Abstract
p24 proteins are a family of type I membrane proteins localized to compartments of the early secretory pathway and to coat protein I (COPI)- and COPII-coated vesicles. They can be classified, by sequence homology, into four subfamilies, named p24α, p24β, p24γ, and p24δ. In contrast to animals and fungi, plants contain only members of the p24β and p24δ subfamilies. It has previously been shown that transiently expressed red fluorescent protein (RFP)–p24δ5 localizes to the endoplasmic reticulum (ER) as a consequence of highly efficient COPI-based recycling from the Golgi apparatus. Using specific antibodies, endogenous p24δ5 has now been localized to the ER and p24β2 to the Golgi apparatus in Arabidopsis root tip cells by immunogold electron microscopy. The relative contributions of the cytosolic tail and the luminal domains to p24δ5 trafficking have also been characterized. It is demonstrated that whereas the dilysine motif in the cytoplasmic tail determines the location of p24δ5 in the early secretory pathway, the luminal domain may contribute to its distribution downstream of the Golgi apparatus. By using knock-out mutants and co-immunoprecipitation experiments, it is shown that p24δ5 and p24β2 interact with each other. Finally, it is shown that p24δ5 and p24β2 exhibit coupled trafficking at the ER–Golgi interface. It is proposed that p24δ5 and p24β2 interact with each other at ER export sites for ER exit and coupled transport to the Golgi apparatus. Once in the Golgi, p24δ5 interacts very efficiently with the COPI machinery for retrograde transport back to the ER.
Keywords: Arabidopsis, coat protein (COP) I, coat protein (COP) II, ER–Golgi transport, p24 proteins, secretory pathway
Introduction
p24 proteins constitute a family of small (20–25 kDa) type I membrane proteins which in mammals and yeast localize to the endoplasmic reticulum (ER), the ER–Golgi intermediate compartment (ERGIC), the cis-Golgi network (CGN), or the Golgi apparatus (Stamnes et al., 1995; Belden and Barlowe, 1996; Blum et al., 1996, 1999; Sohn et al., 1996; Nickel et al., 1997; Rojo et al., 1997; Dominguez et al., 1998; Füllerkrug et al., 1999; Gommel et al., 1999; Emery et al., 2000; Rojo et al., 2000). They are also major constituents of both coat protein I (COPI)- (Stamnes et al., 1995; Sohn et al., 1996; Gommel et al., 1999) and COPII- (Schimmöller et al., 1995; Belden and Barlowe, 1996) coated vesicles. All p24 proteins share the same topology: an N-terminal signal sequence, a large luminal portion, which includes the GOLD (GOLgi Dynamics) and coiled-coil domains, a single transmembrane domain, and a short (12–18 amino acids) cytoplasmic C-terminus (for a review, see Strating and Martens, 2009). The coiled-coil domain of p24 proteins enables intermolecular interactions between copies of the same protein, but also between different p24 proteins (Fullerkrug et al., 1999; Gommel et al., 1999; Marzioch et al. 1999; Jenne et al., 2002). Indeed it has been proposed that oligomerization is required for the proper localization of p24 proteins (Emery et al., 2000; Ciufo and Boyd, 2000). The luminal GOLD domain, present in several proteins involved in Golgi dynamics, is predicted to be involved in specific protein–protein interactions and has been postulated to interact with putative cargo proteins (Anantharaman and Aravind, 2002; Carney and Bowen, 2004).
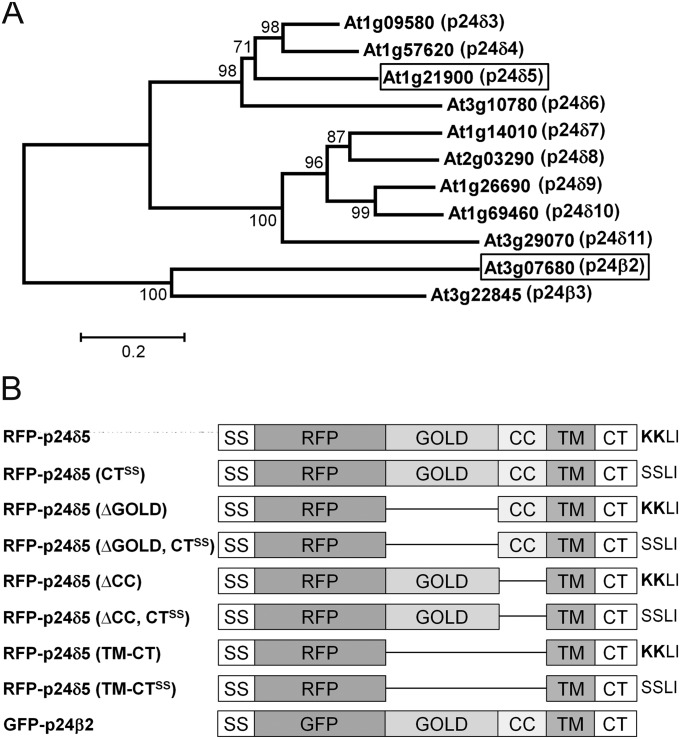
In the past it has been customary in mammalian and yeast p24 research to use individual, sometimes unrelated names for p24 orthologues (e.g. p23, p24, p25, p26, in mammals; Erp1-6p; Emp24p, Erv25p in yeast). However, a more systematic nomenclature has now emerged, first proposed by Dominguez et al. (1998) and recently updated in Strating et al. (2009), which places p24 proteins into one of four subfamilies named p24α, p24β, p24γ, and p24δ. Whereas animals and fungi have representatives in all four subfamilies, plants have only members of the p24δ (nine in Arabidopsis) and the p24β (two in Arabidopsis) subfamilies (Carney and Bowen, 2004). Following this nomenclature, Arabidopsis p24 proteins have been named here with a Greek letter (to identify the subfamily) followed by a number (Fig. 1A). In the case of the delta subfamily, two non-plant members had already been called p24δ1 and 2 (Strating et al., 2009). Therefore, the nine Arabidopsis members were named p24δ3–p24δ11 (in the order they appear in the phylogenetic tree) (Fig. 1A). In the case of the beta subfamily, there was already a p24β1 (Strating et al., 2009), and thus the two Arabidopsis members were named p24β2 and p24β3 (Fig. 1A).
Fig. 1.
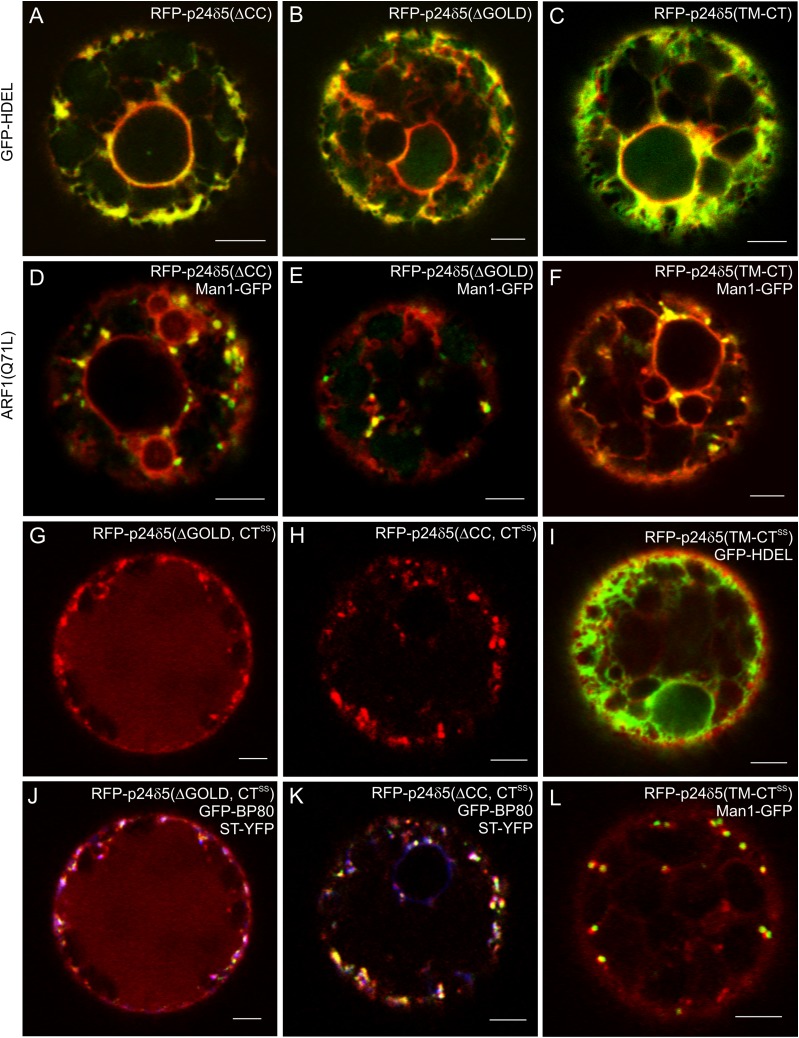
The p24 family in Arabidopsis. (A) A phylogenetic tree containing the δ and β subfamilies of p24 proteins in Arabidopsis. A multiple alignment of the p24 proteins was constructed using ClustalW and the tree was generated from this alignment using the Molecular Evolutionary Genetics Analysis software (MEGA, version 5.03) with the Neighbor–Joining method. The numbers beside the branches represent bootstrap percentage based on 1000 replications. The names assigned to these proteins, following the nomenclature proposed by Dominguez et al. (1998), are shown next to the AGI code. The two proteins analysed in this study, p24δ5 and p24β2, are highlighted. (B) Schematic representation of the RFP–p24δ5 and GFP–p24β2 constructs used in this study, including the different domains of p24δ5 and p24β2. SS, signal sequence; RFP/GFP, red or green fluorescent proteins; GOLD, domain involved in Golgi dynamics (see text for details); CC, coiled-coil domain; TM, transmembrane domain; CT, cytoplasmic tail, which in p24δ5 contains a dilysine (KKXX) motif; CTSS, cytoplasmic tail with the two lysines replaced by serines; ΔGOLD or ΔCC, deletion mutants lacking the GOLD or the CC domains; TM-CT, deletion mutant containing only the transmembrane domain and the cytosolic tail.
Although this classification is based upon sequence alignments for the whole protein, some specific features can be seen in the cytosolic C-terminal tails of the members of the different subfamilies. The cytoplasmic tail of most p24 proteins contains a pair of aromatic residues which has been shown to bind COPII subunits and thus may function as an ER export signal (Dominguez et al., 1998; Contreras et al., 2004b; Aniento et al., 2006). Some p24 proteins also have a canonical dilysine motif which binds COPI and mediates Golgi to ER retrograde transport (Cosson and Letourneur, 1994; Letourneur et al., 1994; Fiedler et al., 1996; Langhans et al., 2008). This is the case for mammalian p25 proteins (p24α1–p24α3), which contain a dilysine motif in the optimal position (-3,-4) for COPI binding (Teasdale and Jackson, 1996; Dominguez et al., 1998). This causes these proteins to locate mainly to the ER or the ERGIC in mammalian cells (Wada et al., 1991; Dominguez et al., 1998; Lavoie et al., 1999; Marzioch et al., 1999; Emery et al., 2000, 2003). Mammalian p23 (p24δ1) has an additional C-terminal residue, which brings the dilysine motif into a suboptimal position for COPI binding, and mainly localizes to the CGN (Nickel et al., 1997; Rojo et al., 1997, 2000; Blum and Lepier, 2008). In mammalian p24 (p24β1), lysines are replaced by arginines (p24), which do not bind coatomer (Cosson and Letourneur, 1994; Contreras et al., 2004a ). As a result, this protein does not localize to the ER but is usually found in the Golgi apparatus (Dominguez et al., 1998; Blum et al., 1999; Gommel et al., 1999; Emery et al., 2000). Interestingly, all Arabidopsis p24 proteins of the delta subfamily (p24δ3–p24δ11) contain a C-terminal tail with the characteristics of mammalian p25 (p24α1–p24α3) proteins, including a dilysine motif in the -3,-4 position and a diaromatic/large hydrophobic motif in the -7,-8 position. One of them, p24δ5, has been shown to localize exclusively to the ER in the steady state (Langhans et al., 2008).
Numerous functions have been ascribed to p24 proteins, including quality control of protein movement through the secretory pathway (Wen and Greenwald, 1999; Belden and Barlowe, 2001), cargo protein selection and packaging into transport vesicles (Schimmöller et al., 1995; Muniz et al., 2000; Takida et al., 2008; Castillon et al., 2011; Fujita et al., 2011), the formation of ER exit sites (ERESs), and the biogenesis and maintenance of the Golgi apparatus (Lavoie et al., 1999; Mitrovic et al., 2008; Koegler et al., 2010). These few examples indicate that p24 proteins are one of the most interesting groups of proteins involved in regulating the structure and function of the organelles of the secretory pathway. A cargo receptor function for p24 proteins has often been suggested (see, for example, Schimmoller et al., 1995), but the identity of putative cargoes has remained elusive. Indeed, only two types of cargoes have been shown to interact with p24 proteins, none of them being a real soluble cargo (as originally postulated). These are glycosylphosphatidylinositol (GPI)-anchored proteins, both in yeast (Schimmoller et al., 1995; Muniz et al., 2000, 2001; Castillon et al., 2011; Fujita et al., 2011) and in mammals (Takida et al. 2008), and G protein-coupled receptors (protease-activated receptors PAR-1 and PAR-2, nucleotide P2Y receptors, and μ-opioid receptors) (Luo et al., 2007, 2011).
Despite the difficulty in the identification of the cargoes, p24 proteins have been shown recently to be involved in a variety of specific functions in animal cells, including early embryonic mouse development (Denzel et al., 2000) and morphogenesis of the mouse embryo and placenta (Jerome-Majewska et al., 2010), insulin biosynthesis and subsequent secretion in pancreatic beta cells (Zhang and Volchuk, 2010), or amyloid precursor metabolism and pathogenesis of Alzheimer disease (Chen et al., 2006; Vetrivel et al., 2007; Hasegawa et al., 2010). In addition, several studies have recently been performed investigating the role of p24 proteins in the secretory pathway of Xenopus laevis (Strating and Martens, 2009; Strating et al., 2010) and Drosophila (Buechling et al., 2011; Port et al., 2011). Therefore, p24 proteins are emerging as very important players in many cellular processes in mammalian cells dependent upon the secretory pathway. In contrast, very little work has been performed on p24 proteins in plants. Only three papers have been published so far: two examining the roles of COPI- and COPII-binding motifs in the cytosolic tail of an Arabidopsis thaliana p24 protein (At1g21900) (here named p24δ5), the other demonstrating that this protein is localized to the ER as a consequence of highly efficient COPI-based recycling from the Golgi apparatus (Contreras et al., 2004a , b ; Langhans et al., 2008). Given the peculiarities of the p24 family and also the different organization of the early secretory pathway in plants, it was deemed necessary to investigate in further detail the localization and trafficking properties of Arabidopsis p24 proteins, as a first step towards their functional characterization.
Following up on previous studies (Contreras et al., 2004a , b ; Langhans et al., 2008), the localization of endogenous p24 proteins in Arabidopsis, including that of a member of each of the two subfamilies (p24δ5 and p24β2), has now been investigated. It was found that p24δ5 localizes mainly to the ER but also to the cis-Golgi, while p24β2 localizes mainly to the Golgi apparatus. In addition the relative contributions of the cytosolic tail and the luminal (GOLD and coiled-coil) domains to p24δ5 trafficking have been characterized. It was found that the cytosolic tail, containing the dilysine motif, is alone sufficient for ER localization at steady state and trafficking of p24δ5 in the early secretory pathway, while the luminal domains may contribute to its transport beyond the Golgi apparatus. Finally, data are presented which suggest that p24δ5 and p24β2 interact with each other and exhibit coupled trafficking at the ER–Golgi interface.
Materials and methods
Plant material
Arabidopsis thaliana ecotype Columbia (Col-0) and T-DNA mutant plants were grown in growth chambers as previously described (Ortiz-Masia et al., 2007). For immunogold electron microscopy, seedlings were grown on Murashige and Skoog (MS) medium containing 0.5% agar, and the roots were harvested after 5 d. To obtain a membrane fraction from Arabidopsis roots, seedlings were grown in liquid MS medium for 15 d. Arabidopsis thaliana cell suspension cultures (LT87) (Axelos et al., 1992) were cultivated as described (Ortiz-Zapater et al., 2006). Tobacco plants (Nicotiana tabacum var. SR1) were grown as described (Pimpl et al., 2006).
Recombinant plasmid production
The coding sequence of green fluorescent protein (GFP)–p24β2 was commercially synthesized de novo (Geneart AG) based on the sequence of GFP and that of the Arabidopsis p24 protein At3g07680 (p24β2). The sequence of the fluorophore is behind the coding sequences of the p24 signal sequence (SS) and the 5' extreme end of the mature p24 coding sequence. The ΔCC, ΔGOLD, ΔCC-CTSS, ΔGOLD-CTSS, TMCT, and TMCT-CTSS mutants of red fluorescent protein (RFP)–p24δ5 were obtained by 3' deletion of RFP–p24δ5 and synthesis of a new sequence (Langhans et al., 2008). The coding sequences of XFP–p24β2/δ5 were cloned into the pBP30 vector (carrying the 35S promoter, Nebenführ et al., 1999) through BglII/NotI.
Plasmids encoding marker proteins were: RFP–p24δ5 and RFP–p24δ5-CTSS (mutant1) (Langhans et al., 2008), ST–yellow fluorescent protein (YFP) (Brandizzi et al., 2002), Man1–RFP and Man1–GFP (Nebenführ et al., 1999), GFP–HDEL (Nebenführ et al., 2000), ARA6–RFP (Ueda et al., 2004), GFP–BP80 (daSilva et al., 2005), and ARF1(Q71L) (Pimpl et al., 2003). All RFP-tagged proteins were tagged with monomeric RFP (mRFP) to prevent oligomerization. Similarly, only mGFP5 was used for GFP-tagged proteins.
Isolation of protoplasts and transient gene expression
Mesophyll protoplasts from N. tabacum var. SR1 leaf cells were isolated and transfected as previously described (Bubeck et al., 2008). Unless otherwise stated, 1–50 μg of plasmid DNA was transfected and expressed for 20 h. Protoplasts from A. thaliana (LT87) cell suspension cultures were isolated as previously described (Axelos et al., 1992).
Generation of antibodies
Rabbit antibodies against different Arabidopsis p24 proteins were generated by Eurogentec (Belgium, http://www.eurogentec.com) using as antigen peptides corresponding either to the N- or to the C-terminus of the indicated proteins: p24δ5-Ct (YLKRYFHKKKLI), p24δ5-Nt (IWLTIPTTGG), p24β2-Nt (IRFVIDREE), and p24β2-Ct (LFERKLGMSRV). Affinity-purified antibodies (Eurogentec) were used for immunogold labelling as well as for covalent binding to magnetic beads in co-immunoprecipitation experiments (see below).
Confocal microscopy and immunofluorescence labelling
Imaging was performed using a Zeiss Axiovert LSM510 Meta confocal laser scanning microscope. At the Metadetector, the main beam splitters (HFT) 458/514 and 488/543 were used. The following fluorophores (excited and emitted by frame switching in the multitracking mode) were used: GFP (488 nm/496–518 nm), YFP (514 nm/529–550 nm), and RFP (543 nm/593–636 nm). To verify the co-localization of fluorescent signals obtained by fast frame switching, parallel observations were performed in the line switching mode. As shown in Supplementary Fig. S8 available at JXB online, there was no difference in the localization of the fluorescent signals, thus eliminating possible false co-localization events caused by the movement of organelles. Post-acquisition image processing was performed using the Zeiss LSM 5 image Browser (4.2.0.121) and CorelDrawX4 (14.0.0.567) or ImageJ (v.1.45m).
Immunogold electron microscopy
Root tips from Arabidopsis were high pressure frozen, freeze substituted, embedded, labelled, and post-stained as described (Bubeck et al., 2008). Affinity-purified antibodies were used at the following dilutions: Ct-p24β2 (1:100); Nt-p24δ5 (1:10); and Ct-p24δ5 (1:10). Micrographs were taken with a JEM1400 transmitting electron microscope operating at 80 kV using a TVIPS F214 digital camera.
Preparation of membrane extracts and co-immunoprecipitation
Membrane fractions were obtained from Arabidopsis cell suspension cultures (LT87) or from Arabidopsis roots. Cells were collected by centrifugation and washed twice in homogenization buffer [50 mM TRIS-HCl, pH 7.5, 0.3 M sucrose, 10 mM KCl, 1 mM dithiothreitol (DTT), 3 mM EDTA, and a cocktail of protease inhibitors (Sigma)]. Pellets were resuspended 1:1 (v/v) in homogenization buffer and cells disrupted by sonication (4×15 s). Cell extracts were separated from unbroken cells by centrifugation (10 min at 2000 g). Membranes were pelleted by centrifugation of the extracts for 1 h at 150 000 g. Arabidopsis roots (from either wild-type or mutant plants) were homogenized in homogenization buffer using a mortar and a pestle, and membrane fractions were obtained as above. Membrane pellets were extracted using a lysis buffer containing 50 mM TRIS-HCl pH 7.5, 150 mM NaCl, 0.5 mM DTT, 0.5% Triton X-100, and a cocktail of protease inhibitors (30 min at 4 °C), and extracts were obtained after centrifugation for 5 min at 10 000 g. Protein extracts were used for SDS–PAGE followed by western blot analysis or for co-immunoprecipitation. The intensity of the bands obtained after western blot was quantified using the Quantity One software (Bio-Rad Laboratories).
Co-immunoprecipitation experiments were performed using magnetic beads (Dynal, Invitrogen), following the recommendations of the manufacturer. Briefly, affinity-purified antibodies were covalently bound to magnetic beads (6 μg of antibody mg−1 Dynabeads). Then 150 μl (1.5 mg) of Dynabeads (with bound antibody) were incubated with 1 ml of protein extracts from membrane fractions obtained from cell suspension cultures (10 mg ml−1) for 2 h at 4 °C. Beads were washed three times with lysis buffer and bound proteins were eluted with elution buffer (buffering salts, pH 2.8, Dynal).
Mutant characterization
Lines (Col-0 background) containing a T-DNA insertion in p24δ4 (SAIL_664_A06, p24δ4-1) and p24δ5 (SALK_016402C, p24δ5-1) were identified from the SALK T-DNA collection (http://signal.salk.edu/cgi-bin/tdnaexpress). They were characterized by PCR as previously described (Ortiz-Masia et al., 2007). The primers used for the p24δ4 mutant (p24δ4-1) were the following: 5′GGATCCACTTAGATCTCCTCAAAATTC3′ and 5′ATACTGTACCATGCGACTCTCGAG3′. The T-DNA left border primer used was 5′TTCATAACCAATCTCGATACAC3′. The primers used for the p25δ5 mutant (p25δ5-1) were 5′GAAGACCATCGTTGTTCTCCGATGGC3′ and 5′TTGGTGATGAAGATTGTTCCC3′. The T-DNA left border and Actin7 (ACT7, At5g09810) primers used were described previously (Ortiz-Masia et al., 2007). Reverse transcription-PCR (RT-PCR) analysis of p24δ4-1 and p24δ5-1 mutants was performed as described (Ortiz-Masia et al., 2007) to show the absence of p24δ4 and p24δ5 mRNA, respectively. The primers used for PCR amplification were the same as above. To generate the double mutant, the homozygous line p24δ4-1 was crossed with the homozygous line p24δ5-1. Individuals homozygous for T-DNA insertion in both p24δ4-1 and p24δ5-1 were identified by PCR with the primers described above.
Results
Localization of endogenous p24 proteins in Arabidopsis
In order to localize endogenous p24 proteins in Arabidopsis, peptide antibodies against two different members of the p24 family were generated. Since previous work (Contreras et al., 2004a , b ; Langhans et al., 2008) was undertaken with p24δ5 (At1g21900) (Fig. 1A), this protein was chosen as one representative of the delta subfamily for antibody generation. Moreover, it has relatively high levels of expression in different tissues, according to gene expression data of public microarray repertoires (Zimmermann et al., 2004). Antibodies were produced using peptides corresponding to both the N- and the C-terminus of p24δ5. In contrast to the N-terminus, the C-terminus of Arabidopsis p24 proteins is very similar among different members of the delta subfamily (Supplementary Fig. S1 at JXB online). In particular, the C-termini of p24δ3, p24δ4, and p24δ6 share 10–11 (out of 12) residues with the C-terminus of p24δ5, while sequence homology is lower in other members of the subfamily. Therefore, C-terminal p24δ5 antibodies may also recognize p24δ3, p24δ4, and p24δ6. According to public microarray databases, p24δ4 has expression levels comparable (although lower) with those of p24δ5, while p24δ6 expression is very low and tissue specific (no data are available for p24δ3). Antibodies against both the N- and the C-terminus of a p24 protein of the beta subfamily, p24β2 (At3g07680), were also generated (Fig. 1A; Supplementary Fig. S1).
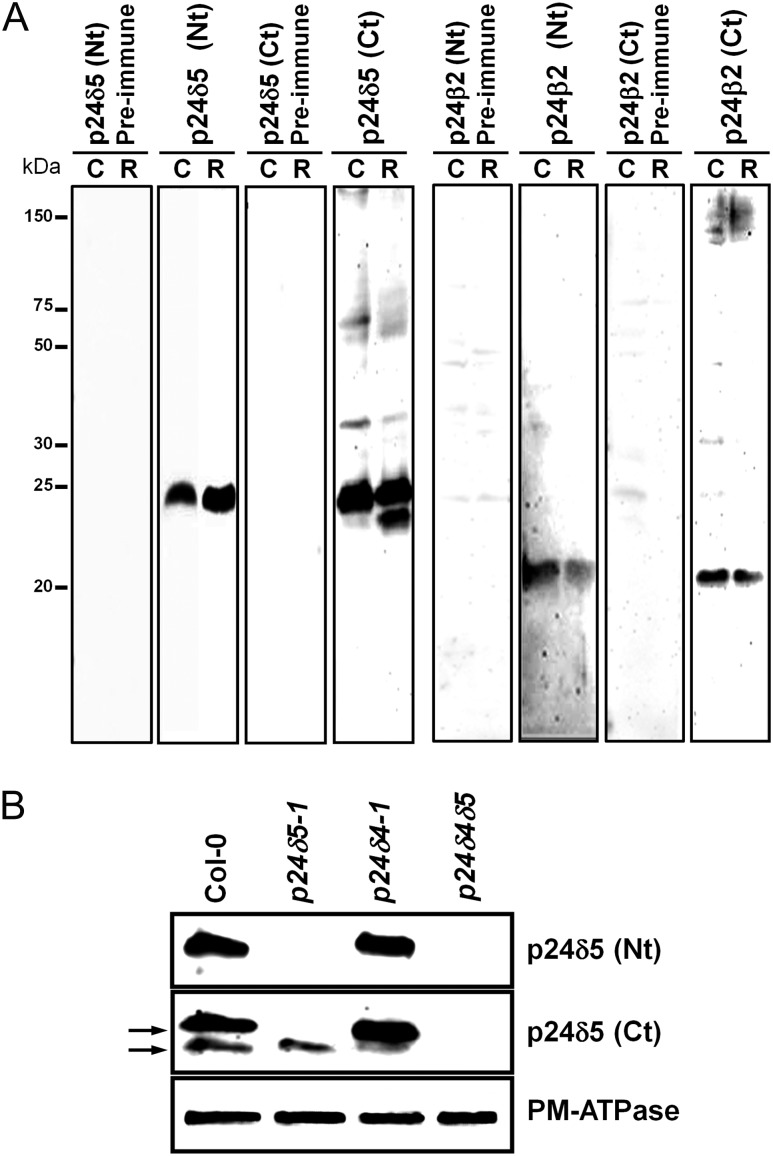
p24 proteins were extracted from membranes of Arabidopsis cell suspension cultures or from Arabidopsis roots. As shown in Fig. 2A, antibodies against the N-terminus of p24δ5 recognized a protein of the expected molecular weight (24 kDa) in membranes from cell suspension cultures as well as in membranes from roots. In the case of p24δ5 C-terminal antibodies, a major band of ∼24 kDa was obtained in membranes from cell suspension cultures, although a double band was detected in root membranes. To characterize the specificity of the antibodies further, available T-DNA insertion mutants for p24δ5 (p24δ5-1) and p24δ4 (p24δ4-1) were used. Both mutants were found to be knock-out mutants. In addition, the p24δ4δ5 double mutant was obtained. Plants from the three lines resembled wild-type plants under standard growth conditions, either on vertical agar plates or on soil (Supplementary Fig. S2 at JXB online). Western blot analysis with the Nt-p24δ5 antibody showed that p24δ5 was absent in root membranes from the p24δ5-1 mutant or from the p24δ4δ5 double mutant, as expected for a p24δ5 knock-out mutant, but was present (at similar levels to those in the wild type) in the p24δ4-1 mutant (Fig. 2B). These mutants were next analysed with the Ct-p24δ5 antibody, which detected two bands in extracts from wild-type root membranes. The upper band should correspond specifically to p24δ5, as it was absent in root membranes from the p24δ5-1 mutant or from the p24δ4δ5 double mutant, confirming the results obtained with the N-terminal antibody, but was present in the p24δ4-1 mutant (Fig. 2B). In contrast, the lower band was still present in the p24δ5-1 mutant but was almost undetectable in the p24δ4δ5 double mutant (Fig. 2B). This suggests that the lower band detected by the Ct-p24δ5 antibody corresponds to a large extent to p24δ4, and that the Ct-p24δ5 antibody does not recognize any other abundant member of the delta subfamily in Arabidopsis roots. Finally, antibodies against both the N- and the C-terminus of p24β2 recognized a protein with an apparent molecular weight of ∼22 kDa (instead of the predicted 24 kDa) (Fig. 2A).
Fig. 2.
Characterization of p24 antibodies. (A) Characterization of antibodies against Arabidopsis p24 proteins. Protein extracts were obtained from membranes of Arabidopsis cell suspension cultures (C) or Arabidopsis roots (R), as described in the Materials and methods, and analysed by SDS–PAGE and western blotting with affinity-purified antibodies against the p24δ5 N-terminus, p24δ5 C-terminus, p24β2 N-terminus, and p24β2 C-terminus, or with the corresponding pre-immune sera. (B) Western blot analysis of membranes from wild-type (Col-0) or p24 knock-out mutants (p24δ5-1, p24δ4-1, or p24δ4δ5) using antibodies against the Nt or the Ct of p24δ5. A 25 μg aliquot of protein was loaded in each lane. Western blot with an antibody against the plasma membrane (PM) ATPase was used as a loading control.
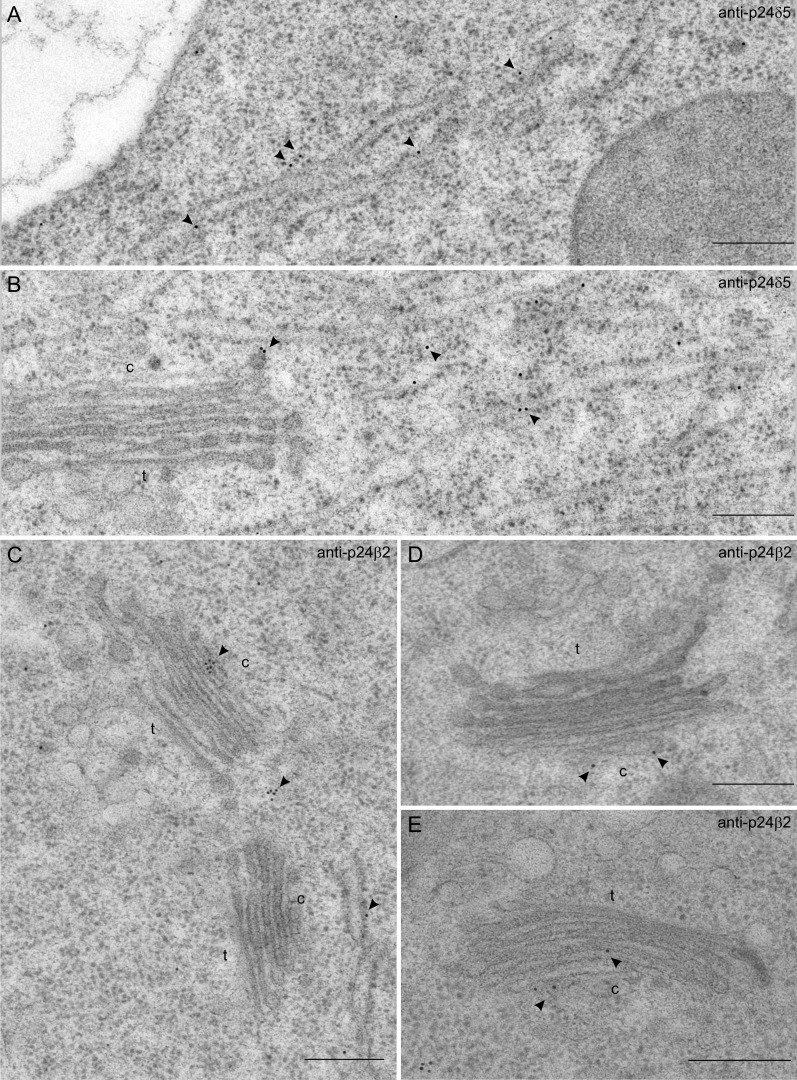
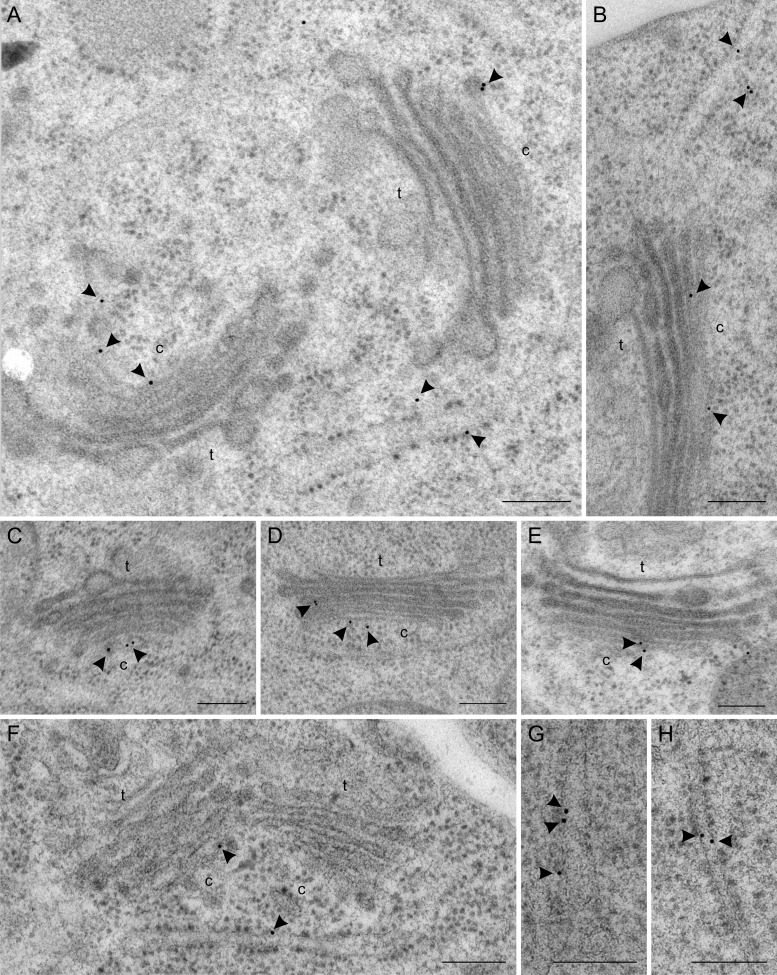
These antibodies were used to localize endogenous p24 proteins by immunogold labelling on sections cut from cryofixed Arabidopsis roots. As shown in Fig. 3A and B, the N-terminal p24δ5 antibody produced a prominent labelling on ER membranes. Occasionally, some labelling was also seen on the cis-Golgi or on putative COPI vesicles (Fig. 3B; see also Table 1). In contrast, the N-terminal p24β2 antibody showed significantly higher labelling at the Golgi apparatus, although some labelling could also be seen at ER membranes (Fig. 3C–E; see also Table 1). The C-terminal p24δ5 antibody was also used to localize endogenous p24δ proteins in p24δ5 (Fig. 4A, C, D) and p24δ4 (Fig. 4B, E) knock-out mutants. As stated above, the Ct-p24δ5 antibody should recognize p24δ5 (in the p24δ4 knock-out mutant) and p24δ4 (in the p24δ5 knock-out mutant). In both mutant lines, labelling was again found at ER membranes, but also a significant labelling at the cis side of the Golgi apparatus. The labelling obtained with the Ct-p24δ5 antibody in both mutant lines was very similar to that observed in wild-type Arabidopsis roots (Fig. 4F–H), suggesting that the localization of endogenous p24δ5 or p24δ4 is not significantly affected by the absence of p24δ4 or p24δ5, respectively.
Fig. 3.
Localization of p24δ5 and p24β2 by immunogold labelling on cryofixed Arabidopsis roots. (A and B) Labelling with antibodies against Nt-p24δ5 at the ER (A and B) and at a putative COPI vesicle (B). (C–E) Labelling with antibodies against Ct-p24β2 at the Golgi (C–E) and at the ER (C). Arrowheads point to gold particles. c, cis-Golgi; t, trans-Golgi. Scale bars=300 nm.
Table 1.
p24 immunogold labelling in wild-type Arabidopsis root cells
| Antibody | Gold particles over ER | Gold particles over Golgi Stack | Gold particles over mitochondria |
| Nt-p24δ5a | 48 | 12 | 6 |
| Ct-p24β2b | 23 | 57 | 7 |
Number of micrographs analysed: 10.
Number of micrographs analysed: 14.
Fig. 4.
Localization of p24δ proteins by immunogold labelling on cryofixed wild-type (Col-0), p24δ5-1, and p24δ4-1 Arabidopsis roots. (A, C, and D) Labelling with Ct-p24δ5 antibodies in roots from the p24δ5-1 mutant. (B and E) Labelling with Ct-p24δ5 antibodies in roots from the p24δ4-1 mutant. (F, G, and H) Labelling with Ct-p24δ5 antibodies in wild-type (Col-0) Arabidopsis roots. Arrowheads point to gold particles. c, cis-Golgi; t, trans-Golgi. Scale bars=300 nm.
The dilysine motif in the cytoplasmic tail of p24δ5 is sufficient for its trafficking in the early secretory pathway
It was previously demonstrated that transiently expressed RFP–p24δ5 shows a uniform signal distribution throughout the ER, both in tobacco mesophyll and in Arabidopsis protoplasts (Langhans et al., 2008), as well as in BY2 cells (Langhans et al., 2008) and in tobacco leaf epidermal cells (Lerich et al., 2011). Whether the dilysine motif at the cytoplasmic tail of p24δ5 is sufficient for ER localization and trafficking in the early secretory pathway, or if the luminal GOLD and coiled-coil domains could also play a role has now been investigated. To this end, RFP–p24δ5 deletion mutants lacking either the GOLD or the coiled-coil domains were prepared (Fig. 1B). These constructs were used for transient expression in tobacco mesophyll protoplasts, since they represent a versatile, well-characterized, and reproducible system for transient expression (Pimpl and Denecke, 2001; da Silva et al., 2006; Ribeiro et al., 2008). Nevertheless, the localization of the different constructs was also confirmed in Arabidopsis protoplasts (see below). As shown in Fig. 5A and B, both deletion mutants localized exclusively to the ER, where they co-localized extensively with the ER marker GFP–HDEL. This suggests that the steady-state localization of RFP–p24δ5 at the ER is not dependent on the GOLD or the coiled-coil domains. A deletion mutant lacking both the GOLD and the coiled-coil domains (and thus having only the transmembrane domain and the cytosolic tail), RFP–p24δ5 (TM-CT) (Fig. 1B), also localized exclusively to the ER (Fig. 5C), indicating that the dilysine motif in the cytosolic tail constitutes the minimal requirement for the steady-state localization of RFP–p24δ5 to the ER. The same ER localization was obtained when these constructs were expressed in Arabidopsis protoplasts (Supplementary Fig. S3A–F at JXB online). Furthermore, this ER localization was not a consequence of overexpression, since all constructs localized to the ER even at very low expression levels (Supplementary Fig. S4). It was also tested whether GOLD or coiled-coil domains were involved in anterograde ER to Golgi transport of RFP–p24δ5. To this end, the three deletion mutants were co-expressed with the ARF1(Q71L) mutant, in order to inhibit retrograde Golgi to ER transport (Langhans et al., 2008). In this case, RFP–p24δ5 deletion mutants still localized to the ER, but also to Golgi-like punctae, where they co-localized with the Golgi marker Man1–GFP (Fig. 5D–F), as was the case for RFP–p24δ5 (Langhans et al., 2008). Therefore, the GOLD and coiled-coil domains do not seem to have an influence on trafficking and localization of RFP–p24δ5 in the early secretory pathway, at least when expressed individually.
Fig. 5.
RFP–p24δ5 deletion mutants localize to the ER but cycle between the ER and the Golgi, whilst mutants lacking the dilysine motif are transported to the pre-vacuolar compartment and the vacuole. (A–L) Transient gene expression in tobacco mesophyll protoplasts. (A–C) RFP–p24δ5 deletion mutants lacking the coiled-coil domain (ΔCC) (A), the GOLD domain (ΔGOLD) (B), or both (TM-CT) (C) show a typical ER pattern and co-localize with the ER marker GFP–HDEL. (D–F) RFP–p24δ5 deletion mutants lacking the coiled-coil domain (ΔCC) (D), the GOLD domain (ΔGOLD) (E), or both (TM-CT) (F) co-localize partially with Man1–GFP in Golgi-like punctae upon ARF1(Q71L) mutant expression. (G and J) An RFP–p24δ5 dilysine mutant lacking the GOLD domain (ΔGOLD, CTSS) shows a prominent vacuolar labelling but it also co-localizes partially in punctae with Golgi (ST–YFP; blue) or PVC (GFP–BP80; green) markers. (H and K) An RFP–p24δ5 dilysine mutant lacking the coiled-coil domain (ΔCC, CTSS) shows a weak vacuolar labelling but it also co-localizes partially in punctae with Golgi (ST–YFP; blue) or PVC (GFP–BP80; green) markers. (I and L) An RFP–p24δ5 dilysine mutant with the transmembrane domain and the cytoplasmic tail (TM-CTSS) co-localizes partially with the ER marker GFP–HDEL and the Golgi marker Man1–GFP. Scale bars=5 μm.
The luminal domain may be involved in transport of p24δ5 beyond the early secretory pathway
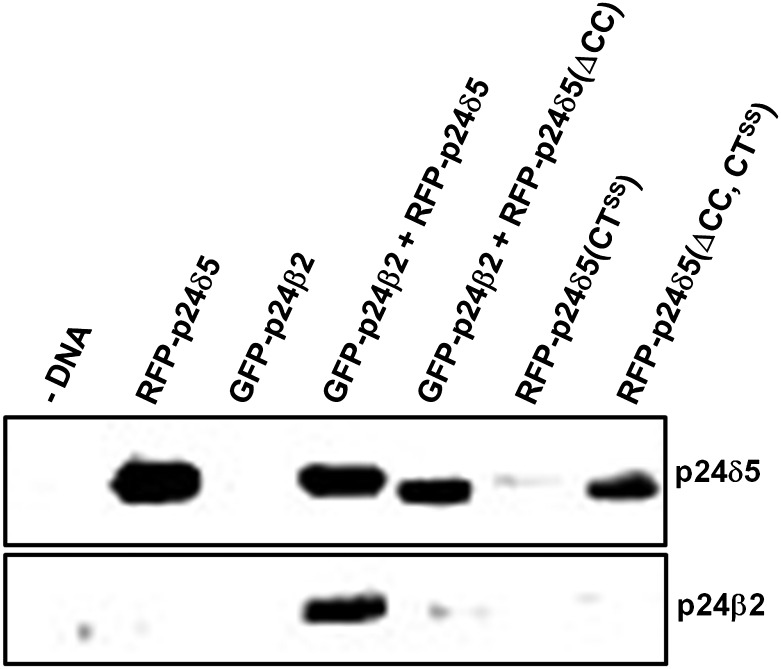
In mammals, the luminal domain of p24 proteins has been proposed to play a role in trafficking of p23 (p24α) to the cell surface (Blum and Lepier, 2008). It was previously found that RFP–p24δ5 mutants lacking the dilysine motif were no longer present at the ER but were transported downstream to the pre-vacuolar compartment (PVC) and the vacuole (Langhans et al., 2008). Therefore, it was decided to investigate whether the GOLD and/or the coiled-coil domains could play a role in RFP–p24δ5 trafficking in the absence of the dilysine motif. To this end, RFP–p24δ5 deletion mutants with a mutated dilysine motif (CTSS) lacking either the GOLD or the coiled-coil domains were prepared (Fig. 1B). As shown in Fig. 5G, H, J, and K, none of these mutants was found at the ER. In contrast, they localized to the Golgi (as shown by co-localization with ST–YFP), the PVC (as shown by co-localization with GFP–BP80), and the vacuole lumen. The dilysine mutant lacking the GOLD domain had a more prominent vacuolar labelling than the dilysine mutant lacking the coiled-coil domain (Fig. 5G, H, J, K). This suggests that the coiled-coil domain facilitates transport of dilysine mutants to the vacuole. To quantify the contribution of the coiled-coil domain and to investigate whether transport of dilysine mutants to the vacuole correlated with their degradation, the levels of the different constructs were analysed by western blotting with an RFP antibody. As shown in Fig. 6, when compared with those of RFP–p24δ5, the levels of RFP–p24δ5 (CTSS) were very low, although the same amounts of DNA were used for transient expression. These data suggest that once synthesized, RFP–p24δ5 (CTSS) is degraded, probably as a consequence of its transport to the vacuole, as had been previously proposed (Langhans et al., 2008). In contrast, the levels of the dilysine mutant lacking the coiled-coil domain [RFP–p24δ5 (ΔCC, CTSS)] were significantly higher, suggesting that a reduced transport to the vacuole may correlate with a reduced degradation (Fig. 6).
Fig. 6.
Biochemical quantitation of the expression of the different constructs. Tobacco mesophyll protoplasts were electroporated in the absence (–DNA) or the presence of 30 μg of plasmid DNAs corresponding to RFP–p24δ5 (and mutant versions) and/or GFP–p24β2. At 24 h post-electroporation, protoplasts were washed, extracted in Laemmli sample buffer, and analysed by SDS–PAGE (12% acrylamide) and western blot analysis with antibodies against RFP (to detect p24δ5 and mutant versions) or the p24β2 C-terminus. Note the lower molecular weight of the RFP–p24δ5 constructs lacking the coiled-coil domain. A 30 μg aliquot of protein was loaded for each of the extracts.
Finally, a dilysine mutant lacking both the GOLD and the coiled-coil domains (RFP–p24δ5, TM-CTSS) was used. As shown in Fig. 5I and L, this was the only dilysine mutant which was partially localized to the ER, showing a partial co-localization with the ER marker GFP–HDEL. It also showed co-localization with the Golgi marker Man1–GFP and eventually with GFP–BP80 (data not shown), but was rarely seen in the vacuole lumen. Altogether, it seems that the absence of the luminal domain impairs trafficking from the ER to the Golgi, and downstream to the PVC and the vacuole. The lower vacuolar labelling (and increased protein levels) obtained in the dilysine mutant lacking the coiled-coil domain suggests that this domain is important in trafficking beyond the Golgi apparatus.
Trafficking properties of a p24 protein of the beta subfamily and coupled transport of Arabidopsis p24 proteins
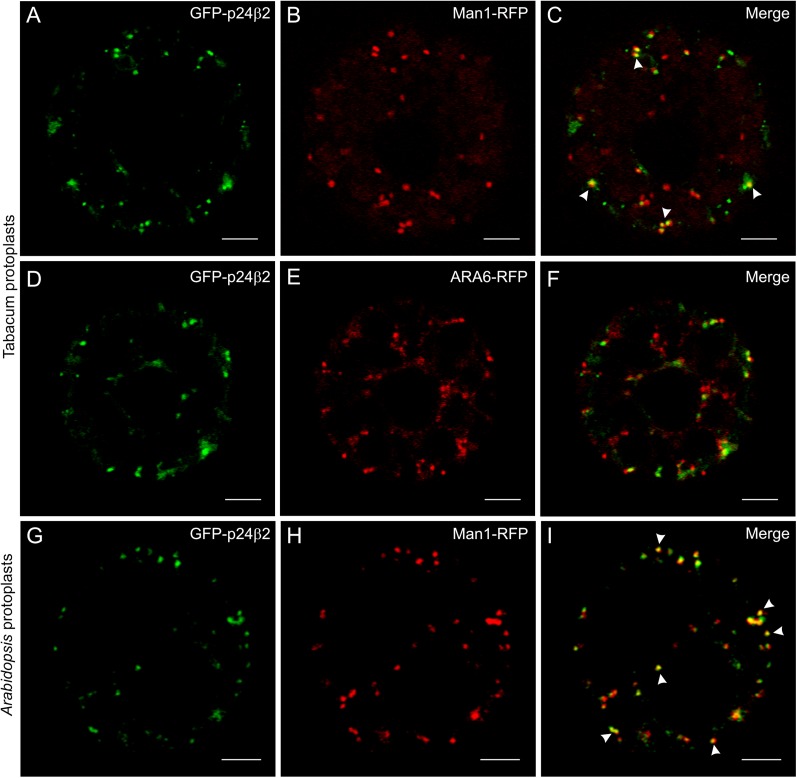
The localization and trafficking properties of a member of the beta subfamily, p24β2 (Fig. 1A), have also been investigated. To this end, a fusion construct similar to the one used to investigate in vivo trafficking and localization of RFP–p24δ5, but with GFP instead of RFP, was prepared (Fig. 1B). Surprisingly, the GFP signal obtained upon expression of GFP–p24β2 appeared to be very low. However, GFP–p24β2 was not present at the ER, in contrast to RFP–p24δ5, but showed a punctate pattern, and it co-localized partially with the Golgi marker Man1–RFP, in both tobacco mesophyll (Fig. 7A–C) and Arabidopsis protoplasts (Fig. 7G–I), but not with the PVC marker ARA6–RFP (Fig. 7D–F).
Fig. 7.
Localization of GFP–p24β2. (A–F) Transient gene expression in tobacco mesophyll protoplasts. (A–C) GFP–p24β2 (A) co-localizes partially with the Golgi marker Man1–RFP (B) in punctate structures (merged image in C). (D–F) GFP–p24β2 (D) punctae do not co-localize with the PVC marker ARA6–RFP (E) (merged image in F). (G–I) Transient gene expression in Arabidopsis protoplasts. GFP–p24β2 (G) co-localizes partially with the Golgi marker Man1–RFP (H) in punctate structures (merged image in I). Arrowheads point to co-localizing signals. Scale bars=5 μm.
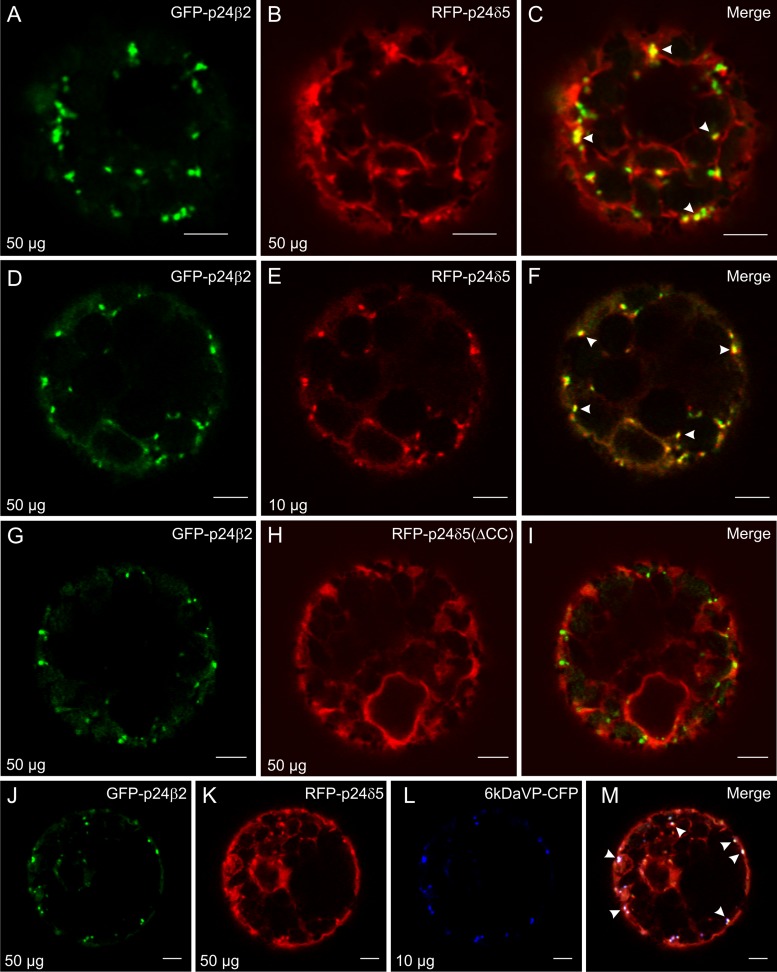
Interestingly, when co-expressed with RFP–p24δ5, the signal of GFP–p24β2 was clearly more intense and localized to punctae. In addition, it was observed that RFP–p24δ5, which showed its typical ER pattern, also localized to the same punctae under these conditions (Fig. 8A–C). An increased ratio in the concentrations of GFP–p24β2 versus RFP–p24δ5 induced a progressive change in the localization of RFP–p24δ5, from its typical reticulate ER pattern to a mostly punctated one, and increased co-localization between both proteins (Fig. 8D–F; Supplementary Fig. S5 at JXB online). This suggests that GFP–p24β2 is able to interact with RFP–p24δ5 and is transported with the latter out of the ER. This co-localization was also observed when both proteins were expressed in Arabidopsis protoplasts (Supplementary Fig. S3G–I).
Fig. 8.
GFP–p24β2 and RFP–p24δ5 co-localize in punctae via the coiled-coil domain. (A–M) Transient gene expression in tobacco mesophyll protoplasts. (A–F) GFP–p24β2 (A and D, 50 μg) and RFP–p24δ5 (B, 50 μg; E, 10 μg) co-localize in punctate structures (see merged images in C and F). (G–I) GFP–p24β2 (G) and RFP–p24δ5 (ΔCC) (H) do not co-localize (see merged image in I). (J–M) GFP–p24β2 (J) and RFP–p24δ5 (K) show extensive co-localization with 6 kDa VP–CFP (L) in punctate structures (see merged images in M). Arrowheads point to co-localizing signals. Scale bars=5 μm.
To test whether p24δ5 interacts with p24β2 via coiled-coil domains, GFP–p24β2 was co-expressed with a deletion mutant of RFP–p24δ5 lacking the coiled-coil domain (Fig. 1B). In this case, almost no co-localization was detected between both proteins (Fig. 8G–I). In addition, the signal of GFP–p24β2 was weak, as it happened in the absence of RFP–p24δ5, and RFP–p24δ5 (ΔCC) showed its typical ER pattern, without punctae, even in the presence of increasing concentrations of GFP–p24β2 (Supplementary Fig. S6 at JXB online). This suggests that both proteins interact with each other via their coiled-coil domain and that this interaction stabilizes overexpressed GFP–p24β2. To quantify whether the protein levels of GFP–p24β2 were indeed dependent on the interaction with RFP–p24δ5, protoplasts expressing these proteins were analysed by western blotting with Ct-p24β2 or RFP antibodies. As shown in Fig. 6, GFP–p24β2 was almost undetectable when expressed alone, but its levels increased enormously upon co-expression with RFP–p24δ5, but not with RFP–p24δ5 (ΔCC). On the other hand, the protein levels of RFP–p24δ5 seem to be independent of the presence of GFP–p24β2 (Fig. 6).
In order to narrow down the identity of the compartment(s) where RFP–p24δ5 and GFP–p24β2 co-localize, RFP–p24δ5 and GFP–p24β2 were co-expressed with 6 kDa VP–CFP (cyan fluorescent protein), a putative COPII/ERES marker (Lerich et al., 2011). As shown in Fig. 8J–M, a high degree of co-localization was observed between the three proteins in punctate structures, suggesting that a fraction of RFP–p24δ5 and GFP–p24β2 is present in COPII-labelled structures.
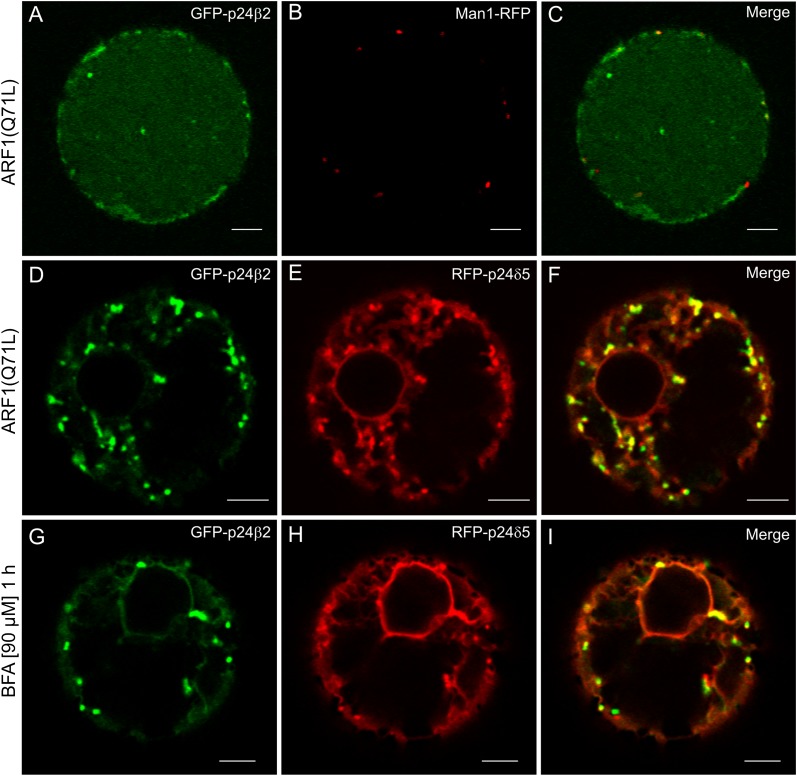
Whether the observed co-localization/interaction between RFP–p24δ5 and GFP–p24β2 correlated with their trafficking was next investigated. To this end, both proteins were co-expressed with the ARF1(Q71L) mutant, to interfere with retrograde Golgi to ER transport. This mutant expression has been shown to redistribute RFP–p24δ5 partially from the ER to Golgi-like punctae (Langhans et al., 2008). However, when GFP–p24β2 was co-expressed with the ARF1(Q71L) mutant, it showed a prominent vacuolar labelling and did not co-localize with ManI–RFP (Fig. 9A–C). This suggests that GFP–p24β2 localizes to the Golgi at steady state but may also cycle between the ER and Golgi. Thus, when retrograde Golgi to ER transport is blocked, the protein is then transported by default to the vacuole. In contrast, when both RFP–p24δ5 and GFP–p24β2 were expressed together under the same conditions [ARF1(Q71L) mutant expression], GFP–p24β2 was not transported to the vacuole but co-localized in punctae with RFP–p24δ5 (Fig. 9D–F). This suggests that RFP–p24δ5 holds back GFP–p24β2 in the ER–Golgi interface. Since both proteins co-localize in punctate structures, it was speculated that some of these punctae may correspond to Golgi stacks. To check this, protoplasts were treated with brefeldin A (BFA). After 1 h treatment, a fraction of both proteins had redistributed to the ER, as is the case for standard Golgi marker proteins, for example ManI–(X)FP, which relocate to the ER under these conditions (Langhans et al., 2011). In contrast, many of the punctate structures were BFA resistant (Fig. 9G–I) and could represent the fraction of both proteins present in COPII-labelled structures. However, it was possible to observe a redistribution of both proteins to the ER after 2 h of BFA treatment (Supplementary Fig. S7 at JXB online). Altogether, these experiments indicate that RFP–p24δ5 and GFP–p24β2 traffic together at the ER–Golgi interface.
Fig. 9.
Coupled trafficking of RFP–p24δ5 and GFP–p24β2 at the ER–Golgi interface. (A–F) Transient gene expression in tobacco mesophyll protoplasts. (A–C) GFP–p24β2 (A) is transported to the vacuole upon co-expression with the ARF1(Q71L) mutant and shows no co-localization with the Golgi marker Man1–RFP (B) (merged image in C). (D–F) GFP–p24β2 (D) is not transported to the vacuole upon co-expression of the ARF1(Q71L) mutant in the presence of RFP–p24δ5 (E) but both proteins co-localize in punctate structures (see merged image in F). (G–I) Treatment with BFA (1 h, 90 μM) after co-expression of GFP–p24β2 (G) and RFP–p24δ5 (H) induces a partial relocalization of both proteins to the ER, but many punctate structures still remain (merged image in I). Scale bars=5 μm.
Interaction between p24 proteins
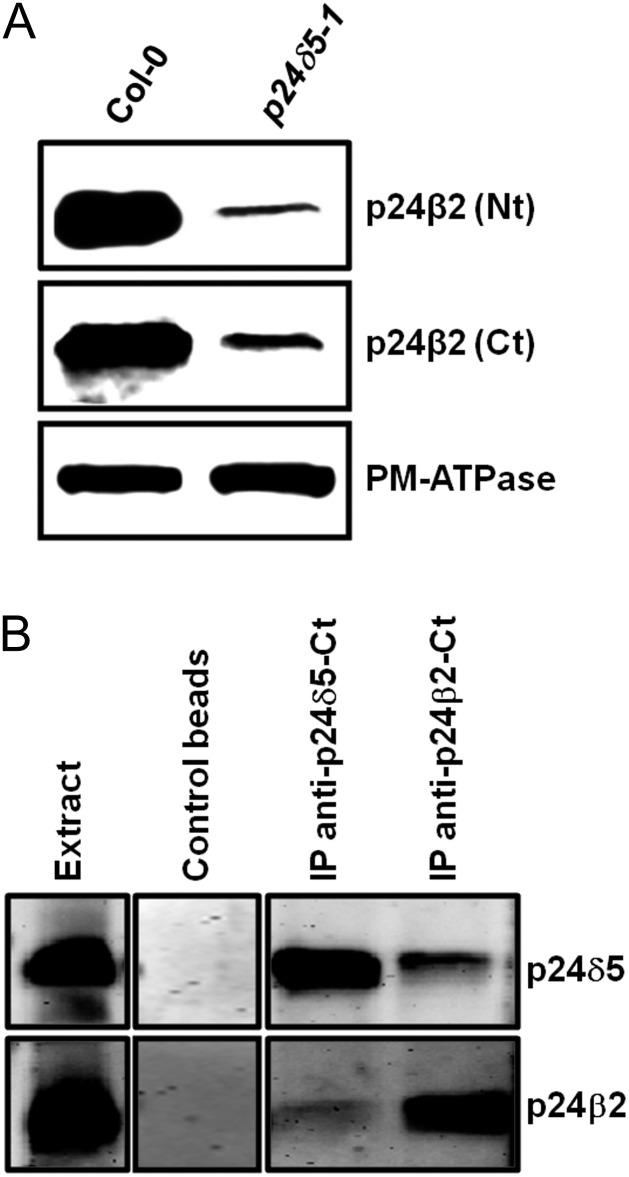
Since the above experiments suggested an interaction between p24δ5 and p24β2, it was decided to investigate this possibility further. Previous studies where a single member of the p24 family has been deleted or knocked-down showed that the protein levels of other family members were reduced, which probably reflects the fact that those p24 proteins interact with each other in hetero-oligomeric complexes (Belden and Barlowe, 1996; Marzioch et al., 1999; Denzel et al., 2000; Vetrivel et al., 2007; Takida et al., 2008; Jerome-Majewska et al., 2010; Koegler et al., 2010; Zhang and Volchuk, 2010). Therefore, the protein levels of p24β2 in a T-DNA mutant lacking p24δ5 (p24δ5-1) were examined. To this end, protein extracts were analysed with Nt- and Ct-p24β2 antibodies. In both cases, it was found that the levels of p24β2 in the p24δ5-1 mutant were drastically reduced (∼70%) when compared with the wild type (Fig. 10A). These results indicate that the protein levels of p24β2 are dependent on p24δ5, suggesting an interaction between these proteins.
Fig. 10.
Interaction between p24 proteins. (A) Western blot analysis showing the levels of p24β2 in membranes from the wild type (Col-0) or the p24δ5 knock-out mutant (p24δ5-1), using antibodies against the Nt or the Ct of p24β2. A 25 μg aliquot of protein was loaded in each lane. Western blot with an antibody against the plasma membrane (PM) ATPase was used as a loading control. (B) Immunoprecipitation of p24δ5 and p24β2 using affinity-purified antibodies against the Ct of both proteins or control beads, followed by SDS–PAGE and western blot with antibodies against p24δ5 (Nt) or p24β2 (Ct). The extract lane contains 20 μg of the membrane proteins used as input for the immunoprecipitation.
To test biochemically for a direct interaction between endogenous p24δ5 and p24β2, co-immunoprecipitation experiments were performed. To this end, affinity-purified antibodies were covalently bound to magnetic beads and were incubated in the presence of protein extracts from membrane fractions, as described in the Materials and methods. Proteins could be immunoprecipitated with the C-terminal antibodies. Under these conditions, the corresponding proteins could be only immunoprecipitated with the C-terminal antibodies. As shown in Fig. 10B, antibodies against the C-terminus of p24δ5 immunoprecipitated p24δ5, as shown by western blot analysis with antibodies against the p24δ5 N-terminus. More importantly, immunoprecipitates containing p24δ5 also contained p24β2. The reverse was also true: antibodies against the C-terminus of p24β2 immunoprecipitated p24β2, and these immunoprecipitates also contained p24δ5 (Fig. 10B). These experiments again suggest that there is a direct interaction between p24δ5 and p24β2.
Discussion
The p24 family in Arabidopsis
In contrast to animals and fungi, plants posses only representatives of the p24δ and the p24β subfamilies (Carney and Bowen, 2004; Strating and Martens, 2009; Strating et al., 2009). The p24α and p24δ subfamilies have a common origin, as is the case for the p24β and p24γ subfamilies. In most vertebrates, the p24α and p24γ subfamilies have expanded, whereas the p24β and p24δ subfamilies each have only a single member. Therefore, only one subfamily from each pair of evolutionarily related subfamilies (p24α/δ and p24β/γ) has expanded. This has led to the suggestion that there is a certain degree of functional redundancy within the two branches, which eliminates the need to expand both subfamilies. The fact that plants have only members of one subfamily in each branch (p24β and p24δ) would be in support of this idea (Carney and Bowen, 2004; Strating and Martens, 2009; Strating et al., 2009). In Arabidopsis, the delta subfamily contains nine different members (which herein have been named p24δ3–p24δ11), while the beta subfamily has only two members (here named p24β2 and p24β3). According to public microarray databases, five of them (p24δ4, p24δ5, p24δ9, p24β2, and p24β3) have high/medium levels of expression in different tissues. In contrast, other members of the delta subfamily have low and tissue-specific expression (Zimmermann et al., 2004). It is thus possible that the first category includes members with a more general/housekeeping function, probably related to the maintenance of the secretory pathway, while the second category may include members with more tissue-specific functions. In this study, the focus was on the first category. To this end, p24δ5 was selected as a representative of the p24δ subfamily and p24β2 as a representative of the p24β subfamily (see Fig. 1A).
To gain some insight into the functions that p24 proteins may have in plants, T-DNA insertion mutants that were available in the SALK collection for p24δ4 and p24δ5 (no T-DNA insertion knock-out mutants for p24β2 are available in mutant collections) were characterized and also a double mutant p24δ4δ5 was generated. The lack of a distinct phenotype in the mutants suggests that p24 proteins of the delta subfamily play redundant functions and thus multiple mutants are being obtained. In any case, the available mutants were useful to characterize the specificity of antibodies and to investigate if the levels of p24 proteins in Arabidopsis were interdependent.
Localization of Arabidopsis p24 proteins
To localize endogenous p24 proteins, specific antibodies against p24δ5 and p24β2 were used. Immunogold labelling on sections from cryofixed samples showed that endogenous p24δ5 localized to ER membranes (as revealed with both Nt- and Ct-p24δ5 antibodies) as well as to the cis side of the Golgi complex (more evident with Ct-p24δ5 antibodies). This localization is consistent with the localization of mammalian p25 (p24α), which is the only p24 family member which, in addition to cis-Golgi labelling also shows extensive ER localization (Wada et al., 1991; Domínguez et al., 1998; Lavoie et al., 1999; Emery et al., 2000, 2003). It is important to emphasize that although p24δ5 belongs to the delta subfamily, its cytosolic tail is reminiscent of that of mammalian p25 (which belongs to the alpha subfamily). This cytosolic tail has been shown to bind COPI with high affinity via a dilysine motif at the -3,-4 position (relative to the cytosolic C-terminus) (Contreras et al., 2004a ) and is important for efficient Golgi to ER retrograde transport (Langhans et al., 2008). In contrast to p24δ5, endogenous p24β2 localizes mainly to the Golgi apparatus. This localization is consistent with the localization of members of the p24β subfamily, both in mammals (Domínguez et al., 1998; Blum et al., 1999; Gommel et al., 1999; Emery et al., 2000) and in yeast (Shimmöller et al., 1995; Belden and Barlowe, 2001).
The localization of endogenous p24 proteins may depend, at least, on the sorting determinants present in the individual proteins as well as on the interactions with other p24 proteins. To investigate these two possibilities, RFP–p24δ5 and deletion mutants were transiently expressed, either individually or in combination with GFP–p24β2.
Sorting determinants in p24δ5
Tagging of p24 proteins with XFP between the signal sequence and the luminal domain produces constructs which faithfully reflect the trafficking dynamics and localization pattern of native p24 proteins (Blum et al., 1999; Barr et al., 2001; Majoul et al., 2001; Gupta and Swarup, 2006; Simpson et al., 2006; Blum and Lepier, 2008). Therefore, as in a previous investigation (Langhans et al., 2008), the same strategy was used for all the constructs used in this study.
First, the relative contribution of the different domains of p24 proteins to their intracellular trafficking and localization was investigated. For this project, p24δ5 was used as being representative of the family, since the sorting determinants in its cytosolic tail have previously been analysed. It was shown that the dilysine motif in the -3,-4 position was the main determinant for its ER localization. Here, the relative contributions of the luminal (GOLD and coiled-coil) domains were determined, using deletion mutants, in the presence or the absence of the dilysine motif in the -3,-4 position. The results show that neither the GOLD nor the coiled-coil domains are necessary for the ER localization of RFP–p24δ5 in the steady state or for the cycling of the protein in the early secretory pathway, at least when expresssed individually. The fact that deletion mutants having only the transmembrane domain and the C-terminal tail localize correctly to the ER, unless the dilysine motif is mutated, strongly implicates the dilysine motif in the cytosolic tail as being the main determinant for the ER localization of RFP–p24δ5, presumably by facilitating its interaction with the COPI machinery. In clear contrast, the luminal domain (in particular the coiled-coil domain) seems to be necessary for transport of p24 dilysine mutants to the vacuole lumen. Since cargoes for plant p24 proteins have not yet been identified (as has been the case for many years for their mammalian and yeast counterparts), it cannot be ruled out that a fraction of p24δ5 (or other p24 proteins) could be involved in the transport of cargo destined for the PVC or the vacuole. Alternatively, transport to the vacuole may simply be a default pathway for membrane proteins in the secretory pathway (Langhans et al., 2008).
The relative contribution of the different p24δ5 domains to trafficking of the p24δ5 protein shown here is consistent with results obtained using similar GFP constructs for p23 (p24δ1) in animal cells. Blum and Lepier (2008) showed that the minimal requirement for p23 cycling within the early secretory pathway was the transmembrane domain and the cytoplasmic tail with an intact KKLIE motif. The luminal domain was expendable for cycling between the ER and the Golgi apparatus, but was necessary for trafficking beyond the Golgi apparatus, in their case to the cell surface. In another study, however, the location of a p23/p24 dimer was shown to be independent of the KKLIE motif in p23 but instead required the coiled-coil domains in both proteins (Emery et al., 2000). It has to be noted that, in contrast to p23 (p24δ1), p24δ5 has a p25 (p24α)-like C-terminal tail, with high affinity for COPI (Contreras et al., 2004a ), which is responsible for its efficient Golgi to ER recycling (Langhans et al., 2008). Therefore, it is likely that the sorting information contained in the cytosolic tail of RFP–p24δ5 is sufficient for its steady-state distribution in the ER. This scenario has also been proposed for mammalian p25, which normally resides in the ER, even when co-expressed with p23, p24, and p26 (Emery et al., 2000).
Interactions between p24 proteins and coupled transport of p24 family members
Shuttling of p24 proteins in the ER–Golgi interface has been proposed to depend on interactions with other p24 family members (Dominguez et al., 1998; Füllekrug et al., 1999; Emery et al., 2000). Early experiments concluded that p24 proteins form heterotetrameric complexes via their coiled-coil domains with one member of each subfamily, both in animals (p23, p24, p25, and p27) (Fullerkrug et al., 1999) and in yeast (Erv25p, Emp24p, Erp1p, and Erp2p) (Marzioch et al., 1999). However, the pattern of interdependence between different p24 members seen in yeast raises the possibility that the postulated p24 tetramer is a double dimer (Ciufo and Boyd, 2000). Moreover, the studies of Jenne et al. (2002) have shown that p24 proteins occur mostly as monomers and dimers of various compositions, depending on their subcellular location. In most cases, the best characterized dimers are the ones formed between one member of the p24δ subfamily and one member of the p24β subfamily. In yeast, Erv25 (p24δ subfamily) and Emp24 (p24β subfamily) have been shown to form a complex which is efficiently incorporated into ER-derived COPII vesicles and can exit the ER without the presence of Erp1 and Erp2, putative components of the yeast tetrameric p24 complex (Belden and Barlowe, 1996, 2001). In animals, a complex is formed between p23 (p24δ subfamily) and p24 (p24β subfamily) which can also exit the ER to be transported to the Golgi apparatus (Gommel et al., 1999; Emery et al., 2000). In both cases, retrograde transport of these complexes was postulated to depend on the dilysine motif present in the p24δ members: Erv25 in yeast (Belden and Barlowe, 2001) and p23 in animals (Gommel et al., 1999).
Several lines of evidence suggest that Arabidopsis p24δ5 and p24β2 interact with each other, and that this interaction is mediated by the coiled-coil domain. First, the protein levels of p24β2 seem to be dependent on p24δ5: a knock-out mutant lacking p24δ5 showed a drastic reduction in the levels of p24β2. This is consistent with previous reports in yeast and mammalian cells showing that depletion of one member of the p24 family affects the protein levels of other family members, suggesting interaction between those proteins (Belden and Barlowe, 1996; Marzioch et al., 1999; Denzel et al., 2000; Vetrivel et al., 2007; Takida et al., 2008; Jerome-Majewska et al., 2010; Koegler et al., 2010; Zhang and Volchuk, 2010). Secondly, our co-immunoprecipitation data indicate that endogenous p24δ5 and p24β2 interact with each other: immunoprecipitation with p24δ5 antibodies is able to co-immunoprecipitate p24β2, and vice versa.
Transiently expressed proteins also seem to interact with each other. When expressed individually, GFP–p24β2 seems to be unstable, since the protein is hardly detectable either by confocal laser scanning microcopy or after western blot analysis. However, the levels of GFP–p24β2 increase enormously upon co-expression with RFP–p24δ5, an effect which is not seen when RFP–p24δ5 lacks the coiled-coil domain. This suggests that the interaction with p24δ5, via the coiled-coil domains, stabilizes p24β2. In contrast, transiently expressed RFP–p24δ5 seems to be stable in the absence of co-expressed GFP–p24β2, probably as a consequence of its predominant localization to the ER. In addition, the co-expression of GFP–p24β2 changes the pattern of RFP–p24δ5 from its typical reticulate ER pattern to punctae where both proteins co-localize. This does not occur when p24δ5 lacks the coiled-coil domain.
Finally, the two proteins seem to travel together along the early secretory pathway. RFP–p24δ5 and GFP–p24β2 may be efficiently incorporated into ERESs, as suggested by their significant co-localization with the COPII marker 6 kDa VP–CFP, for transport to the Golgi. When retrograde Golgi to ER transport is inhibited by expression of the ARF1(Q71L) mutant, GFP–p24β2 is transported downstream to the vacuole, where it appears to be degraded. In contrast, when RFP–p24δ5 is co-expressed with GFP–p24β2, the latter is not transported to the vacuole, but both proteins co-localize to punctate structures. This suggests that RFP–p24δ5 is able to hold GFP–p24β2 at the ER–Golgi interface, and possibly to mediate coupled trafficking of both proteins back to the ER. Therefore, it is possible that the steady-state localization found for endogenous proteins and their stability reflects the tight balance between the levels of these two proteins (or of other members of the family).
In summary, the results presented herein are consistent with a coupled trafficking of both proteins at the ER–Golgi interface. p24δ5 and p24β2 could interact with each other at ERESs for ER exit and coupled transport to the Golgi apparatus. Once in the Golgi, p24δ5 could interact very efficiently with the COPI machinery for retrograde transport back to the ER.
Addendum
While this manuscript was undergoing review, a paper was published showing trafficking and localization of p24 proteins in Arabidopsis (Chen et al., 2012), which overlaps with some of the results presented herein. Using transient expression in tobacco leaf epidermal cells, these authors propose a subclass-specific localization for Arabidopsis p24 proteins: p24δ1a–d (p24δ3–6 in this study) are found to localize exclusively to the ER, p24δ2a–d (p24δ7–11 in this study) localize to ER and Golgi, and p24β proteins localize exclusively to the Golgi apparatus. As representatives of each subclass for trafficking studies, they chose p24δ1d (p24δ6 in this study), p24δ2d (p24δ10 in this study), and p24β1 (p24β2 in this study). Surprisingly, these authors maintain that the steady-state localization of these proteins is not dependent on the position of the XFP tag. There are also significant discrepancies with the results presented here, which it is felt could be explained by this fact: p24 proteins have a very short cytosolic C-terminal tail, which contains the sorting signals for COPI/COPII binding. In the case of p24δ proteins, these signals consist of a dilysine motif at the -3,-4 position and a diaromatic motif at the -7,-8 position. Indeed, it has been shown that the position of these signals (with respect to the C-terminus) is important for optimal binding of COP subunits (Teasdale and Jackson, 1996). Accordingly, it is difficult to imagine that the presence of an XFP molecule at the C-terminus has no influence in trafficking and localization of the fusion proteins, even if they might be still partially functional. This is the reason why p24–XFP constructs used in mammalian research (Blum et al., 1996; Majoul et al., 2001; Simpson et al., 2006; Blum and Lepier, 2008), as well as in a previous paper on plant p24 proteins (Langhans et al., 2008) and in this study, have the XFP tag at the luminal N-terminus (immediately behind the signal sequence). The most striking differences between both studies can be summarized as follows. First, although transiently expressed RFP–p24δ5 localizes exclusively to the ER (as do all the members of the p24δ1 subfamily in the study by Chen et al.), endogenous p24δ5 (and possibly p24δ4) localizes to the ER but also to the cis-Golgi, suggesting that the steady-state localization of endogenous p24 proteins and their stability reflect the tight balance between the levels of the different members of the family. In this respect, the present study represents the first report on the localization of endogenous p24 proteins, through immunogold electron microscopy, in Arabidopsis.
Secondly, when RFP–p24δ1d (p24δ6 in this study) was co-expressed with p24β1–YFP (Ct-fusion) (p24β2 in this study), there was no change in the localization of RFP–p24δ1d. This is in marked contrast to the present co-expression data showing that GFP–p24β2 (Nt-fusion) changes the localization of RFP–p24δ5 (a close homologue of p24δ1d) and this requires the coiled-coil domain. Thirdly, Chen et al. showed that when p24β1–YFP (Ct-fusion) (p24β2 in this study) was co-expressed with the ARF1(Q71L) mutant, to interfere with Golgi to ER transport, the protein was still Golgi localized and only occasionally was found at the PVC (never at the vacuole). In the present study, when this protein was co-expressed with the ARF1 mutant, it was transported to the vacuole, unless it was co-expressed with RFP–p24δ5. This suggests that RFP–p24δ5 holds GFP–p24β2 at the ER–Golgi interface and may explain why transiently expressed GFP–p24β2 is not stable in the absence of RFP–p24δ5. Finally, the present study shows the first biochemical demonstration of interactions between Arabidopsis p24 proteins. In summary, while both studies are somehow complementary, there are a number of discrepancies which need to be resolved.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Nt- and Ct-sequences of Arabidopsis p24 proteins.
Figure S2. Characterization of T-DNA insertion mutants.
Figure S3. Localization of RFP–p24δ5 and deletion mutants and co-localization between GFP–p24β2 and RFP–p24δ5 or RFP–p24δ5(ΔCC) in Arabidopsis protoplasts.
Figure S4. RFP–p24δ5 and RFP–p24δ5 (ΔCC) localize to the ER at different expression levels.
Figure S5. Co-expression of RFP–p24δ5 and different DNA concentrations of GFP–p24β2.
Figure S6. Co-expression of RFP–p24δ5(ΔCC) and different DNA concentrations of GFP–p24β2.
Figure S7. A 2 h BFA treatment redistributes RFP–p24δ5 and GFP–p24β2 to the ER.
Figure S8. A comparison between frame scan and line scan modes for co-localization studies.
Supplementary Material
Acknowledgments
The 6 kDa VP–CFP plasmid was kindly provided by Dr Aiming Wang (Southern Crop Protection and Food Research Centre, AAFC, Canada). We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, the greenhouse section of SCSIE (University of Valencia), and Pilar Selvi and Barbara Jesenofsky for excellent technical assistance.
This work was supported by the Ministerio de Ciencia e Innovación [grant number BFU2009-07039 to F.A.] and the Deutsche Forschungsgemeinschaft [grant number RO 440/14-1 to D.G.R.]. J.C.M. was a recipient of fellowships from Generalitat Valenciana (Geronimo Forteza Program) and from the Ministerio de Educación y Ciencia (FPU Program).
References
- Anantharaman V, Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-5-research0023. research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniento F, Matsuoka K, Robinson DG. ER-to-Golgi transport: the COPII-pathway. In: Robinson DG, editor. The plant endoplasmic reticulum. Berlin: Springer-Verlag; 2006. pp. 99–124. [Google Scholar]
- Axelos M, Curie C, Bardet C, Lescure B. A protocol for transient expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiology and Biochemistry. 1992;30:123–128. [Google Scholar]
- Barr FA, Preisinger C, Kopajtich R, Körner R. Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. Journal of Cell Biology. 2001;155:885–891. doi: 10.1083/jcb.200108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. Journal of Biological Chemistry. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. Journal of Biological Chemistry. 2001;276:43040–43048. doi: 10.1074/jbc.M108113200. [DOI] [PubMed] [Google Scholar]
- Blum R, Feick P, Puype M, Vandekerckhove J, Klengel R, Nastainczyk W, Schulz I. Tmp21 and p24A, two type I proteins enriched in pancreatic microsomal membranes, are members of a protein family involved in vesicular trafficking. Journal of Biological Chemistry. 1996;271:17183–17189. doi: 10.1074/jbc.271.29.17183. [DOI] [PubMed] [Google Scholar]
- Blum R, Lepier A. The luminal domain of p23 (Tmp21) plays a critical role in p23 cell surface trafficking. Traffic. 2008;9:1530–1550. doi: 10.1111/j.1600-0854.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- Blum R, Pfeiffer F, Feick P, Nastainczyk W, Kohler B, Schafer KH, Schulz I. Intracellular localization and in vivo trafficking of p24A and p23. Journal of Cell Science. 1999;112:537–548. doi: 10.1242/jcs.112.4.537. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N. The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. The Plant Cell. 2002;14:1077–1092. doi: 10.1105/tpc.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck J, Scheuring D, Hummel E, Langhans M, Viotti C, Foresti O, Denecke J, Banfield DK, Robinson DG. The syntaxins SYP31 and SYP81 control ER–Golgi trafficking in the plant secretory pathway. Traffic. 2008;9:1629–1652. doi: 10.1111/j.1600-0854.2008.00803.x. [DOI] [PubMed] [Google Scholar]
- Buechling T, Chaudhary V, Spirohn K, Weiss M, Boutros M. p24 proteins are required for secretion of Wnt ligands. EMBO Reports. 2011;12:1265–1272. doi: 10.1038/embor.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney GE, Bowen NJ. p24 proteins, intracellular trafficking, and behaviour: Drosophila melanogaster provides insights and opportunities. Biology of the Cell. 2004;96:271–278. doi: 10.1016/j.biolcel.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muñiz M. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Molecular Biology of the Cell. 2011;22:2924–2936. doi: 10.1091/mbc.E11-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hasegawa H, Schmitt-Ulms G, et al. TMP21 is a presenilin complex component that modulates γ-secretase but not ϵ-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- Chen J, Qi X, Zheng H. Subclass-specific localization and trafficking of Arabidopsis p24 proteins in the ER–Golgi interface. Traffic. 2012;13:400–415. doi: 10.1111/j.1600-0854.2011.01317.x. [DOI] [PubMed] [Google Scholar]
- Ciufo LF, Boyd A. Identification of a lumenal sequence specifying the assembly of Emp24p into p24 complexes in the yeast secretory pathway. Journal of Biological Chemistry. 2000;275:8382–8388. doi: 10.1074/jbc.275.12.8382. [DOI] [PubMed] [Google Scholar]
- Contreras I, Ortiz-Zapater E, Aniento F. Sorting signals in the cytosolic tail of membrane proteins involved in the interaction with plant ARF1 and coatomer. The Plant Journal. 2004a;38:685–698. doi: 10.1111/j.1365-313X.2004.02075.x. [DOI] [PubMed] [Google Scholar]
- Contreras I, Yang Y, Robinson DG, Aniento F. Plant COPI and COPII coat proteins show a differential affinity for p24 cytosolic tails. Plant Cell Physiology. 2004b;45:1779–1786. doi: 10.1093/pcp/pch200. [DOI] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- DaSilva LL, Foresti O, Denecke J. Targeting of the plant vacuolar sorting receptor BP80 is dependent on multiple sorting signals in the cytosolic tail. The Plant Cell. 2006;18:1477–1497. doi: 10.1105/tpc.105.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaSilva LL, Taylor JP, Hadlington JL, Hanton SL, Snowden CJ, Fox SJ, Foresti O, Brandizzi F, Denecke J. Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. The Plant Cell. 2005;17:132–148. doi: 10.1105/tpc.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzel A, Otto F, Girod A, Pepperkok R, Watson R, Rosewell I, Bergeron JJ, Solari RC, Owen MJ. The p24 family member p23 is required for early embryonic development. Current Biology. 2000;10:55–58. doi: 10.1016/s0960-9822(99)00266-3. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Dejgaard K, Fullekrug J, Dahan S, Fazel A, Paccaud JP, Thomas DY, Bergeron JJ, Nilsson T. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COPI and COPII coatomer. Journal of Cell Biology. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G, Parton RG, Rojo M, Gruenberg J. The trans-membrane protein p25 forms highly specialized domains that regulate membrane composition and dynamics. Journal of Cell Science. 2003;116:4821–4832. doi: 10.1242/jcs.00802. [DOI] [PubMed] [Google Scholar]
- Emery G, Rojo M, Gruenberg J. Coupled transport of p24 family members. Journal of Cell Science. 2000;113:2507–2516. doi: 10.1242/jcs.113.13.2507. [DOI] [PubMed] [Google Scholar]
- Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- Fujita M, Watanabe R, Jaensch N, Romanova-Michaelides M, Satoh T, Kato M, Riezman H, Yamaguchi Y, Maeda Y, Kinoshita T. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. Journal of Cell Biology. 2011;194:61–75. doi: 10.1083/jcb.201012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllerkrug J, Suganuma T, Tang BL, Hong W, Storrie B, Nilsson T. Localization and recycling of gp27 (hp24g3): complex formation with other p24 family members. Molecular Biology of the Cell. 1999;10:1939–1955. doi: 10.1091/mbc.10.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommel D, Orci L, Emig EM, Hannah MJ, Ravazzola M, Nickel W, Helms JB, Wieland FT, Sohn K. p24 and p23, the major transmembrane proteins of COPI-coated vesicles, form hetero-oligomeric complexes and cycle between organelles of the early secretory pathway. FEBS Letters. 1999;447:179–185. doi: 10.1016/s0014-5793(99)00246-x. [DOI] [PubMed] [Google Scholar]
- Gupta V, Swarup G. Evidence for a role of transmembrane protein p25 in localization of protein tyrosine phosphatase TC48 to the ER. Journal of Cell Science. 2006;119:1703–1714. doi: 10.1242/jcs.02885. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Liu L, Nishimura M. Dilysine retrieval signal-containing p24 proteins collaborate in inhibiting γ-cleavage of amyloid precursor protein. Journal of Neurochemistry. 2010;115:771–781. doi: 10.1111/j.1471-4159.2010.06977.x. [DOI] [PubMed] [Google Scholar]
- Jenne N, Frey K, Brügger B, Wieland FT. Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. Journal of Biological Chemistry. 2002;277:46504–46511. doi: 10.1074/jbc.M206989200. [DOI] [PubMed] [Google Scholar]
- Jerome-Majewska LA, Achkar T, Luo L, Lupu F, Lacy E. The trafficking protein Tmed2/p24beta(1) is required for morphogenesis of the mouse embryo and placenta. Developmental Biology. 2010;341:154–166. doi: 10.1016/j.ydbio.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegler E, Bonnon C, Waldmeier L, Mitrovic S, Halbeisen R, Hauri HP. p28, a novel ERGIC/cis Golgi protein, required for Golgi ribbon formation. Traffic. 2010;11:70–89. doi: 10.1111/j.1600-0854.2009.01009.x. [DOI] [PubMed] [Google Scholar]
- Langhans M, Förster S, Helmchen G, Robinson DG. Differential effects of the brefeldin A analogue (6R)-hydroxy-BFA in tobacco and Arabidopsis. Journal of Experimental Botany. 2011;62:2949–2957. doi: 10.1093/jxb/err007. [DOI] [PubMed] [Google Scholar]
- Langhans M, Marcote MJ, Pimpl P, Virgili-López G, Robinson DG, Aniento F. In vivo trafficking and localization of p24 proteins in plant cells. Traffic. 2008;9:770–785. doi: 10.1111/j.1600-0854.2008.00719.x. [DOI] [PubMed] [Google Scholar]
- Lavoie C, Paiement J, Dominguez M, Roy L, Dahan S, Gushue JN, Bergeron JJ. Roles for alpha(2)p24 and COPI in endoplasmic reticulum cargo exit site formation. Journal of Cell Biology. 1999;146:285–300. doi: 10.1083/jcb.146.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerich A, Langhans M, Sturm S, Robinson DG. Is the 6kDa tobacco etch viral protein a bona fide ERES marker? Journal of Experimental Botany. 2011;62:5013–5023. doi: 10.1093/jxb/err200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Luo W, Wang Y, Reiser G. Proteinase-activated receptors, nucleotide P2Y receptors, and μ-opioid receptor-1B are under the control of the type I transmembrane proteins p23 and p24A in post-Golgi trafficking. Journal of Neurochemistry. 2011;117:71–81. doi: 10.1111/j.1471-4159.2011.07173.x. [DOI] [PubMed] [Google Scholar]
- Luo W, Wang Y, Reiser G. p24A, a type I transmembrane protein, controls ARF1-dependent resensitization of protease-activated receptor-2 by influence on receptor trafficking. Journal of Biological Chemistry. 2007;282:30246–30255. doi: 10.1074/jbc.M703205200. [DOI] [PubMed] [Google Scholar]
- Majoul I, Straub M, Hell SW, Duden R, Söling HD. KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Developmental Cell. 2001;1:139–153. doi: 10.1016/s1534-5807(01)00004-1. [DOI] [PubMed] [Google Scholar]
- Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, Solari RC, Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Molecular Biology of the Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic S, Ben-Tekaya H, Koegler E, Gruenberg J, Hauri HP. The cargo receptors Surf4, endoplasmic reticulum–Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Molecular Biology of the Cell. 2008;19:1976–1990. doi: 10.1091/mbc.E07-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz M, Morsomme P, Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Muñiz M, Nuoffer C, Hauri HP, Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. Journal of Cell Biology. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Frohlick JA, Staehelin LA. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiology. 2000;124:135–151. doi: 10.1104/pp.124.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiology. 1999;121:1127–1142. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W, Sohn K, Bunning C, Wieland F. p23, a major COPI-vesicle membrane protein, constitutively cycles through the early secretory pathway. Proceedings of the National Academy of Sciences, USA. 1997;94:11393–11398. doi: 10.1073/pnas.94.21.11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Masia D, Perez-Amador MA, Carbonell J, Marcote MJ. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Letters. 2007;581:1834–1840. doi: 10.1016/j.febslet.2007.03.075. [DOI] [PubMed] [Google Scholar]
- Ortiz-Zapater E, Soriano-Ortega E, Marcote MJ, Ortiz-Masiá D, Aniento F. Trafficking of the human transferrin receptor in plant cells: effects of tyrphostin A23 and brefeldin A. The Plant Journal. 2006;48:757–770. doi: 10.1111/j.1365-313X.2006.02909.x. [DOI] [PubMed] [Google Scholar]
- Pimpl P, Denecke J. Protein–protein interactions in the secretory pathway, a growing demand for experimental approaches in vivo . Plant Molecular Biology. 2002;50:887–902. doi: 10.1023/a:1021266320877. [DOI] [PubMed] [Google Scholar]
- Pimpl P, Hanton SL, Taylor JP, Pinto-DaSilva LL, Denecke J. The GTPase ARF1p controls the sequence-specific vacuolar sorting route to the lytic vacuole. The Plant Cell. 2003;15:1242–1256. doi: 10.1105/tpc.010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P, Taylor JP, Snowden C, Hillmer S, Robinson DG, Denecke J. Golgi-mediated vacuolar sorting of the endoplasmic reticulum chaperone BiP may play an active role in quality control within the secretory pathway. The Plant Cell. 2006;18:198–211. doi: 10.1105/tpc.105.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Hausmann G, Basler K. A genome-wide RNA interference screen uncovers two p24 proteins as regulators of Wingless secretion. EMBO Reports. 2011;12:1144–1152. doi: 10.1038/embor.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro D, Foresti O, Denecke J, Wellink J, Goldbach R, Kormelink RJ. Tomato spotted wilt virus glycoproteins induce the formation of endoplasmic reticulum- and Golgi-derived pleomorphic membrane structures in plant cells. Journal of General Virology. 2008;89:1811–1818. doi: 10.1099/vir.0.2008/001164-0. [DOI] [PubMed] [Google Scholar]
- Rojo M, Emery G, Marjomäki V, McDowall A, Parton RG, Gruenberg J. The transmembrane protein p23 contributes to the organization of the Golgi apparatus. Journal of Cell Science. 2000;113:1043–1057. doi: 10.1242/jcs.113.6.1043. [DOI] [PubMed] [Google Scholar]
- Rojo M, Peperkok R, Emery G, Kellner R, Stang E, Parton RG, Gruenberg J. Involvement of the transmembrane protein p23 in biosynthetic protein transport. Journal of Cell Biology. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmöller F, Singer-Krüger B, Schröder S, Krüger U, Barlowe C, Riezman H. The absence of Em24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO Journal. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JC, Nilsson T, Pepperkok R. Biogenesis of tubular ER-to-Golgi transport intermediates. Molecular Biology of the Cell. 2006;17:723–737. doi: 10.1091/mbc.E05-06-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K, Orci L, Ravazzola M, Amherdt M, Brunner M, Kahn RA, Rothman JE. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. Journal of Cell Biology. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes MA, Craighead MW, Hoe MH, Lampen N, Geromanos S, Tempst P, Rothman JE. An integral membrane components of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proceedings of the National Academy of Sciences, USA. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating JR, Martens GJ. The p24 family and selective transport processes at the ER–Golgi interface. Biology of the Cell. 2009;101:495–509. doi: 10.1042/BC20080233. [DOI] [PubMed] [Google Scholar]
- Strating JR, van Bakel NH, Leunissen JA, Martens GJ. A comprehensive overview of the vertebrate p24 family: identification of a novel tissue-specifically expressed member. Molecular Biology of Evolution. 2009;26:1707–1714. doi: 10.1093/molbev/msp099. [DOI] [PubMed] [Google Scholar]
- Takida S, Maeda Y, Kinoshita T. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochemical Journal. 2008;409:555–562. doi: 10.1042/BJ20070234. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annual Review of Cell and Developmental Biology. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A. Functional differentiation of endosomes in Arabidopsis cells. The Plant Journal. 2004;40:783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, Gong P, Bowen JW, et al. Dual roles of the transmembrane protein p23/TMP21 in the modulation of amyloid precursor protein metabolism. Molecular Neurodegeneration. 2007;2:4. doi: 10.1186/1750-1326-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJ, 2nd, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJ. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. Journal of Biological Chemistry. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Wen C, Greenwald I. p24 proteins and quality control of LIN-12 and GLP-1 trafficking in Caenorhabditis elegans . Journal of Cell Biology. 1999;145:1165–1175. doi: 10.1083/jcb.145.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Volchuk A. p24 family type 1 transmembrane proteins are required for insulin biosynthesis and secretion in pancreatic beta-cells. FEBS Letters. 2010;584:2298–2304. doi: 10.1016/j.febslet.2010.03.041. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.