Background: The oxyR and recG genes appeared to be located within the same operon in many bacteria.
Results: OxyR and RecG might work together to control the expression of oxidative stress-related genes.
Conclusion: RecG might be required for the induction of the OxyR regulon by unwinding palindromic DNA for transcription.
Significance: This provides novel insights into the oxidative stress transcriptional machinery in Pseudomonas.
Keywords: DNA Helicase, Gene Regulation, Oxidative Stress, Pseudomonas, Transcription Promoter, OxyR, RecG
Abstract
The oxyR gene appears to reside in an operon with the recG helicase gene in many bacteria, including pathogenic Pseudomonas aeruginosa and Pseudomonas putida. Analysis of P. putida transcriptomes shows that many OxyR-controlled genes are regulated by the ATP-dependent RecG helicase and that RecG alone modulates the expression of many genes. We found that purified RecG binds to the promoters of many OxyR-controlled genes and that expression of these genes was not induced under conditions of oxidative stress in recG mutants of P. aeruginosa, P. putida, and Escherichia coli. In vitro data revealed that promoters containing palindromic sequences are essential for RecG binding and that single-strand binding proteins and ATP are also needed for RecG to promote transcription, whereas a magnesium ion has the opposite effect. The OxyR tetramer preferentially binds to promoters after RecG has generated linear DNA in the presence of ATP; otherwise, the OxyR dimer has higher affinity. This study provides new insights into the mechanism of bacterial transcription by demonstrating that RecG might be required for the induction of the OxyR regulon by unwinding palindromic DNA for transcription. This work describes a novel bacterial transcriptional function by RecG helicase with OxyR and may provide new targets for controlling Pseudomonas species pathogen.
Introduction
OxyR regulates many genes involved in defense against hydrogen peroxide (H2O2)2-induced oxidative stress in Escherichia coli and Salmonella enterica (1–3). E. coli genes regulated by OxyR include trxB (encoding a thioredoxin reductase), katG (encoding hydroperoxidase I), gorA (encoding a glutathione reductase), and ahpCF (encoding an alkyl hydroperoxide reductase). Structural and biochemical studies have identified the molecular mechanism underlying OxyR activation in E. coli (3, 4). OxyR is redox-sensitive and forms an intramolecular disulfide bond in the presence of H2O2. This oxidized form of OxyR acts as a transcriptional activator by binding to its cognate consensus promoter, which contains palindromic sequences. Interestingly, the OxyR dimer has higher affinity for its in vitro binding sites than the tetramer, despite the fact that the OxyR tetramer is considered the active form. However, the reason for this observation is not fully understood (3).
Little is known about the oxidative stress response in Pseudomonas putida (an organism that is very abundant in soils) as transcriptome analysis under oxidative stress has not been reported for this species. Additionally, the function of P. putida OxyR is not well characterized. The oxyR gene appears to reside in an operon with the recG helicase gene in many bacteria, including P. putida and pathogenic Pseudomonas aeruginosa; however, only the DNA repair function of RecG has been examined under conditions of oxidative stress (5). RecG is a helicase (6), but its function in conjunction with OxyR has never been addressed.
DNA helicases are motor proteins that transiently catalyze the unwinding of stable duplex DNA molecules using ATP hydrolysis as the energy source (6, 7). They play an essential role in nearly all aspects of nucleic acid metabolism such as DNA replication, repair, recombination, and transcription (5, 6). The RecG protein of E. coli is a double-stranded DNA helicase that targets a variety of branched DNA substrates, including Holliday junctions, three-strand junctions, and various loop structures (5, 8, 9). RecG homologues are found in most bacterial species (5) and play important roles in the control of chromosome replication and segregation (5, 10). It has been suggested that some helicases may sense redox status directly (11, 12). Therefore, the functions of helicases in bacteria may be quite diverse.
Our data show that RecG directly binds to palindromic sequences of OxyR binding sites and is required for OxyR-mediated activation of many oxidative stress defense genes. RecG alone also influences the expression of many genes whose promoters exhibit palindromic features. Furthermore, we demonstrate that RecG requires Mg2+ and ATP for its function. By identifying a novel mechanism of RecG action, we provide a new perspective on mechanisms of bacterial transcription.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids are shown in supplemental Table S1. The mutant strains of P. aeruginosa are purchased from the Washington University Genome Center. The mutant strains of E. coli K-12 were purchased from NBRP-E. coli at NIG (Japan). Antibiotics (kanamycin, 100 μg/ml; rifampicin, 200 μg/ml) were added where necessary. The open reading frames (ORFs) of the oxyR and recG genes were PCR-amplified using OxyR-OE F/OxyR-OE R and RecG-OE F/RecG-OE R primer pairs, respectively. The amplified fragments, harboring the oxyR and recG genes, were cloned into the BamHI/HindIII sites of pET-28a(+), yielding pET-oxyR and pET-recG. pET-oxyR and pET-recG were transformed into E. coli BL21(DE3) cells via electroporation. E. coli BL21(DE3) cells were grown with moderate shaking at various temperatures in 2-YT medium 2×yeast extract-Tryptone (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl per liter of deionized water) supplemented with kanamycin (100 μg/ml). The cells were grown to mid-log phase (A600 of ∼0.5) at 37 °C with aeration and then induced by adding 0.25 mm isopropyl thio-d-galactoside for 5–7 h at 30 °C. Wild-type P. putida cells were cultured in Luria-Bertani medium (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter of deionized water) at 30 °C with aeration for transcriptome analysis. All chemicals were acquired from Sigma unless otherwise stated.
Microarray Experiments
The cells were grown to early exponential phase (A600 of ∼0.2) at 30 °C with aeration. The cells were then treated with oxidative stress agents (methyl viologen (paraquat), 0.5 mm; cumen hydroperoxide, 3 mm) for 10 min. Total RNA was isolated using an RNeasy mini kit (Qiagen) in accordance with the manufacturer's recommendations. The integrity of the bacterial total RNA was assessed by capillary electrophoresis using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) and further purified using an RNeasy mini kit (Qiagen). cDNA probes for cDNA microarray analysis were prepared by reverse transcription of total RNA (50 μg) in the presence of aminoallyl-dUTP and 6 μg of random primers (Invitrogen) for 3 h. The cDNA probes were cleaned up using a Microcon YM-30 column (Millipore, Bedford, MA) followed by coupling to Cy3 dye (for the reference) or Cy5 dye (for the test sample) (Amersham Biosciences Pharmacia). The dried Cy3 or Cy5-labeled cDNA probes were then resuspended in hybridization buffer containing 30% formamide (v/v), 5× saline-sodium citrate, 0.1% SDS (w/v), and 0.1 mg/ml salmon sperm DNA. The Cy3- or Cy5-labeled cDNA probes were mixed together and hybridized to a microarray slide. The hybridization images on the slides were scanned using an Axon 4000B (Axon Instrument, Union City, CA) and analyzed with GenePix Pro software (version 3.0, Axon Instrument) to determine the gene expression ratios (control versus test sample). Gene expression log 2 expression ratios were normalized using GenePix Pro software (version 3.0, Axon Instrument). The microarray data were deposited in the NCBI GEO site (under accession nos. GSE34409 and GSE34410).
Northern Blot Analysis and Quantitative Real-time RT-PCR (qRT-PCR)
Total RNA was isolated from 5 ml of exponentially growing cells using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. RNA concentrations were estimated using absorbance at 260 nm. Samples of total RNA (5 μg) were run on denaturing agarose gels containing 0.25 m formaldehyde, and the gels were stained with ethidium bromide to visualize 23 S and 16 S rRNA. The fractionated RNA was transferred to nylon membranes (Schleicher & Schuell) using a Turboblotter (Schleicher & Schuell). The mRNA levels were determined by hybridizing the membrane with a gene-specific, 32P-labeled probe (Takara) prepared by PCR amplification with their respective primer pair as indicated in supplemental Table S1. Autoradiography was conducted using an IP plate (Fujifilm) and a Multiplex Bio-Imaging system (Fujifilm). For qRT-PCR, total RNA was isolated from 5 ml of exponentially growing cells using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Ten micrograms of total RNA were treated with DNase I for 1 h at 37 °C. cDNAs were synthesized from DNase-treated total RNA to obtain first strand cDNA suitable for PCR amplification by using RevertAid H Minus first strand cDNA synthesis kit (Fermentas). cDNAs were synthesized with the primer pair, trxB F/trxB R. qRT-PCR was performed using the iCycler iQ real-time PCR detection system (Bio-Rad). cDNA was produced from the same RNA used for RT-PCR. For real-time RT-PCR, 1 μl of template cDNA, 5 pmol of primers, 0.5× SYBR Green and 1 unit of Taq polymerase (Fermentas) were used. Fluorescence was measured at the end of each 72 °C incubation and analyzed with iCycler iQ software (version 3.0). Melting curve analysis (60–95 °C in 0.5 °C increments) was performed to ensure PCR specificity. For quantification, the 16 S rRNA gene was used to obtain reference expression data. Four independent experiments were performed, and means and S.D. are shown.
Protein Purification
All purification steps were conducted at 4 °C using an FPLC system (AKTA FPLC, Unicorn 4.0, Amersham Biosciences). E. coli cell pellets were resuspended in buffer A (50 mm Tris-Cl and 1 mm dithiothreitol, pH 7.5) and disrupted via sonication. After the removal of cell debris by 30 min of centrifugation at 14,000 × g, the soluble fraction was loaded onto an anion exchange column (1 ml, DEAE-cellulose, Amersham Biosciences) equilibrated with buffer A, and the proteins were eluted with a 20-ml linear gradient of 0–1 m NaCl in buffer A (pH 7.5). The fractions (1.0-ml each) were collected and concentrated by ultrafiltration in a Centricon (2 ml of YM-10, Amicon). The concentrates were then applied to a nickel-nitrilotriacetic acid column (1 ml, His-trap, Amersham Biosciences), equilibrated with binding buffer (20 mm sodium phosphate, 0.5 m NaCl, 40 mm imidazole, pH 7.4), and the proteins were eluted with 15 ml of elution buffer (20 mm sodium phosphate, 0.5 m NaCl, 250 mm imidazole, pH 7.4). The fractions were dialyzed via ultrafiltration in a Centricon chamber (2 ml of YM-10, Amicon) and stored at −80 °C in 10% glycerol. SDS-PAGE (10%) was used to track the progress of OxyR and RecG purification, and the gel was stained using Coomassie Blue G-250.
DNA Unwinding Assay
Sets of junction and duplex DNA were constructed using oligonucleotides as described previously (13). The assays for DNA unwinding by RecG were performed at 30 °C in 20 mm Tris-HCl (pH 7.5), 2 mm dithiothreitol, 100 μg/ml bovine serum albumin, and the indicated concentrations of RecG, single-strand binding protein (SSB), ATP, and MgCl2 and 0.1 nm DNA. The optimum MgCl2 and ATP concentrations were estimated in 20-μl reaction volumes. The reactions were started by the addition of RecG and, after 30 min, the reactions were stopped by adding 5 μl of 100 mm Tris-HCl (pH 7.5), 2.5% (w/v) SDS, 200 mm EDTA, 10 mg/ml proteinase K and incubating at 37 °C for 10 min. The samples were analyzed by electrophoresis on 10% polyacrylamide gels.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was conducted as described previously (14). The ptrxB DNA probes were generated using the trxB pro full, trxB pro-1 R, trxB pro-2 F, trxB pro-3 F, trxB pro-4 F, and trxB pro-5 R primers (supplemental Table S1). Also, DNA probes for other genes were produced using their respective primer pairs (supplemental Table S1). The reaction mixture (final volume, 20 μl), containing the ptrxB probe, purified OxyR, and RecG proteins (0–0.2 μg), SSB (Promega, 0.05–0.1 μg), loading buffer, and poly dI-dC (1 μg) in binding buffer (10 mm Tris, pH 7.5, 2 mm MgCl2, 10% (v/v) glycerol, and 75 mm KCl), was incubated for 30 min at 4 °C. The resulting complexes were then analyzed by electrophoresis on 4.5% polyacrylamide gels for 2 h at 120 V. Autoradiography was conducted using an IP plate (Fujifilm) and a Multiplex Bio-Imaging System (Fujifilm). Plasmid EMSA was performed under the same conditions, except for agarose gel electrophoresis. The sample mixtures were analyzed by electrophoresis on 1.5% agarose gels for 1 h at 135 V. Agarose gels were stained with EtBr (1 μg/ml) for 15 min and then destained with distilled water for 15 min.
Nuclease Sensitivity Assay
T7 endonuclease I (New England Biolabs) was performed at room temperature for 1 h in the appropriate buffers. Ten units of T7 endonuclease were used to digest 1 μg of plasmid. After digestion, the plasmid DNA was subjected to 0.8% agarose gel electrophoresis. To map the cleavage sites, nuclease-digested plasmids were purified, digested with EcoRI, and then subjected to 2% agarose gel electrophoresis. The fragments were purified and sequenced using internal primers.
In Vitro Transcription Assay
In vitro run-off transcription assays were performed using a modified version of a previously reported method (15). Briefly, RNAP (1.5 pmol) was incubated at 30 °C for 5 min in 15 μl of transcription buffer (40 mm Tris, pH 7.9, 0.5 mm MgCl2, 0.6 mm EDTA, 0.4 mm potassium phosphate, 1.5 mm DTT, 0.25 mg/ml BSA, and 20% (v/v) glycerol) with 0.15 pmol of template DNA. RNA synthesis was initiated by the addition of 3 μl of substrate mixture containing 0.5 μCi [α-32P]CTP (1000 Ci/mmol) and 0.4 mm each of UTP, ATP, and GTP. After incubating for 30 min at room temperature, the reaction was terminated by adding 50 μl of stop solution (375 mm sodium acetate, pH 5.2, 15 mm EDTA, 0.15% SDS, and 0.1 mg/ml salmon sperm DNA). Transcripts were precipitated with ethanol, resuspended in formamide sample buffer (80% (v/v) formamide, 8% glycerol, 0.1% SDS, 8 mm EDTA, 0.01% bromphenol blue, and 0.01% xylene cyanol) and analyzed on a 10% polyacrylamide gel containing 7 m urea.
RESULTS
Transcriptome Analyses of P. putida and recG Mutant under Oxidative Stress
Transcriptome analysis of P. putida under oxidative stress has never been reported. Superoxide-generating methyl viologen (paraquat) drastically altered the expression profile, which was categorized into different functional groups in our microarray study (data not shown, supplemental Table S2). The microarray results showed that trxB, lsfA, katA, ahpC, ahpF, and PP2441 genes induced under paraquat oxidative stress conditions (supplemental Table S2). Microarray analysis was also conducted under stress conditions induced by cumen hydroperoxide (CHP) to characterize peroxide-mediated oxidative stress in P. putida (data not shown, supplemental Table S3). Studies with H2O2 have been reported in Pseudomonas species (16) but not with organic peroxide. Many oxidative stress defense-related genes (e.g. lsfA, katA, sodA, ohr, ahpC, ahpF, trx-2, and PP2441) were highly up-regulated under CHP oxidative stress (supplemental Table S3). Eleven genes were highly induced by both paraquat and CHP treatment (supplemental Tables S2 and S3), suggesting that they play important roles in defense against oxidative stress in P. putida. These genes include the oxidative stress-related genes ahpC, ahpF, katA, lsfA, and PP3639, the metabolism-related genes aceA and PP3832, a transporter gene cysP, and a regulatory gene rpoX. Of the identified oxidative stress-related genes, ahpC, ahpF, and katA are induced under oxidative stress in P. aeruginosa (16).
OxyR and SoxRS regulate many genes involved in oxidative stress defense in E. coli and Salmonella typhimurium in response to H2O2 and superoxide, respectively (17). Therefore, we examined the expression of genes from the oxyR and soxRS regulons of E. coli in our P. putida microarray after the induction of oxidative stress (supplemental Tables S4 and S5). The gene expressions of E. coli oxyR and soxRS regulon in P. putida seem to be different from those of E. coli. Some E. coli oxyR regulon genes were induced by CHP, but the expression of soxRS regulon genes absolutely did not match with the soxRS regulon in P. putida under paraquat treatment. This is consistent with reports indicating that E. coli-SoxR homologues function differently in Pseudomonas species (18). It is worth noting that several paraquat-induced genes, including trxB (encoding a thioredoxin reductase), belong to the H2O2-induced OxyR regulon in E. coli (3, 17).
Interestingly, the oxyR and recG genes appeared to be located within the same operon in Pseudomonas and other types of bacteria (supplemental Fig. S1) and were co-expressed as an operon in P. putida and P. aeruginosa (supplemental Fig. S1C) (16). Thus, we sought to understand why the recG gene belongs to the same operon as the oxyR regulator. We hypothesized that recG may be involved in the expression of oxyR regulon genes. To test this, transcriptome analyses using the recG mutant strain were conducted in the presence or absence of oxidative stress. When we compared the transcriptome data of wild-type bacteria with those of recG mutants under oxidative stress, almost all of the genes in the wild type that were up-regulated by oxidative stress were not induced in the recG mutant strain under paraquat stress (Table 1). This suggests that that OxyR and RecG may work together to control the expression of many oxidative stress-related genes in P. putida.
TABLE 1.
List of genes whose expression was reduced among PQ-induced genes in P. putida recG mutant
| PP no. | WT+PQa | ΔrecG+PQ | ΔrecG/WT | Palindoromeb | Description |
|---|---|---|---|---|---|
| PP2439 (ahpC) | 17.39 ± 1.09 | 1.17 ± 0.05 | 0.46 ± 0.02 | + | Alkyl hydroperoxide reductase, C subunit |
| PP0481 (katA) | 14.83 ± 1.12 | 1.04 ± 0.02 | 0.48 ± 0.01 | + | Catalase |
| PP2334 | 12.13 ± 0.09 | 1.34 ± 0.17 | 0.94 ± 0.05 | + | Carboxyvinyl-carboxyphosphonate |
| PP2440 (ahpF) | 10.27 ± 1.11 | 1.84 ± 0.06 | 0.41 ± 0.07 | ahpC operon | Alkyl hydroperoxide reductase, F subunit |
| PP0786 (trxB) | 6.92 ± 0.11 | 1.27 ± 0.13 | 0.50 ± 0.02 | + | Thioredoxin reductase |
| PP2339 (acnB) | 5.62 ± 0.09 | 1.43 ± 0.13 | 0.50 ± 0.08 | + | Aconitate hydratase 2 |
| PP4403 (bkdB) | 4.96 ± 0.04 | 1.12 ± 0.43 | 0.97 ± 0.01 | + | 2-Oxoisovalerate dehydrogenase, lipoamide |
| PP5215 (trx-2) | 3.41 ± 0.33 | 0.40 ± 0.05 | 0.95 ± 0.09 | + | Thioredoxin |
| PP5172 | 3.41 ± 0.33 | 0.40 ± 0.05 | 0.99 ± 0.01 | + | Conserved hypothetical protein |
| PP0951 (rpoX) | 3.41 ± 0.41 | 1.38 ± 0.12 | 1.24 ± 0.03 | + | Sigma54 modulation protein |
| PP4621 (hmgA) | 3.39 ± 0.09 | 1.02 ± 0.09 | 1.07 ± 0.02 | + | Homogentisate 1,2-dioxygenase |
| PP0234 (oprE) | 3.27 ± 0.77 | 1.19 ± 0.19 | 0.95 ± 0.02 | + | Porin E |
| PP1206 (oprD) | 3.18 ± 0.11 | 1.02 ± 0.07 | 0.47 ± 0.01 | - | Porin D |
| PP0222 | 3.14 ± 0.35 | 1.05 ± 0.28 | 0.93 ± 0.03 | + | Monooxygenase, DszA family |
| PP2335 | 3.07 ± 0.12 | 1.15 ± 0.04 | 0.95 ± 0.03 | + | Methylcitrate synthase, putative |
| PP0170 | 3.05 ± 0.01 | 1.13 ± 0.30 | 0.96 ± 0.04 | + | ABC transporter, periplasmic binding protein |
| PP4194 (gltA) | 2.97 ± 0.22 | 1.34 ± 0.11 | 0.96 ± 0.13 | + | Citrate synthase |
| PP0883 | 2.89 ± 0.12 | 0.93 ± 0.05 | 0.46 ± 0.06 | + | Porin, putative |
| PP0238 (ssuD) | 2.87 ± 0.08 | 1.17 ± 0.21 | 0.99 ± 0.01 | + | Organosulfonate monooxygenase |
| PP2441 | 2.79 ± 0.11 | 1.23 ± 0.00 | 0.94 ± 0.02 | + | Hypothetical protein |
| PP5278 | 2.77 ± 0.29 | 1.03 ± 0.07 | 0.98 ± 0.01 | + | Aldehyde dehydrogenase family protein |
| PP3148 | 2.77 ± 0.19 | 1.04 ± 0.28 | 0.50 ± 0.04 | − | Glutamine synthetase, putative |
| PP0235 (lsfA) | 2.73 ± 0.09 | 1.31 ± 0.09 | 0.99 ± 0.01 | + | Antioxidant protein LsfA |
| PP3122 | 2.73 ± 0.17 | 0.94 ± 0.24 | 1.00 ± 0.01 | − | CoA-transferase, subunit A, putative |
| PP0885 | 2.69 ± 0.11 | 0.99 ± 0.09 | 0.95 ± 0.06 | − | Dipeptide ABC transporter, periplasmic |
| PP4402 (bkdA2) | 2.69 ± 0.08 | 0.99 ± 0.08 | 0.96 ± 0.01 | + | 2-Oxoisovalerate dehydrogenase, β subunit |
| PP4064 (ivd) | 2.66 ± 0.07 | 0.97 ± 0.21 | 0.50 ± 0.09 | + | Isovaleryl-CoA dehydrogenase |
| PP1071 | 2.62 ± 0.02 | 0.98 ± 0.15 | 0.95 ± 0.05 | + | Amino acid ABC transporter, periplasmic amino acid |
| PP5171 (cysP) | 2.58 ± 0.09 | 0.98 ± 0.15 | 0.49 ± 0.00 | + | Sulfate ABC transporter, periplasmic |
| PP3123 | 2.57 ± 0.12 | 0.98 ± 0.15 | 0.97 ± 0.02 | + | CoA-transferase, subunit B, putative |
| PP0223 | 2.50 ± 0.09 | 1.18 ± 0.04 | 1.04 ± 0.02 | + | Monooxygenase, DszC family |
| PP2333 | 2.33 ± 0.01 | 1.12 ± 0.40 | 1.01 ± 0.00 | − | Transcriptional regulator, GntR family |
| PP0252 (hslO) | 2.28 ± 0.02 | 1.15 ± 0.04 | 1.07 ± 0.06 | + | Chaperonin, 33 kDa |
| PP4620 | 2.27 ± 0.12 | 0.95 ± 0.12 | 0.97 ± 0.01 | + | Fumarylacetoacetase |
| PP2337 | 2.17 ± 0.07 | 1.09 ± 0.23 | 0.96 ± 0.02 | + | Conserved hypothetical protein |
| PP4185 (sucD) | 2.16 ± 0.01 | 1.26 ± 0.26 | 0.50 ± 0.01 | − | Succinyl-CoA synthetase, α subunit |
a Two independent experiments were performed using microarray analysis and the average values with standard deviation are shown. Genes whose expressions were changed under paraquat (PQ) compared with wild-type in contrast to PQ.
b Palindromic sites were determined using a palindrome search program, and the maximum number of mismatches allowed within palindromic sites was two bases. The plus (+) and minus (−) mean the present and absent of a palindromic site, respectively.
RecG and OxyR Co-regulate trxB Gene in P. putida
The trxB and gor genes were chosen for further analysis because their gene expression was regulated by OxyR under peroxide-induced oxidative stress in E. coli (1). Thioredoxin reductase, encoded by the trxB gene, and glutathione reductase, encoded by the gor gene, are reducing agents used for defense against oxidative stress in bacteria (1, 17); however, expression of trxB and gor has not been studied in P. putida. In contrast to results obtained with E. coli, trxB gene expression was induced to a greater extent by paraquat-induced stress than by CHP-induced stress (supplemental Table S4), whereas no change was observed in gor expression (data not shown). We verified the microarray data by performing Northern blot analysis of trxB and gor gene expression under various oxidative stress conditions (data not shown). We then examined trxB gene regulation in response to oxidative stress in P. putida. Expression of trxB increased in response to superoxide-generating materials such as paraquat and menadione but not to agents that induce peroxide-mediated oxidative stress (Fig. 1A). Expression of gor barely increased under the different oxidative stress conditions (data not shown). qRT-PCR and Northern blot analyses confirmed that the expression of trxB was induced maximally after 10 min of treatment with 0.5 mm of the superoxide oxidative stress inducer, paraquat, compared with H2O2 treatment (Fig. 1B). The P. putida trxB mutants were sensitive to superoxide oxidative stress-inducing agents such as paraquat and menadione (data not shown), and their growth rate (0.56 ± 0.08 h−1) was reduced profoundly compared with that of the wild type (1.03 ± 0.16 h−1). However, the motility of the wild type and mutant strains was similar (data not shown).
FIGURE 1.
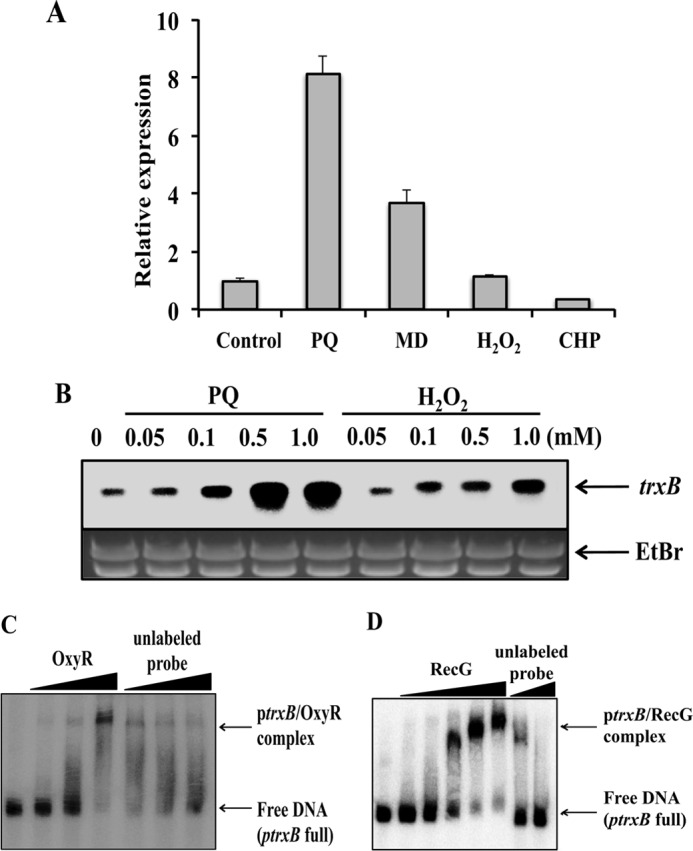
Analysis of trxB gene expression under various oxidative stress conditions. A, qRT-PCR (graph) analyses of trxB gene expression. Four qRT-PCR experiments were performed, and means and S.D. are shown. C, no treatment; PQ, methyl viologen (paraquat; 0.5 mm); MD, menadione (1.0 mm); H2O2, hydrogen peroxide (1.0 mm); and CHP (3 mm) for 10 min. B, Northern blot analysis shows that increasing concentrations of paraquat and H2O2 induced trxB gene expression. The ethidium bromide-stained (EtBr) gel prior to blotting demonstrated consistent loading in all lanes. C, in vitro EMSA analysis. Lanes 1–4, increasing amounts of purified OxyR (0, 0.05, 0.1, and 0.2 μg). Lanes 5–7, labeled as unlabeled probe, represent increasing amounts of unlabeled trxB (full) promoter (0.3, 0.7, and 1.2 μm unlabeled probe), whereas the amount of OxyR was fixed at 0.2 μg. D, in vitro EMSA analysis. Lanes 1–6 represent increasing amounts of purified RecG (0, 0.02, 0.05, 0.10, 0.15, and 0.20 μg). Lanes 7 and 8, labeled as unlabeled probe, correspond to increasing amounts of unlabeled trxB (full) promoter (0.4 and 1.2 μm unlabeled probe), whereas the amount of RecG was fixed at 0.2 μg. At least three independent experiments were performed in all experiments.
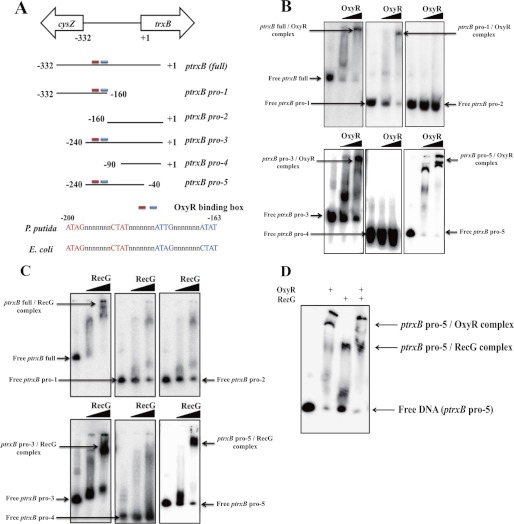
EMSA using purified proteins showed that both OxyR and RecG bound to the trxB promoter region (ptrxB (full) region) (Fig. 1, C and D). We performed EMSA using various fragments of the trxB promoter (ptrxB (full), ptrxB pro-1, ptrxB pro-2, ptrxB pro-3, ptrxB pro-4, and ptrxB pro-5) (Fig. 2). The OxyR-binding site was already characterized by previous study on E. coli OxyR function (1, 17). Our data showed that OxyR bound to the −200 to −163 promoter region (Fig. 2B) and RecG bound to the −200 to −90 promoter region of the trxB in P. putida (Fig. 2C). Binding studies using a mixture of RecG and OxyR proteins showed that the both proteins could bind to the trxB gene promoter region (ptrxB pro-5) (Fig. 2D). Expression of the trxB gene in the recG mutants was not induced under conditions of oxidative stress (Fig. 3A), indicating that RecG may be important for regulating trxB gene expression within the oxyR regulon.
FIGURE 2.
A, schematic representation of the trxB promoter region that are used for EMSA in our OxyR binding studies. B, in vitro EMSA using OxyR on various the trxB promoter regions. The concentration of purified OxyR was used in 0.05 and 0.2 μg. C, in vitro EMSA using RecG on various the trxB promoter regions. The concentration of purified RecG was used in 0.05 and 0.2 μg. D, in vitro EMSA using OxyR and RecG on the ptrxB-pro5 promoter regions. The concentrations of purified OxyR and RecG were fixed in 0.2 μg. At least three independent experiments were performed in all experiments.
FIGURE 3.

Effect of OxyR and RecG on the transcriptional regulation of the oxyR regulon. A, expression analysis for oxidative stress-related genes under oxidative conditions in P. putida wild-type and recG gene mutant cells. C, no treatment; PQ, methyl viologen (paraquat; 0.5 mm) for 10 min. The EtBr-stained gel prior to blotting demonstrated consistent loading in all lanes. B, in vitro EMSA analysis. Purified OxyR and RecG (both 0.2 μg) were used. Autoradiography was conducted using an IP plate (Fujifilm) and a Multiplex Bio-Imaging system (Fujifilm). At least three independent experiments were performed in all experiments.
RecG Regulates Other oxyR Regulon Genes
We further analyzed seven genes that putatively belong to the E. coli oxyR regulon and also have promoters that contain an OxyR-binding consensus sequence (supplemental Fig. S2A). Interestingly, the expression of five of these genes (katA, ahpC, trx-2, hslO, and PP0877) was reduced in the recG mutant under oxidative stress (Fig. 3A). The expression of katA and the ahpC-ahpF operon in the recG mutant was significantly reduced even under untreated conditions (Fig. 3A); whereas the expression of trxB, trx-2, hslO, and PP0877 was reduced under both untreated and treated conditions (Fig. 3A). Purified RecG and OxyR both bound to the promoters of these genes (Fig. 3B), suggesting that RecG also regulates other OxyR-regulated genes.
OxyR-binding Sites Generate Cruciform DNA Structures
In this study, we show that RecG functions as a helicase and can unwind four and three junction DNAs, but it cannot unwind duplex DNA structure (supplemental Fig. S3). Because RecG functions as a helicase and binds to Holliday junctions, R-loops, D-loops, and cruciform DNA structures (8, 9), we examined whether the promoter region of the trxB gene forms non-linear DNA structures in vitro, which can be recognized by RecG (supplemental Fig. S2, A–C). OxyR-binding sites contain palindromic sequences that bind the OxyR protein in the form of a tetramer (3, 4). Two OxyR dimers bind to similar palindromic sites, resulting in tetramer formation (3). Interestingly, we discovered that the OxyR-binding sequences might form a cruciform DNA structure (supplemental Fig. S2, B, red box, and C). EMSA was then performed using various fragments of the trxB gene promoter in the presence or absence of the OxyR-binding sites (Fig. 4, A and B). When one palindromic OxyR-binding site was truncated, RecG was unable to bind to the trxB promoter (Fig. 4B); however, the OxyR dimer, but not the tetramer, might be able to still bind to the truncated site, which was judged by the EMSA pattern (Fig. 4B, second panel). Because cruciform DNA structures commonly contain single strand DNA regions, we examined the effect of adding purified SSB from E. coli to our EMSA assays. The data showed that SSB also bound to the trxB gene promoter (Fig. 4C). Helicase RecG binds to the C-terminal region of SSB (19, 20), which was verified in P. putida using EMSA. SSB might facilitate the binding of RecG to the trxB promoter (Fig. 4D). In a previous study, it was known that the C terminus of SSB could bind to some helicases such as RecG (20), and SSB acts as a DNA maintenance hub at active chromosomal forks for stabilization of DNA replication. In the binding studies described earlier, an excess of RecG protein was used (Figs. 1D and 3B); if a low concentration of RecG (0.05 μg) was used, no band shift was observed (Fig. 4D, left panel). However, in the presence of SSB, only a low concentration of RecG protein was required to bind to the promoter (Fig. 4, D, right panel, and E). Also, if OxyR-binding sites were truncated, both RecG and SSB could not bind to the trxB promoter (Fig. 4F).
FIGURE 4.
Studies on the trxB gene promoter region. A, schematic representation of the trxB promoter region, which were used for EMSA in the presence or absence of the OxyR-binding sequence. B, in vitro EMSA analysis showed that OxyR and RecG proteins bind to various regions of the trxB promoter in P. putida. A schematic representation of the truncated promoter is shown in A. Purified OxyR and RecG (both 0.2 μg) were used. C, in vitro EMSA analysis with SSB protein and ptrxB pro-5. Lanes 1–6 represent increasing amounts of purified SSB (0, 0.01, 0.02, 0.05, 0.08, and 0.10 μg). Lanes 7 and 8 correspond to 2.0 and 4.0 mg, respectively, of unlabeled trxB promoter in the presence of 0.1 μg of SSB. D and E, assessment of SSB and RecG binding to the ptrxB pro-5 promoter using in vitro EMSA analysis. D, lanes 1–5 represent increasing amounts of purified RecG (0, 0.01, 0.02, 0.04, and 0.05 μg). Lanes 6–9 correspond to increasing amounts of SSB (0.01, 0.02, 0.04, and 0.05 μg), whereas the amount of RecG was fixed at 0.05 μg. E, lanes 1–4 represent increasing amounts of purified RecG (0, 0.02, 0.05, and 0.10 μg). Lanes 5–7 correspond to increasing amounts of RecG (0.02, 0.05, and 0.10 μg), whereas the amount of SSB was fixed at 0.05 μg. F, assessment of SSB and RecG binding to the ptrxB pro-5 promoter in the presence or absence of the OxyR-binding sequence using in vitro EMSA. RecG (0.05 μg) and SSB (0.05 μg) were used for EMSA. At least three independent experiments were performed in all experiments.
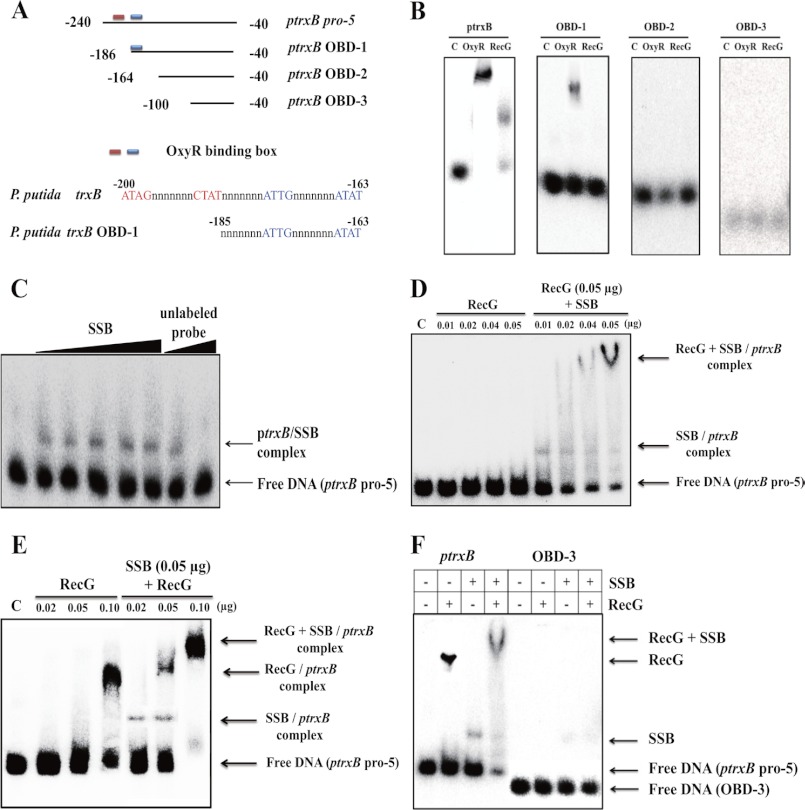
To analyze the DNA structure of the trxB promoter region, various DNA fragments of the trxB promoter regions were used for EMSA using SSB proteins (supplemental Fig. S4A). An upstream 20-nucleotide from pro-5 was truncated in the pro-6 fragment, and a downstream 110-nucleotide from pro-6 was truncated in pro-7. The OxyR-binding sites of pro-6 and pro-7 are present at the edge of these fragments. The OxyR-binding sites of promoter-1 and promoter-2 are located at the center of the fragments. Interestingly, SSB bound to the pro-5 region, but not to OBD-1, OBD-2, and OBD-3 (supplemental Fig. S4B). Therefore, the OxyR-binding sites are necessary for binding of SSB. More importantly, SSB could not bind to the pro-6 and pro-7 regions (supplemental Fig. S4, A and B), which suggests that the OxyR-binding sites at the edges of the DNA may not form precise cruciform DNA structures. The location of the OxyR-binding sites in the linear DNA fragment is very important for the proper formation of cruciform DNA structures. Subsequently, SSB bound to the long promoter regions (promoter-1 and promoter-2) that had OxyR-binding sites at their centers. Interestingly, the EMSA data from promoter-1 and promoter-2 showed a number of band shifts (supplemental Fig. S4B), suggesting that the promoter-1 and promoter-2 regions may contain different cruciform structures. The sub 2, 3, and 4 DNA fragments were generated by nucleotide cascade substitutions within the OxyR-binding sites (supplemental Fig. S4C); therefore, the sub 2, 3, and 4 regions may not form proper DNA structures. Indeed, SSB did not bind to the sub 2, sub 3, and sub 4 regions (supplemental Fig. S4D).
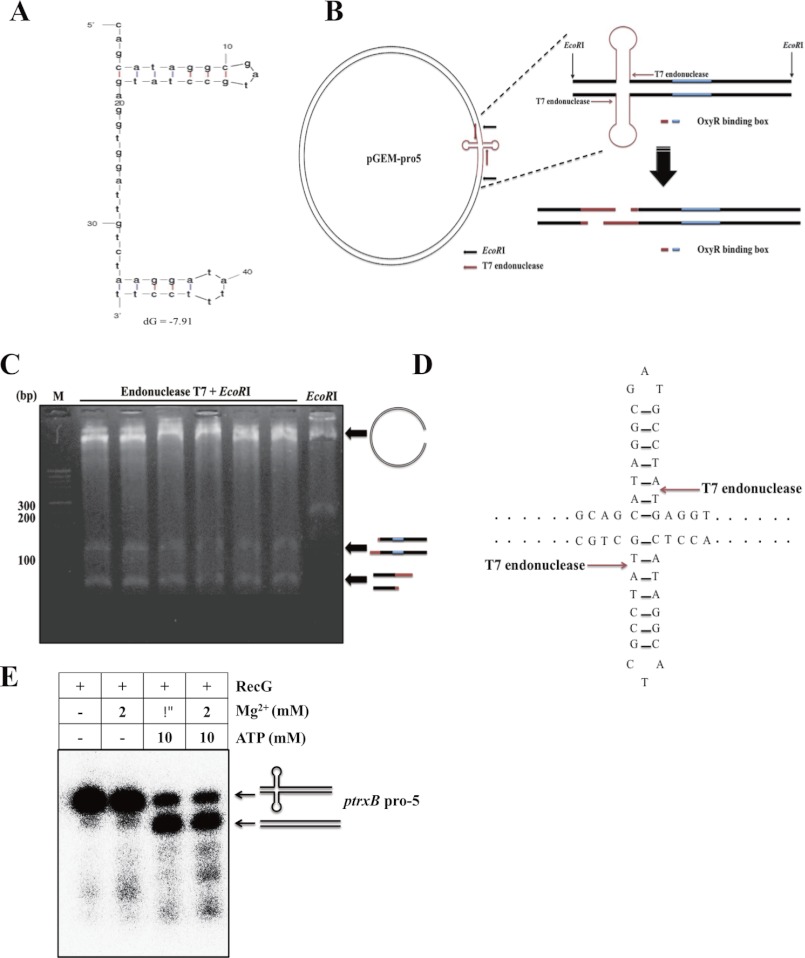
The presence of cruciform DNA structures was further examined using T7 endonuclease I, which cleaves cruciform structures (13). We mapped the T7 endonuclease I cleavage sites using restriction enzyme digestion and sequencing of the DNA fragments (13). First, we predicted the most thermodynamically stable DNA structure for the trxB promoter region using the mfold program (13). We found that the OxyR-binding sites generated a main hairpin DNA structure within the trxB promoter region (Fig. 5A). When incubated with T7 endonuclease I, a considerable amount of the pGEM-pro5 vector harboring the trxB promoter was digested into linear fragments. To further map the cleavage sites, we performed sequence analysis of each nuclease-cleaved fragment (Fig. 5, B and C). Two EcoRI-T7 endonuclease fragments were sequenced, and the cleavage sites were localized to one position (Fig. 5, C and D). By contrast, EcoRI-treated samples generated only one fragment (apart from the vector backbone, Fig. 5D; last lane). Taken together, these data suggest that OxyR-binding sites may generate cruciform DNA structures within linear DNA.
FIGURE 5.
Identification of cruciform DNA structure in the trxB promoter region using T7 endonuclease I. A, prediction of thermodynamic stable cruciform DNA structure using the OxyR binding sequence using mfold program. B, the arrow indicates the cleavage site by T7 endonuclease I. Schematic representations of EcoRI plus T7 endonuclease cleavage sites mapping on the trxB promoter region. C, the plasmid containing the trxB promoter region (pGEM-pro5) was digested with EcoRI T7 endonuclease and was subjected to agarose gel (2.0%) electrophoresis. Lane 1, DNA ladder; lanes 2–7, EcoRI+T7 endonuclease-treated samples; lane 8, EcoRI-treated sample. D, the fragments were sequenced, and results were represented on the cruciform DNA structure using arrows mark. E, helicase unwind assay was performed with RecG, Mg2+, and ATP in the trxB gene promoter region (ptrxB pro-5). The concentrations of purified RecG were fixed in 0.2 μg. At least two independent experiments were performed in all experiments.
Both Mg2+ and ATP Affect Expression of OxyR-regulated Genes
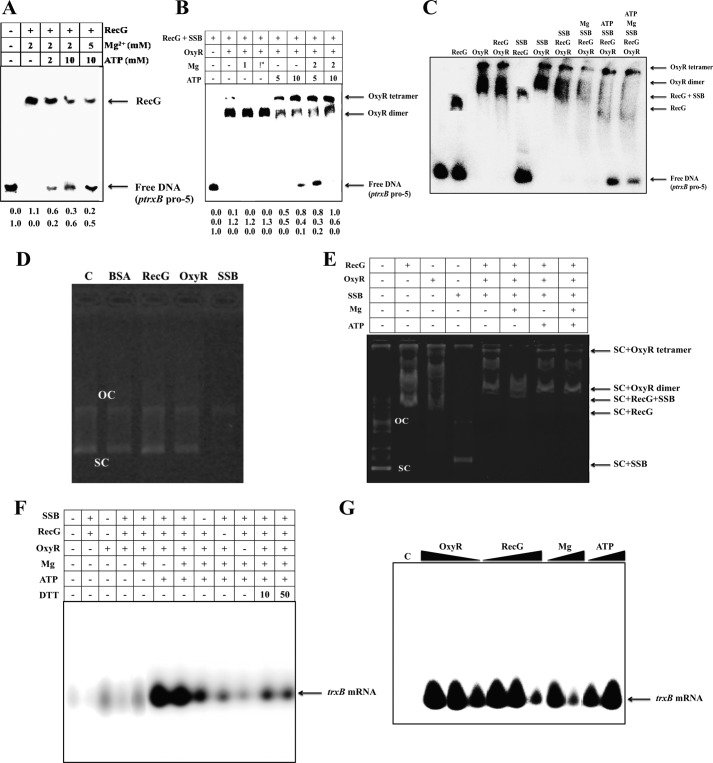
RecG function is modulated by Mg2+ and ATP (21, 22). Mg2+ stabilizes cruciform structures (23), whereas ATP is used as an energy source for unwinding by RecG helicase (22). Therefore, depending on the Mg2+:ATP ratio, cruciform DNA structures can be altered by RecG helicase (22). We found that addition of Mg2+ induced strong binding of RecG to the trxB promoter, whereas the addition of ATP weakened it (Fig. 6A). When the Mg2+:ATP molar ratio was changed to 1:5, RecG binding to the promoter region decreased because RecG used the ATP to create linear DNA. This ratio is the typical molar ratio of mobile cruciform DNA (22, 24), and the result was confirmed using a helicase unwinding assay (Fig. 5E).
FIGURE 6.
The effect of SSB, Mg2+, and ATP on the regulatory mechanism of RecG and OxyR. A, EMSA analysis showing that MgCl2 and ATP affected the binding of RecG to the trxB gene promoter (ptrxB-pro5). Binding affinity was monitored using the indicated concentrations of ATP and MgCl2. RecG concentration was fixed at 0.05 μg. B, in vitro EMSA analysis demonstrated that OxyR protein binding was dependent upon the concentration (mm) of MgCl2 and ATP. RecG, SSB, and OxyR concentrations were added at 0.02 μg, 0.01 μg, and 0.2 μg, respectively. The numbers below each lane show the intensity of the band relative to the intensity of control band (lane 1), which were analyzed by ProXPRESS 2D (PerkinElmer Life Science) and Total Lab software (version 2.0, Nonlinear Dynamics, BioSystematica). C, assessment of the binding of various components to the trxB promoter using in vitro EMSA. RecG, SSB, OxyR, MgCl2, and ATP concentrations were added at 0.1 μg, 0.1 μg, 0.2 μg, 2 mm, and 10 mm, respectively. D, control EMSA experiment using pGEM-T easy plasmid. The reaction mixture (20 μl final volume), containing the pGEM-T easy, purified OxyR and RecG proteins (both 0.1 μg), SSB (Promega, 0.1 μg), BSA (0.5 μg), loading buffer, and poly dI-dC (1 μg) in binding buffer (10 mm Tris, pH 7.5, 2 mm MgCl2, 10% (v/v) glycerol, and 75 mm KCl) was incubated for 30 min at 4 °C. E, plasmid EMSA analysis performed with 2 mm MgCl2, 10 mm ATP, SSB, RecG, and OxyR. RecG, SSB, and OxyR concentrations were added at 0.2, 0.05, and 0.2 μg, respectively. SC and OC refer to the supercoiled plasmid and open circular plasmid, respectively. F, effect of various components on runoff transcription assay from promoters of the trxB genes. An autoradiogram of denaturing 10% PAGE of radiolabeled RNA products is shown. Transcription reactions were conducted at 30 °C for 30 min under standard reaction conditions (see “Experimental Procedures”). RecG, SSB, OxyR, MgCl2, and ATP concentrations were added at 0.1 μg, 0.1 μg, 0.2 μg, 2 mm, and 10 mm, respectively. DTT concentrations were added at 10 and 50 mm. G, a runoff transcription assay was performed with various concentrations of RecG, OxyR, SSB, MgCl2, and ATP. The amount of reaction components was the same as in F, except for the following: lanes 1–4 represent decreasing amounts of purified OxyR (0, 0.10, 0.05, and 0.01 μg); lanes 5–7 represent decreasing amounts of purified RecG (0.05, 0.1, and 0.2 μg); lanes 8 and 9 represent increasing amounts of MgCl2 (5 and 10 mm); and lanes 10 and 11 represent increasing amounts of ATP (20 and 50 mm). At least three independent experiments were performed in all experiments.
DNA binding studies were performed using RecG, OxyR, SSB, Mg2+, and ATP, and the results showed that binding of the OxyR dimer was stronger than that of the OxyR tetramer. In the presence of Mg2+, the OxyR dimer bound to the promoter more strongly than the OxyR tetramer (Fig. 6, B, lanes 3 and 4, and C), perhaps due to the tight cruciform DNA structures formed. However, when ATP was added to the reaction, the OxyR tetramer bound to the promoter region more strongly than the OxyR dimer. These data suggest that RecG, along with SSB, binds to the cruciform DNA structure of the oxyR regulon promoters, and RecG then creates linear DNA by hydrolyzing ATP. Subsequently, the OxyR tetramer binds to the linearized DNA, and oxidized OxyR induces expression of the oxyR regulon.
When size exclusion chromatography was performed using FPLC, the purified OxyR was eluted predominantly as a tetramer (data not shown). It has been known that the active form of OxyR is homotetrameric, consisting of a dimer where each dimer is made up of 34-kDa monomers (25). Many regulatory proteins such as CbnR and DntR form the tetramer as a dimer of dimers (26). In Fig. 6B, RecG and SSB concentrations were 5- and 10-fold less than those in the Fig. 6C so that binding of those two proteins were not detected in the first lane of Fig. 6B in the presence of RecG plus SSB. When high concentration of OxyR was added (the second lane in Fig. 6B), two different sizes of bands were observed, which might be a dimer and a tetramer of OxyR bound to the DNA. We cannot rule out the possibility that different forms of DNAs and different oligomeric OxyR proteins might exist in our experiments because there were more than two OxyR-DNA bands in many EMSA experiments (Figs. 1, C, and 2, B and D). When ATP was added, RecG was released, and more OxyR tetramer-bound DNA was formed. We speculated that more free-DNA became available because more OxyR proteins were bound to the less amounts of DNA (Fig. 6B, lane 7).
DNA binding studies using a supercoiled plasmid harboring the trxB promoter (pGEM-pro5) supported this conclusion (Fig. 6, D and E). In the presence of Mg2+, the OxyR dimer bound to the promoter more strongly than the OxyR tetramer (Fig. 6E, lane 6). The reverse occurred when ATP was added to the reaction (Fig. 6E, lane 7). By contrast, control samples containing vector alone did not show a shift under the same conditions (Fig. 6D). We performed the in vitro transcription analysis for investigation of transcriptional effect of SSB, RecG, and ATP (15). Data obtained from the in vitro transcription assay were consistent with the EMSA data (Fig. 6F), which showed that presence and absence of those components changed OxyR-dependent in vitro transcription. Highest in vitro transcription was observed only in the presence of all three components (Fig. 6F, lanes 6 and 7). In addition, in vitro transcription decreased when an excess of DTT was used (Fig. 6F, lanes 11 and 12). We also observed that when the amount of OxyR used in the reaction decreased, or the amount of RecG or Mg2+ increased, gene expression was reduced, whereas increasing the amount of ATP had the opposite effect (Fig. 6F). 0.25 mm Mg2+ was in the in vitro transcription reaction because the transcription buffer (0.5 mm MgCl2) was diluted when reaction mixture was prepared. Extra 2 mm Mg2+ was added to see the effect of Mg2+ stabilization for cruciform DNA. However, many in vitro transcription assays used 0.25–5 mm Mg2+ concentration for RNA polymerase activity so that dramatic effect of Mg2+ addition was not observed in the Fig. 6F (lanes 6 and 7), but when more Mg2+ (10 mm) was added to the assay (Fig. 6G), transcriptional activity decreased because Mg2+ stabilized the cruciform DNA.
RecG Acts as Global Regulator
To examine whether recG acts as a global regulator, we performed transcriptome analysis using the recG mutant under oxidative stress. Almost all genes that were induced by oxidative stress in wild-type cells were down-regulated in the recG mutant under oxidative stress (Table 1). RecG deletion could reduce expression of some genes in the absence of oxidative stress. When paraquat- and CHP-induced genes (39 and 60, respectively) were analyzed, 13 of 39 and 8 of 60 genes were reduced in the recG mutant without oxidative stress (supplemental Tables S2 and S3, ΔrecG/WT column). Among 51 paraquat and 54 CHP down-regulated genes, only three and nine genes were affected by RecG deletion in the absence of oxidative stress, respectively. No paraquat-down-regulated genes were induced in the recG mutant. Thus, RecG can modulate gene expression even in the absence of oxidative stress. The presence of palindromic sequences within their promoters was also examined because RecG acts as a regulator of loop or cruciform DNA structures. Of the 36 genes whose expression was reduced in the recG mutant under paraquat treatment, 30 contained a palindromic site within their promoter regions (representatives are shown in supplemental Fig. S5). Notably, the oxidative stress-related genes ahpC, katA, trx-2, hslO, and lsfA, which are induced by oxidative stress and contain the OxyR-binding sequence, had palindromic sequences in their promoter regions. Other oxidative stress-related genes that also contain a palindromic sequence include the following: cysP, a transporter related gene induced in response to chromate-oxidative stress in Shewanella oneidensis (27); rpoX, which has a similar function as rpoS and regulates various genes under oxidative stress in Vibrio alginolyticus (28); acnB, which encodes an aconitase and is sensitive to oxidative stress in E. coli (29); and bkdB, which is annotated as the branched-chain α-keto acid dehydrogenase subunit, E2, and is induced by cold stress in Bacillus subtilis (30). We next compared the transcriptomes of the recG mutant and wild type strains in the absence of oxidative stress and found that the expression of >99 genes (including the previously mentioned oxyR-regulated genes) in the recG mutant were reduced 2-fold compared with that in the wild type (supplemental Table S6). Therefore, RecG may induce structural changes in DNA regardless of OxyR function and appears to be involved in global bacterial transcription.
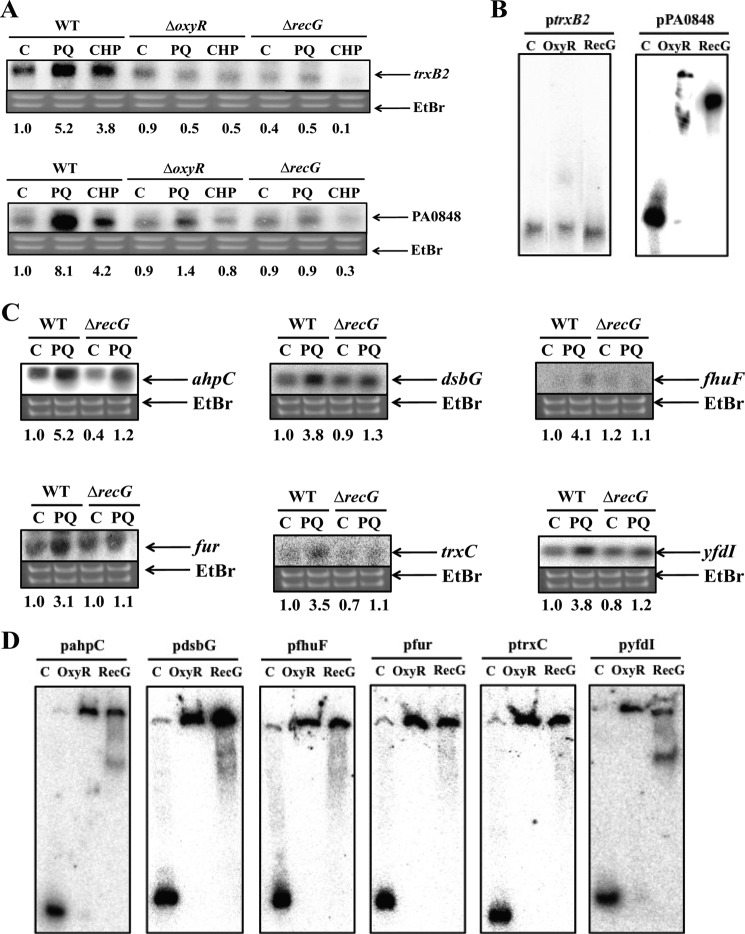
RecG Is Important for Gene Expression in P. aeruginosa and E. coli
The P. putida oxyR gene mutant appears to be lethal, as we were unable to generate this mutant successfully, despite several attempts. Additionally, oxyR deletion mutants of P. putida have not been reported, with the exception of a single amino acid mutant (31). Therefore, P. aeruginosa was used to study the in vivo role of OxyR in Pseudomonas. The oxyR-recG operon of P. aeruginosa is similar to that of P. putida; however, there are two trxB genes in the P. aeruginosa genome, one of which (trxB2) has a sequence similar to that of the trxB gene of P. putida. The trxB2 gene is located next to the PA0848 gene, which, together, may comprise an operon in P. aeruginosa. Additionally, an oxyR box is present in the PA0848 promoter. Interestingly, the expression of these genes was severely reduced in oxyR and recG mutants in P. aeruginosa (Fig. 7A). To investigate whether OxyR and RecG bind to the promoters of these two genes, EMSA was performed using purified OxyR and RecG from P. putida. The results showed that both proteins bound to the PA0848 promoter (Fig. 7B) of P. aeruginoas. The PA0848 and trxB2 genes exist in an operon, and the promoter region exists in front of the PA0848 gene. Furthermore, OxyR and RecG bound to the promoters of OxyR-regulated genes of E. coli (i.e. ahpC, dsbG, fhuF, fur, trxC, and yfdI) and regulated their expression (Fig. 7, C and D).
FIGURE 7.
Investigation of whether RecG functions as global regulator. A, expression analysis of oxidative stress-related genes from supplemental Fig. S3A under oxidative conditions in P. aeruginosa. PA0848 and trxB2 genes belong to the same operon in P. aeruginosa. An image of the ethidium bromide-stained gel was used to verify consistent loading. Cells were treated for 10 min with methyl viologen (paraquat; PQ; 0.5 mm) or CHP (3 mm), or untreated (C). The EtBr gel prior to blotting demonstrated consistent loading in all lanes. The numbers below each lane show the intensity of the band relative to the intensity of control band (lane 1), which were analyzed by ProXPRESS 2D (PerkinElmer Life Science) and Total Lab software (version 2.0, Nonlinear Dynamics, BioSystematica). B, in vitro EMSA showed that OxyR and RecG bound to the promoter of the PA0848-trxB2 operon in P. aeruginosa. Purified OxyR and RecG (0.2 μg of each) were used. C, expression analysis of oxidative stress-related genes under oxidative conditions in E. coli. An image of the ethidium bromide-stained gel was used to verify consistent loading. Cells were treated for 10 min with C (no treatment); PQ (methyl viologen (paraquat); 0.5 mm). The numbers below each lane show the intensity of the band relative to the intensity of control band (lane 1), which were analyzed by ProXPRESS 2D (Perkin Elmer Life Science) and Total Lab software (version 2.0, Nonlinear Dynamics, BioSystematica). The EtBr gel prior to blotting demonstrated consistent loading in all lanes. D, in vitro EMSA analysis showed that OxyR and RecG bound to the promoters of ahpC, dsbG, fhuF, fur, trxC, and yfdI of E. coli. Purified OxyR and RecG (0.2 μg) were used. At least three independent experiments were performed in all experiments.
DISCUSSION
Although the function of RecG in the recombination and repair of DNA has been studied (5), we demonstrated for the first time that RecG also plays an important role in bacterial transcription by binding to promoters containing palindromic sequences. This function is specific to RecG because other helicases (dinG, PA3272, rep, ruvA, and recQ) and the recJ gene, encoding a nuclease, which is known to be involved in RecQ helicase activity of P. aeruginosa, did not influence gene expression of OxyR-regulated genes (supplemental Fig. S6). Our microarray data also suggest that RecG may function alone in transcriptional regulation. Genes whose expression levels were significantly reduced in the recG mutant were classified based on COG function (data not shown, supplemental Table S6). Thus, the unwinding function of RecG may be required for transcription of these regions.
Cruciform formation within palindromic regions of linear DNA carries an energy cost because bases must to be unpaired to initiate extrusion of the cruciform arms (9, 10). Thus, cruciform DNA will branch-migrate back to form duplex linear stable molecules. Cruciform DNA formation can be driven by negative supercoiling, but their formation in linear DNA is highly unlikely. However, our data showed that the trxB promoter generated cruciform DNA within linear DNA if the DNA regions next to the palindromic regions were long enough (supplemental Fig. S4). Although the binding site for SSB is 35 or 65 nucleotides, depending on the salt conditions, other results suggest that SSB can bind to a 21-nucleotide region in S. enterica DNA (depending on the salt concentration) (32). The EMSAs were conducted under low-salt conditions, which would have allowed SSB to bind to such small nucleotide regions. Also, other studies indicate that extrusion of linear DNA generates hairpin and cruciform structures (33, 34). Although little is known about the physical DNA structure of the trxB-like promoters, the data provided evidence that SSB may bind to trxB promoters in both linear and supercoiled DNA.
The data also showed that the pattern of transcriptional expression affected by RecG varied depending on the location of the cruciform DNA. The promoter region of the trxB gene produces cruciform DNA within OxyR-binding sites, but the palindromic sequences of the ahpC and katA genes are located upstream of the OxyR-binding sites. Although expression of the ahpC and katA genes was reduced overall in the recG mutant compared with that in wild-type cells, expression of these genes was still induced in response to oxidative stress (Fig. 3A). Thus, we speculate that RecG influences DNA stability, which, in turn, may affect the expression of ahpC and katA regardless of oxidative stress. Because RecG does not directly affect the OxyR-binding sites of the ahpC and katA genes, unlike the trxB gene, the tetramer form of OxyR may still be functional at these sites. Other oxyR- regulated genes (trx-2, hslO, and trxB) contain cruciform DNA within their OxyR-binding sites. The data suggest that the function of RecG may correlate with the position of the cruciform DNA on the target gene.
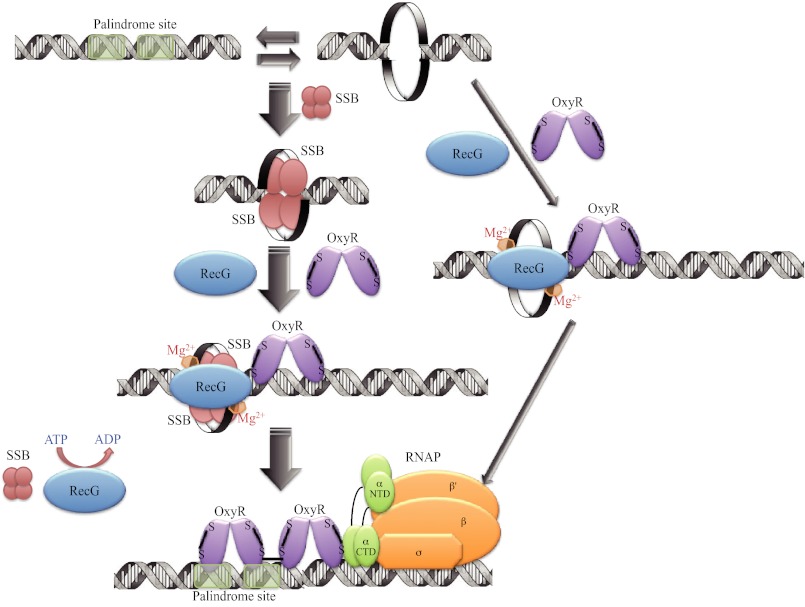
We also found that OxyR senses superoxide but not hydrogen peroxide. The mechanism of OxyR activation may vary in different types of bacteria. In the case of Porphyromonas gingivalis, the OxyR regulator does not sense hydrogen peroxide but activates various oxidative stress-related genes (35). Additionally, OxyR functions as a negative regulator in some types of bacteria (36). Previous studies have shown that the OxyR dimer has stronger DNA binding affinity than the tetramer (3, 4). Although the observed interactions with the tetramer appear to be weak and may require other proteins to be stabilized in solution, the tetramer is still required for the activation of OxyR-regulated genes. The data indicate that DNA is linearized by RecG helicase and that the addition of SSB and ATP provides stability for the binding of the OxyR tetramer, which then promotes full activation of OxyR-regulated genes (Fig. 8). Taken together, the results reveal a novel mechanism underlying RecG-mediated regulation of transcription in Pseudomonas species and demonstrate that the OxyR tetramer may preferentially bind to its cognate palindromic promoter after RecG generates linear DNA.
FIGURE 8.
Model of RecG helicase function during bacterial transcription. RecG helicase, in conjunction with SSB and ATP, linearizes hairpin DNA. When oxidized, the OxyR tetramer stably binds to the linearized promoter and activates transcription. The thickness of the arrows represents the preferences of potential helicase function. RNAP, RNA polymerase enzyme.
Supplementary Material
This work was supported by the National Research Foundation Grant funded by the Korean government (Ministry of Education, Science and Technology) (2012-0005277).

This article contains supplemental Tables S1–S6 and Figs. S1–S6.
- H2O2
- hydrogen peroxide
- CHP
- cumen hydroperoxide
- EMSA
- electrophoretic mobility shift assay
- OBD
- OxyR-binding site deletion
- SSB
- single stranded binding protein
- qRT-PCR
- quantitative real-time RT-PCR
- NBRP
- National BioResource Project
- NIG
- National Institute of Genetics.
REFERENCES
- 1. Imlay J. A. (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee C., Lee S. M., Mukhopadhyay P., Kim S. J., Lee S. C., Ahn W. S., Yu M. H., Storz G., Ryu S. E. (2004) Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 4. Zaim J., Kierzek A. M. (2003) The structure of full-length LysR-type transcriptional regulators. Modeling of the full-length OxyR transcription factor dimer. Nucleic Acids Res. 31, 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rudolph C. J., Upton A. L., Briggs G. S., Lloyd R. G. (2010) Is RecG a general guardian of the bacterial genome? DNA Repair 9, 210–223 [DOI] [PubMed] [Google Scholar]
- 6. Sharples G. J., Ingleston S. M., Lloyd R. G. (1999) Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J. Bacteriol. 181, 5543–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall M. C., Matson S. W. (1999) Helicase motifs: The engine that powers DNA unwinding. Mol. Microbiol. 34, 867–877 [DOI] [PubMed] [Google Scholar]
- 8. Gregg A. V., McGlynn P., Jaktaji R. P., Lloyd R. G. (2002) Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 9, 241–251 [DOI] [PubMed] [Google Scholar]
- 9. Briggs G. S., Mahdi A. A., Wen Q., Lloyd R. G. (2005) DNA binding by the substrate specificity (wedge) domain of RecG helicase suggests a role in processivity. J. Biol. Chem. 280, 13921–13927 [DOI] [PubMed] [Google Scholar]
- 10. Rudolph C. J., Upton A. L., Harris L., Lloyd R. G. (2009) Pathological replication in cells lacking RecG DNA translocase. Mol. Microbiol. 73, 352–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ren B., Duan X., Ding H. (2009) Redox control of the DNA damage-inducible protein DinG helicase activity via its iron-sulfur cluster. J. Biol. Chem. 284, 4829–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Voloshin O. N., Camerini-Otero R. D. (2007) The DinG protein from Escherichia coli is a structure-specific helicase. J. Biol. Chem. 282, 18437–18447 [DOI] [PubMed] [Google Scholar]
- 13. Kurahashi H., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B. S., Toda T. (2004) Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 279, 35377–35383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yeom S., Yeom J., Park W. (2010) Molecular characterization of FinR, a novel redox-sensing transcriptional regulator in Pseudomonas putida KT2440. Microbiology 156, 1487–1496 [DOI] [PubMed] [Google Scholar]
- 15. Kang J. G., Hahn M. Y., Ishihama A., Roe J. H. (1997) Identification of σ factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2). Nucleic Acids Res. 25, 2566–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ochsner U. A., Vasil M. L., Alsabbagh E., Parvatiyar K., Hassett D. J. (2000) Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182, 4533–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imlay J. A. (2006) Iron-sulfur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 [DOI] [PubMed] [Google Scholar]
- 18. Park W., Peña-Llopis S., Lee Y., Demple B. (2006) Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli. Biochem. Biophys. Res. Commun. 341, 51–56 [DOI] [PubMed] [Google Scholar]
- 19. Cadman C. J., McGlynn P. (2004) PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 32, 6378–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costes A., Lecointe F., McGovern S., Quevillon-Cheruel S., Polard P. (2010) The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet. 6, e1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singleton M. R., Scaife S., Wigley D. B. (2001) Structural analysis of DNA replication fork reversal by RecG. Cell 107, 79–89 [DOI] [PubMed] [Google Scholar]
- 22. McGlynn P., Lloyd R. G. (1999) RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 27, 3049–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vologodskaia M. Y., Vologodskii A. V. (1999) Effect of magnesium on cruciform extrusion in supercoiled DNA. J. Mol. Biol. 289, 851–859 [DOI] [PubMed] [Google Scholar]
- 24. Shida T., Iwasaki H., Saito A., Kyogoku Y., Shinagawa H. (1996) Analysis of substrate specificity of the RuvC holliday junction resolvase with synthetic Holliday junctions. J. Biol. Chem. 271, 26105–26109 [DOI] [PubMed] [Google Scholar]
- 25. Knapp G. S., Tsai J. W., Hu J. C. (2009) The oligomerization of OxyR in Escherichia coli. Protein Sci. 18, 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeller T., Klug G. (2004) Detoxification of hydrogen peroxide and expression of catalase genes in Rhodobacter. Microbiology 150, 3451–3462 [DOI] [PubMed] [Google Scholar]
- 27. Brown S. D., Thompson M. R., Verberkmoes N. C., Chourey K., Shah M., Zhou J., Hettich R. L., Thompson D. K. (2006) Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol. Cell Proteomics 5, 1054–1071 [DOI] [PubMed] [Google Scholar]
- 28. Zhao J. J., Chen C., Zhang L. P., Hu C. Q. (2009) Cloning, identification, and characterization of the rpoS-like σ factor rpoX from Vibrio alginolyticus. J. Biomed. Biotechnol. 2009, 126986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar J. K., Tabor S., Richardson C. C. (2004) Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 3759–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaan T., Homuth G., Mäder U., Bandow J., Schweder T. (2002) Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 148, 3441–3455 [DOI] [PubMed] [Google Scholar]
- 31. Hishinuma S., Ohtsu I., Fujimura M., Fukumori F. (2008) OxyR is involved in the expression of thioredoxin reductase TrxB in Pseudomonas putida. FEMS Microbiol. Lett. 289, 138–145 [DOI] [PubMed] [Google Scholar]
- 32. Huang Y. H., Lee Y. L., Huang C. Y. (2011) Characterization of a single-stranded DNA binding protein from Salmonella enterica serovar Typhimurium LT2. Protein J. 30, 102–108 [DOI] [PubMed] [Google Scholar]
- 33. Taylor A. F., Smith G. R. (1992) RecBCD enzyme is altered upon cutting DNA at a Chi recombination hotspot. Proc. Natl. Acad. Sci. U.S.A. 89, 5226–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dai X., Rothman-Denes L. B. (1998) Sequence and DNA structural determinants of N4 virion RNA polymerase-promoter recognition. Genes Dev. 12, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohara N., Kikuchi Y., Shoji M., Naito M., Nakayama K. (2006) Superoxide dismutase-encoding gene of the obligate anaerobe Porphyromonas gingivalis is regulated by the redox-sensing transcription activator OxyR. Microbiology 152, 955–966 [DOI] [PubMed] [Google Scholar]
- 36. Panmanee W., Hassett D. J. (2009) Differential roles of OxyR-controlled antioxidant enzymes alkyl hydroperoxide reductase (AhpCF) and catalase (KatB) in the protection of Pseudomonas aeruginosa against hydrogen peroxide in biofilm versus planktonic culture. FEMS Microbiol. Lett. 295, 238–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.