durme J., Timmerman E., Goethals M., Schymkowitz J., Rousseau F., Vandekerckhove J., Gevaert K. (2011) Redox proteomics of protein-bound methionine oxidation. Mol. Cell. Proteomics 10, M110.00686640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marino S. M., Gladyshev V. N. (2010) Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 404, 902–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kochańczyk M. (2011) Prediction of functionally important residues in globular proteins from unusual central distances of amino acids. BMC Struct. Biol. 11, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levine R. L., Mosoni L., Berlett B. S., Stadtman E. R. (1996) Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 15036–15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beal J. L., Foster S. B., Ashby M. T. (2009) Hypochlorous acid reacts with the N-terminal methionines of proteins to give dehydromethionine, a potential biomarker for neutrophil-induced oxidative stress. Biochemistry 48, 11142–11148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Achilli C., Ciana A., Rossi A., Balduini C., Minetti G. (2008) Neutrophil granulocytes uniquely express, among human blood cells, high levels of methionine sulfoxide reductase enzymes. J. Leukoc. Biol. 83, 181–189 [DOI] [PubMed] [Google Scholar]
- 45. Rosen H., Klebanoff S. J., Wang Y., Brot N., Heinecke J. W., Fu X. (2009) Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc. Natl. Acad. Sci. U.S.A. 106, 18686–18691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) Comparative protein structure modeling with MODELLER. Curr. Protoc. Bioinformatics Chapter 5, Unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.