Abstract
Branching morphogenesis is a crucial developmental process in which vertebrate organs generate extensive epithelial surface area while retaining a compact size. In the vertebrate submandibular salivary gland, branching morphogenesis is crucial for generation of the large surface area necessary to produce sufficient saliva. However, in many salivary gland diseases, saliva-producing acinar cells are destroyed, resulting in dry mouth and secondary health conditions. Systems-based approaches can provide insights into understanding salivary gland development, function, and disease. The traditional approach to understanding these processes is identification of molecular signals using reductionist approaches; we review current progress with such methods in understanding salivary gland development. Taking a more global approach, multiple groups are currently profiling the transcriptome, the proteome, and other “omes” in both developing mouse tissues and in human patient samples. Computational methods have been successful in deciphering large data sets, and mathematical models are starting to make predictions regarding the contribution of molecules to the physical processes of morphogenesis and of cellular function. A challenge for the future will be to establish comprehensive, publicly accessible salivary gland databases spanning the full range of genes and proteins; plans are underway to provide these resources to researchers in centralized repositories. The greatest challenge for the future will be to develop realistic models that integrate multiple types of data to both describe and predict embryonic development and human disease.
Keywords: Salivary gland, Systems biology, Morphogenesis, Cell signaling, Computational models
Introduction
Every organ in the body begins embryonic development as a primitive precursor that undergoes organogenesis to acquire the morphology needed for optimal biological function. Embryonic development of secretory and absorptive organs including the salivary glands, kidneys, lungs, mammary glands, prostate, and lacrimal glands depend on a process known as branching morphogenesis [1]. Branching morphogenesis allows organs to maximize epithelial surface area while efficiently minimizing empty volume within the tissue.
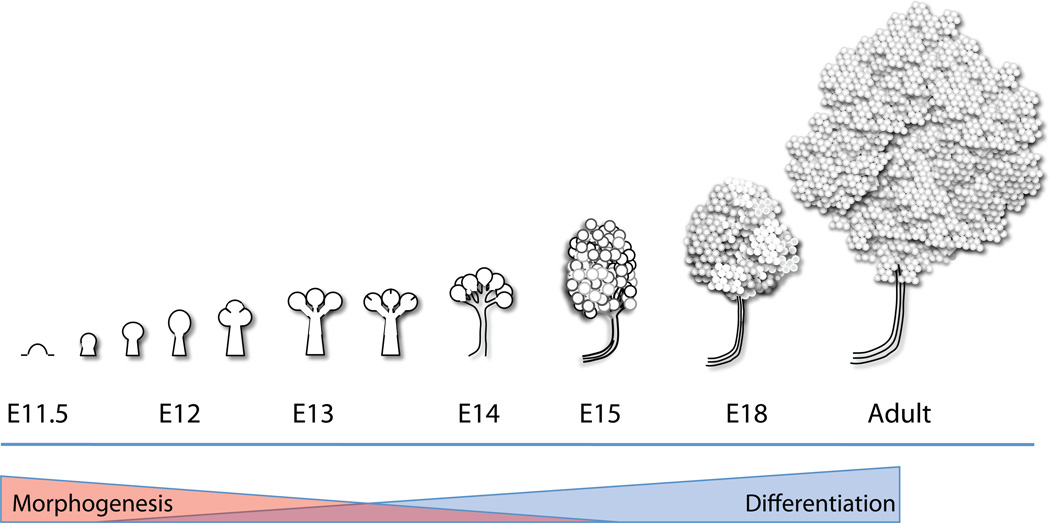
Animal models that have provided information relevant to human salivary gland development include the mouse, rat, and the fruit fly, Drosophila melanogaster. While studies in Drosophila have provided significant insight into tube formation, studies in mouse and rat have provided significant insight into the process of branching morphogenesis. The mouse submandibular salivary gland begins its development at embryonic day 11.5 (E11.5) (with the day of coital plug discovery defined as E0) as a thickening of the primitive oral epithelium that grows into the first branchial (mandibular) arch mesenchyme to form the solid epithelial placode (reviewed in [2, 3]). This placode protrudes into the mesenchyme by E12, forming a single, solid mass of cells connected to the tongue epithelium by a stalk of immature duct epithelial cells. By E12.5, indentations termed clefts start to form on the surface of the epithelial bud accompanied by alterations in the basement membrane. The clefts progress to separate the primary bud into multiple buds [4, 5]. As cleft progression proceeds, the epithelium proliferates, and the base of the cleft becomes the nascent ductal structure. This process of salivary branching morphogenesis (reviewed in [4]) is repeated multiple times over the course of several days, continually increasing the complexity of the organ (Figure 1). By late E14, the simple one-bud one-duct salivary gland has both grown and branched significantly, and the main duct begins to lumenize. The end buds undergo reorganization and begin to form acini – the main secretory units of the salivary gland. By E15–16, lumenization of the main secretory duct is nearly complete, and by E17, the acini complete lumenization, so that the gland has a continuous network of lumenized ducts connecting the acini to the oral cavity [2, 4]. Both nerves and blood vessels populate the gland in association with the branching epithelium. Cellular differentiation occurs in parallel with branching morphogenesis through pathways that are at least partially independent [5] as the glands continue to mature after birth.
Figure 1. Overview of salivary gland development.
The submandibular salivary gland undergoes branching morphogenesis, beginning as a protrusion from the oral epithelium at E11 that forms a solid primary bud on a stalk by E12. The primary bud undergoes an iterative pattern of cleft formation followed by duct outgrowth and bud expansion to produce the acini and ducts of the adult gland. As the process of morphogenesis attenuates during development, cell differentiation increases to produce the complex assembly of cells that comprise the adult gland. Not drawn to scale, and some details are excluded for simplicity.
In the adult, salivary gland function is complex, involving the contribution of multiple epithelial cell types to make saliva, and regulation of exocrine secretion by signals originating outside the gland. Saliva production begins with the secretion of incomplete saliva by submandibular acinar cells, including contributions from both serous and mucous acinar cells. This primary fluid is modified by ductal cells as it is transported to the oral cavity, where it mixes with secretions from other salivary glands to generate whole saliva. Salivary gland secretion is regulated externally, with salivary output controlled by both parasympathetic and sympathetic innervation [6].
Adult salivary glands can be affected by infection, inflammation, autoimmune disease, and tumorigenesis. Infections of the salivary gland, or siladenitis, can occur from bacterial, viral, fungal, or other causes. Sjogren’s syndrome (SS) is a complex systemic autoimmune disease affecting primarily the salivary glands and the lacrimal glands with an elusive etiology [7, 8]. Some cases of Sjogren’s disease may develop following infection. During the slow course of disease development, the glandular tissues are infiltrated by CD4+ T lymphocytes. Autoimmunity develops with 90% of patients expressing antibodies targeting autoantigens [9]. There is also typically increased B cell activation and progression to a non-Hodgkin’s, mucosa-associated lymphoid tissue (MALT) lymphoma in 5% of patients [10]. SS is classified as primary or secondary, with secondary rheumatoid diseases being associated with other systemic autoimmune diseases [8]. The etiology of SS is poorly understood, and it is difficult to diagnose until it is in its later stages due to many factors, including a lack of molecular markers.
Primary tumors of the major salivary glands are relatively rare, accounting for 11% of oropharyngeal neoplasms in the U.S. [11]. A distinguishing feature of salivary gland neoplasms is that they are highly histologically variable due to the significant variance in their cellular origins, and presumably variable etiology. As a result, histological classification of salivary gland tumors is challenging and subject to misdiagnosis by non-experts [12, 13]. Although potentially invaluable for tumor classification, molecular signatures for salivary tumors are lacking.
Other than malignant cancers, the most significant problems for patients with salivary gland disease result from loss of function. Hyposalivation, or decreased saliva production, occurs in both primary and secondary SS, and also following radiation therapy for head and neck cancers. Loss of salivary flow results in “dry mouth” and affects additional processes, such as mastication and swallowing; secondary oral disease states can subsequently develop, including sialadenitis, dental caries, periodontal disease, and persistent oral infections [14]. As a result of these secondary conditions, loss of saliva leads to a general decline in the quality of life for such patients. The molecular basis for hyposalivation in patients suffering from SS and the mechanism of the selective destruction of salivary acinar cells leading to hyposalivation as a result of radiation therapy remain poorly understood, and there are few satisfactory therapeutic options for these patients.
There are multiple challenges currently facing salivary gland research encompassing a wide range of topics from understanding development to identifying the causes of human disease. During development, it is still not understood what forces change tissue shape, what cells contribute to these forces, and what molecular pathways direct these changes. Even less is understood regarding the molecular pathways controlling cellular differentiation into the multiple cell types comprising a gland. While many details regarding cell physiology of adult acinar and ductal cells have been identified, an integrated, universally accepted model of saliva secretion at the molecular level still remains to be developed. In salivary gland disease, there is a lack of understanding of the molecular events leading to hyposalivation and to progression of salivary neoplasms. A common feature of all of these questions involves the need to understand comprehensively the changes in tissue behavior from the organ to the molecular level. Integration of all molecular interactions and cellular processes that is coordinated in all cell types results in branching morphogenesis and differentiation (Figure 2); whereas defects at one or more of these levels can produce disease. Consequently, understanding disease pathogenesis and developing rational therapies will require an understanding of the emergent properties of these interactions, which is the strength of systems biology.
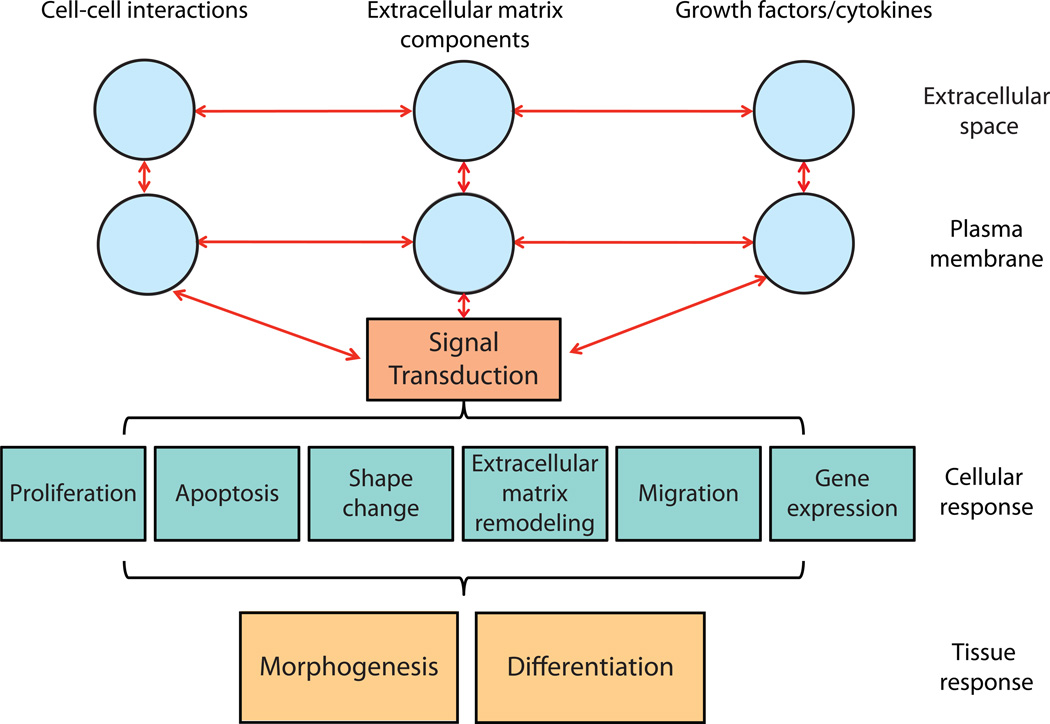
Figure 2. Signaling hierarchy controlling branching morphogenesis and cellular differentiation.
Extracellular interactions between cells, between cells and the ECM, and with growth factors and cytokines activate cytoplasmic signaling pathways that alter cell behavior. The combination of these cellular behaviors results in changes at the tissue level: morphogenesis and differentiation. The goal of systems biology is to understand all of the inputs contributing to tissue and organ-level changes so that they can be synthesized into a systems-level understanding of organ biology.
Here we will review the use of systems-based approaches to understand salivary gland development, function, and dysfunction (see Figure 3). First we will review initial progress using reductionist approaches towards understanding aspects of salivary gland development. Next we will overview research in which profiling methods have been used to characterize mRNA, miRNA, and proteins in both tissue and in saliva. Then we will discuss how a major tool of systems biology, mathematical models, are currently being utilized to investigate specific issues in salivary biology. In order for such mathematical models to achieve the complexity required to model biological events, they will need to encompass biological processes at multiple scales, analogous to modeling accomplished for vulval development in Caenorhabditis elegans [15] and for vertebrate limb development [16]. Additionally, much more comprehensive molecular detail than is currently available will be required, and it must be accessible to both researchers and computational scientists. As we describe later in this review, new advances and ongoing projects supported primarily by the National Institute of Dental and Craniofacial Research (NIDCR) will soon provide extensive salivary gland systems information for the research community.
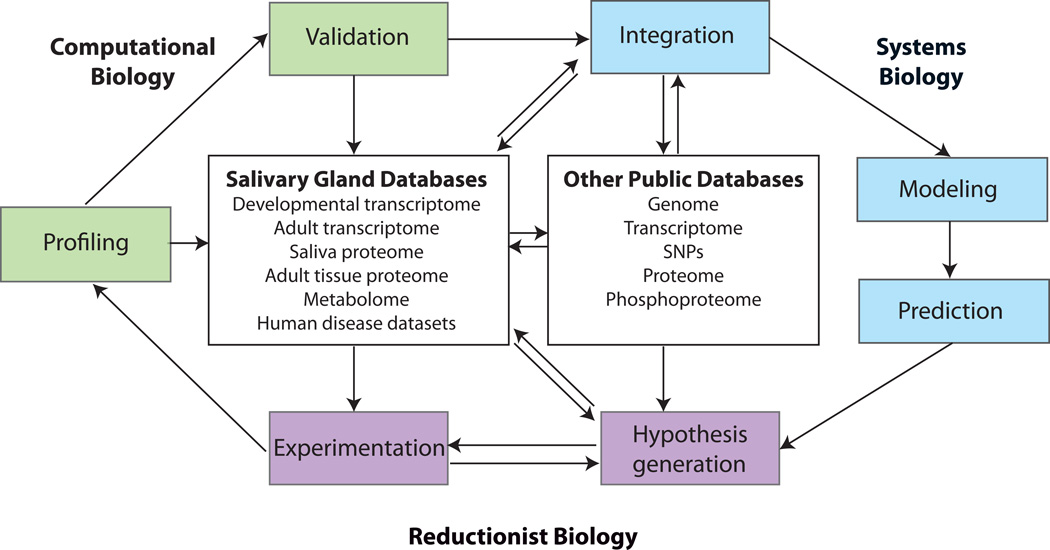
Figure 3. Schematic diagram illustrating how systems biology can be integrated into research projects.
Reductionist approaches utilize experimentation to test hypotheses, which can lead to systems approaches through profiling methods. Profiling data can be validated using traditional and/or computational methods and can become part of public databases. Individual profiling data can also be computationally integrated with public datasets, both salivary gland-specific and general, to transition into the realm of systems biology. Systems biology includes computational modeling of various types and ultimately prediction, which can lead to further hypothesis generation and experimentation.
REDUCTIONIST APPROACHES TO UNDERSTAND BRANCHING
Traditionally, cell and developmental biologists have taken a reductionist approach to understanding the development and function of salivary glands. This approach – rooted in the scientific method itself – has provided useful information regarding linear signaling pathways. Genetic ablation studies have provided significant insights into growth factors involved in early development. Studies of the epidermal growth factor receptor 1 (EGFR1) knockout mouse showed reduced branching in the submandibular gland [17]. Strikingly, examination of the fibroblast growth factor 10 (FGF10) and FGF receptor 2 (FGFR2) isoform IIIb [18–20] knockout mice showed development of only a rudimentary submandibular primary bud [21], as did the FGF8 [22] and sonic hedgehog (Shh) knockout mice [23]. Many studies have used ex vivo embryonic organ cultures to identify specific molecular mechanisms responsible for these phenotypes. First established in the 1950’s [24–27], the embryonic submandibular gland organ culture system has provided researchers with a 3D experimental system that can be used to ask in-depth, complex questions about the control of morphogenesis. Figure 4 shows an overview of a subset of the signaling pathways experimentally verified to control salivary gland development.
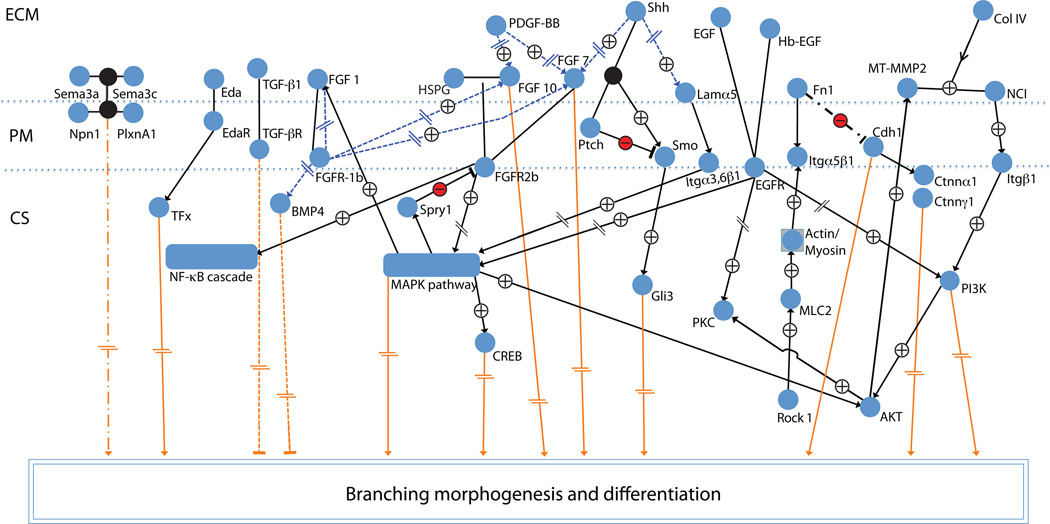
Figure 4. Cellular signaling map of embryonic salivary gland development.
A simplified overview of major signaling pathways known to control salivary gland development based on experimental studies. Slash dot slash: known effect, but pathway not identified/described yet; orange lines/arrows: pathways that affect morphogenesis/differentiation; plus sign: positive effect (activation); minus: negative effect (inhibition); interrupted lines: intermediate steps omitted; blue dotted lines and arrows: affect expression; open arrows: protein is modified (proteolysis); blunt line (inverted T): inhibition; and circular edge rectangles: signaling modules. See [66] for a detailed computational model of signaling pathways.
To identify signaling pathways involved in morphogenesis, protein function and mRNA expression can be perturbed by various means in ex vivo organ cultures. Function-blocking antibodies were used to identify a function for many molecules in branching morphogenesis, including the transmembrane receptors integrin α6 (a laminin receptor) [28] and integrin α5 and the integrin α5β1 ligand, the extracellular matrix glycoprotein fibronectin [29]. A combination of function blocking antibodies to FGF1, FGF7, and FGF10 were necessary to inhibit branching of organ cultures [30], pointing to a function for multiple FGFs in salivary gland development. A role for the EGFR tyrosine kinase [31] and downstream effector Erk1/2 [32] was initially demonstrated using pharmacological inhibitors of these proteins. Phosphatidylinositol 3’-kinase (PI3K) emerged as a kinase important in cleft formation in a subsequent study to screen multiple pharmacological inhibitors of kinases and phosphatases [33] and as an EGFR effector [33, 34]. Other cytoplasmic signaling proteins, phospholipase C gamma1 (PLCγ1) and protein kinase C (PKC), were identified as additional mediators of EGFR signaling [34]. More recently, antisense RNAs were used to identify functions for FGF receptor 1 (FGFR1) [35] and semaphorin 3A and 3B receptors plexin A2 and D1 [36] in branching morphogenesis. Small inhibitory RNAs (siRNAs) were first shown to be useful in salivary gland organ cultures by selectively knocking down gene function in a study that identified a role for fibronectin in branching morphogenesis [29] and have since become an important tool in ex vivo organ culture studies.
Recent studies have combined multiple techniques to manipulate protein function and expression to identify complex relationships between known molecules in the non-linear signaling network controlling salivary gland development. Rebustini et al. recently identified a function for the protease MT2-MMP in integrating basement membrane dynamics, integrin signaling, and growth factor activation in the control of epithelial proliferation and branching morphogenesis. MT2-MMP cleaves collagen IV in the basement membrane and releases NC1 (non-collagenous domain 1). The released NC1 domain then interacts with β1 integrin to regulate PI3K signaling, which in turn upregulates heparin-binding EGF (HB-EGF) expression, HB-EGF-induced MT2-MMP expression, and FGFR2b expression to increase epithelial proliferation. Downstream of β1 integrin signaling, collagen IV is induced by upregulation of Col4a2 gene expression [37]. The relationship between platelet-derived growth factor (PDGF) signaling and FGF signaling was identified using purified PDGF ligand, siRNAs targeting Pdgfa and Pdgfb, and inhibitors of receptor function. Using these tools, PDGF-AA was identified as regulator of FGF7 and 10 signaling [38]. FGF7 and 10 signaling, in turn, was found to potentiate cells to respond to EGFR1 signaling in the early stages of development [39]. A recent study identified an upstream regulator of fibronectin signaling to be the Rho-associated, coiled-coil containing protein kinase 1 (ROCK I), which regulates the transition from cleft initiation to cleft progression through myosin light chain phosphorylation and a subsequent defect in fibronectin assembly [40]. Fibronectin stimulates cell proliferation [40] and negatively regulates E-cadherin localization at cell-cell membranes [29], wheras E-cadherin is implicated in regulation of ductal lumen formation and acinar cell fate [41]. Although these types of reductionist approaches have been successful in starting to build an “interactome” of salivary gland signaling pathways controlling development, progress could be accelerated by identification of the entire ensemble of molecules.
GLOBAL PROFILING APPROACHES
Transcriptome profiling
Gene profiling has become a powerful tool for researchers studying both developmental processes and human disease to identify the transcriptome. One of the first salivary gland mRNA profiling studies was performed using mouse submandibular/sublingual glands harvested from embryos at E14, E17, postnatal day 1 (D1) and D5, and from 3 month-old adult mice [35]. Transcript expression in both salivary glands at each time point was compared using filter arrays containing 1176 known genes. This array analysis identified FGFR1 as a gene important in submandibular gland development. The finding that FGFR1 directly up-regulates itself, Ffg1, Fgf3 and Bmp7 and that it indirectly down-regulates Fgf7, Fgf10, and Bmp4 was identified using these data, antisense fgfr1 knockdown, and SU5042 to regulate FGFR1 activity [35]. Gene profiling studies have also been performed using human salivary gland tissue. Biopsy tissue from human minor salivary glands was evaluated by gene profiling methods to compare gene expression between primary Sjogren’s syndrome patients and normal controls [42]. Activation of interferon pathways was detected in the tissue of the primary Sjogren’s syndrome patients with 23 genes upregulated relative to normal controls.
A fundamental question in branching morphogenesis involves identifying the signals for cleft formation. Since the existing whole-organ gene profiling methods destroy cellular context, a new methodology was required to address the hypothesis that gene expression in the cleft region is different than in the end buds where clefts are not forming. Sakai and colleagues searched for mRNA differences in these regions using laser capture microdissection. From thin sections of salivary gland tissue, a laser capture microscope was used to outline and isolate the region of the epithelium adjacent to the cleft, the region at the distal tip of the end bud, and their corresponding mesenchymal regions. Because only small amounts of tissue could be obtained, a modified serial analysis of gene expression (SAGE) technique was developed that utilizes a two-cycle amplification step with the high fidelity T7 polymerase prior to generation of the SAGE libraries (T7-SAGE) [43]. Preliminary analysis of SAGE data from this study was confirmed by qPCR, and in situ hybridization revealed that fibronectin was expressed at higher levels in the epithelial cells immediately adjacent to the cleft than in the end bud region [44]. Subsequent studies identified a critical function for fibronectin in branching morphogenesis [29]. A second gene differentially expressed in cleft epithelial cells compared to buds was the protease inhibitor TIMP3 [44]. The gene knockout of TIMP3 in mice displays defects in branching morphogenesis accompanied by increased degradation of fibronectin [45]; the authors suggest that the defective morphogenesis is due to enhanced proteolytic activity degrading fibronectin. Spatial profiling of the transcriptome within specific subsets of cells will likely become increasingly important in future studies of developmental processes and human disease.
miRNA profiling
In recent years, the importance of RNA interference pathways in regulation of gene expression has been appreciated. That microRNAs (miRNAs) control multiple developmental processes and can be misregulated in disease is now clear. Profiling of miRNAs expressed in salivary gland development has been accomplished at E15.5, postnatal day 0 (P0), P5, and P25 [46] with many candidate miRNAs identified. miRNAs have also been discovered to exist in human saliva. In a recent study, miRNA profiles were compared between normal and oral squamous cell carcinoma patients. Two miRNAs are present at lower levels in saliva of cancer patients versus normal matched control saliva [47]. Analysis of the function of miRNAs in both development and disease will likely be a key component of many future studies.
Proteomic profiling
Proteomic approaches have also been applied to the study of salivary glands, the most notable study being a comprehensive catalog of the contents of human saliva – the salivary proteome – prepared by a team of researchers from five US universities [70]. This study identified 917 submandibular/sublingual proteins and 914 parotid proteins, of which 252 are submandibular/sublingual specific and 249 are parotid specific and are primarily extracellular or secretory in nature. Saliva has been demonstrated to provide many insights into normal and disease functions [72]. The salivary proteome is now available as a tool for identification and diagnosis of both systemic and salivary gland diseases. In a study comparing whole saliva from normal and Sjogren’s syndrome patients, proteins, peptides, and mRNAs differentially expressed in these populations were identified that may be informative for detection of Sjogren’s syndrome [71]. Since proteomic profiling has demonstrated its utility, it is likely that future developmental studies will employ these methods as protein detection methods become increasingly sensitive.
MATHEMATICAL MODELING AS A SYSTEMS ANALYSIS TOOL
Since profiling approaches generate large data sets, computational algorithms for integration and mathematical models are needed to build hypotheses from these data sets. Mathematical models can be generated from different types of datasets. Both traditional bottom-up methods that start with detailed data and create a model with broader applicability and top-down approaches that start with general phenomena and build models to attempt to explain the underlying complexity have been developed to address specific aspects of salivary gland biology.
Cell physiology models
The classical approach towards mathematical modeling of biological systems is the bottom-up approach that seeks to create a predictive model by combining all of the known parts. There is an extensive literature on the study of saliva secretion using primary parotid cells (reviewed in [48]), which are similar to the serous acinar cells in the submandibular gland in that they secrete the watery, proteinaceous component of the saliva. Since the precise mechanisms through which paracrine secretion occurs is not entirely clear, a mathematical model was developed to investigate the nature of the inputs inducing fluid flow. Based on measured ion concentrations, estimations of ion movement using four transporter and channel proteins, and models of calcium dynamics, a physiological model was created for parotid cell fluid secretion in a single cell [49]. Using this tool, oscillations of calcium ions (Ca++) were identified as a regulator of fluid flow, with the critical factor predicted to be the total Ca++ concentration rather than the oscillation frequency. To validate the model, Ca++ concentrations and oscillations were measured in primary parotid cells and found to substantiate the model’s predictions [49]. Future work will expand the model to create a multicellular one that can make predictions regarding primary saliva secretion.
Physical models of branching morphogenesis
Mathematical models can also attempt to model complex physical events for which all the inputs are not known to identify the critical components, which is the basis of a top-down type of modeling approach. Mathematical modelers have attempted to describe branching morphogenesis in terms of a quantitative physical mathematical model. In the first study, the epithelial and mesenchymal cells were both assumed to be “Stokes fluids” separated by an interface meant to represent the basement membrane and modeled in 2D [50]. A subsequent model incorporated three additional levels of complexity: 1) the 3D aspect of the tissue, 2) the contractile ability of the mesenchymal cells, and 3) the migratory tendencies of the epithelial cells [51] based on cell migration movements published in a separate study [52]. In both models, all other parameters for which a specific value was needed were estimated from studies in other embryonic tissues. The new 3D model better approximated the properties of the mesenchymal compartment by embedding cells within an extracellular matrix by using a mixture model rather than a Stokes fluid model. This new model – in which properties of both the epithelial and the mesenchymal cells are more realistically represented – has provided hypotheses that can be tested ex vivo. Another approach, in which changes in tissue shape are monitored using an image-based, quantitative global cell detection tool known as cell-graphs [53], may improve future models by more accurately modeling behavior of both epithelial and mesenchymal cells [54]. Future models will likely integrate with biological data to incorporate additional levels of complexity to move closer towards an understanding of the forces controlling branching morphogenesis.
Studies have integrated live-cell imaging data with mathematical modeling to better understand the mechanical forces at work in tube formation in Drosophila. Using 2-photon confocal imaging to examine defective cell movements in ribbon (rib) mutants vs. wild type Drosophila, Cheshire and colleagues identified slow and incomplete luminal morphogenesis and were able to accurately measure changes in cell shapes [55]. Since rib is a regulator of crumbs (crb), an apical polarity protein, and moesin (moe), a member of the Ezrin-Radixin-Moesin (ERM) family, which regulates cytoskeletal and membrane binding, the authors hypothesized that rib mutants were resistant to apical deformation. To test their hypothesis, the authors developed a finite element-based model where the tube was modeled as an isotropic and incompressible hyperelastic material. From the model, the authors surmised that the defective morphogenesis of the rib mutants was likely due to a three- to five-fold increased apical stiffness and two-fold increased effective apical viscosity [55]. This type of coupling of biological image data with mathematical modeling will likely lead to better understanding of the physical nature of other morphogenic events.
Signaling network models
In most morphogenetic events, substantially more information is available regarding the identity of the molecular players involved in these processes than is known regarding the physical forces at work. However, few studies have examined the relationship between all of the players using network-based modeling. The first studies to address salivary gland development using network-based modeling were performed by Jaskoll and Melnick. In one study, they noted the relevance of Waddington’s “epigenetic landscape” [56] as a description of the non-linearity of signal transduction during embryonic development [57]. The authors suggest that neural net type modeling could be applicable for modeling salivary development. In another study where neural network modeling was employed, the authors investigated a function for the transcription factor NF-kB in developing salivary glands. Both cDNA microarrays containing 1176 genes and proteomic profiles using 2D gels (600 proteins) were used to profile downstream targets of NF-kB in ex vivo organ cultures. From these data, a computational algorithm was used to perform probabilistic neural network analysis to predict likely effectors of NF-kB activity in developing E15 salivary glands [58]. Upstream activators of NF-kB in the salivary gland were not known at the time of this study. However, once Ectodysplasin (Eda), a mesenchyme-produced ligand for the epithelial Eda receptor (Edar) [59], was identified as a critical growth factor controlling salivary gland development and a regulator of NF-kB in the salivary gland [60], a second systems-based study was performed to determine if canonical NF-kB signaling is sufficient to account for the complex Eda Tabby (EdaTa) phenotype [66]. To generate a predictive model, a subset of genes to be included in the model were selected from published reductionist studies. Relevant data regarding the associated pathways and interactions were extracted from publicly available databases including KEGG [61], OMIM [62], GeneCards [63], iHOP [64], and BioModels[65], and imported into ProcessDB (Integrative Bioinformatics). To generate a mechanistic kinetic framework of pathway interactions, a combination of bottom-up and top-down approaches was developed from the mined data that included 5 cellular locations (nucleus, cytoplasm, secretory pathway, plasma membrane, and extracellular space), 217 processes (binding, chemical reaction, or transport) and 138 states (a molecule or complex in a physical space). Unexpectedly, the resulting model predicted that NF-kB is not a significant mediator of EdaR signaling, which was confirmed with real-time PCR. However, many genes were changed in EdaTa mice, including Edar, Fgf8, Shh, Egf, Tfga, and Egfr, but the model predicted that the likely cause of these expression changes was an unidentified transcription factor (TFx). Based on further in silico analysis, the authors postulate that the transcription factor may be C/EBP [66]. These studies, the first to perform quantitative systems analysis in the context of salivary gland development, are an initial step towards achieving a comprehensive understanding of salivary gland development.
Computational analysis in the study of Sjogren’s syndrome
Systems biology-based approaches are beginning to be utilized as a tool to better understand complex salivary gland diseases. Hu et al. used a systems approach to identify new disease-hub genes, which they define as promising targets for therapeutic intervention and diagnosis of SS [67,71]. This study compared parotid tissue from three classes of patients: patients with primary SS, patients with primary SS associated with mucosa-associated lymphoid tissue (MALT) lymphoma, and patients without primary SS (non-primary SS controls). Microarray profiling to examine gene expression and proteomic analysis to examine protein expression were performed on all samples, and weighted gene-co-expression network analysis was performed on these data to identify disease-hub genes. Computational analysis has also been employed in the study of animal models of SS. In one study, the widely used nonobese diabetic (NOD) mouse model of SS [68] was compared with the parent line, Balb/C, in terms of biomarker expression, autoantibody production, glandular inflammation, and saliva production [69]. Principal component analysis was used to identify significant positive and negative correlations within the data. Interestingly, in this study each biomarker typically associated exclusively with only one of the other parameters. These data indicate that SS disease progression may not follow a linear trajectory, even within this animal model. In humans, the disease etiology is likely to be significantly more complex and variable.
CURRENT AND FUTURE CENTRALIZED DATA REPOSITORIES
The study of salivary gland development and function employing systems biology approaches is still in its infancy, and more tools are required. A comprehensive repository for databases will facilitate research in the field. Several ambitious projects are underway to provide such information for salivary gland embryonic development and adult functions. They range from the identification and characterization of genes and proteins expressed at the various stages of salivary gland development to characterizations of the components of human saliva and saliva-producing tissue in health and disease. The NIDCR has recognized that to accelerate progress in salivary gland research, the generation and accessibility of such databases is of high importance, and it is supporting new programs and consortia to make them available to researchers.
One new consortium is providing novel systems information about saliva, the product of salivary glands. The Salivaomics Knowledge Base (SKB) (http://hspp.dent.ucla.edu/skb.html) provides a convenient central repository for multiple saliva-relevant databases. This multi-group project supported by the NIDCR aims to generate a comprehensive, centralized tool to study ‘omics’ of adult salivary glands. It contains the consortium project cataloging the saliva proteome [70], the saliva transcriptome, and also includes a clinical sample database. The SKB plans to cover comprehensively all aspects of saliva and saliva-producing cells for clinical applications to better understand salivary gland diseases, as well as diseases that can be tracked or diagnosed through saliva. The SKB includes multiple independent databases, including transcriptome comparisons of gene expression profiles of patients that are already publicly available. It also aims to incorporate salivary gland metabolomics, microRNA [47], and microbe profiles into its central repository. When complete, it will offer a central portal to tools, databases, and scientific findings to help characterize oral diseases by saliva analysis (http://www.skb.ucla.edu/).
An NIDCR intramural project specifically focused on providing gene expression profiles spanning the developmental stages of salivary gland development is being completed. This Gene Expression Atlas of Salivary Gland Development will consist of two parts, one focusing on whole-gland or whole-tissue characterization of gene expression within a full range of stages of gland development, and the other focusing on tissue site-specific expression of genes within selected developmental stages.
The Hoffman group will provide a new, more complete systems-based analysis of salivary gland development than previously published [35] by profiling mRNA expression in both submandibular and sublingual glands using arrays providing whole genome coverage. The tissue samples used will span time-points from the initial bud stage, at which the glands can first be identified, through adult. Three independent replicate datasets will be merged in the final database.
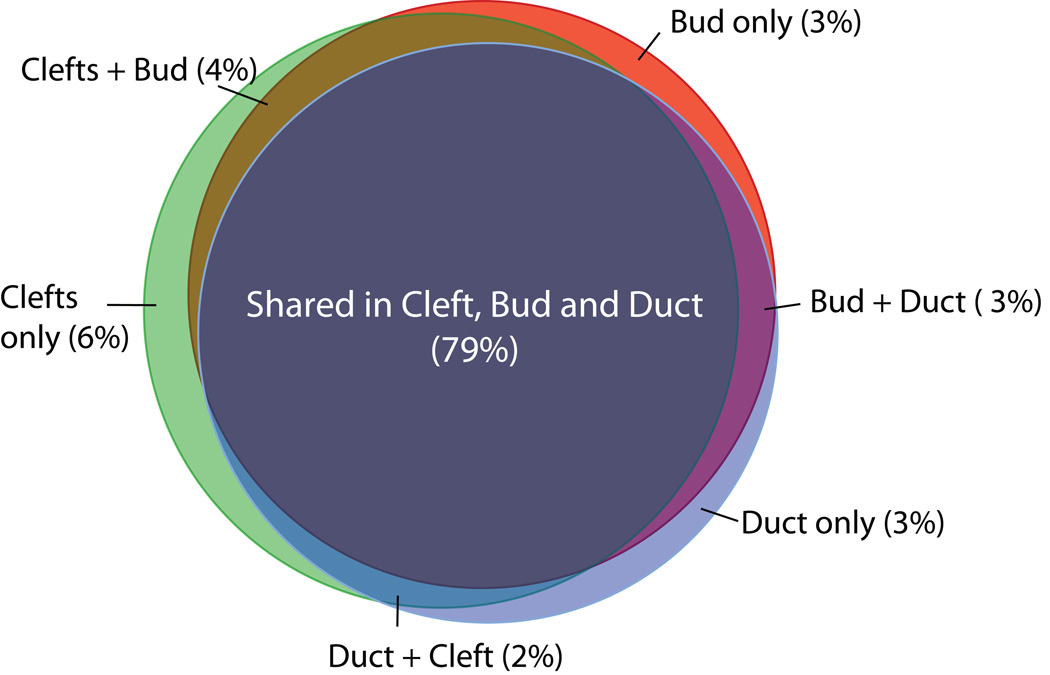
The other major program conducted by Musselmann and co-workers uses laser microdissection to generate gene expression data for a series of specialized epithelial sites in developing salivary glands. Figure 5 summarizes preliminary data on shared and unique gene expression profiles in developing epithelial cells from cleft, bud, and duct regions. NIDCR plans to complete initial analyses and open the entire dataset to the research community in March 2010.
Figure 5. Venn diagram of genes expressed in epithelial cells from cleft, bud, and duct regions of E12.5 mouse salivary glands.
Note the extensive overlaps of gene expression between regions, as well as some differentially expressed genes. Based on preliminary analyses of microarray data involving >20,000 genes; numbers in parentheses indicate the percentage of total unique genes in that category.
NIDCR’s FaceBase Consortium was launched in October 2009 as a major program to provide a centralized, comprehensive source of systems data for all craniofacial tissues. This ambitious multi-institution program will provide a centralized database that will consolidate information on a series of ongoing and new projects in order to provide researchers with crucial systems information on the genes and molecules expressed during normal craniofacial and oral development, as well as for defects in craniofacial disorders. The FaceBase program should soon provide a centralized portal for researchers interested in a variety of specialized projects and research questions in oral and craniofacial biology, including systems analysis of salivary gland development.
Conclusions and Outlook
Our understanding of the molecular basis of salivary gland branching morphogenesis has progressed significantly in the past few years, but a systems-level understanding of salivary gland development is still far from a reality. Many databases and tools are currently on the horizon that will help to bring us closer to this goal in the near future. These databases will profile mRNA expression within the organ over a developmental time-course and even profile gene expression within specific subpopulations of cells. Other databases will focus on various aspects of human disease and profile mRNA, miRNA, protein, cellular metabolism, and microbe-related expression to complement existing transcriptional and proteomic databases already available to the research community. Once complete, these resources will facilitate new computational models of developing salivary glands and human disease conditions. Integration of data from multiple sources will be a challenge inherent in utilizing the massive datasets that will become available.
To make progress towards a comprehensive understanding of the development and function of the salivary gland will require more than just cataloging all of the events and molecules. Many researchers will need to consider integrating systems-based computational approaches into their work to identify the critical players. From the point of view of an individual researcher, the large datasets from “-omics” can be quite daunting. The enormity of the datasets as well as the lack of tools to access and integrate these data into their own work poses a challenge for researchers. Continued development of tools to make data accessible and integration possible for researchers and clinicians alike will be needed to make the transition into systems thinking. Interdisciplinary collaborations will continue to be important to bridge the gaps between diverse fields of inquiry so that every researcher does not need to become a systems or computational biologist. Not every researcher will want to use a purely systems-based approach for their work, and nor should they. The necessity of reductionist approaches for testing hypotheses generated from systems biology methods will still persist. Nevertheless, the generation of salivary-specific “-omics” data sets along with new integration and modeling tools generated by computer scientists will likely encourage more researchers to enter into the realm of salivary gland systems biology.
There is much to be gained from the application of systems biology-based approaches to the study of salivary gland development and disease. Because salivary gland development is such an intricate and complex process, it is unlikely that it will be possible to understand the non-linear integration of all important events and molecules without the assistance of mathematical models to both integrate observations and identify new testable hypotheses. Salivary diseases are equally or more complex than the development of the organ itself. SS appears to be a disease with potentially multiple causes and a non-linear progression. Modeling tools could provide significant insight and hypothesis generation to understand the complex relationships between observed manifestations of the disease. Salivary gland cancers appear to be equally as complex and difficult to detect at early stages. Computational approaches could be useful for identifying biomarkers for early diagnosis of all salivary gland diseases. Future disease models could provide an opportunity for earlier disease diagnosis, selection of appropriate therapeutic options, and provide accurate prognoses.
Acknowledgements
The authors thank Dr. Michael Gerdes for useful discussions and critical reading of the manuscript. This work was sponsored in part by the NIH/NIDCR by grants RO1DE019244, R21DE019197 and RC1DE020402 (to M.L.) and NIDCR Intramural support (to K.M.Y.).
Footnotes
Further Reading
Joyce AR, Palsson BO. The model organism as a system: integrating 'omics' data sets. Nature Reviews 2006 Mar;7(3):198–210.
Martin M-S, Iain DCF, Frederick K. Multiscale modeling for biologists. Wiley Interdisciplinary Reviews: Systems Biology and Medicine 2009;1(1):4–14.
References
- 1.Davies J, editor. Branching Morphogenesis. 1 ed. Springer; 2005. [Google Scholar]
- 2.Tucker AS. Salivary gland development. Seminars in cell & developmental biology. 2007 Apr;18(2):237–244. doi: 10.1016/j.semcdb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Jaskoll T, Melnick M. Embryonic Salivary Gland Branching Morphogenesis. In: Davies JA, editor. Branching Morphogenesis. New York: Landes Bioscience/Springer; 2005. pp. 160–175. [Google Scholar]
- 4.Patel VN, Rebustini IT, Hoffman MP. Salivary gland branching morphogenesis. Differentiation; research in biological diversity. 2006 Sep;74(7):349–364. doi: 10.1111/j.1432-0436.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 5.Cutler LS. The dependent and independent relationships between cytodifferentiation and morphogenesis in developing salivary gland secretory cells. The Anatomical record. 1980 Mar;196(3):341–347. doi: 10.1002/ar.1091960310. [DOI] [PubMed] [Google Scholar]
- 6.Proctor GB, Carpenter GH. Regulation of salivary gland function by autonomic nerves. Auton Neurosci. 2007 Apr 30;133(1):3–18. doi: 10.1016/j.autneu.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Lee BH, Tudares MA, Nguyen CQ. Sjogren's syndrome: an old tale with a new twist. Archivum immunologiae et therapiae experimentalis. 2009 Jan-Feb;57(1):57–66. doi: 10.1007/s00005-009-0002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaleu N, Jonsson R, Koller MM. Sjogren's syndrome. European journal of oral sciences. 2005 Apr;113(2):101–113. doi: 10.1111/j.1600-0722.2004.00183.x. [DOI] [PubMed] [Google Scholar]
- 9.Mathews SA, Kurien BT, Scofield RH. Oral manifestations of Sjogren's syndrome. J Dent Res. 2008 Apr;87(4):308–318. doi: 10.1177/154405910808700411. [DOI] [PubMed] [Google Scholar]
- 10.Kovacs L, Szodoray P, Kiss E. Secondary tumours in Sjogren's syndrome. Autoimmunity reviews. 2009 Jul 12; doi: 10.1016/j.autrev.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Boukheris H, Curtis RE, Land CE, Dores GM. Incidence of carcinoma of the major salivary glands according to the WHO classification, 1992 to 2006: a population-based study in the United States. Cancer Epidemiol Biomarkers Prev. 2009 Nov;18(11):2899–2906. doi: 10.1158/1055-9965.EPI-09-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speight PM, Barrett AW. Salivary gland tumours. Oral diseases. 2002 Sep;8(5):229–240. doi: 10.1034/j.1601-0825.2002.02870.x. [DOI] [PubMed] [Google Scholar]
- 13.Speight PM, Barrett AW. Prognostic factors in malignant tumours of the salivary glands. The British journal of oral & maxillofacial surgery. 2009 Dec;47(8):587–593. doi: 10.1016/j.bjoms.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Napenas JJ, Brennan MT, Fox PC. Diagnosis and treatment of xerostomia (dry mouth) Odontology / the Society of the Nippon Dental University. 2009 Jul;97(2):76–83. doi: 10.1007/s10266-008-0099-7. [DOI] [PubMed] [Google Scholar]
- 15.Kam N, Kugler H, Marelly R, Appleby L, Fisher J, Pnueli A, et al. A scenario-based approach to modeling development: a prototype model of C. elegans vulval fate specification. Dev Biol. 2008 Nov 1;323(1):1–5. doi: 10.1016/j.ydbio.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman SA, Christley S, Glimm T, Hentschel HG, Kazmierczak B, Zhang YT, et al. Multiscale models for vertebrate limb development. Current topics in developmental biology. 2008;81:311–340. doi: 10.1016/S0070-2153(07)81011-8. [DOI] [PubMed] [Google Scholar]
- 17.Jaskoll T, Melnick M. Submandibular gland morphogenesis: stage-specific expression of TGF-alpha/EGF, IGF, TGF-beta, TNF, and IL-6 signal transduction in normal embryonic mice and the phenotypic effects of TGF-beta2, TGF-beta3, and EGF-r null mutations. The Anatomical record. 1999 Nov 1;256(3):252–268. doi: 10.1002/(SICI)1097-0185(19991101)256:3<252::AID-AR5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes & development. 1998 Oct 15;12(20):3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, et al. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000 Nov 2;277(3):643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 20.De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development (Cambridge, England) 2000 Feb;127(3):483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 21.Jaskoll T, Abichaker G, Witcher D, Sala FG, Bellusci S, Hajihosseini MK, et al. FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC developmental biology. 2005;5:11. doi: 10.1186/1471-213X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaskoll T, Witcher D, Toreno L, Bringas P, Moon AM, Melnick M. FGF8 dose-dependent regulation of embryonic submandibular salivary gland morphogenesis. Dev Biol. 2004 Apr 15;268(2):457–469. doi: 10.1016/j.ydbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Jaskoll T, Leo T, Witcher D, Ormestad M, Astorga J, Bringas P, Jr, et al. Sonic hedgehog signaling plays an essential role during embryonic salivary gland epithelial branching morphogenesis. Dev Dyn. 2004 Apr;229(4):722–732. doi: 10.1002/dvdy.10472. [DOI] [PubMed] [Google Scholar]
- 24.Grobstein C. Morphogenetic interaction between embryonic mouse tissues separated by a membrane filter. Nature. 1953 Nov 7;172(4384):869–870. doi: 10.1038/172869a0. [DOI] [PubMed] [Google Scholar]
- 25.Grobstein C. Inductive epitheliomesenchymal interaction in cultured organ rudiments of the mouse. Science (New York, NY. 1953 Jul 10;118(3054):52–55. doi: 10.1126/science.118.3054.52. [DOI] [PubMed] [Google Scholar]
- 26.Borghese E. The development in vitro of the submandibular and sublingual glands of Mus musculus. Journal of anatomy. 1950 Jul;84(3):287–302. [PMC free article] [PubMed] [Google Scholar]
- 27.Borghese E. Explantation experiments on the influence of the connective tissue capsule on the development of the epithelial part of the submandibular gland of Mus musculus. Journal of anatomy. 1950 Jul;84(3):303–318. [PMC free article] [PubMed] [Google Scholar]
- 28.Kadoya Y, Kadoya K, Durbeej M, Holmvall K, Sorokin L, Ekblom P. Antibodies against domain E3 of laminin-1 and integrin alpha 6 subunit perturb branching epithelial morphogenesis of submandibular gland, but by different modes. The Journal of cell biology. 1995 Apr;129(2):521–534. doi: 10.1083/jcb.129.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003 Jun 19;423(6942):876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 30.Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, et al. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development (Cambridge, England) 2005 Mar;132(6):1223–1234. doi: 10.1242/dev.01690. [DOI] [PubMed] [Google Scholar]
- 31.Kashimata M, Gresik EW. Epidermal growth factor system is a physiological regulator of development of the mouse fetal submandibular gland and regulates expression of the alpha6-integrin subunit. Dev Dyn. 1997 Feb;208(2):149–161. doi: 10.1002/(SICI)1097-0177(199702)208:2<149::AID-AJA2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 32.Kashimata M, Sayeed S, Ka A, Onetti-Muda A, Sakagami H, Faraggiana T, et al. The ERK-1/2 signaling pathway is involved in the stimulation of branching morphogenesis of fetal mouse submandibular glands by EGF. Dev Biol. 2000 Apr 15;220(2):183–196. doi: 10.1006/dbio.2000.9639. [DOI] [PubMed] [Google Scholar]
- 33.Larsen M, Hoffman MP, Sakai T, Neibaur JC, Mitchell JM, Yamada KM. Role of PI 3-kinase and PIP3 in submandibular gland branching morphogenesis. Dev Biol. 2003 Mar 1;255(1):178–191. doi: 10.1016/S0012-1606(02)00047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koyama N, Kashimata M, Sakashita H, Sakagami H, Gresik EW. EGF-stimulated signaling by means of PI3K, PLCgamma1, and PKC isozymes regulates branching morphogenesis of the fetal mouse submandibular gland. Dev Dyn. 2003 Jun;227(2):216–226. doi: 10.1002/dvdy.10309. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman MP, Kidder BL, Steinberg ZL, Lakhani S, Ho S, Kleinman HK, et al. Gene expression profiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF-dependent mechanisms. Development (Cambridge, England) 2002 Dec;129(24):5767–5778. doi: 10.1242/dev.00172. [DOI] [PubMed] [Google Scholar]
- 36.Chung L, Yang TL, Huang HR, Hsu SM, Cheng HJ, Huang PH. Semaphorin signaling facilitates cleft formation in the developing salivary gland. Development (Cambridge, England) 2007 Aug;134(16):2935–2945. doi: 10.1242/dev.005066. [DOI] [PubMed] [Google Scholar]
- 37.Rebustini IT, Myers C, Lassiter KS, Surmak A, Szabova L, Holmbeck K, Pedchenko V, Hudson BG, Hoffman MP. MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev Cell. 2009 Oct;17(4):482–493. doi: 10.1016/j.devcel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto S, Fukumoto E, Yoshizaki K, Iwamoto T, Yamada A, Tanaka K, et al. Platelet-derived growth factor receptor regulates salivary gland morphogenesis via fibroblast growth factor expression. J Biol Chem. 2008 Aug 22;283(34):23139–23149. doi: 10.1074/jbc.M710308200. [DOI] [PubMed] [Google Scholar]
- 39.Nitta M, Kume T, Nogawa H. FGF alters epithelial competence for EGF at the initiation of branching morphogenesis of mouse submandibular gland. Dev Dyn. 2009 Feb;238(2):315–323. doi: 10.1002/dvdy.21780. [DOI] [PubMed] [Google Scholar]
- 40.Daley WP, Gulfo KM, Sequeira SJ, Larsen M. Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev Biol. 2009 Oct 3; doi: 10.1016/j.ydbio.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker JL, Menko AS, Khalil S, Rebustini I, Hoffman MP, Kreidberg JA, et al. Diverse roles of E-cadherin in the morphogenesis of the submandibular gland: insights into the formation of acinar and ductal structures. Dev Dyn. 2008 Nov;237(11):3128–3141. doi: 10.1002/dvdy.21717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottenberg JE, Cagnard N, Lucchesi C, Letourneur F, Mistou S, Lazure T, et al. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjogren's syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2006 Feb 21;103(8):2770–2775. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai T, Larsen M, Yamada KM. Microanalysis of gene expression in tissues using T7- SAGE: serial analysis of gene expression after high-fidelity T7-based RNA amplification. Chapter 19. Current protocols in cell biology / editorial board, Juan S Bonifacino. 2002 Nov; doi: 10.1002/0471143030.cb1903s16. [et al Unit 19 13. [DOI] [PubMed] [Google Scholar]
- 44.Sakai T, Larsen M, Yamada KM. Morphogenesis and branching of salivary glands: Characterization of new matrix and signaling regulators. Oral Biosci Med. 2005;2:105–113. [Google Scholar]
- 45.Gill SE, Pape MC, Leco KJ. Tissue inhibitor of metalloproteinases 3 regulates extracellular matrix--cell signaling during bronchiole branching morphogenesis. Dev Biol. 2006 Oct 15;298(2):540–554. doi: 10.1016/j.ydbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Jevnaker AM, Osmundsen H. MicroRNA expression profiling of the developing murine molar tooth germ and the developing murine submandibular salivary gland. Archives of oral biology. 2008 Jul;53(7):629–645. doi: 10.1016/j.archoralbio.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009 Sep 1;15(17):5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner RJ, Sugiya H. Understanding salivary fluid and protein secretion. Oral diseases. 2002 Jan;8(1):3–11. doi: 10.1034/j.1601-0825.2002.10815.x. [DOI] [PubMed] [Google Scholar]
- 49.Gin E, Crampin EJ, Brown DA, Shuttleworth TJ, Yule DI, Sneyd J. A mathematical model of fluid secretion from a parotid acinar cell. Journal of theoretical biology. 2007 Sep 7;248(1):64–80. doi: 10.1016/j.jtbi.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lubkin SR, Li Z. Force and deformation on branching rudiments: cleaving between hypotheses. Biomechanics and modeling in mechanobiology. 2002 Jun;1(1):5–16. doi: 10.1007/s10237-002-0001-4. [DOI] [PubMed] [Google Scholar]
- 51.Wan X, Li Z, Lubkin SR. Mechanics of mesenchymal contribution to clefting force in branching morphogenesis. Biomechanics and modeling in mechanobiology. 2008 Oct;7(5):417–426. doi: 10.1007/s10237-007-0105-y. [DOI] [PubMed] [Google Scholar]
- 52.Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. Journal of cell science. 2006 Aug 15;119(Pt 16):3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- 53.Demir C, Gultekin SH, Yener B. Augmented cell-graphs for automated cancer diagnosis. Bioinformatics (Oxford, England) 2005 Sep 1;21(Suppl 2):ii7–ii12. doi: 10.1093/bioinformatics/bti1100. [DOI] [PubMed] [Google Scholar]
- 54.Bilgin CC, Ray S, Daley WP, Baydil B, Sequeira SJ, Yener B, et al. Cell-graph modeling of salivary gland branching morphogenesis. Proceedings of the IEEE International Symposium on Biomedical Imaging: from nano to micro. 2010 [Google Scholar]
- 55.Cheshire AM, Kerman BE, Zipfel WR, Spector AA, Andrew DJ. Kinetic and mechanical analysis of live tube morphogenesis. Dev Dyn. 2008 Oct;237(10):2874–2888. doi: 10.1002/dvdy.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waddington CH. The strategy of the genes. London: George Allen & Unwin; 1957. pp. 11–58. [Google Scholar]
- 57.Melnick M, Jaskoll T. Mouse submandibular gland morphogenesis: a paradigm for embryonic signal processing. Crit Rev Oral Biol Med. 2000;11(2):199–215. doi: 10.1177/10454411000110020401. [DOI] [PubMed] [Google Scholar]
- 58.Melnick M, Chen H, Min Zhou Y, Jaskoll T. The functional genomic response of developing embryonic submandibular glands to NF-kappa B inhibition. BMC developmental biology. 2001;1:15. doi: 10.1186/1471-213X-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pispa J, Mikkola ML, Mustonen T, Thesleff I. Ectodysplasin, Edar and TNFRSF19 are expressed in complementary and overlapping patterns during mouse embryogenesis. Gene Expr Patterns. 2003 Oct;3(5):675–679. doi: 10.1016/s1567-133x(03)00092-9. [DOI] [PubMed] [Google Scholar]
- 60.Jaskoll T, Zhou YM, Trump G, Melnick M. Ectodysplasin receptor-mediated signaling is essential for embryonic submandibular salivary gland development. Anat Rec A Discov Mol Cell Evol Biol. 2003 Apr;271(2):322–331. doi: 10.1002/ar.a.10045. [DOI] [PubMed] [Google Scholar]
- 61.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research. 1999 Jan 1;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM) Human mutation. 2000;15(1):57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 63.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics (Oxford, England) 1998;14(8):656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- 64.Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics (Oxford, England) 2005 Sep 1;21(Suppl 2):ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 65.Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic acids research. 2006 Jan 1;34(Database issue):D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melnick M, Phair RD, Lapidot SA, Jaskoll T. Salivary gland branching morphogenesis: a quantitative systems analysis of the Eda/Edar/NFkappaB paradigm. BMC developmental biology. 2009;9:32. doi: 10.1186/1471-213X-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu S, Zhou M, Jiang J, Wang J, Elashoff D, Gorr S, et al. Systems biology analysis of Sjogren's syndrome and mucosa-associated lymphoid tissue lymphoma in parotid glands. Arthritis and rheumatism. 2009 Jan;60(1):81–92. doi: 10.1002/art.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makino S, Kunimoto K, Muraoka Y, Mizushima Y, Katagiri K, Tochino Y. Breeding of a non-obese, diabetic strain of mice. Jikken dobutsu. 1980 Jan;29(1):1–13. doi: 10.1538/expanim1978.29.1_1. [DOI] [PubMed] [Google Scholar]
- 69.Delaleu N, Immervoll H, Cornelius J, Jonsson R. Biomarker profiles in serum and saliva of experimental Sjogren's syndrome: associations with specific autoimmune manifestations. Arthritis research & therapy. 2008;10(1):R22. doi: 10.1186/ar2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. Journal of proteome research. 2008 May;7(5):1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, et al. Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis and rheumatism. 2007 Nov;56(11):3588–3600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. American journal of dentistry. 2009 Aug;22(4):241–248. [PMC free article] [PubMed] [Google Scholar]