Abstract
Type-1 interferon (IFN)-mediated responses are a crucial first line of defense against viral infections and are critical for generating both innate and adaptive immunity. Therefore, viruses have necessarily evolved mechanisms to impede the IFN response. HSV-2 was found to completely abolish type-1 IFN-mediated signaling via multiple STAT2-associated mechanisms. Although the extent and kinetics of this inactivation were indistinguishable between the various cell-lines examined, there were distinct differences in the mechanisms HSV-2 employed to subvert IFN-signaling amongst the cell-lines. These mechanistic differences could be segregated into two categories dependent on the phase of the HSV replicative cycle that was responsible for this inhibition: 1) early phase-inhibited cells which exhibited abrogation of IFN-signaling prior to viral DNA replication; 2) late phase-inhibited cells where early phase inhibition mechanisms were not functional, but viral functions expressed following DNA replication compensated for their ineffectiveness. In early phase-inhibited cells, HSV-2 infection targeted STAT2 protein for proteosomal degradation and prevented de novo expression of STAT2 by degrading its mRNA. In contrast, HSV-2 infected late phase-inhibited cells exhibited no apparent changes in STAT2 transcript or protein levels. However, in these cells STAT2 was not activated by phosphorylation and failed to translocate to the cell nucleus, thereby preventing transactivation of antiviral genes. In primary human fibroblasts, HSV-2 failed to fully degrade STAT2 and therefore, both early and late phase mechanisms functioned cooperatively to subvert IFN-mediated antiviral gene expression. Taken together, these results indicate the importance that HSV-2 has assigned to STAT2, investing significant genomic currency throughout its replicative lifecycle for continuous targeted destruction and inhibition of this protein.
Keywords: HERPESVIRUS 2, HUMAN, HERPES SIMPLEX, STAT2 TRANSCRIPTION FACTOR, INTERFERON TYPE I, HOST-PATHOGEN INTERACTIONS, IMMUNE EVASION
1. Introduction
Herpes simplex virus types 1 (HSV-1) and 2 (HSV-2) are two closely related members of the alphaherpesvirus family that share a nearly identical set of approximately 80 open reading frames. These genes are expressed during the HSV replicative cycle in a triphasic sequential and coordinately regulated temporal cascade of gene expression (Honess and Roizman, 1974; Honess and Roizman, 1975; Roizman et al., 1975): 1) immediate early (IE) genes are expressed immediately upon deposition of viral DNA into the nucleus (Honess and Roizman, 1974; Honess and Roizman, 1975); 2) IE proteins launch the expression of early (E) genes that initiate viral DNA replication and subsequently elicit late (L) gene expression (Honess and Roizman, 1974); 3) the L genes consist of proteins that are either structural in nature or function to mediate capsid assembly, virion maturation, and viral egress (Honess and Roizman, 1974). In order to control viral infections, hosts have evolved cell-intrinsic mechanisms that attempt to neutralize processes within each phase of this replicative cycle, thereby suppressing efficient viral replication until innate and adaptive responses can effectively respond (Darnell, 1997; Garcia-Sastre and Biron, 2006; Levy and Garcia-Sastre, 2001; Platanias and Fish, 1999; Samuel, 2001; Stark et al., 1998).
The type-I interferon (IFN) response is a critical determinant for cell-intrinsic control of viral infections (Darnell, 1997; Garcia-Sastre and Biron, 2006; Platanias and Fish, 1999; Samuel, 2001; Stark et al., 1998), and therefore, occlusion or deletion of IFN-associated pathways can result in severe disease following infection (Conrady et al., 2011; Garcia-Sastre and Biron, 2006; Leib et al., 1999; Pasieka et al., 2008). Type-I IFNs mediate the antiviral response by inducing a JAK-STAT signaling cascade that results in tyrosine phosphorylation, cytoplasmic hetero-oligomerization, and nuclear translocation of the IFN-stimulated gene factor 3 (ISGF3) transcription factor (Horvath, 2004; Horvath and Darnell, 1997; Improta et al., 1994; Kessler et al., 1990; Platanias et al., 1994; Schindler et al., 1992). ISGF3 consists of three subunits: signal transducers and activators of transcription 1 (STAT1); STAT2; and interferon regulatory factor 9 (IRF9) (Horvath and Darnell, 1997; Improta et al., 1994; Schindler et al., 1992). Of these subunits, STAT2 is unique and critical to the type-I IFN signaling pathways; whereas, STAT1 functions both in type I and type II IFN signaling (Horvath and Darnell, 1997). IFN activation of the ISGF3 complex culminates in its binding to IFN-stimulated responsive elements (ISREs) within target gene promoters and the subsequent transactivation of hundreds of interferon stimulated genes (ISGs) (Bandyopadhyay et al., 1995; Darnell, 1997; Horvath and Darnell, 1997; Levy and Garcia-Sastre, 2001; Platanias and Fish, 1999; Samuel, 2001; Stark et al., 1998). ISG expression mediates many antiviral effects, including the inhibition of viral protein translation, genomic replication, viral egress, and cell-to-cell spread (Darnell, 1997; Mott et al., 2009; Pierce et al., 2005; Samuel, 2001). In addition, ISG expression facilitates the recognition of virally infected cells by the innate and adaptive immune response (Gallucci et al., 1999; Honda et al., 2005; Le Bon et al., 2001; Le Bon and Tough, 2002).
HSV is able to establish its characteristic lifelong infection, at least in part, by evading or subverting host antiviral defenses via specific virus-encoded countermeasures (Chee and Roizman, 2004; Duerst and Morrison, 2003; Melroe et al., 2004; Paladino and Mossman, 2009; Peng et al., 2009; Yokota et al., 2001). Several HSV-1 proteins have been shown to antagonize type-I IFN induced antiviral responses: 1) HSV-1 ICP0 functions as an ubiquitin ligase and targets specific IFN-associated cellular antiviral proteins (e.g. PML and Sp100) for proteosomal degradation (Boutell et al., 2002; Chelbi-Alix and de The, 1999; Hagglund and Roizman, 2002; Hagglund et al., 2002). 2) ICP0 inhibits IRF3- and IRF7-mediated induction of type-I IFN and ISG expression (Harle et al., 2002; Mossman et al., 2000; Mossman and Smiley, 2002; Paladino et al., 2010). 3) γ34.5 and Us11 block host-initiated shutdown of protein translation (Cassady et al., 1998; Chou and Roizman, 1994; He et al., 1997; Leib et al., 2000; Mossman and Smiley, 2002; Pasieka et al., 2006). 4) HSV-1 ICP27 can decrease IFN-activated STAT1 phosphorylation and partially block STAT1 translocation to cell nuclei (Johnson and Knipe, 2010; Johnson et al., 2008). 5) The HSV encoded RNase, virion host shutoff (VHS) protein, degrades cellular transcripts and thereby prevents expression of IFN-associated antiviral genes (Chee and Roizman, 2004; Duerst and Morrison, 2004; Murphy et al., 2003; Narita et al., 1998; Su et al., 1993). Mutant viruses that specify deletions in these genes exhibit increased sensitivity to IFNs and are highly attenuated in mouse models (Duerst and Morrison, 2003; Halford et al., 2010; Korom et al., 2008; Leib et al., 1999). Although a paucity of direct mechanistic studies exist for HSV-2, genetic mapping and pathogenic studies have indicated that the HSV-2 VHS protein is vital for regulating type-I IFN responses, and therefore, deletion of VHS profoundly attenuates HSV-2 in vivo (Murphy et al., 2003; Narita et al., 1998; Su et al., 1993).
In the present study, the ability of HSV-2 to interfere with IFN-mediated signaling and transactivation of antiviral gene expression was examined. As has been shown for HSV-1, IFN-mediated expression of ISGs was inhibited following HSV-2 infection of normal primary adult human dermal fibroblasts. However, in examining the mechanisms HSV-2 employs to interfere with activation of ISG expression, an intriguing cell-line dependent phenomena was identified that took advantage of peculiarities inherent to the established transformed cell-lines and enabled the visualization of previously masked late-replicative phase-mediated inhibitory events. Similar to what has been observed for HSV-1 (Chee and Roizman, 2004; Yokota et al., 2001), we found that in some cell-lines HSV-2 inhibition of type-I IFN signaling events could be accounted for by virus-mediated loss of STAT2 protein. In these cells, multiple complementary HSV-2 early replicative phase mechanisms were required to fully extinguish STAT2 protein levels. However, despite HSV-2 inhibiting signaling in all cell-lines examined, STAT2 protein expression was not altered in some cell-lines by HSV-2 infection. This finding permitted the unmasking of late replicative phase STAT2-associated events that can function cooperatively to ablate type-I IFN signaling. Specifically, in cells where HSV-2 did not deplete STAT2 protein levels, IFN treatment failed to activate STAT2 phosphorylation, although STAT1 phosphorylation was unaffected. Inhibition of STAT2 activation permitted its retention in the cell cytoplasm and abolished its translocation to cell nuclei. In primary cells, HSV-2 infection failed to fully degrade cellular STAT2, indicating that both early and late replicative phase mechanisms are likely required for full modulation of IFN-mediated signaling in the host. The findings described herein demonstrate that HSV-2 specifies multiple complementary mechanisms throughout its replicative lifecycle that can compensate for incomplete functioning of one mechanism or differences between cells in order to facilitate complete ablation of IFN signaling.
2. Materials and methods
2.1. Cells and viruses
Vero, C33A, HEK293, and HeLa cells were initially acquired from ATCC (Manassas, VA) and maintained in DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 5% FCS (Hyclone). The HEK-Blue IFN α/β (293α/β) cell-line, which is engineered to express additional STAT2 protein and permits the detection of bioactive type-I interferons, was purchased from Invivogen (San Diego, CA). Normal adult human dermal fibroblasts (HDFa) were obtained from Invitrogen and maintained in Medium 106 supplemented with low serum growth supplement (Invitrogen). The HSV-1 and HSV-2 G strain viruses were obtained from ATCC, propagated in Vero cells and stored as infectious cell preparations at −80 °C.
2.2. Reagents and inhibition of viral and cellular processes
Aliquots of recombinant human IFNβ and human IFNγ (PBL Biomedical Laboratories) were stored at −80 °C and diluted to the indicated Units/ml (U/ml) just prior to use. To inhibit viral DNA replication and late gene expression, cells were treated with vehicle, acyclovir (300μM; Sigma Chemical) or phosphonoacetic acid (PAA; 400μg/ml; Sigma Chemical) 30 minutes prior to infection and treatment was maintained until cells were processed at the indicated time points (Knopf, 1987; Pierce et al., 2005). To study the effect of cellular proteasome inhibition on HSV-mediated protein degradation, 293A cells were pre-treated with the proteasome inhibitors, Z-Leu-Leu-Leu (10μM; Sigma Chemical) and Clastolactacystin β-lactone (5μM; Sigma Chemical), for 10h prior to HSV-2 infection. Proteosome inhibitors were maintained throughout the infection, and cells were harvested at the indicated time points post infection. In order to determine the relative stability of the cellular STAT2 protein, cells were treated with transcriptional (actinomycin D; 2μg/ml; Sigma Chemical) and translational (cyclohexamide; 25μg/ml; Sigma Chemical) inhibitors to prevent de novo expression of STAT2 proteins. 293A cell lysates were collected at the indicated time points and analyzed by western for the relative levels of STAT2.
2.3. Effects of HSV-2 infection on IFN-mediated signaling in primary HDFa cells
HDFa cells were grown in T75s and were either mock-infected or infected with HSV-1 or HSV-2 (MOI of 5). At 16 hours post infection (hpi), cells were mock-treated or treated with 300U/ml of IFNβ and incubated at 37°C for 8 additional hours. Cells were subsequently harvested and processed for western blot analysis of ISG expression, degradation of ISGF3-associated proteins, and inhibition of STAT phosphorylation.
2.4. Reverse transcription and semi-quantitative RT-PCR
5×105 cells per well in 6 well plates were either mock-infected or infected with HSV-2 at an MOI of 10. Total RNA was isolated from the cells after 0, 4, 8, and 16 hpi using the Ambion RNAqueous-4PCR kit. 1 μg of RNA from each sample was reverse transcribed using Ambion RETROscript and oligo dT. To detect relative expression of specific genes, 2 μl of cDNA was PCR-amplified for 25 cycles using the following primer sets: STAT2: Forward-GTGCAGCTGATCCTGAAAGG, Reverse-GCTCATACTAGGGACGGGAA; STAT1: Forward-CCAGGCTCTTGATTTCATGC, Reverse-AATTCTGGAAAACGCCCAG; IRF9: Forward- TTCTGTGGAGTATGGCTGGAG, Reverse- GAGCTCTTCAGAACCGCCTA; Actin: Forward-CCTTGCACATGCCGGAG, Reverse-GCACAGAGCCTCGCCTT. 20μl of each PCR amplified product was analyzed on 2% high resolution agarose gels and visualized via ethidium bromide staining.
2.5. Promoter-reporter plasmids and luciferase assays
The induction of IFN-inducible gene expression was determined by a luciferase reporter assay. The IFNβ inducible pISRE-TA-Luc, the IFNγ inducible pGAS-TA-Luc, the control pTA-Luc promoter-reporter vectors, as well as the pSEAP2 normalization vector were obtained from Clontech. 2× 105 cells seeded in 12 well plates were transfected with 0.1μg of the specific promoter-reporter vector, 0.6μg of pSEAP2 (as a transfection normalizing control) and 0.9μg of pCMVsport-βgal vector to maintain a consistent amount of transfected DNA (1.6μg) using Lipofectamine 2000 (Invitrogen). 24 hours post transfection, cells were either mock- or HSV-2-infected (MOI=10). For studies that assessed the effects of inhibiting viral replication and late gene expression on promoter activation, PAA (400μg/ml) or acyclovir (300μM) was added 30 minutes prior to infection and was maintained until lysis. At the indicated time points (0, 4, 8, and 16h) post infection 300U/ml IFNβ (PBL Interferon Source) was added. 3h post treatment, supernatants were collected for determination of SEAP levels and cells were lysed in Luciferase Passive Lysis Buffer (Promega) for quantification of luciferase levels. SEAP levels were utilized to normalize transfection efficiency and were determined according to the manufacturer’s directions by conversion of the Quantiblue substrate (Invivogen). Experiments were performed in triplicate and reported as mean +/− standard deviation.
2.6. STAT-specific 3′ UTR plasmids and inhibition assays
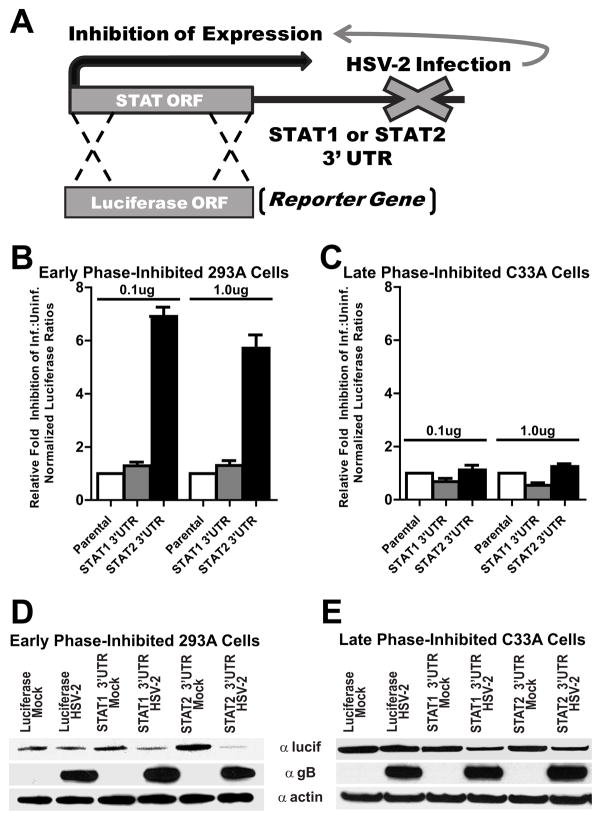
Luciferase-based reporter gene constructs that specified the 3′ untranslated region (3′UTR) of either STAT1 or STAT2 were generated by cloning the respective 3′UTR immediately downstream of a constitutively expressed firefly luciferase open reading frame as depicted in Figure 4A. 3′UTR luciferase reporter plasmids, or the parental control vector that contained no 3′UTR, were transfected with Lipofectamine 2000 (Invitrogen) either into 293A cells or C33A cells in a 12 well plate with the indicated amount of plasmid (0.1ug or 1ug). A constitutively expressed renilla luciferase was co-transfected as a normalizing control. Transfected cells were subsequently mock- infected or infected with HSV-2. At 16 hpi cells were lysed in passive lysis buffer (Promega) and analyzed via a dual luciferase assay (Promega) on a Berthold FB12 luminometer for the levels of Firefly and Renilla luciferase. Firefly luciferase activity for each sample was normalized to renilla luciferase levels and the ratio of infected normalized luciferase activity: uninfected normalized luciferase activity was determined for each plasmid. Subsequently, the value for the parental luciferase vector was arbitrarily set to 1.0. Relative fold inhibition for each of the STAT 3′UTR’s was calculated by dividing the infected:uninfected value for the specific 3′UTR by the parental vector infected:uninfected value. Experiments were carried out in triplicate and reported as mean +/− standard deviation.
Figure 4.
Cell-line dependent analysis of the influence HSV-2 infection has on the STAT2 3′UTR. (A) Diagram of the 3′UTR indicator vector constructs that specify the 3′UTR of either STAT1 or STAT2 cloned downstream of a luciferase reporter open reading frame. Luciferase assays were performed following transfection of either 0.1ug or 1.0 ug into the indicated cell lines. HSV-2-mediated inhibition through the 3′UTR is measured by normalized luciferase activity in infected cells relative to the activity of the construct in uninfected cells. The control parental luciferase vector with no 3′UTR was set to an arbitrary value of 1 and the fold inhibition of each of the 3′UTR constructs was determined relative to this value. (B) In early phase-inhibited 293A cells, HSV-2 infection inhibits luciferase activity of transcripts that specify the STAT2 3′UTR (black bars), but not the STAT1 3′UTR (gray bars). (C) In late phase-inhibited C33A cells, HSV-2 infection does not affect the relative luciferase activity from transcripts specifying either the STAT1 or STAT2 3′UTR. (D & E) The relative effect of HSV-2 infection on luciferase expression was determined for transcripts that specified no 3′ UTR (Luciferase) or either the STAT1 3′UTR or STAT2 3′UTR in early phase-inhibited 293A or late phase-inhibited C33A cells.
To examine the effect of HSV-2 infection on translation of luciferase mRNAs that specified either the STAT1 or STAT2 3′UTR, each of the above plasmids were transfected into either 293A or C33A cells and, 8h later, subsequently mock-infected or infected with HSV-2 (MOI of 5). At 16 hpi, cells were harvested and prepared for western blot analysis.
2.7. Cellular protein extraction, SDS-PAGE and western blot analysis
Cells were infected with HSV-2 at an MOI of 10 and lysates were prepared at the indicated time points. Total cell lysates were prepared in mammalian protein extraction buffer (Pierce Chemical) supplemented with 0.1% SDS. For differential cytoplasmic versus nuclear protein extractions, lysates were prepared using an NE-PER nuclear and cytoplasmic protein extraction kit (Pierce) according to the manufacturer’s instructions. All lysis buffers were supplemented with complete protease inhibitor cocktail (Roche, San Francisco, CA) and phosphatase inhibitor cocktails I and II (Sigma). Cell lysate preparations were clarified by centrifugation, normalized for protein concentration (80 μg/well), and prepared for SDS-PAGE analysis in NuPage LDS sample loading buffer containing NuPage sample reducing agent (Invitrogen). Samples were separated on NuPage 4–12% Bis-Tris gradient gels and transferred to nitrocellulose membranes for immunoblotting (Sanchez et al., 2012). Blots were blocked with 5% nonfat dry milk for 1h and probed overnight at 4 C with the indicated primary antibody at the following dilutions: α-STAT2, 1:250 (Upstate, BK502 ), α-STAT1, 1:1000 (BD, Cat:610186), α-ISGF3γ (αIRF9), 1:250 (BD, Cat:610285), α-PO4STAT2, 1:350 (BD, pY 690), α-PO4STAT1, 1:1000 (Cell Signaling, Tyr701), α-Actin, 1:5000 (Sigma, AC15), α-HSV1/2 gB, 1:5000 (Abcam, 10B7); α-luciferase, 1:500 (Santa Cruz), α-ISG15, 1:1000 (Cell Signaling, 22D2); α-Mx1/2/3, 1:1000 (Santa Cruz). Proteins were visualized by chemiluminescence using HRP conjugated anti-rabbit (1:200,000) or anti-mouse (1:50,000) secondary antibodies and Femto Supersignal chemiluminescent detection (Pierce Chemical). All antibody dilutions and washes were performed in TBS-0.05% Tween 20.
2.8. Indirect immunofluorescent microscopy
293α/β cells were grown on collagen coated glass coverslips overnight prior to infection. Cells were either mock-treated or treated with 300μM acyclovir and subsequently infected with HSV-2 at an MOI of 5. 12h later cells were incubated with vehicle or IFNβ (1000 U/ml) for 30 minutes followed by fixation/permeabilization with ice-cold 100% methanol (−20 °C) for 10 min. Cells were rehydrated in TBS for 5 min, treated with Image-it FX signal enhancer (Invitrogen), and blocked in TBS with 3% BSA/5% normal goat serum for 1h. Slides were incubated overnight at 4 °C with the primary antibodies (α-STAT2, 1:150 or α-PO4STAT2, 1:150) diluted in TBS with 1% BSA. Cells were subsequently incubated with secondary Alexafluor 488 diluted 1:750 in TBS with 0.1% BSA and Hoeschst stain (Invitrogen). All washes were done in TBS with 0.05% Tween 20. Coverslips were mounted with Prolong-Antifade Gold and visualized with a Zeiss Axio Observer Z1 inverted microscope (Sanchez et al., 2012).
3. Results
3.1. HSV-2 inhibits IFN-mediated induction of ISGs in primary human dermal fibroblasts
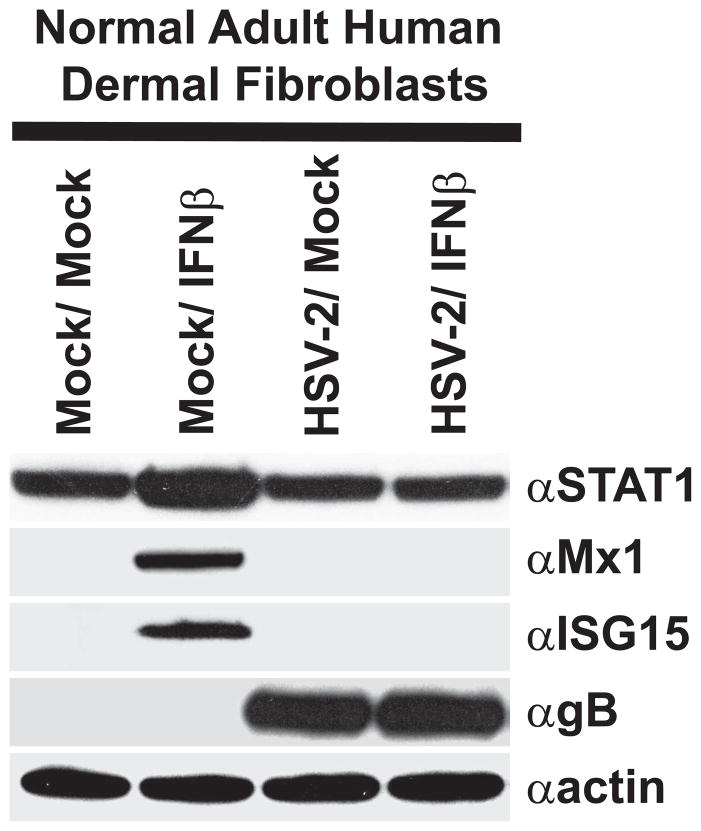
In cultured cells, Herpes simplex viruses are somewhat resistant to the antiviral effects of type-I IFN treatment (Kramer et al., 1983). IFNs facilitate inhibition of viral replication and viral protein translation through the transactivation of numerous ISGs. Therefore, the ability of HSV-2 to inhibit IFN-mediated induction of ISG expression was examined following infection of primary human dermal fibroblasts. Treatment of uninfected HDFa’s with IFNβ upregulated STAT1 expression, a component of the IFN signaling cascade, and induced expression of the cellular ISGs, Mx1 and ISG15 (Fig. 1). In contrast, in HSV-2 infected cells IFNβ treatment was unable to transactivate expression of either Mx1 or ISG15 and did not upregulate STAT1 (Fig. 1). This data suggests that HSV-2 encodes at least one mechanism for subversion of IFN-mediated induction of cell-intrinsic antiviral pathways.
Figure 1.
HSV-2 infection abrogates IFN-mediated induction of ISG expression in primary HDFa cells. Western blot analysis of ISG expression in HDFa cells that were mock-infected or infected with HSV-2 and subsequently mock-treated or treated with IFNβ. Blots were probed with antibodies to STAT1, Mx1, ISG15, HSV1/2 gB, or actin.
3.2. HSV-2 exhibits cell-line dependent differential inhibition of type-I IFN signaling
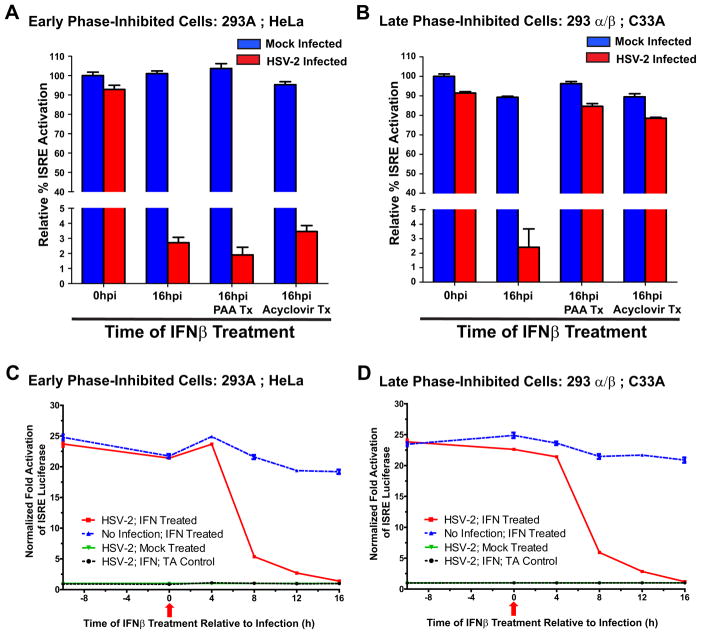
HSV-1 has previously been shown to facilitate IFN resistance by occluding type-1 IFN signaling pathways (Chee and Roizman, 2004; Melroe et al., 2004; Yokota et al., 2001). Therefore, the ability of HSV-2 to inhibit IFN–mediated JAK-STAT signaling and thereby transactivation of antiviral ISG expression was examined in a number of transformed cell-lines. All cell-lines infected with HSV-2 exhibited a marked decrease at 16 hpi in their ability to activate IFN-mediated transcriptional activation of the type-I IFN-dependent ISRE promoter (Figs. 2A & B). However, depending on the cell-line infected, a difference in the replicative phase in which HSV-2 inhibits the IFN signaling cascade was observed. In 293A and HeLa cells, inhibition of HSV-2 replication by either PAA or acyclovir did not affect HSV-2’s ability to abrogate IFN signaling (Fig. 2A). Since both PAA and acyclovir inhibit viral DNA replication and thereby late viral gene expression (Knopf, 1987; Pierce et al., 2005), this data suggests that early viral proteins, or “leaky-late” viral proteins, are fully capable of inhibiting IFN signaling in these cell-lines (hereafter designated as early phase-inhibited cells). In contrast, treatment of HSV-2 infected 293α/β or C33A cells with PAA or acyclovir abolished the ability of HSV-2 to occlude IFN-mediated signaling (Fig. 2B), indicating that early viral gene expression is not sufficient for subverting IFN signaling in these cell-lines (hereafter designated late phase-inhibited cells). Therefore, late viral gene products or late-initiated cellular events must compensate for these inadequacies. Despite the distinct differences in the HSV-2 replicative phase that mediated inhibition of IFN signaling, there were no apparent differences between cell-lines in the kinetics with which HSV-2 inhibited IFN signaling (Figs. 2C & D). All cell-lines examined demonstrated a precipitous inhibition of IFN signaling between 4 and 8 hpi with almost complete abolition of signaling by 16 hpi. Taken together, this data suggests that HSV-2 appears to affect IFN-mediated actions through distinctly different, but compensatory mechanisms and that HSV-2 encodes the ability to affect IFN signaling pathways both prior to and following viral DNA replication.
Figure 2.
HSV-2 specifies mechanisms that inhibit type-I IFN signaling both early and late in the replicative cycle. (A & B) ISRE promoter-reporter analysis of HSV-2 mediated inhibition of type-I IFN signaling in mock- (blue bars) or HSV-2- (red bars) infected 293A and HeLa (early phase-inhibited) cells (A) or 293α/β and C33A (late phase-inhibited) cells (B). Treatment of various HSV-2 infected cell-lines with the viral DNA replication inhibitors PAA or acyclovir segregates the replicative phase that HSV-2 utilizes to mediate abrogation of IFN signaling. (C & D) Kinetics of HSV-2 inhibition of IFN-mediated activation of the ISRE promoter in early phase-inhibited (C) and late phase-inhibited (D) cells. Red arrow indicates the time of HSV-2 infection (0h) relative to addition of IFNβ at −8, 0, 4, 8, 12, and 16h.
3.3. HSV-2 selectively destabilizes STAT2 transcripts in a cell-dependent manner
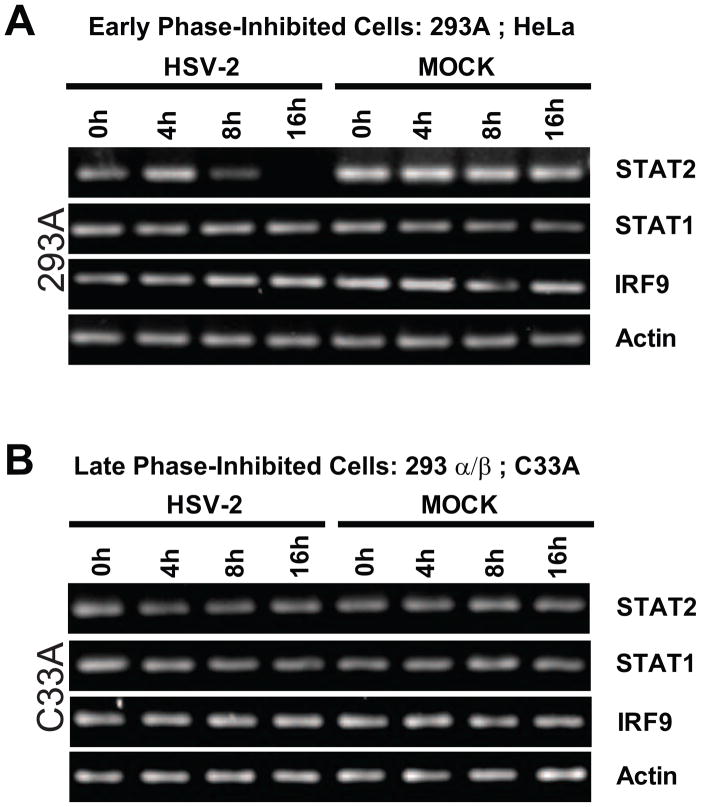
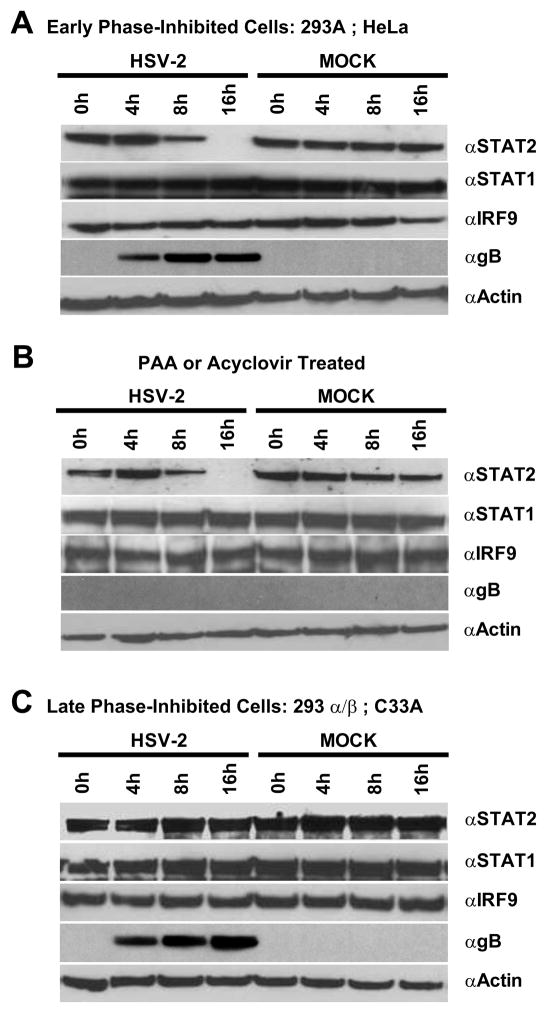
It has been demonstrated previously for HSV-1 that the UL41/VHS gene contributes to inhibition of IFN-mediated signaling pathways (Chee and Roizman, 2004). Given VHS’s role as an mRNA-specific RNase that accelerates degradation of host transcripts (Elgadi et al., 1999; Feng et al., 2005; Taddeo et al., 2004), the relative levels of transcripts for each member of the ISGF3 complex were analyzed at various time points following HSV-2 infection (Fig. 3). In early phase-inhibited 293A and HeLa cell-lines, STAT2 transcripts were significantly decreased by 8 hpi and undetectable by 16 hpi (Fig. 3A). These time points coincided with HSV-mediated inhibition of IFN signaling (Fig. 2C). By comparison, relative levels of STAT1 and IRF9 transcripts did not appear affected by HSV-2 infection at any time points examined in these cells. In contrast, late phase-inhibited 293α/β or C33A cells exhibited no apparent change in IRF9, STAT1, or STAT2 transcript levels (Fig. 3B). This data indicates that STAT2 transcripts are selectively targeted for degradation in HSV-2 infected cells that are sensitive to early phase inhibition, but are unaffected in late phase-inhibited cells, where HSV-2’s early phase inhibition mechanism does not appear to function.
Figure 3.
Semi-quantitative RT-PCR of cellular transcripts for the members of the IFN-associated ISGF3 complex (STAT2, STAT1 and IRF9) in mock- or HSV-2-infected early phase-inhibited (A) and late phase-inhibited (B) cells at 0, 4, 8, and 16 hpi.
3.4. HSV-2 infection affects STAT2 transcripts through their 3′UTR in a cell-dependent manner
The 3′UTR of specific cellular transcripts has been shown to be important for mRNA stability, as well as for regulating mRNA translation (Khabar and Young, 2007). The STAT2 transcript consists of a relatively large 3′UTR region that could serve as a potential target for HSV-mediated initiation of mRNA degradation or inhibition of protein expression. In order to assess if HSV-2 infection affected transcripts that specified the 3′UTR of STAT2, a 3′UTR luciferase reporter assay was employed (Fig. 4). Plasmids were constructed with the STAT1 or STAT2 3′UTRs cloned downstream of a parental plasmid that specified a constitutively expressed firefly luciferase open reading frame (Fig. 4A). Luciferase activity was assayed and the relative fold inhibition of luciferase activity following HSV-2 infection was determined. In early phase sensitive 293A cells, HSV-2 infection significantly inhibited luciferase activity of constructs containing the STAT2 3′UTR. However, HSV-2 infection did not significantly influence the relative activity of constructs specifying either the STAT1 3′UTR or the parental luciferase (Fig. 4B). In contrast, in late phase-inhibited C33A cells HSV-2 infection exhibited no substantial relative effect on luciferase activity for any of the constructs, including the STAT2 3′UTR (Fig. 4C). The effects of HSV-2 infection were further explored by examining luciferase protein expression from transcripts that specified either the STAT1 or STAT2 3′UTR. In the absence of any 3′UTR, HSV-2 infection had no effect on relative luciferase protein levels in either early phase- or late phase-inhibited cells (Figs. 4D & E). Following HSV-2 infection of both early and late phase-inhibited cells, luciferase protein levels from STAT1 3′UTR transcripts were only slightly, and to a similar extent, decreased (Figs. 4D & E). In uninfected early phase-inhibited 293A cells, luciferase protein expression levels were relatively higher in the STAT2 3′UTR mock-infected cells. However, in agreement with the luciferase activity data, HSV-2 infection significantly decreased the levels of translated luciferase protein from transcripts containing the STAT2 3′UTR (Fig. 4D). In contrast, in late phase-inhibited cells, HSV-2 infection only slightly decreased luciferase protein levels from transcripts that specified the STAT2 3′UTR in a manner nearly analogous to what was seen for transcripts with the STAT1 3′UTR (Fig. 4E). Therefore, HSV-2 infection has a more profound effect on the production of proteins produced from transcripts that specify the 3′UTR of STAT2 in early phase-inhibited than HSV does in late phase-inhibited cells.
3.5. HSV-mediated degradation of STAT2 transcripts is not solely responsible for alteration of STAT2 protein levels in early phase-inhibited cells
To assess if the specific degradation of STAT2 mRNA beginning at 8 hpi affected protein production, protein levels of STAT2, STAT1, and IRF9 were examined at the indicated time points post infection (Fig. 5). In concordance with transcript levels, STAT2 protein was markedly reduced 8 hpi and completely absent by 16 hpi in HSV-2 infected early phase-inhibited cells; whereas neither STAT1 nor IRF9 protein levels were affected (Fig. 5A). Likewise, in early phase-inhibited cells, abrogation of DNA replication and late gene expression by PAA treatment resulted in a comparable loss of STAT2 expression starting at 8 hpi (Fig. 5B), indicating that a late gene product was not responsible for the absence of STAT2 in these cells. Similar results were obtained with acyclovir treatment (data not shown). In contrast, HSV-2 infection of late phase-inhibited cells did not significantly affect IRF9, STAT1, or STAT2 levels, suggesting that an alternative mechanism must account for HSV-mediated inhibition of type-I interferon signaling pathways in these cell-lines.
Figure 5.
HSV-2 infection specifically affects STAT2 protein levels in early phase-inhibited (A) but not late phase-inhibited (C) 293α/β cells. HSV-2 infected 293A cells were analyzed by western blot for levels of STAT1, STAT2, IRF9, HSV gB and actin expression at 0, 4, 8, and 16 hpi. (B) In early phase-inhibited 293A cells, abrogation of HSV-2 DNA replication by PAA does not affect HSV-2-mediated loss of STAT2 expression. The same results were observed with acyclovir treatment (data not shown). (C) In late phase-inhibited 293α/β cells, STAT2 protein levels are unaffected by HSV-2 infection.
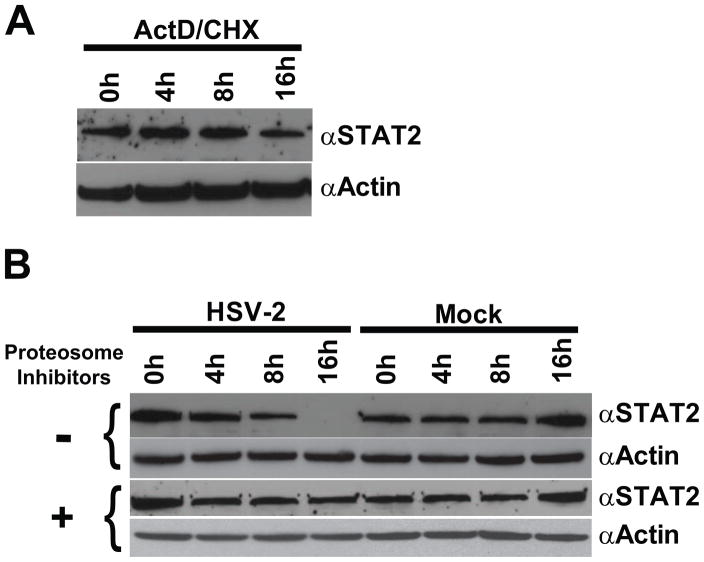
Although these protein results paralleled the cell-dependent manner of HSV-mediated degradation of STAT2 transcripts, it has previously been reported that STAT2 is an exceptionally stable protein with a half-life of greater than 24 hours (Le et al., 2008; Lee et al., 1997). In agreement with these findings, inhibition of transcription and translation by a combination of actinomycin D and cyclohexamide showed only minimal effects in 293A cells on STAT2 protein levels by 16 h post treatment (Fig 6A). Given the stability of the STAT2 protein, HSV-mediated loss of STAT2 mRNA beginning at 8 hpi could not fully account for the concomitant loss of STAT2 protein. HSV has been shown to target specific cellular proteins for proteosomal degradation (Boutell et al., 2002; Chelbi-Alix and de The, 1999; Hagglund and Roizman, 2002; Hagglund et al., 2002). To assess if proteosomal degradation is responsible for the absence of STAT2 in HSV-2 infected early phase sensitive cells, 293A cells were treated with proteasome inhibitors, infected with HSV-2, and STAT2 protein levels were assessed at the indicated time points (Fig. 6B). Treatment with proteasome inhibitors completely abrogated changes in STAT2 protein levels following HSV-2 infection. Taken together, these results indicate that in early phase-inhibited cells HSV-2 targets both STAT2 mRNA and protein via complementary approaches in order to achieve inhibition of STAT2-mediated IFN signaling.
Figure 6.
Cellular STAT2 has an exceedingly stable half-life and is therefore targeted for proteosomal degradation in HSV-2 infected cells. (A) Assessment of the stability of cellular STAT2 in 293A cells following Actinomycin D (ActD) and Cyclohexamide (CHX) inhibition of de novo STAT2 protein expression. (B) Western analysis of HSV-2’s ability to alter cellular STAT2 protein levels in the presence (+) or absence (−) of proteosome inhibitors.
3.6. HSV-2 prevents type-I IFN mediated phosphorylation of STAT2, but not STAT1, in late phase-inhibited cells
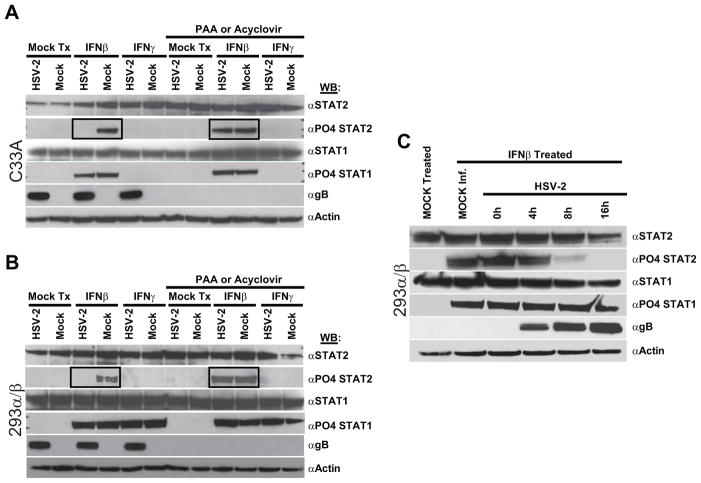
The mechanisms by which HSV-2 infection mediates subversion of IFN signaling during late phases of replication were next examined. As shown previously, HSV-2 infection did not alter STAT1 or STAT2 expression levels in late phase-inhibited C33A (Fig. 7A: αSTAT1; αSTAT2) or 293α/β (Fig. 7B) cells. Upon interaction with their cognate receptors, type-I IFNs stimulate phosphorylation of STAT1 and STAT2; whereas, type-II IFN (IFNγ) stimulates phosphorylation of only STAT1 (Kessler et al., 1990; Platanias et al., 1994; Uddin et al., 1995). To examine if HSV-2 inhibited IFN signaling via abrogation of STAT activation, cells were mock-treated, treated with type-I IFNβ, or treated with type-II IFNγ and examined for activation of STAT1 or STAT2 by phosphorylation. As expected, mock-infected cells treated with IFNβ exhibited phosphorylation of STAT1 and STAT2; however, only STAT1 was phosphorylated in IFNβ treated HSV-2 infected cells (Fig 7A and B: IFNβ), indicating that HSV-2 specifically inhibited the phosphorylation of STAT2 but not STAT1. IFNγ treatment of 293α/β cells resulted in phosphorylation of STAT1, but not STAT2, irrespective of HSV-2 infection (Fig. 7B:IFNγ), indicating that HSV-2 would not subvert type-II interferon responses through inhibition of STAT1 phosphorylation. As has been shown previously, C33A cells do not respond to IFNγ and therefore did not exhibit any STAT1 phosphorylation upon IFNγ treatment (Fig 7A: IFNγ). Phosphorylation of STAT2 following IFNβ stimulation could be re-established in HSV-2 infected C33A or 293α/β cells if they were treated with either PAA or acyclovir, suggesting that HSV-2-mediated inhibition of STAT2 activation could account for the absence of IFN signaling in late phase-inhibited cells. Examination of the kinetics of HSV-2 mediated inhibition of STAT2 phosphorylation indicated that HSV-2 occludes STAT2 phosphorylation with identical kinetics to its inhibition of IFN signaling. Phosphorylation of STAT2 was drastically inhibited at 8 hpi and completely inhibited by 16 hpi (Fig. 7C). STAT1 phosphorylation was not affected throughout the time course of the experiment.
Figure 7.
In HSV-2 infected late phase-inhibited cells, which do not degrade cellular STAT2 protein, HSV-2 infection inhibits STAT2 phosphorylation. C33A (A) or 293α/β (B) cells were mock- or HSV-2-infected and at 16 hpi were mock-treated (Mock Tx) or treated with IFNβ or IFNγ and subsequently analyzed by western for phosphorylation of STAT1 and STAT2. Parallel experiments were conducted in the presence of the viral DNA replication inhibitor PAA. Identical results were observed with acyclovir treatment (data not shown). (C) The kinetics of HSV-2 inhibition of IFNβ-mediated STAT2 phosphorylation was examined by treating HSV-2 infected cells at 0, 4, 8, and 16 hpi.
3.7. A late HSV-2 viral event inhibits type-I IFN-mediated translocation of STAT2 from the cytoplasm to the nucleus
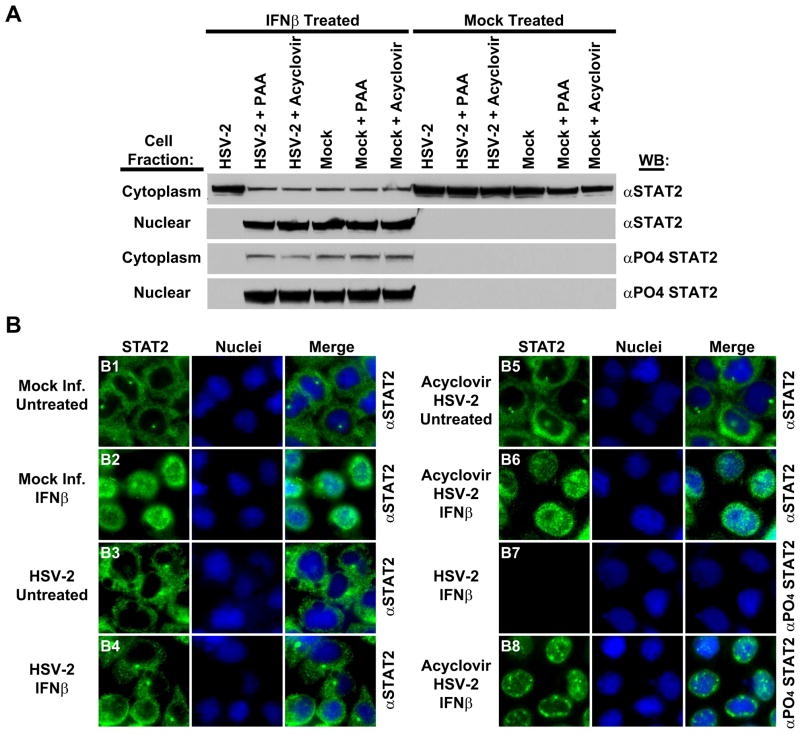
Latent STAT2 resides within the cytoplasm of cells until activated by stimuli. Upon phosphorylation, STAT2 translocates from the cytoplasm to the nucleus where it functions as a tripartite complex with STAT1 and IRF9 to initiate transactivation of ISGs (Horvath and Darnell, 1997; Kessler et al., 1990; Platanias et al., 1994). To examine cellular localization of STAT2 in HSV-2 infected late phase-inhibited cells, STAT2 translocation was ascertained by both cell fractionation (Fig. 8A) and immunofluorescent localization (Fig. 8B). In the absence of IFN treatment, STAT2 was not phosphorylated and could be found only in the cytoplasm of cells, irrespective of HSV-2 infection or other treatments (Fig. 8A: Mock Treated; Fig. 8B: B1; B3; B5). Treatment of mock-infected cells with IFNβ resulted in STAT2 phosphorylation and translocation to the nucleus (Fig. 8A: IFNβ Treated; Mock Infected; Fig. 8B2). In contrast, STAT2 was not phosphorylated and was localized only to the cytoplasm of HSV-2 infected IFNβ treated cells (Fig. 8A: IFNβ Treated; Fig. 8B: B4;B7). IFNβ stimulation of HSV-2 infected cells treated with PAA or acyclovir resulted in nuclear accumulation of phosphorylated STAT2 (Fig. 8A: IFNβ (Acyclovir; PAA); Fig. 8B: B6; B8), indicating further that the inhibition of STAT2 phosphorylation by HSV-2 is essential for control of IFN signaling in late phase-inhibited cells.
Figure 8.
HSV-2 late phase inhibition of STAT2 phosphorylation abrogates translocation of STAT2 from the cytoplasm to the nucleus. (A) Mock-infected or HSV-2-infected 293α/β cells were mock-treated or treated with IFNβ, separated into cytoplasmic and nuclear fractions, and analyzed by western for STAT2 (αSTAT2) and phosphorylated STAT2 (αPO4 STAT2) proteins. Parallel experiments were conducted in the presence of the viral DNA replication inhibitors PAA or acyclovir. (B) Immunofluorescence analysis of STAT2 (green) and phosphorylated STAT2 (green) 293α/β cellular localization under the indicated conditions. Nuclei are demarcated in blue by Hoescht staining.
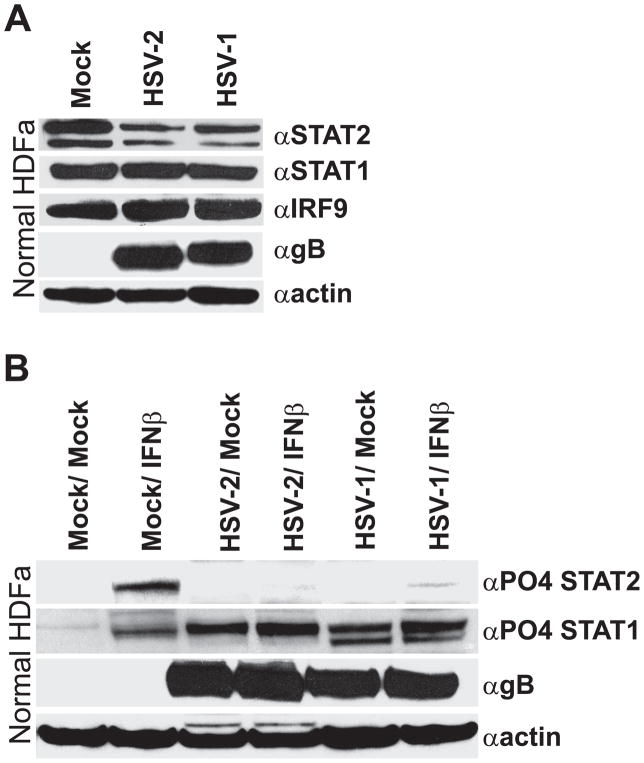
3.8. In primary HDFa cells, HSV-2 does not completely degrade STAT2, but compensates by inhibiting STAT2 phosphorylation
Permanently transformed cell-lines frequently have a number of peculiarities with regards to IFN signaling. Although these peculiarities enabled often masked late phase mechanisms to be revealed, the processes in these cells may not be indicative of what occurs in more normal cells. Similar to transformed cell-lines, in primary HDFa’s HSV-2 did not affect either STAT1 or IRF9 protein levels (Fig. 9A). However, unlike transformed cell-lines both HSV-1 and -2 infection mediated partial degradation of STAT2, reducing its levels but not completely ablating its presence (Fig. 9A). Because HSV-2 completely inhibited ISG expression in these cells (Fig. 1), the ability of HSV-2 to occlude phosphorylation of the remaining STAT2 was examined (Fig. 9B). Treatment of mock-infected HDFa cells with IFNβ induced the phosphorylation of both STAT1 and STAT2. However, detectable STAT2 phosphorylation was absent in HSV-2 infected HDFa cells and was significantly reduced in HSV-1 infected HDFa cells. Unlike transformed cells, an apparent phosphorylated STAT1 species was present following HSV-1 and HSV-2 infection, irrespective of if they had been treated with IFNβ. Taken together these results indicate that herpes simplex viruses utilize multiple complementary and compensatory approaches to completely modulate IFN signaling and subsequent expression of antiviral ISGs.
Figure 9.
In primary HDFa cells, HSV-1 and HSV-2 infection induces only partial but specific loss of STAT2 expression, but compensates by inhibiting STAT2 phosphorylation. (A) Western blot analysis of HDFa cells either mock infected or infected with HSV-2 or HSV-1. To determine relative levels of protein expression, blots were probed with antibodies to STAT2, STAT1, IRF9, HSV1/2 gB, or actin. (B) The ability of IFNβ to induce STAT1 or STAT2 phosphorylation in mock infected or in the context of an HSV-1 or HSV-2 infection was determined by western blot analysis.
4. Discussion
Type-I IFN-mediated responses are a crucial first line of defense against viral infections and are critical for generating both innate and adaptive immunity (Gallucci et al., 1999; Honda et al., 2005; Le Bon et al., 2001; Le Bon and Tough, 2002). Therefore, viruses have necessarily evolved mechanisms to impede IFN-induced expression of antiviral genes (Brukman and Enquist, 2006; Chee and Roizman, 2004; Duerst and Morrison, 2003; Garcia-Sastre and Biron, 2006; Hahm et al., 2005; Melroe et al., 2004; Yokota et al., 2001). In the present study, we examined the effects of HSV-2 infection on type-I IFN signaling in various cell-lines and discovered that HSV-2 abolished IFNβ signaling and induction of ISG expression in all cell-lines examined. Although the extent and kinetics of this inactivation was indistinguishable between the various cell-lines, distinct differences in the mechanisms HSV-2 employed to subvert IFN-signaling in a given cell-line were observed. These differences were cell-line dependent and could be segregated into two categories: 1) early replicative phase mechanisms that abrogated IFN signaling prior to DNA replication; 2) late replicative phase mechanisms that compensated for an ineffectual early phase response and were functional following viral DNA replication. However, in primary human dermal fibroblasts, both mechanisms cooperated to ensure complete inhibition of IFN-mediated ISG expression.
In transformed cell-lines that exhibited inhibition of type-I IFN signaling via an early replicative phase-inhibited process, the defining phenotype that could account for inhibition of IFN signaling is a complete loss of STAT2 expression. In these cells, STAT2 protein loss resulted from a complexly orchestrated series of events: 1) HSV-2 infection initiated the degradation of STAT2 transcripts, but did not affect either STAT1 or IRF9 mRNAs; 2) In cells that exhibited early replicative phase inhibition of IFN signaling, HSV-2 infection altered either translation or mRNA stability of transcripts that specified the 3′UTR of STAT2 more so than it affected transcripts with no 3′UTR or the 3′UTR of STAT1; 3) Nascent STAT2 proteins were targeted for cellular proteosomal degradation. It has previously been demonstrated that STAT2 protein levels are significantly decreased following infection of cells with HSV-1 (Chee and Roizman, 2004; Yokota et al., 2001). VHS was shown to be at least partially responsible for this effect, since cells infected with viruses that specified a VHS deletion did not exhibit the same degree of STAT2 disappearance as wild-type HSV-1 infected cells (Chee and Roizman, 2004). VHS is a virally encoded RNase that degrades both cellular and viral mRNAs (Elgadi et al., 1999; Feng et al., 2005; Taddeo et al., 2004) and has been shown to selectively target some specific cellular mRNAs through their 3′ UTR (Esclatine et al., 2004a; Esclatine et al., 2004b). The finding that cellular STAT2 transcripts are degraded following HSV-2 infection is in agreement with the role VHS may play in facilitating the disappearance of STAT2 protein from these cells. Furthermore, it may account mechanistically for observations that HSV-2 viruses that are deleted in VHS exhibit increased sensitivity to type-I IFNs and are severely attenuated in vivo (Duerst and Morrison, 2004; Korom et al., 2008; Murphy et al., 2003). However, our findings highlight that VHS-mediated degradation of STAT2 mRNA cannot fully account for the complete loss of cellular STAT2 protein in these cells by 16 hpi. As has been reported previously in other cell-lines (Le et al., 2008; Lee et al., 1997), STAT2 was exceedingly stable and possessed a long half-life in uninfected early phase-inhibited cells. In order to circumvent this issue, HSV-2 infection facilitated the proteosomal-dependent degradation of STAT2. The preferential targeting of STAT2 for degradation is not unique to HSV. Human parainfluenza virus 2 blocks IFN signaling by inducing proteosomal degradation of STAT2, but not STAT1, through interactions with its V protein (Parisien et al., 2002a; Parisien et al., 2001; Parisien et al., 2002b; Ulane and Horvath, 2002). HSV-2 encodes an ubiquitin ligase, ICP0, that has been shown to target other cellular proteins for proteosomal degradation (Boutell et al., 2002; Chelbi-Alix and de The, 1999; Hagglund and Roizman, 2002; Hagglund et al., 2002), and it is therefore possible that ICP0 may mediate the observed loss of STAT2 protein. In this regard, VHS and ICP0 would serve complementary functions that work in concert to prevent de novo expression of STAT2 protein via mRNA degradation and to destroy nascent STAT2 protein through targeted proteosomal degradation.
Because STAT2 is completely degraded in many transformed cell-lines, the downstream effects of HSV-2 on STAT2 could not be readily visualized. However, the finding that STAT2 expression was not affected in all HSV-2 infected cells enabled the unmasking of HSV-2 late replicative phase-mediated mechanisms of IFN signaling inhibition. Although the extent and kinetics of HSV-2 abrogation of IFN signaling were indistinguishable between cell-lines, there were distinct differences in the mechanisms utilized for late replicative phase inhibition. In HSV-2 infected late replicative phase-inhibited cells, STAT2 phosphorylation and subsequent translocation to cell nuclei was completely abolished. IFN-mediated STAT2 phosphorylation and nuclear translocation could be restored by treating infected cells with viral DNA replication inhibitors, suggesting that either late viral proteins or events initiated by HSV-2 replication block STAT2 phosphorylation. HSV-2 may specifically target STAT2 phosphorylation either by directly blocking its phosphorylation or by activating a phosphatase that can actively remove the phosphate modifications. Phosphorylated STAT2 was also not detected in infected cells treated with phosphatase inhibitors prior to infection, indicating that phosphate removal of activated STAT2 by cellular phosphatases may not be the primary mechanism initiated by HSV-2 to preclude STAT2 phosphorylation (data not shown). Therefore, it is likely that HSV-2 initiates events to inhibit the direct phosphorylation of STAT2. In addition to HSV-encoded late viral proteins that may abrogate STAT2 phosphorylation, another possibility may be that HSV-2 replication induces cellular proteins that ultimately inhibit STAT2 phosphorylation. In this regard, HSV-1 has been shown to upregulate suppressors of cytokine signaling 1 and 3 (SOCS1 & 3) expression following infection (Frey et al., 2009; Yokota et al., 2005; Yokota et al., 2004). Cellular SOCS proteins regulate type-I IFN signaling pathways by binding JAKs and thereby prevent tyrosine phosphorylation of STAT proteins (Akhtar and Benveniste, 2011). Like HSV-1, HIV-1 Tat has been shown to upregulate SOCS3 expression. Furthermore, the Tat-induced expression of SOCS3 prevents STAT2 tyrosine phosphorylation and type-I IFN signaling (Akhtar et al., 2010). It remains to be determined if a viral protein or a cellular protein accounts for the absence of STAT2 phosphorylation following IFN treatment.
Collectively, the data indicates that HSV-2 has evolved mechanisms that can abolish type-I IFN signaling throughout each replicative phase of its lifecycle. As with HSV-1, HSV-2 inhibition of type-I IFN signaling occurs through a variety of complementary and compensatory approaches. Early in infection, HSV-2 degrades IFN signaling-associated mRNAs, such as STAT2, and complements that approach with the targeted proteosomal destruction of STAT2 protein. Following viral DNA replication, mechanisms such as VHS-dependent degradation of RNA have been shown to be less efficient due to inactivation of VHS-associated RNase activities through interactions with VP16 (Lam et al., 1996; Smibert et al., 1994). As VHS-dependent RNA degradation mechanisms abate in the late replicative phases, HSV-2 may compensate for low levels of STAT2 expression by initiating events that inhibit STAT2 phosphorylation and/or translocation to cell nuclei. Additionally, if HSV-2 encounters cells or cell-types where one mechanism does not efficiently function, it has a redundant capacity to inhibit type-I IFN-mediated antiviral gene expression. Both early and late mechanisms, each in of themselves, appear capable of inhibiting type-I IFN signaling and induction of ISG expression with the same kinetics and effectiveness. HSV-2, like HSV-1, most likely interferes with type-I IFN signaling by additional mechanisms that are not STAT2- or ISGF3-associated. Combined, these mechanisms appear to function cooperatively to completely ablate type-I IFN signaling at all replicative phases and enable efficient HSV-2 gene expression, viral genomic replication and cell-to-cell spread. These complementary and compensatory mechanisms were shown to be especially important in primary human fibroblasts. HSV-2 compensated for its inability to fully degrade STAT2 by complementing partial degradation with abolishment of STAT2 phosphorylation. This finding emphasizes the importance that these multiple mechanisms play for complete control of IFN-mediated antiviral responses in the host.
Highlights.
HSV-2 targeted cellular STAT2 to interfere with type-I interferon signaling.
HSV-2 specified both early and late phase mechanisms to inhibit interferon signaling.
Early HSV-2 mechanisms resulted in the absence of STAT2 from infected cells.
The absence of STAT2 required multiple complementary early phase mechanisms.
Late HSV-2 mechanisms inhibited STAT2 phosphorylation and translocation to nuclei.
Acknowledgments
The authors thank Dr. Augusto Ochoa and the Stanley S. Scott Cancer Center for critical financial and professional development support. This work was supported by a Louisiana Board of Regents Research Competitiveness Award LEQSF-RD-A-13 and by the National Institutes of Health from the National Center for Research Resources (P20RR021970) and the National Institute of General Medical Sciences (P20GM103501). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources, the National Institute of General Medical Sciences, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar LN, Benveniste EN. Viral exploitation of host SOCS protein functions. J Virol. 2011;85(5):1912–1921. doi: 10.1128/JVI.01857-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar LN, Qin H, Muldowney MT, Yanagisawa LL, Kutsch O, Clements JE, Benveniste EN. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J Immunol. 2010;185(4):2393–2404. doi: 10.4049/jimmunol.0903563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay SK, Leonard GT, Jr, Bandyopadhyay T, Stark GR, Sen GC. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270(33):19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76(2):841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukman A, Enquist LW. Suppression of the interferon-mediated innate immune response by pseudorabies virus. J Virol. 2006;80(13):6345–6356. doi: 10.1128/JVI.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassady KA, Gross M, Roizman B. The herpes simplex virus US11 protein effectively compensates for the gamma1(34.5) gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J Virol. 1998;72(11):8620–8626. doi: 10.1128/jvi.72.11.8620-8626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee AV, Roizman B. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J Virol. 2004;78(8):4185–4196. doi: 10.1128/JVI.78.8.4185-4196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix MK, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18(4):935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- Chou J, Roizman B. Herpes simplex virus 1 gamma(1)34.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci U S A. 1994;91(12):5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady CD, Halford WP, Carr DJ. Loss of the type I interferon pathway increases vulnerability of mice to genital herpes simplex virus 2 infection. J Virol. 2011;85(4):1625–1633. doi: 10.1128/JVI.01715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277(5332):1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Duerst RJ, Morrison LA. Innate immunity to herpes simplex virus type 2. Viral Immunol. 2003;16(4):475–490. doi: 10.1089/088282403771926300. [DOI] [PubMed] [Google Scholar]
- Duerst RJ, Morrison LA. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology. 2004;322(1):158–167. doi: 10.1016/j.virol.2004.01.019. [DOI] [PubMed] [Google Scholar]
- Elgadi MM, Hayes CE, Smiley JR. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol. 1999;73(9):7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esclatine A, Taddeo B, Evans L, Roizman B. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc Natl Acad Sci U S A. 2004a;101(10):3603–3608. doi: 10.1073/pnas.0400354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esclatine A, Taddeo B, Roizman B. The UL41 protein of herpes simplex virus mediates selective stabilization or degradation of cellular mRNAs. Proc Natl Acad Sci U S A. 2004b;101(52):18165–18170. doi: 10.1073/pnas.0408272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Everly DN, Jr, Read GS. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J Virol. 2005;79(15):9651–9664. doi: 10.1128/JVI.79.15.9651-9664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey KG, Ahmed CM, Dabelic R, Jager LD, Noon-Song EN, Haider SM, Johnson HM, Bigley NJ. HSV-1-induced SOCS-1 expression in keratinocytes: use of a SOCS-1 antagonist to block a novel mechanism of viral immune evasion. J Immunol. 2009;183(2):1253–1262. doi: 10.4049/jimmunol.0900570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci S, Lolkema M, Matzinger P. Natural adjuvants: endogenous activators of dendritic cells. Nat Med. 1999;5(11):1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312(5775):879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Hagglund R, Roizman B. Characterization of the novel E3 ubiquitin ligase encoded in exon 3 of herpes simplex virus-1-infected cell protein 0. Proc Natl Acad Sci U S A. 2002;99(12):7889–7894. doi: 10.1073/pnas.122246999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagglund R, Van Sant C, Lopez P, Roizman B. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2002;99(2):631–636. doi: 10.1073/pnas.022531599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22(2):247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Halford WP, Puschel R, Rakowski B. Herpes simplex virus 2 ICP0 mutant viruses are avirulent and immunogenic: implications for a genital herpes vaccine. PLoS One. 2010;5(8):e12251. doi: 10.1371/journal.pone.0012251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harle P, Sainz B, Jr, Carr DJ, Halford WP. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology. 2002;293(2):295–304. doi: 10.1006/viro.2001.1280. [DOI] [PubMed] [Google Scholar]
- He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A. 1997;94(3):843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis. I Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci U S A. 1975;72(4):1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CM. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur J Biochem. 2004;271(23–24):4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- Horvath CM, Darnell JE. The state of the STATs: recent developments in the study of signal transduction to the nucleus. Curr Opin Cell Biol. 1997;9(2):233–239. doi: 10.1016/s0955-0674(97)80067-1. [DOI] [PubMed] [Google Scholar]
- Improta T, Schindler C, Horvath CM, Kerr IM, Stark GR, Darnell JE., Jr Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc Natl Acad Sci U S A. 1994;91(11):4776–4780. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Knipe DM. Herpes simplex virus-1 infection causes the secretion of a type I interferon-antagonizing protein and inhibits signaling at or before Jak-1 activation. Virology. 2010;396(1):21–29. doi: 10.1016/j.virol.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Song B, Knipe DM. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology. 2008;374(2):487–494. doi: 10.1016/j.virol.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler DS, Veals SA, Fu XY, Levy DE. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 1990;4(10):1753–1765. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- Khabar KS, Young HA. Post-transcriptional control of the interferon system. Biochimie. 2007;89(6–7):761–769. doi: 10.1016/j.biochi.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf CW. The herpes simplex virus type 1 DNA polymerase gene: site of phosphonoacetic acid resistance mutation in strain Angelotti is highly conserved. J Gen Virol. 1987;68 ( Pt 5):1429–1433. doi: 10.1099/0022-1317-68-5-1429. [DOI] [PubMed] [Google Scholar]
- Korom M, Wylie KM, Morrison LA. Selective ablation of virion host shutoff protein RNase activity attenuates herpes simplex virus 2 in mice. J Virol. 2008;82(7):3642–3653. doi: 10.1128/JVI.02409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MJ, Dennin R, Kramer C, Jones G, Connell E, Rolon N, Gruarin A, Kale R, Trown PW. Cell and virus sensitivity studies with recombinant human alpha interferons. J Interferon Res. 1983;3(4):425–435. doi: 10.1089/jir.1983.3.425. [DOI] [PubMed] [Google Scholar]
- Lam Q, Smibert CA, Koop KE, Lavery C, Capone JP, Weinheimer SP, Smiley JR. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15(10):2575–2581. [PMC free article] [PubMed] [Google Scholar]
- Le Bon A, Schiavoni G, D’Agostino G, Gresser I, Belardelli F, Tough DF. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14(4):461–470. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14(4):432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- Le VT, Trilling M, Wilborn M, Hengel H, Zimmermann A. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J Gen Virol. 2008;89(Pt 10):2416–2426. doi: 10.1099/vir.0.2008/001669-0. [DOI] [PubMed] [Google Scholar]
- Lee CK, Bluyssen HA, Levy DE. Regulation of interferon-alpha responsiveness by the duration of Janus kinase activity. J Biol Chem. 1997;272(35):21872–21877. doi: 10.1074/jbc.272.35.21872. [DOI] [PubMed] [Google Scholar]
- Leib DA, Harrison TE, Laslo KM, Machalek MA, Moorman NJ, Virgin HW. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J Exp Med. 1999;189(4):663–672. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib DA, Machalek MA, Williams BR, Silverman RH, Virgin HW. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc Natl Acad Sci U S A. 2000;97(11):6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Garcia-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12(2–3):143–156. doi: 10.1016/s1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- Melroe GT, DeLuca NA, Knipe DM. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol. 2004;78(16):8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman KL, Saffran HA, Smiley JR. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J Virol. 2000;74(4):2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman KL, Smiley JR. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J Virol. 2002;76(4):1995–1998. doi: 10.1128/JVI.76.4.1995-1998.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott KR, Underhill D, Wechsler SL, Town T, Ghiasi H. A role for the JAK-STAT1 pathway in blocking replication of HSV-1 in dendritic cells and macrophages. Virol J. 2009;6:56. doi: 10.1186/1743-422X-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JA, Duerst RJ, Smith TJ, Morrison LA. Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J Virol. 2003;77(17):9337–9345. doi: 10.1128/JVI.77.17.9337-9345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Ando Y, Soushi S, Kurata T, Arao Y. The BglII-N fragment of herpes simplex virus type 2 contains a region responsible for resistance to antiviral effects of interferon. J Gen Virol. 1998;79 ( Pt 3):565–572. doi: 10.1099/0022-1317-79-3-565. [DOI] [PubMed] [Google Scholar]
- Paladino P, Collins SE, Mossman KL. Cellular localization of the herpes simplex virus ICP0 protein dictates its ability to block IRF3-mediated innate immune responses. PLoS One. 2010;5(4):e10428. doi: 10.1371/journal.pone.0010428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino P, Mossman KL. Mechanisms employed by herpes simplex virus 1 to inhibit the interferon response. J Interferon Cytokine Res. 2009;29(9):599–607. doi: 10.1089/jir.2009.0074. [DOI] [PubMed] [Google Scholar]
- Parisien JP, Lau JF, Horvath CM. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J Virol. 2002a;76(13):6435–6441. doi: 10.1128/JVI.76.13.6435-6441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien JP, Lau JF, Rodriguez JJ, Sullivan BM, Moscona A, Parks GD, Lamb RA, Horvath CM. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology. 2001;283(2):230–239. doi: 10.1006/viro.2001.0856. [DOI] [PubMed] [Google Scholar]
- Parisien JP, Lau JF, Rodriguez JJ, Ulane CM, Horvath CM. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J Virol. 2002b;76(9):4190–4198. doi: 10.1128/JVI.76.9.4190-4198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasieka TJ, Baas T, Carter VS, Proll SC, Katze MG, Leib DA. Functional genomic analysis of herpes simplex virus type 1 counteraction of the host innate response. J Virol. 2006;80(15):7600–7612. doi: 10.1128/JVI.00333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasieka TJ, Lu B, Leib DA. Enhanced pathogenesis of an attenuated herpes simplex virus for mice lacking Stat1. J Virol. 2008;82(12):6052–6055. doi: 10.1128/JVI.00297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Zhu J, Klock A, Phasouk K, Huang ML, Koelle DM, Wald A, Corey L. Evasion of the mucosal innate immune system by herpes simplex virus type 2. J Virol. 2009;83(23):12559–12568. doi: 10.1128/JVI.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AT, DeSalvo J, Foster TP, Kosinski A, Weller SK, Halford WP. Beta interferon and gamma interferon synergize to block viral DNA and virion synthesis in herpes simplex virus-infected cells. J Gen Virol. 2005;86(Pt 9):2421–2432. doi: 10.1099/vir.0.80979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC, Fish EN. Signaling pathways activated by interferons. Exp Hematol. 1999;27(11):1583–1592. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- Platanias LC, Uddin S, Colamonici OR. Tyrosine phosphorylation of the alpha and beta subunits of the type I interferon receptor. Interferon-beta selectively induces tyrosine phosphorylation of an alpha subunit-associated protein. J Biol Chem. 1994;269(27):17761–17764. [PubMed] [Google Scholar]
- Roizman B, Kozak M, Honess RW, Hayward G. Regulation of herpesvirus macromolecular synthesis: evidence for multilevel regulation of herpes simplex 1 RNA and protein synthesis. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):687–701. doi: 10.1101/sqb.1974.039.01.083. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez RL, Ramsay AJ, Foster TP. Efficient generation and rapid isolation via stoplight recombination of Herpes simplex viruses expressing model antigenic and immunological epitopes. J Virol Methods. 2012;179(1):116–126. doi: 10.1016/j.jviromet.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A. 1992;89(16):7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Popova B, Xiao P, Capone JP, Smiley JR. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J Virol. 1994;68(4):2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Su YH, Oakes JE, Lausch RN. Mapping the genetic region coding for herpes simplex virus resistance to mouse interferon alpha/beta. J Gen Virol. 1993;74 ( Pt 11):2325–2332. doi: 10.1099/0022-1317-74-11-2325. [DOI] [PubMed] [Google Scholar]
- Taddeo B, Esclatine A, Roizman B. Post-transcriptional processing of cellular RNAs in herpes simplex virus-infected cells. Biochem Soc Trans. 2004;32(Pt 5):697–701. doi: 10.1042/BST0320697. [DOI] [PubMed] [Google Scholar]
- Uddin S, Chamdin A, Platanias LC. Interaction of the transcriptional activator Stat-2 with the type I interferon receptor. J Biol Chem. 1995;270(42):24627–24630. doi: 10.1074/jbc.270.42.24627. [DOI] [PubMed] [Google Scholar]
- Ulane CM, Horvath CM. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304(2):160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- Yokota S, Yokosawa N, Kubota T, Suzutani T, Yoshida I, Miura S, Jimbow K, Fujii N. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phosphorylation of STATs and janus kinases during an early infection stage. Virology. 2001;286(1):119–124. doi: 10.1006/viro.2001.0941. [DOI] [PubMed] [Google Scholar]
- Yokota S, Yokosawa N, Okabayashi T, Suzutani T, Fujii N. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 confers efficient viral replication. Virology. 2005;338(1):173–181. doi: 10.1016/j.virol.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Yokota S, Yokosawa N, Okabayashi T, Suzutani T, Miura S, Jimbow K, Fujii N. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 contributes to inhibition of the interferon signaling pathway. J Virol. 2004;78(12):6282–6286. doi: 10.1128/JVI.78.12.6282-6286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]